Abstract
The inhibition of male-specific lethal 2 (msl-2) mRNA translation by the RNA-binding protein sex-lethal (SXL) is an essential regulatory step for X-chromosome dosage compensation in Drosophila melanogaster. The mammalian upstream of N-ras (UNR) protein has been implicated in the regulation of mRNA stability and internal ribosome entry site (IRES)-dependent mRNA translation. Here we have identified the Drosophila homolog of mammalian UNR as a cofactor required for SXL-mediated repression of msl-2 translation. UNR interacts with SXL, a female-specific protein. Although UNR is present in both male and female flies, binding of SXL to uridine-rich sequences in the 3′ untranslated region (UTR) of msl-2 mRNA recruits UNR to adjacent regulatory sequences, thereby conferring a sex-specific function to UNR. These data identify a novel regulator of dosage compensation in Drosophila that acts coordinately with SXL in translational control.
Keywords: msl-2, SXL, UNR, dosage compensation, translational control
Translational control of gene expression is an important mode of regulation in numerous biological settings. For example, during embryonic development translational control plays a critical role in the establishment of the embryo axes, the specification of cell fates, oocyte maturation, sex determination, and dosage compensation (for review, see Kuersten and Goodwin 2003; Wilhelm and Smibert 2005). Dosage compensation is the process that equalizes the expression of X-linked genes in those organisms in which males and females contain a different number of X chromosomes. In Drosophila melanogaster, dosage compensation is achieved by increasing the transcriptional output of the single male X chromosome by approximately twofold (for review, see Gilfillan et al. 2004; Hamada et al. 2005; Straub et al. 2005). This process requires the activity of a ribonucleoprotein (RNP) assembly known as the dosage compensation complex (DCC) or male-specific lethal (MSL) complex. The DCC fails to assemble in female flies primarily because of the lack of one of its subunits, the protein MSL-2. Synthesis of MSL-2 is prevented by the female-specific RNA-binding protein sex-lethal (dSXL) via a dual mechanism that involves the splicing inhibition of a facultative 5′ untranslated region (UTR) intron in msl-2 pre-mRNA and the translational repression of the msl-2 transcript (Bashaw and Baker 1997; Kelley et al. 1997; Gebauer et al. 1998). Studies using a Drosophila cell-free translation system have been instrumental in deciphering the molecular mechanism of msl-2 translational regulation. Translational repression of msl-2 mRNA requires dSXL binding to stretches of uridines present in the 5′ and 3′ UTRs of the transcript. dSXL achieves tight repression by blocking two consecutive steps of translation initiation: dSXL bound to the 3′ UTR inhibits the initial binding of the small ribosomal subunit to msl-2 mRNA, while 5′-UTR-bound dSXL inhibits scanning of those subunits that have escaped the first control (Beckmann et al. 2005). Mutational analysis has shown that the sequences adjacent to the relevant SXL-binding sites in the 3′ UTR are also required for efficient repression, and could serve as binding sites for putative corepressors (Gebauer et al. 2003; Grskovic et al. 2003).
Mammalian upstream of N-ras (UNR) is a cytoplasmic protein essential for development, as deletion of the UNR promoter leads to embryonic lethality (Boussadia et al. 1997). UNR contains five cold-shock domains (CSD) (Jacquemin-Sablon et al. 1994), a five-stranded β-barrel fold with conserved RNP1 and RNP2 motifs that mediates binding to single-stranded nucleic acids (Ermolenko and Makhatadze 2002). Proteins of the CSD family perform a variety of functions and are thought to act as RNA chaperones, promoting a linear conformation of the mRNA (Graumann and Mahariel 1998). In humans, UNR has been implicated in the destabilization of c-fos mRNA (Chang et al. 2004) and the activation of translation driven by the IRESs of several transcripts, including c-myc (Evans et al. 2003), rhinovirus (Hunt et al. 1999), poliovirus (Boussadia et al. 2003), PITSLRE protein kinase (Tinton et al. 2005), and the proapoptotic factor Apaf-1 (Mitchell et al. 2003).
Here we show that Drosophila UNR (dUNR) is required for translational repression of msl-2 mRNA. dUNR is recruited to the 3′ UTR of msl-2 mRNA by dSXL. Consistent with this, although dUNR is present in both sexes, it associates with msl-2 mRNA only in female flies. dUNR-depleted extracts fail to support translational repression by dSXL, while restoring dUNR levels reinstates translation inhibition. These data indicate that dUNR is a corepressor of msl-2 mRNA translation and identify a novel regulator of dosage compensation in Drosophila.
Results
Identification of Drosophila UNR as a dSXL-associated protein
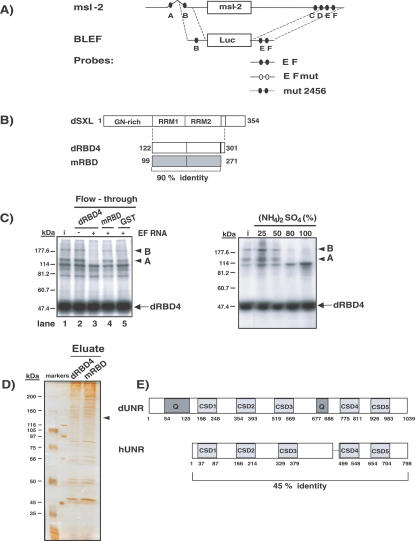
Binding of dSXL to uridine stretches in the 5′ and 3′ UTRs of msl-2 mRNA is necessary for translational repression (Fig. 1A, sites A-F). The minimal sequences required for translation inhibition by dSXL consist of SXL-binding site B in the 5′ UTR and sites E and F in the 3′ UTR (Fig. 1A, BLEF mRNA). Nucleotides adjacent to sites E and F are also required and interact with two high-molecular-weight polypeptides that could be potential corepressors (Gebauer et al. 2003; Grskovic et al. 2003), which we hereafter refer to as A and B. A fragment of dSXL containing the RNA-binding domains followed by seven amino acids exerts full translational repression activity and coimmunoprecipitates with polypeptides A and B (Fig. 1B, dRBD4; Grskovic et al. 2003). However, the equivalent fragment of the SXL homolog from Musca domestica (mRBD), despite sharing 90% identity, is unable to interact with these polypeptides and cannot repress translation (Grskovic et al. 2003). We therefore reasoned that the putative corepressors should be retained in a dRBD4 affinity chromatography column, but should flow through an mRBD column.
Figure 1.
