Abstract
Sodalis glossinidius is a maternally transmitted endosymbiont of tsetse flies (Glossina spp.), an insect of medical and veterinary significance. Analysis of the complete sequence of Sodalis' chromosome (4,171,146 bp, encoding 2,432 protein coding sequences) indicates a reduced coding capacity of 51%. Furthermore, the chromosome contains 972 pseudogenes, an inordinately high number compared with that of other bacterial species. A high proportion of these pseudogenes are homologs of known proteins that function either in defense or in the transport and metabolism of carbohydrates and inorganic ions, suggesting Sodalis' degenerative adaptations to the immunity and restricted nutritional status of the host. Sodalis possesses three chromosomal symbiosis regions (SSR): SSR-1, SSR-2, and SSR-3, with gene inventories similar to the Type-III secretion system (TTSS) ysa from Yersinia enterolitica and SPI-1 and SPI-2 from Salmonella, respectively. While core components of the needle structure have been conserved, some of the effectors and regulators typically associated with these systems in pathogenic microbes are modified or eliminated in Sodalis. Analysis of SSR-specific invA transcript abundance in Sodalis during host development indicates that the individual symbiosis regions may exhibit different temporal expression profiles. In addition, the Sodalis chromosome encodes a complete flagella structure, key components of which are expressed in immature host developmental stages. These features may be important for the transmission and establishment of symbiont infections in the intra-uterine progeny. The data suggest that Sodalis represents an evolutionary intermediate transitioning from a free-living to a mutualistic lifestyle.
Through diverse processes and complex interactions, beneficial microbes contribute to the health, evolution, and diversity of ecosystems. Despite their wide occurrence, insights into the genomic and functional aspects of their biology are only recently accumulating. Members of the Class Insecta are among the taxa that have succeeded in often resource-limited and inhospitable ecological terrains, due in part to fitness advantages granted by symbiosis. Tsetse flies (Diptera: Glossinidia), vectors of pathogenic African trypanosomes that cause sleeping sickness in humans and nagana in animals, rely on symbiotic flora for important physiological processes such as fecundity.
Sodalis glossinidius is one of three vertically transmitted microbial symbionts in tsetse (Dale and Maudlin 1999; Aksoy 2000). This enteric organism resides primarily intra- and extracellularly in the host midgut, but can also be detected in hemolymph (Cheng and Aksoy 1999). Sodalis infections have been implicated in enhancing trypanosome susceptibility of the tsetse host (Welburn and Maudlin 1999), and in one field study, parasite-infected populations were found to carry greater symbiont densities (Maudlin et al. 1990). Identical 16S rDNA sequences obtained from Sodalis from different tsetse host species implied its relatively recent association with the tsetse host (Aksoy 1995; Chen et al. 1999).
Three additional lines of evidence support Sodalis' recent divergence from a free-living ancestor. The first is the ability to successfully cultivate Sodalis on insect cells (Welburn et al. 1987) and subsequently in cell-free medium (Beard et al. 1993). This is in contrast to most other symbiotic microbes, which have been difficult to culture from animals, presumably due to their long evolutionary histories in unique host niches (Amann et al. 1995). Second, the Sodalis chromosome, which was initially predicted to be 2.1 Mb by using pulsed-field gel electrophoresis analysis (Akman et al. 2001), is significantly larger than the genomes of insect obligate mutualists (Wernegreen 2002). Finally, heterologous hybridization analysis of Sodalis DNA to Escherichia coli-K12 macroarrays revealed the presence of >1800 putatively functional orthologs in this genome (Akman et al. 2001), indicative of a large functional repertoire.
Despite its recent association, Sodalis does not exhibit a pathogenic association with its tsetse host. In fact, in experiments where Sodalis was selectively eliminated from tsetse by using the sugar analog and antibiotic streptozotocin, host longevity was significantly reduced (Dale and Welburn 2001). This implies a mutualistic function for this symbiosis in tsetse. Furthermore, a quantitative analysis of Sodalis numbers during host development indicates that they are closely monitored for homeostasis (S. Aksoy, unpubl.).
Unlike the diverse microbial flora harbored by animals with multiple diets, analysis to date indicates that tsetse flies are colonized by a small number of symbiotic microbes. This phenomenon is likely a consequence of the highly sterile nature of tsetse's vertebrate blood-specific diet, upon which it depends during all developmental stages. In addition to Sodalis, tsetse flies harbor the intracellular obligate mutualist Wigglesworthia glossinidia, which has coevolved with its tsetse host for >80 Myr (Chen et al. 1999). Many tsetse populations have also been invaded by an organism related to parasitic Wolbachia pipientis (Cheng et al. 2000). While both Wigglesworthia and Sodalis take advantage of the viviparous reproductive mode of tsetse for transmission into the intrauterine larva via the mother's milk (Ma and Denlinger 1974; Cheng and Aksoy 1999), Wolbachia utilizes a transovarial route (Cheng et al. 2000).