Purification of Drosophila UNR. (A)
Schematic representation of the msl-2 mRNA and the RNA constructs used in this study. msl-2 mRNA contains SXL-binding sites (A-F, filled ovals) in its 5′ UTR (626 nucleotides [nt]) and 3′ UTR (1047 nt). BLEF mRNA harbors the minimal msl-2 sequences required for translational repression, consisting of 69 nt in the 5′ UTR containing site B, and 46 nt in the 3′ UTR containing sites E and F, fused to the Firefly luciferase ORF (Gebauer et al. 2003). Probes used for UV-cross-link and gel mobility-shift assays are also depicted (see Materials and Methods for their detailed sequences). (B) Scheme of the Drosophila SXL protein (dSXL) and its derivatives. dRBD4 is a deletion derivative fully functional in translational repression; mRBD is the equivalent fragment of the SXL homolog from Musca domestica, which shows no translational repression ability (Grskovic et al. 2003). Amino acid numbers are indicated, as well as the percentage of identity between the Drosophila and Musca SXL fragments. (C, left panel) Retention of putative corepressors in SXL columns. Embryo extract was loaded on glutathione-Sepharose columns containing either GST-dRBD4, GST-mRBD, or GST alone, in the presence or absence of the EF RNA fragment. Retention of the polypeptides A and B was tested by analyzing their presence in the column flowthrough using UV-cross-link to radiolabeled EF RNA and coimmunoprecipitation with dRBD4, as previously described (Grskovic et al. 2003). (i) Input. (C, right panel) Enrichment of the putative corepressors by ammonium sulfate precipitation. Drosophila embryo extracts were subjected to precipitation by saturation with (NH4)2SO4. The polypeptides A and B (arrowheads) were detected by the cross-link-IP assay described above. (D) Enrichment of an ∼130-kDa protein in the dRBD4 column eluate. Proteins present in the eluates from dRBD4 and mRBD columns were separated in a 10% acrylamide gel and silver stained (see Results for further experimental details). The putative ∼130-kDa corepressor is indicated with an arrow. (E) Schematic diagram of Drosophila and human UNR proteins (dUNR and hUNR, respectively). dUNR contains five CSDs and two glutamine-rich regions (Q). The amino acid numbers and the identity between the two UNR proteins are shown.
Because dSXL does not repress translation of msl-2 mRNA when tethered via a heterologous RNA-binding domain to a noncognate sequence in the 3′ UTR (Grskovic et al. 2003), we considered the possibility that stable dSXL/corepressor complex formation required the presence of the msl-2 EF RNA fragment (containing the binding sites for dSXL and polypeptides A and B). Overnight (0-12 h) embryo extract was loaded on glutathione-Sepharose columns containing GST-dRBD4, GST-mRBD, or GST, in the presence or absence of the EF RNA (Fig. 1C, left panel). Retention of polypeptides A and B in the column was evaluated by determining their presence in the column flowthrough using UV cross-linking to radiolabeled EF RNA and coimmunoprecipitation with dRBD4, as previously described (Grskovic et al. 2003). The results show that polypeptides A and B (Fig. 1C, labeled by arrowheads) are not retained in the dRBD4 column lacking EF RNA, as evidenced by their presence in the column flowthrough (Fig. 1C, lane 2). However, addition of EF RNA results in retention of the polypeptides (Fig. 1C, lane 3). mRBD or GST columns containing EF RNA do not retain the polypeptides (Fig. 1C, lanes 4,5), indicating that retention in the dRBD4 column is specific of the presence of the residues that mediate repression. The results also suggest that the formation of a repressive complex containing dSXL and putative corepressors requires dSXL-binding to the EF sites.
For preparative-scale purification, polypeptides A and B were enriched by ammonium sulfate precipitation of the embryo extract, prior to column chromatography (Fig. 1C, right panel). A 25% ammonium sulfate cut showed approximately fourfold enrichment, and was loaded onto GST-dRBD4 and GST-mRBD columns saturated with EF RNA (see Materials and Methods for details). After extensive washing, the bound proteins were eluted by digestion with TEV protease, which cleaves after the GST moiety, and were separated by SDS-PAGE and silver stained. An ∼130-kDa protein present in the GST-dRBD4 eluate was absent from the mRBD eluate (Fig. 1D, arrowhead). This protein was sequenced by MALDI-TOF mass spectrometry. The sequence of 12 peptides of eight to 16 amino acids was obtained, all of which matched to the Drosophila gene CG7015 of unknown function located on chromosome 3L. A homology search showed that the ∼130-kDa protein is the likely Drosophila homolog of mammalian UNR, showing an overall identity of 45% with its human counter-part (Fig. 1E). Drosophila UNR (dUNR) is a protein of 1039 amino acids containing five CSDs and two glutamine-rich regions. Although the predicted molecular weight of dUNR is 116 kDa, the highly basic N terminus is likely to account for its apparent slower mobility in SDS-PAGE (data not shown).
dUNR interacts with msl-2 mRNA
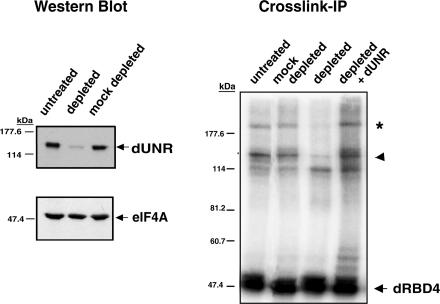
The size of dUNR suggested that it could correspond to polypeptide A. The species that we refer to as polypeptide B has been shown to be a complex of dSXL and an unidentified protein, which is not resolved by the RNase T1 treatment of the immunoprecipitation (IP) assay (Grskovic et al. 2003). To test whether dUNR corresponded to polypeptides A or B, we raised antibodies against the C terminus of dUNR (amino acids 396-1039) and used them to deplete Drosophila embryo extracts. Mock depletion with preimmune serum was carried out in parallel as a control. Western blot analysis showed that the extent of depletion exceeded 95% (Fig. 2, left panel). Depletion was specific because the translation initiation factor eIF4A, used as an internal control, was not depleted when α-dUNR antibodies were used, and there was no depletion of dUNR in mock-depleted extracts.
Figure 2.
dUNR interacts with msl-2 mRNA. Antibodies against dUNR were generated and used to deplete Drosophila embryo extracts. (Left panel) The extent of dUNR depletion was assessed by Western blot using eIF4A as a specificity control. (Right panel) The presence of the polypeptides A and B in the depleted extracts was tested by cross-link-IP, as indicated in the legend of Figure 1. The assay was performed in the absence or presence of 100 ng of purified recombinant dUNR. The arrowhead indicates the position of dUNR, while the asterisk indicates a complex between dUNR and dRBD4.