Here we report on the genome sequence of Sodalis, the first from an insect mutualist symbiont. We present characteristics of the Sodalis genome that represent the dynamic process of transitioning from a free-living to a mutualistic lifestyle. Furthermore, we discuss aspects of an active gene decay process and functional adaptations that enable the persistence of a successful symbiotic relationship.
Results and Discussion
The Sodalis proteome and its comparison with closely related bacteria
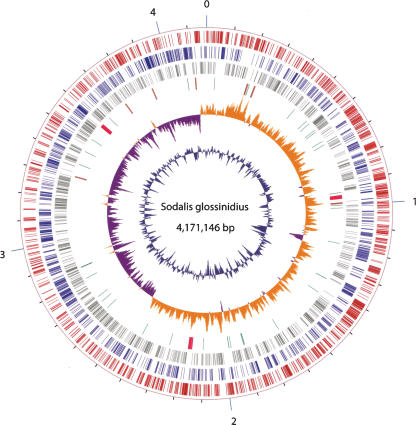
The Sodalis genome consists of one circular chromosome of 4,171,146 bp with an average G+C content of 54.7%. In addition, Sodalis has three extrachromosomal plasmids designated pSG1, pSG2, and pSG3, as well as a phage, ΦSG1. The chromosome exhibits a GC skew pattern typical of prokaryotic genomes that have two major shifts, one near the origin and one near the terminus of replication, with dnaA assigned as base pair 1 of the chromosome (Fig. 1). Of the 2432 putative protein-coding sequences (CDSs) annotated on the chromosome, 1465 (60%) are assigned to putative functions on the basis of homology to other known proteins, 484 (20%) are conserved in several bacteria but have unknown functions, 262 (11%) are phage related, and 221 (9%) have no homology to entries in the public databases. In addition to these CDSs, the chromosome also contains 972 pseudogenes distributed throughout the chromosome (Fig. 1). These segments are likely rendered nonfunctional by virtue of the accumulation of various mutations compared with corresponding functional homologs. The predicted CDSs cover only 51% of the chromosome, making this genome one of the least dense bacterial genomes in terms of coding capacity. The general features of the Sodalis chromosome and the associated plasmids are summarized in Table 1.
Figure 1.
Circular representation of the Sodalis glossinidius chromosome. The outer scale (indicated by numbers 1–4) is shown in megabases. From the outside in: circle 1, positive strand CDSs (red); circle 2, negative strand CDSs (blue); circle 3, pseudogenes (gray); circle 4, tRNA genes (green), rRNA genes (brown), genes for type III secretion systems (pink); circle 5, GC skew ([G-C]/[G+C]; khaki indicates values >0; purple indicates values <0); and circle 6, G+C content (higher values outward).
Table 1.
General features of the Sodalis genome
| Chromosome | pSG1 | pSG2 | pSG3 | |
|---|---|---|---|---|
| Size (bp) | 4,171,146 | 83,306 | 27,240 | 10,810 |
| GC content (%) | 54.7 | 48.9 | 44.7 | 47.8 |
| Predicted CDS | 2432 | 54 | 23 | 7 |
| Assigned function | 1465 | 23 | 7 | 1 |
| Conserved hypothetical | 484 | 6 | 5 | 0 |
| Unknown function | 221 | 25 | 11 | 4 |
| Phage related | 262 | 0 | 0 | 2 |
| Coding density (%) | 50.9 | 41.5 | 48.1 | 25 |
| Average CDS size (bp) | 873 | 641 | 569 | 386 |
| Pseudogenes | 972 | 23 | 5 | 3 |
| tRNA gene | 69 | 0 | 0 | 0 |
| rrn operon | 7 | 0 | 0 | 0 |
| 16S-23S-5S rRNA | 6 | — | — | — |
| 16S-23S-5S-5S rRNA | 1 | — | — | — |
Phylogenetic analysis of Sodalis from tsetse species in different subgenera has indicated a single highly related lineage in Enterobacteriacaea, affirming its recent establishment in tsetse (Chen et al. 1999; Charles et al. 2001). Comparison of 25 ribosomal proteins from sequenced organisms in Enterobacteriaceae indicates that Sodalis is closely related to E. coli K-12, Salmonella, Yersinia, and Photorhabdus (Supplemental Fig. 1A,B). This relationship is also supported by the synteny of orthologous genes examined between Sodalis and Salmonella and between Sodalis and Yersinia (Supplemental Fig. 2).
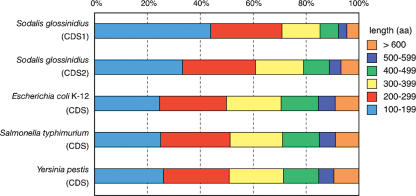
When compared with the proteomes of related free-living microbes E. coli K-12, Salmonella, and Yersinia, the size distribution of Sodalis' CDSs indicates that its genome encodes a lower percentage of large protein products (Fig. 2). These data suggest that the Sodalis genome contains a significant number of fragmented CDSs or pseudogenes. Alignment of these pseudogenes with their functional homologs validated this hypothesis. Most of the pseudogenes were found to be generated by substitutions, insertions, or deletions of single nucleotides or large segments of DNA or by the incorporation of insertion elements. The majority of Sodalis' pseudogenes were created when a functional CDS was divided into more than three fragments, or through the loss of a large segment of DNA. Furthermore, we identified several loci with functional homologs in other species, but those in Sodalis were divided into two fragments due to a single mutation. These loci were considered pseudogenes if the CDS was less than half the length of its functional homolog in related species. On the other hand, if the CDS was greater than half the length of its functional homolog, the loci were not considered pseudogenes. Hence, some of Sodalis' assigned CDSs may in fact be pseudogenes, and future functional studies will be required to determine the precise composition of Sodalis' proteome. Based on this definition of a pseudogene, we mapped 972 loci rendered nonfunctional on the Sodalis chromosome (and some in the three plasmids) (Table 1).
Figure 2.
CDSs in each size category, represented as a percentage of the total number of CDSs present (the legend indicates the size, in amino acids, of the corresponding encoded putative proteins). For each indicated bacteria, CDSs encoding putative proteins >100 amino acids were counted and used in this analysis. S. glossinidius CDS1 indicates the number of total CDSs in each category, while CDS2 excludes the pseudogenes assigned in this study.
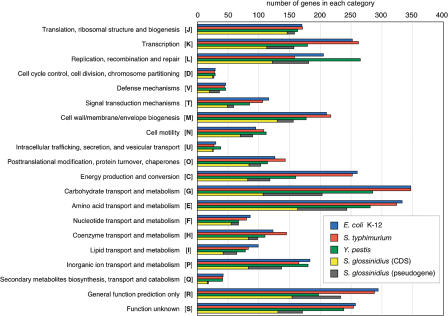
Sodalis' low gene density is further demonstrated when its putative protein products are categorized into clusters of orthologous groups (COGs) and compared with those from closely related bacteria (Fig. 3). This low coding density (51%) is equivalent to that of the obligate intracellular human parasite Mycobacterium leprae (50%) (Cole et al. 2001). Mycobacterium, which transmits horizontally, may have discarded genes that can be compensated for by a host-dependent parasitic lifestyle, while Sodalis, which transmits vertically, may have lost unnecessary genes during adaptation to a symbiotic lifestyle that corresponds to tsetse's restricted nutritional ecology. In addition to gene loss, Sodalis' 972 pseudogenes are significantly more than those found in related free-living species: 204 in Salmonella typhi CT18 (Parkhill et al. 2001a), 149 in Yersinia pestis CO92 (Parkhill et al. 2001b), and 157 in Photorhabdus luminescens (Duchaud et al. 2003). Eighty percent of the pseudogenes can be assigned putative functions based on their sequence homology to known genes. The three COGs with high numbers of pseudogenes (≥40%) are defense mechanisms, carbohydrate transport and metabolism, and inorganic ion transport and metabolism (Fig. 3; Supplemental Fig. 3). The functional reduction of metabolic pathways may again reflect Sodalis' adaptation to its hematophogous host's restricted nutritional requirements. Closely related bacterial species Y. pestis and Salmonella typhimurium have retained larger gene inventories and greater phenotypic plasticity. The enteropathogenic bacteria opportunistically colonize different hosts with broad dietary intakes and hence need to be able to metabolize a variety of different compounds.
Figure 3.
Comparative analysis of the number of genes present in each functional category as defined by the COG database for S. glossinidius, E. coli K-12, S. typhimurium, and Y. pestis. The number of genes in each COG category in Sodalis rendered nonfunctional (pseudogene) is denoted by the gray shaded area.