Cross-link-IP assays in depleted extracts showed that polypeptides A and B were absent from the dUNR-depleted extract, while they remained in the mock-depleted extract (Fig. 2, right panel). Significantly, both polypeptides were recovered upon addition of recombinant full-length dUNR to the depleted extract. These data show that polypeptide A is dUNR, while polypeptide B likely corresponds to a complex between dUNR and dSXL.
dUNR is a cytoplasmic protein present throughout development
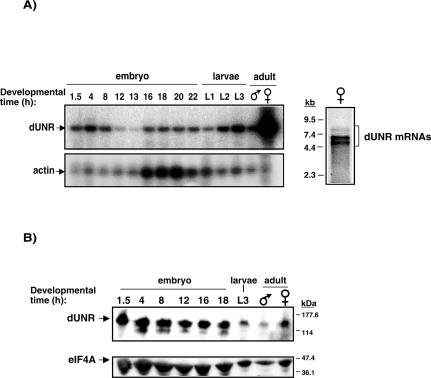
Dosage compensation is believed to operate in most—if not all—tissues and throughout development (Belote 1983). Therefore, if dUNR is required for the regulation of dosage compensation, its expression should be ubiquitous. To determine the distribution of dUNR during development, we performed semiquantitative RT-PCR analysis on total RNA samples obtained from embryos and larvae at different developmental stages, as well as adult flies. Coamplification of actin was carried as an internal control. Figure 3A shows that dUNR mRNA is present at all developmental stages, although it is strikingly more abundant in adult females. These results are consistent with those of Arbeitman et al. (2002) obtained using microarrays (http://genome.med.yale.edu/Lifecycle/query_gen.php?input1=CG7015).
Figure 3.
Developmental distribution of dUNR. (A, left panel) Semiquantitative RT-PCR analysis of dUNR mRNA during development. Amplification of actin mRNA within the same reactions was performed as a control. Amplified fragments were visualized by Southern blot. (A, right panel) Northern blot of female RNA using the dUNR full-length ORF as a probe. Molecular weight markers are indicated on the left. (B) Western blot of dUNR during Drosophila development. The amount of eIF4A was monitored as a loading control.
Northern blot of female fly RNA revealed the existence of three mRNAs that hybridize with the dUNR probe, probably resulting from differential pre-mRNA processing (Fig. 3A, right panel). Although dUNR mRNA is more abundant in females, Western blot analysis showed that comparable levels of dUNR protein are observed in both sexes, suggesting the existence of a sex-specific mechanism that equalizes dUNR expression (Fig. 3B, cf. dUNR levels and those of the loading control, eIF4A; see also Fig. 6C, below).
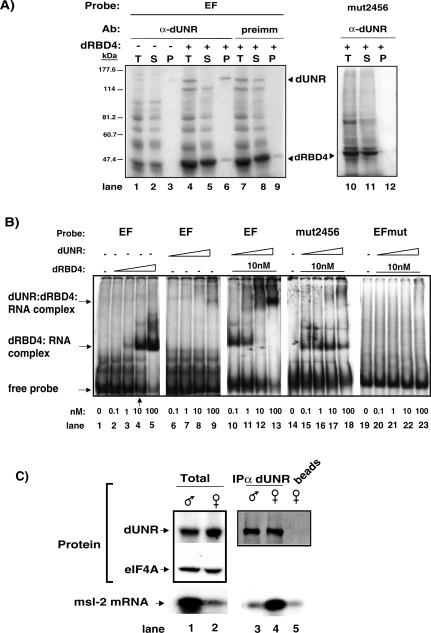
Figure 6.
Binding of dUNR to msl-2 mRNA requires dSXL. (A) Cross-link of endogenous dUNR to 32P-labeled EF RNA, and IP with α-dUNR antibodies. The total set of cross-linked proteins (T), as well as the supernatant (S) and pellet (P) fractions of the IP are shown. In some tubes, dRBD4 was added to the reaction before cross-linking. Precipitation with preimmune serum, and cross-linking with radiolabeled mut2456 RNA (lacking dUNR-binding sites) were carried as negative controls. The positions of dRBD4 and dUNR are indicated. (B) Gel mobility-shift assays of radiolabeled wild-type EF RNA or derivatives lacking either dSXL-binding sites (EFmut) or dUNR-binding sites (mut2456), in the absence or presence of increasing amounts of recombinant dUNR and dRBD4 proteins. The positions of the different complexes are indicated. (C) dUNR interacts with msl-2 mRNA in female, but not male, flies. (Upper panel) Western blot of dUNR in total extracts from adult male and female flies (lanes 1,2), and after IP with α-dUNR serum (lanes 3,4) or beads alone (lane 5). The amount of eIF-4A was measured as a loading control. (Lower panel) Semiquantitative RT-PCR analysis of msl-2 mRNA from total (1 μg) male and female RNA (lanes 1,2) and equivalent volumes of RNA extracted from the IP shown in the upper panel (lanes 3-5). Amplified fragments were visualized by Southern blot.
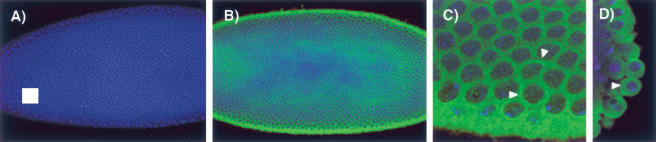
To determine the localization of dUNR within the cell, immunostaining of Drosophila embryos at the blastoderm stage was performed (Fig. 4). While staining with the preimmune serum failed to show any signal, staining with α-dUNR antibodies showed strong cytoplasmic staining (Fig. 4A,B, respectively). At higher resolution, a concentration of dUNR around the nucleus was observed, both in somites and in pole cells (Fig. 4C,D).
Figure 4.
Immunostaining of dUNR. Blastoderm-stage Drosophila embryos were incubated with preimmune serum (A) or α-dUNR serum (B-D) (green). DNA was stained using TOPRO3 (blue). Identical exposures were done for all pictures. The arrowhead indicates the stronger fluorescence intensity around the nucleus in somites (C) and pole cells (D).
dUNR interacts with dSXL
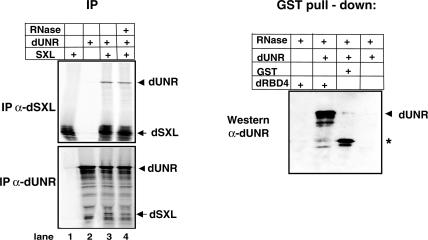
We next tested whether dUNR interacted with dSXL. We first used coimmunoprecipitation of 35S-labeled dSXL and dUNR translated in rabbit reticulocyte lysates with either α-dUNR or α-dSXL antibodies. These two antibodies did not show cross-reactivity, indicating that IP was specific (Fig. 5, left panel, lanes 1,2). However, when the two proteins were mixed, α-dSXL antibodies immunoprecipitated dUNR and, conversely, α-dUNR antibodies immunoprecipitated dSXL (Fig. 5, lane 3). Interestingly, coimmunoprecipitation of the two proteins persisted in the presence of RNase A (Fig. 5, lane 4), implying that the interaction between dUNR and dSXL may occur in the absence of RNA. To further substantiate this finding, we performed a GST pull-down assay using purified recombinant dUNR and either GST-dRBD4 or GST as a control, in the presence of RNase A. Consistent with the coimmunoprecipitation data, the GST pull-down assay showed specific retention of dUNR in the GST-dRBD4 column, indicating that dUNR interacts with dSXL directly (Fig. 5, right panel). We conclude that dUNR and dSXL interact with each other. While this interaction can occur in the absence of RNA, stable complex formation in the competitive conditions of the Drosophila extract probably requires msl-2 mRNA, thus explaining the results of Figure 1C.
Figure 5.
dUNR interacts with dSXL. (Left panel) Coimmunoprecipitation of in vitro-translated dUNR and dSXL proteins with either α-dSXL or α-dUNR antibodies, in the absence or presence of RNase A. (Right panel) Pull-down of purified recombinant dUNR with glutathione-agarose beads containing either GST or GST-dRBD4. The assay was performed in the presence of RNase A. The position of dUNR is indicated. The asterisk denotes a bacterial contaminant present in the GST preparation that cross-reacts with the α-dUNR antibody.
dUNR binding to msl-2 mRNA requires dSXL
We next tested the interaction of endogenous dUNR with the relevant sequences of msl-2 mRNA by using UV-cross-link to radiolabeled EF RNA and IP with α-dUNR antibodies. In the absence of dRBD4, dUNR cross-linked poorly to a wild-type substrate, as only a weak signal was detected in the total (T) and pellet (P) fractions of the IP (Fig. 6A, lanes 1,3). Addition of dRBD4 dramatically improved dUNR cross-linking to the RNA, as the cross-linked polypeptide was precipitated with α-dUNR antibodies (Fig. 6A, lanes 4-6). The signal remained in the supernatant when the preimmune serum was used, indicating that the IP was specific (Fig. 6A, lanes 7-9). Endogenous dUNR is, indeed, likely to bind to the sequences adjacent to the dSXL-binding sites in the 3′ UTR of msl-2, because dUNR cross-linking was lost when these sequences were mutated (Fig. 6A, lanes 10-12; see Fig. 1A and Materials and Methods for a description of the probes). These data suggest that dUNR associates with msl-2 mRNA in a dSXL-dependent manner.
To confirm this observation, the binding of purified recombinant dRBD4 and dUNR to EF RNA was analyzed by gel mobility-shift assays. As previously reported, dRBD4 bound to EF RNA with an apparent Kd of ∼10 nM (Fig. 6B, lanes 1-5; Gebauer et al. 1998; Grskovic et al. 2003). Similar concentrations of dUNR failed to bind to the RNA (Fig. 6B, lanes 6-9). However, in the presence of 10 nM dRBD4, a supershift was observed upon titration of dUNR (Fig. 6B, lanes 10-13). Consistent with the supershift being due to dUNR binding to the RNA, it was not observed with a probe containing mutated dUNR-binding sites (Fig. 6B, lanes 14-18, mut2456). Importantly, a probe with mutated dSXL-binding sites (EFmut) failed to support both dSXL and dUNR binding, implying that dSXL binding to msl-2 mRNA is a prerequisite for dUNR binding (Fig. 6B, lanes 19-23).
A prediction from these results is that the interaction between dUNR and msl-2 mRNA should only be detected in female flies, which contain dSXL. To test this prediction, dUNR was immunoprecipitated from either adult male or female extracts, and msl-2 mRNA was amplified by RT-PCR from the RNAs present in the immunoprecipitate. Figure 6C (lanes 1,2) shows that, while the amount of dUNR in extracts from both sexes was comparable, msl-2 mRNA was more abundant in male flies, as previously reported (Zhou et al. 1995; Kelley et al. 1997). Despite its relative abundance, only background levels of msl-2 mRNA were detected in dUNR immunoprecipitates obtained from male flies (Fig. 6C, cf. lanes 3 and 5). In contrast, msl-2 mRNA was readily detected in dUNR immunoprecipitates obtained from female flies (Fig. 6C, lane 4). Taken together, these results show that dUNR binds to msl-2 mRNA in female, but not male, flies and that dUNR binding to msl-2 mRNA requires dSXL interaction with the adjacent uridine-rich sequences.
dUNR is required for translational repression of msl-2 mRNA
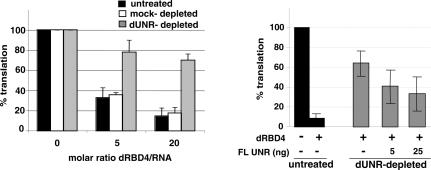
To evaluate the relevance of the dSXL:dUNR interaction for msl-2 mRNA translational regulation, we established dUNR-depleted extracts that were competent for translation. We then tested the ability of dSXL to inhibit the translation of a reporter mRNA containing the minimal regulatory sequences of msl-2 mRNA (BLEF mRNA) (see Fig. 1A) in the depleted extracts. While addition of dSXL to untreated extracts inhibited BLEF mRNA translation by ∼10-fold, immunodepletion of dUNR largely impaired translational repression by dSXL (Fig. 7, left panel, cf. black and gray bars). Furthermore, the response to dSXL was unmodified in mock-depleted extracts (white bars). Significantly, addition of recombinant dUNR to the depleted extracts partially restored dSXL-mediated translational repression (Fig. 7, right panel). We conclude that dUNR is required for dSXL-mediated translational repression of msl-2 mRNA.
Figure 7.
Drosophila UNR is required for translational repression of msl-2 mRNA. (Left panel) BLEF mRNA was incubated in typical translation reactions containing increasing amounts of dRBD4 and either untreated (black bars), dUNR-depleted (gray bars), or mock-depleted (white bars) extracts. Firefly luciferase values were corrected for cotranslated Renilla expression and plotted against the molar ratio dRBD4/RNA. The activity obtained in the absence of recombinant protein was taken as 100%. (Right panel) Translation inhibition by dSXL is restored upon addition of recombinant dUNR. Translation reactions were assembled with dUNR-depleted extracts in the absence or presence of increasing amounts of recombinant dUNR. Untreated extracts were used as a reference (black bars). Where indicated, 20-fold molar excess of dRBD4 over reporter BLEF RNA was used.
Discussion
Inhibition of msl-2 expression is essential for development of female flies, as forced expression of MSL-2 causes the assembly of the DCC on both X chromosomes and lethality (Kelley and Kuroda 1995). Here we show that the Drosophila homolog of mammalian UNR is necessary to inhibit msl-2 expression. dUNR is recruited to the 3′ UTR of msl-2 mRNA by dSXL, a female-specific protein, and plays an essential role in repressing its translation. Our data are fully consistent with those of Duncan et al. (2006), showing that dUNR associates to the 3′ UTR of msl-2 mRNA in female cells and is necessary to repress msl-2 translation in vivo. Together, our data identify dUNR as a regulator of dosage compensation.
In vitro selection experiments (SELEX) indicate that hUNR binds to purine-rich regions in the mRNA, with the consensus sequences (A/G)5AAGUA/G or (A/G)8AACG and an apparent dissociation constant (Kd) of ∼10 nM (Triqueneaux et al. 1999). Although dUNR also recognizes purine-rich sequences in the 3′ UTR of msl-2 mRNA that fall within these consensus, it does so with a very poor affinity (Fig. 6B), a situation reminiscent to that of the bacterial cold-shock proteins (Ermolenko and Makhatadze 2002). Binding of dUNR to msl-2 mRNA requires the binding of dSXL (Fig. 6). The observation that msl-2 RNA fragments containing mutated dSXL-binding sites, but wild-type dUNR-binding sites, do not bind to either of the two proteins suggests that dSXL does not simply induce a conformational change in dUNR that allows it to bind RNA (Fig. 6B, EFmut). Rather, dSXL recruits dUNR to bind in close proximity in the 3′ UTR of msl-2 mRNA. Stable recruitment of dUNR requires the interaction of dUNR with both dSXL and msl-2 mRNA, as supported by the following evidence. First, dUNR is not retained in the dRBD4 column unless this column is saturated with msl-2 mRNA (Fig. 1C). Second, dUNR is not retained in the mRBD column despite the presence of msl-2 mRNA (Fig. 1C). Third, no complex formation is observed in a gel mobility-shift assay when the dUNR-binding sites are mutated (Fig. 6B, mut2456). Fourth, msl-2 mRNA and dUNR do not interact in male flies, which lack dSXL (Fig. 6C). Nevertheless, dUNR and dSXL can interact directly in vitro (Fig. 5). Addition of EF RNA does not improve this interaction, and addition of embryo extract actually competes it (data not shown). These results suggest that, although the interaction of dSXL and dUNR can occur directly, the interaction with msl-2 mRNA stabilizes the complex in the competitive conditions of the extract.
As mentioned above, dUNR protein and msl-2 mRNA do not interact in male flies despite their relative abundance (Fig. 6C). In addition, supplementing cytoplasmic embryo extracts with dSXL—a primarily nuclear protein (Bopp et al. 1991)—promotes dUNR association with msl-2 mRNA (Fig. 6A), and translational repression by dUNR is only observed in dSXL-containing cells (Duncan et al. 2006). These data suggest that the interaction of dUNR with msl-2 mRNA is mediated by dSXL in vivo, and imply that dSXL is the critical determinant for the formation of a repressive complex on the 3′ UTR of msl-2 mRNA (Fig. 8). In this scenario, dSXL conveys a sex-specific function to dUNR. The stepwise assembly of a translation inhibitory complex on msl-2 mRNA is reminiscent of Drosophila hunchback. The 3′ UTR of maternal hunchback mRNA is bound by Pumilio (Pum), and this event triggers the sequential recruitment of Nanos (Nos) and Brain tumor (Brat), which ultimately results in the translational repression of hunchback mRNA (Sonoda and Wharton 1999, 2001). Sequential binding of dSXL and dUNR to msl-2 mRNA could result from their respective subcellular locations: While dSXL is nuclear and associates with msl-2 pre-mRNA, dUNR is primarily—if not exclusively—cytoplasmic (Fig. 4). Interestingly, dUNR accumulates at the nuclear periphery, which perhaps reflects or ensures the rapid formation of repressive complexes as msl-2 and probably other mRNAs are exported to the cytoplasm. Additionally, accumulation around the nucleus could reflect the association of dUNR with the endoplasmic reticulum, as reported for mammalian UNR (Jacquemin-Sablon et al. 1994).
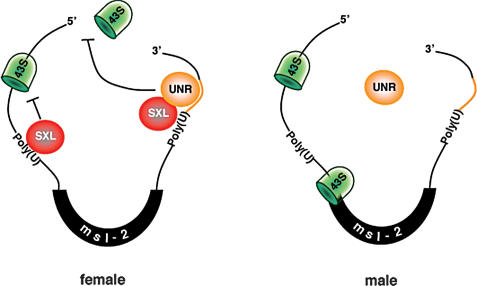
Figure 8.
Model for the role of Drosophila UNR in the regulation of msl-2 mRNA translation. The female-specific protein SXL binds to poly(U) stretches in the 5′ and 3′ UTRs of msl-2 mRNA. SXL bound to the 3′ UTR recruits UNR to adjacent purine-rich sequences. The SXL/UNR complex inhibits the association of the 43S ribosomal complex with the 5′ end of the mRNA, thereby repressing translation. SXL bound to the 5′ UTR inhibits the scanning of ribosomal complexes that may have escaped the UNR block (Beckmann et al. 2005). In male flies, which lack SXL, UNR does not bind to the 3′ UTR of msl-2 mRNA and translation proceeds.
Dosage compensation is believed to function from the blastoderm stage (Rastelli et al. 1995; Franke et al. 1996). As expected for a protein involved in the regulation of dosage compensation, dUNR is present throughout development (Fig. 3). Curiously, although dUNR mRNA is dramatically more abundant in female flies, this difference is compensated at the protein level, suggesting the existence of sex-specific mechanisms to modulate dUNR expression (Figs. 3, 6C). Indeed, the amount of dUNR might need to be tightly controlled. Overexpression of mammalian UNR leads to cell death (Cornelis et al. 2005), and our preliminary data suggest that substantial overexpression of dUNR results in lethality of both male and female flies (S. Patalano and F. Gebauer, unpubl.). Three forms of dUNR mRNA can be detected in Drosophila (Fig. 3A). Several mRNAs have also been detected in mammals (Ferrer et al. 1999), consistent with the observation of three alternative polyadenylation sites of the hUNR gene (Jeffers et al. 1990) and alternative splicing of the hUNR pre-mRNA (Boussadia et al. 1993). These data suggest that the different dUNR mRNAs arise by alternative processing, although the significance of this observation remains to be explored.
The role of UNR in Drosophila contrasts with the known functions of UNR in mammals. hUNR is part of a complex assembled on the coding region of c-fos mRNA that is involved in the deadenylation-dependent destabilization of this transcript (Grosset et al. 2000). The interaction of hUNR with PABP within this complex is believed to bridge the complex to the poly(A) tail, although the mechanism by which the complex influences deadenylation is unknown (Chang et al. 2004). In Drosophila, the steady-state levels of msl-2 mRNA are, indeed, lower in females (Fig. 6C; Zhou et al. 1995; Kelley et al. 1997). However, we could not observe any effect of dUNR and dSXL on msl-2 mRNA stability in our translation assays (Gebauer et al. 1998; data not shown). hUNR also binds to the IRES elements in the 5′ UTRs of several transcripts and activates their translation. In the best understood example, that of Apaf-1 mRNA, hUNR induces a conformational change in the IRES that makes it accessible for binding of PTB, a positive regulator of Apaf-1 translation (Mitchell et al. 2003). Contrary to hUNR, dUNR binds to the 3′ UTR of msl-2 mRNA and represses its translation (Figs. 6, 7; see also Duncan et al. 2006). Nonetheless, the underlying effects of dUNR binding may be similar if dUNR acts as an RNA or RNP chaperone to facilitate an RNA conformation, or the assembly of repressive factors, that inhibit translation.
Translation of msl-2 occurs via a cap-dependent mechanism (Gebauer et al. 2003). Cap-dependent translation initiation involves the recruitment of 43S ribosomal complexes (molecular assemblies of the 40S ribosomal subunit with a set of translation initiation factors and the initiator tRNA) to the cap structure at the 5′ end of the mRNA. Translation inhibition mediated by the 3′ UTR of msl-2 results from a block of 43S ribosomal recruitment (Beckmann et al. 2005). However, translational repression of msl-2 mRNA by dSXL can occur in the absence of a cap structure and a poly(A) tail (Gebauer et al. 1999, 2003). Understanding how 43S recruitment is affected by dSXL without the involvement of the cap is, indeed, intriguing. A possibility is that, as with mammalian UNR, dUNR interacts with PABP (Chang et al. 2004). PABP could, in turn, exert a poly(A)- and cap-independent effect on translation (Kahvejian et al. 2005). Certainly, the mapping of dUNR domains relevant for translational control and the identification of dedicated factors that interact with dUNR are likely to provide insights into this mechanism of translation regulation that is key to control dosage compensation in Drosophila.
Materials and methods
Sequences
The RNA probes used in this study contained the following relevant sequences: EF, UUUUUUUGAGCAUGAAUUUUUUUGAGCACGUGAACCUAGGAUUAAG; EFmut, CUCUCUCUAGCAUGAACUCUCUCUAGCACGUGAACCUAGGAUUAAG; and mut2456, UUUUUUUGCUCUCUCAUUUUUUUGCUCUCUCUCUCUCUCUCUCUCU.
Purification of dUNR
Ten milliliters of Drosophila 0-12-h embryo extract (155 mg of total protein) were precipitated by saturation with ammonium sulfate according to the protocol described by Englard and Sefert (1990). Precipitates were resuspended in 0.56 mL of buffer DE (10 mM HEPES at pH 7.5, 5 mM DTT) supplemented with protease inhibitor cocktail (Boehringer), and tested for the presence of the putative corepressors by UV-cross-link to radiolabeled EF RNA and coimmunoprecipitation with dRBD4 using α-dSXL antiserum.
GST-dRBD4 and GST-mRBD columns were prepared as follows: Four-hundred micrograms of the respective recombinant proteins were incubated with 100 μg of EF RNA on ice for 20 min. Four-hundred microliters of a (1:1) slurry of glutathione-Sepharose beads previously equilibrated in buffer T [30 mM HEPES at pH 8.0, 0.6 mM Mg(OAc)2, 80 mM KOAc] were added, and the suspension was incubated for 20 min at 4°C. The suspension was loaded on a column and washed with 5 volumes of buffer T supplemented with 3 mg/mL heparin (buffer TH). Two-hundred-ten microliters of the 25% ammonium sulfate cut (1 mg of protein) were adjusted to buffer T and loaded four times over either the GST-dRBD4 or the GST-mRBD columns. After washing three times with 5 column volumes of buffer TH, 30 μg of TEV protease were added in a buffer containing 30 mM HEPES (pH 8.0), 1 mM EDTA, and 2 mM DTT, and the columns were incubated for 1 h at 30°C. The columns were spun to obtain the eluates, and the proteins were separated in an 8% acrylamide gel and visualized by silver staining.
DNA constructs and antibody generation
The N-terminal fragment of dUNR (nucleotides 1-1190) was amplified from embryo RNA using oligonucleotides designed to introduce a Flag tag at the N terminus, and cloned into pGEM-T (Promega). The C-terminal fragment of dUNR (nucleotides 1148-3120) was obtained from the Berkeley Drosophila Genome Project (clone LD3025) and was also subcloned into pGEM-T. Flag-tagged full-length dUNR cDNA (FL) was obtained by subcloning the inserts of these plasmids into the NdeI and XhoI sites of the Escherichia coli expression vector pET-15b (Novagen).
The C-terminal fragment of dUNR (amino acids 396-1039) was also subcloned into the vector pET-15b, expressed as a His-tagged fusion and used to generate polyclonal antibodies in rabbits. The α-dUNR serum was characterized by Western blot and immunoprecipitation of recombinant dUNR and Drosophila embryo extracts.
Protein expression and purification
dSXL derivatives were expressed in E. coli as N-terminal GST-tagged fusions and purified as described previously (Grskovic et al. 2003). His-tagged, full-length dUNR was purified following the pET system user's manual (Novagen). Proteins were dialyzed against buffer D (20 mM HEPES at pH 8.0, 20% glycerol, 1 mM DTT, 0.01% NP-40, 0.2 mM EDTA).
In vitro transcription and translation
mRNAs were synthesized as described previously (Gebauer et al. 1999). All mRNAs contained a 5′ 7mGpppG cap and a poly(A) tail of 73 residues.
In vitro translation reactions in Drosophila embryo extracts were performed in a final volume of 10 μL as described (Gebauer et al. 1999). BLEF and Renilla luciferase mRNAs were added at a final concentration of 20 ng/μL and 10 ng/μL, respectively. The translation efficiency was determined by measuring the luciferase activity using the Dual Luciferase Assay System (Promega), and Firefly luciferase values were corrected for Renilla expression.
UV-cross-link and IP
UV-cross-link and IP were performed essentially as described (Grskovic et al. 2003). Briefly, 32P-labeled EF RNA or its mutated derivatives were incubated in a typical translation reaction containing 50 ng of dRBD4. After 30 min of incubation at 25°C, reactions were cross-linked at 254 nm and 600 mJ on ice, and digested with RNase T1 or A. Proteins were immunoprecipitated with α-dSXL or α-dUNR antibodies, or with dUNR preimmune serum bound to protein A-Sepharose beads. Unless otherwise indicated, proteins were separated by 8% SDS-PAGE and visualized using a PhosphorImager.
For interaction assays, full-length dSXL and dUNR were expressed as 35S-labeled proteins using the TnT expression system (Promega) following the instructions of the manufacturer. Samples were mixed, treated with 10 μg of RNase A for 30 min at 25°C and immunoprecipitated as indicated above.
The IP and selection of RNA shown in Figure 6C were performed as follows. Male or female protein extract (1.5 mg) were incubated with 50 μL of protein A beads either alone or coupled with α-dUNR antibodies for 1 h at 4°C. The beads were extensively washed, and the RNA was extracted from the immunoprecipitates using the Trizol reagent (Invitrogen) and precipitated. Samples were resuspended in H2O, and the DNA contaminant was removed using Nucleospin RNA II columns (Macherey-Nagel). Equivalent sample volumes were used for RT-PCR using oligonucleotides that amplified positions 813-969 of the msl-2 3′ UTR. Amplified fragments were separated on a 2% agarose gel, transferred to a Hybond-N+ membrane (Amersham), and visualized by Southern blot using a labeled oligonucleotide as a probe.
GST pull-down
GST pull-down assays were carried out in a volume of 20 μL containing 1 μg of GST or GST-dRBD4 and 2 μg of dUNR in buffer D (20 mM HEPES at pH 8.0, 20% glycerol, 1 mM DTT, 0.01% NP-40, 0.2 mM EDTA). After incubation at 25°C for 30 min, 10 μg of RNase A were added and the incubation was extended for 30 min. Ten microliters of a (1:1) slurry of glutathione-agarose beads (Sigma) and 30 μL of capture buffer (600 mM NaCl, 20 mM HEPES at pH 8.0, 100 mM KOAc, 0.2 mM EDTA, 0.02% NP-40) were added, and the mixture was incubated for 2 h at 4°C on a rotating wheel. The beads were washed four times with 1 mL of capture buffer, and the retained proteins were fractionated on an 8% SDS-polyacrylamide gel.
Northern blot analysis and RT-PCR
Total RNA was extracted from wild-type Drosophila staged animals using the Trizol reagent. Poly(A)+-enriched RNA was obtained using oligo(dT)-cellulose beads. For Northern blots, 8 μg of poly(A)+-enriched RNA were resolved in a 1% agarose denaturing gel, transferred, and hybridized as described (Church and Gilbert 1984) using a random-primed probe against the full-length dUNR ORF. RT-PCR was performed from 1.5 μg of total RNA using AMV retro-transcriptase and the PCR Master Mix (Promega) with oligonucleotides complementary to positions 2420 and 3120 of the dUNR ORF. Actin mRNA was coamplified as an internal control. Amplified fragments were separated on a 2% agarose gel and visualized by Southern blot using labeled oligonucleotides as probes.
Western blot
Protein preparations were obtained from wild-type Drosophila staged animals, and 80 μg were separated by 8% SDS-PAGE and transferred to a PVDF membrane. α-dUNR and α-eIF4A (a kind gift of G. Hernández, Institute for Biochemistry and Molecular Biology, University of Bern, Bern, Switzerland) sera were used at 1:500 and 1:2500 dilutions, respectively, and proteins were visualized by chemiluminescence (ECL; Amersham).
Gel mobility-shift assays
32P-labeled EF RNA or its mutated derivatives were incubated with increasing amounts of recombinant dRBD4 and/or dUNR, and processed as described previously (Valcarcel et al. 1993).
Immunodepletions
To obtain the depleted extracts used in Figure 2, protein A-Sepharose beads were blocked with BSA and covalently coupled α-dUNR antiserum. Beads were incubated with Drosophila embryo extracts at a (1:2) volume ratio of beads to extract for 1 h at 4°C. The supernatant was recovered and the procedure was repeated with fresh beads for a total of six times. The resulting depleted extracts were competent for cross-link-IP assays, although translationally inactive.
To obtain translationally active depleted extracts, magnetic protein A-Dynabeads (Biotech) were used. Three-hundred microliters of magnetic beads were blocked with 1.7 mg of Drosophila embryo extract for 1 h at room temperature. Beads were washed with a buffer containing 10 mM HEPES (pH 8.0) and 8% glycerol and coupled to 120 μg of purified α-dUNR IgGs. Two rounds of depletion using a (1:1) ratio of beads to extract were performed by incubation for 1 h at 4°C. The efficiency of depletion was monitored by Western blot.
Immunostaining
Immunostaining was performed following fixation with formaldehyde as described (Rotwell and Sullivan 2000). α-dUNR serum was used at a 1:100 dilution. As a secondary, anti-rabbit IgG coupled to Alexa 488 (Molecular Probes) was used at a dilution of 1:500. DNA was stained with TOPRO3 (Molecular Probes), and embryos were visualized using a Leica SP2 confocal microscope.
Acknowledgments
We thank Greco Hernández for the α-eIF4A antibody, Juan Valcárcel for the α-dSXL antibody, Alain Debec and Salud Llamazares for advise on immunostaining, and members of the Hentze laboratory for reagents and discussions. We also thank Juan Valcárcel, Raúl Méndez, Rafael Cuesta, Marica Grskovic, Matthias W. Hentze, and Kent Duncan for carefully reading this manuscript. I.A. was supported by a FI-IQUC fellowship from the Generalitat of Catalunya. This work was supported by grant BMC2003-04108 from the Spanish Ministry of Science and Technology.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.371906.
References
- Arbeitman M.N., Furlong, E.E., Imam, F., Johnson, E., Null, B.H., Baker, B.S., Krasnow, M.A., Scott, M.P., Davis, R.W., and White, K.P. 2002. Science 297: 2270-2275. [DOI] [PubMed] [Google Scholar]
- Bashaw G.J. and Baker, B.S. 1997. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell 89: 789-798. [DOI] [PubMed] [Google Scholar]
- Beckmann K., Grskovic, M., Gebauer, F., and Hentze, M.W. 2005. A dual inhibitory mechanism for ribosomal 43S complex recruitment and scanning restricts msl-2 mRNA translation for dosage compensation in Drosophila. Cell 122: 529-540. [DOI] [PubMed] [Google Scholar]
- Belote J.M. 1983. Male-specific lethal mutations of Drosophila melanogaster. II. Parameters of gene action during male development. Genetics 105: 881-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp D., Bell, L.R., Cline, T.W., and Schedl, P. 1991. Developmental distribution of female-specific Sex-lethal proteins in Drosophila melanogaster. Genes & Dev. 5: 403-415. [DOI] [PubMed] [Google Scholar]
- Boussadia O., Jacquemin-Sablon, H., and Dautry, F. 1993. Exon skipping in the expression of the gene immediately upstream of N-ras (unr/NRU). Biochim. Biophys. Acta 1172: 64-72. [DOI] [PubMed] [Google Scholar]
- Boussadia O., Amiot, F., Cases, S., Triqueneaux, G., Jacquemin-Sablon, H., and Dautry, F. 1997. Transcription of unr (upstream of N-ras) down-modulates N-ras expression in vivo. FEBS Lett. 420: 20-24. [DOI] [PubMed] [Google Scholar]
- Boussadia O., Niepmann, M., Creancier, L., Prats, A.C., Dautry, F., and Jacquemin-Sablon, H. 2003. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 77: 3353-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.C., Yamashita, A., Chen, C.Y., Yamashita, Y., Zhu, W., Durdan, S., Kahvejian, A., Sonenberg, N., and Shyu, A.B. 2004. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes & Dev. 18: 2010-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G.M. and Gilbert, W. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. 81: 1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis S., Tinton, S.A., Schepens, B., Bruynooghe, Y., and Beyaert, R. 2005. UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein. Nucleic Acids Res. 33: 3095-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K., Grskovic, M., Strein, C., Beckmann, K., Niggeweg, R., Abaza, I., Gebauer, F., Wilm, M., and Hentze, M.W. 2006. Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: Translational repression for dosage compensation. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Englard S. and Seifter, S. 1990. Precipitation techniques. Methods Enzym. 182: 285-300. [DOI] [PubMed] [Google Scholar]
- Ermolenko D.N. and Makhatadze, G.I. 2002. Bacterial cold-shock proteins. Cell. Mol. Life Sci. 59: 1902-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J.R., Mitchell, S.A., Spriggs, K.A., Ostrowski, J., Bomsztyk, K., Ostarek, D., and Willis, A.E. 2003. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene 22: 8012-8020. [DOI] [PubMed] [Google Scholar]
- Ferrer N., Garcia-Espana, A., Jeffers, M., and Pellicer, A. 1999. The unr gene: Evolutionary considerations and nucleic acid-binding properties of its long isoform product. DNA Cell Biol. 18: 209-218. [DOI] [PubMed] [Google Scholar]
- Franke A., Dernburg, A., Bashaw, G.J., and Baker, B.S. 1996. Evidence that MSL-mediated dosage compensation in Drosophila begins at blastoderm. Development 122: 2751-2760. [DOI] [PubMed] [Google Scholar]
- Gebauer F., Merendino, L., Hentze, M., and Valcarcel, J. 1998. The Drosophila splicing regulator sex-lethal directly inhibits translation of male-specific-lethal 2 mRNA. RNA 4: 142-150. [PMC free article] [PubMed] [Google Scholar]
- Gebauer F., Corona, D., Preiss, T., Becker, P., and Hentze, M. 1999. Translational control of dosage compensation in Drosophila by Sex-lethal: Cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A). EMBO J. 18: 6146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F., Gskovic, M., and Hentze, M. 2003. Drosophila Sex-lethal inhibits the stable association if the 40S ribosomal subunit with msl-2 mRNA. Mol. Cell 11: 1397-1404. [DOI] [PubMed] [Google Scholar]
- Gilfillan G.D., Dahlsveen, I.K., and Becker, P.B. 2004. Lifting a chromosome: Dosage compensation in Drosophila melanogaster. FEBS Lett. 567: 8-14. [DOI] [PubMed] [Google Scholar]
- Graumann P.L. and Marahiel, M.A. 1998. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 23: 286-290. [DOI] [PubMed] [Google Scholar]
- Grosset C., Chen, C.Y., Xu, N., Sonenberg, N., Jacquemin-Sablon, H., and Shyu, A.B. 2000. A mechanism for translationally coupled mRNA turnover: Interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103: 29-40. [DOI] [PubMed] [Google Scholar]
- Grskovic M., Hentze, M.W., and Gebauer, F. 2003. A co-repressor assembly nucleated by Sex-lethal in the 3′UTR mediates translational control of Drosophila msl-2 mRNA. EMBO J. 22: 5571-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada F.N., Park, P.J., Gordadze, P.R., and Kuroda, M.I. 2005. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes & Dev. 19: 2289-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt S.L., Hsuan, J.J., Totty, N., and Jackson, R.J. 1999. Unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes & Dev. 13: 437-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemin-Sablon H., Triqueneaux, G., Deschamps, S., le Maire, M., Doniger, J., and Dautry, F. 1994. Nucleic acid binding and intracellular localization of unr, a protein with five cold shock domains. Nucleic Acids Res. 22: 2643-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers M., Paciucci, R., and Pellicer, A. 1990. Characterization of unr; a gene closely linked to N-ras. Nucleic Acids Res. 18: 4891-4899. [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A., Svitkin, Y.V., Sukarieh, R., M'Boutchou, M.N., and Sonenberg, N. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes & Dev. 19: 104-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R.L. and Kuroda, M. 1995. Equality for X chromosomes. Science 270: 1607-1610. [DOI] [PubMed] [Google Scholar]
- Kelley R., Wang, J., Bell, L., and Kuroda, M. 1997. Sex-lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387: 195-199. [DOI] [PubMed] [Google Scholar]
- Kuersten S. and Goodwin, E.B. 2003. The power of the 3′ UTR: Translational control and development. Nat. Rev. Genet. 4: 626-637. [DOI] [PubMed] [Google Scholar]
- Mitchell S.A., Spriggs, K.A., Coldwell, M.J., Jackson, R.J., and Willis, A.E. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11: 757-771. [DOI] [PubMed] [Google Scholar]
- Rastelli L., Richman, R., and Kuroda, M.I. 1995. The dosage compensation regulators MLE, MSL-1 and MSL-2 are interdependent since early embryogenesis in Drosophila. Mech. Dev. 53: 223-233. [DOI] [PubMed] [Google Scholar]
- Rotwell W.F. and Sullivan, W. 2000. Fluorescent analysis of Drosophila embryos. In Drosophila protocols (eds. W. Sullivan et al.), pp. 141-157. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sonoda J. and Wharton, R.P. 1999. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes & Dev. 13: 2704-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001. Drosophila Brain tumor is a translational repressor. Genes & Dev. 15: 762-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T., Gilfillan, G.D., Maier, V.K., and Becker, P.B. 2005. The Drosophila MSL complex activates the transcription of target genes. Genes & Dev. 19: 2284-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinton S.A., Schepens, B., Bruynooghe, Y., Beyaert, R., and Cornelis, S. 2005. Regulation of the cell-cycle-dependent internal ribosome entry site of the PITSLRE protein kinase: Roles of Unr (upstream of N-ras) protein and phosphorylated translation initiation factor eIF-2α. Biochem. J. 385: 155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triqueneaux G., Velten, M., Franzon, P., Dautry, F., and Jacquemin-Sablon, H. 1999. RNA binding specificity of UNR, a protein with five cold shock domains. Nucleic Acids Res. 27: 1926-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcarcel J., Singh, R., Zamore, P.D., and Green, M.R. 1993. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 362: 171-175. [DOI] [PubMed] [Google Scholar]
- Wilhelm J.E. and Smibert, C.A. 2005. Mechanisms of translational regulation in Drosophila. Biol. Cell 97: 235-252. [DOI] [PubMed] [Google Scholar]
- Zhou S., Yang, Y., Scott, M.J., Pannuti, A., Fehr, K.C., Eisen, A., Koonin, E.V., Fouts, D.L., Wrightsman, R., Manning, J.E., et al. 1995. Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. EMBO J. 14: 2884-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]