Biosynthetic capabilities of Sodalis reflect adaptations to host environment
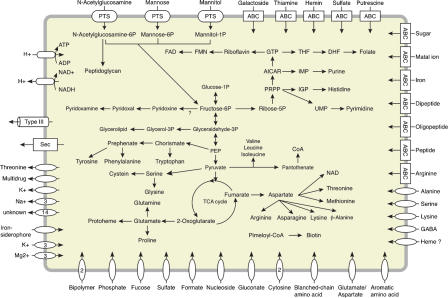
Sodalis has apparently retained many of the capabilities of free-living bacteria, including functional pathways for glycolysis, gluconeogenesis, the tricarboxylic acid (TCA) cycle, and pentose phosphate pathway (Fig. 4). However, in comparison to related free-living Enterobacteriacea, Sodalis has a very inactive biochemical profile (Dale and Maudlin 1999). The COG most strongly affected by gene degradation is that of carbohydrate transport and metabolism (Fig. 3; Supplemental Fig. 3). The Sodalis genome encodes only three intact phosphoenolpyruvate-carbohydrate phosphotransferase systems to import N-acetylglucosamine (SG0859), mannose (SG1325-SG1327), and mannitol (SG0014). Also absent are genes encoding glycolytic enzymes such as galactosidase and glucosidase, which may be dispensable given the low carbohydrate content of vertebrate blood. The Sodalis chromosome is predicted to encode components of pathways required for the synthesis of all amino acids except alanine, while many of the genes required for amino acid degradation are missing. Most genes involved in energy production and conversion via anaerobic respiration, such as nitrite and fumarate reductase, glycerol-3-phosphate dehydrogenase, and formate dehydrogenase, have been eroded or lost. Furthermore, Sodalis is apparently unable to ferment anaerobic lactate and butyrate and does not have a functional glyoxylate bypass. In general, Sodalis seems to have retained synthetic rather than degradative pathways.
Figure 4.
Overview of Sodalis's general metabolic capabilities deduced from chromosomally located genes and their putative functions.
Sodalis' gene set responsible for altering its cellular behavior in response to environmental cues is highly streamlined. Its genome has retained only five sigma factors (the primary RpoD, RpoE, RpoH, RpoN, and FliA) and eight signal transduction systems, in contrast to the human commensal microbe Bacteroides thetaiotaomicron, which has 50 ECF-type sigma factors and 32 one-component systems (Xu et al. 2003). Analysis of this small repertoire indicates Sodalis may be able to respond to acidic pH, anaerobic growth conditions, and fluctuating iron levels via PmrAB (Hacker and Kilama 1974); nitrogen-limiting conditions via NtrBC; and oxidative/osmotic stresses via BarA, a protein that can also regulate biofilm formation (Sahu et al. 2003). In the iron-rich tsetse midgut environment, Sodalis apparently relies on the direct transport of heme/hemoglobin across the cytoplasmic membrane via a specific ABC transporter (SG1538-SG1540, homologous to HutB/HemU/HmuV of Yersinia enterolitica) in a TonB-dependent manner (SG1381). However, a heavy metal–specific two-component system adjacent to this locus has been rendered nonfunctional. Sodalis can also utilize a TonB-independent iron transport system (SG1516-SG1519), homologous to Salmonella SitABCD, which has been implicated in virulence (Zhou et al. 1999). The ferrous iron transporter FeoA is intact, although FeoB is degraded and the global regulator Fur (ferric uptake regulator) is missing. In addition, plasmid pSG1 carries a cluster of genes similar to an achromobactin biosynthetic pathway (acsABCD, yhcA, lysA, and acr: SGP1_0039–SGP1_0045) and uptake and transport system (cbrABCD: SGP1_0035-SGP1_0038), thus suggesting that Sodalis may also utilize an achromobactin-related siderophore system. In the cell, excess ferrous ions can be oxidized to the ferric state and stored via a ferritin-like protein (SG1275) or bacterioferritin (SG2280) to avoid toxicity.
Analysis of genes from the defense mechanisms COG indicates that Sodalis' need for multidrug resistance genes may have been eliminated, possibly because it resides in such a specialized environment. For example, β-lactamase and its regulator, as well as multidrug efflux transporters, appear to be nonfunctional (Fig. 3; Supplemental Fig. 3). Despite its existence in the hemolymph and gut, the presence of Sodalis apparently does not invoke either a systemic or epithelial immune response in the fly (Hao et al. 2001). This may be due to its truncated lipopolysaccharide (LPS) structure, which lacks an O-antigen, the major effector protein in Gram-negative bacteria recognized by the insect immune system. In addition, in vitro experiments have shown that in comparison to E. coli-K12, Sodalis exhibits a significantly higher resistance to the bacteriocidal actions of several tsetse immune effectors, in particular the antimicrobial peptides Attacin and Diptericin (Hao et al. 2001; Hu and Aksoy 2005). The Sodalis genome encodes a catalase, Mg-superoxide dismutase, and two peroxidases, as well as homologs of various stress response regulators (CspA1, CspC, ClpAB, HtpG, HslUV, HtpX, GrpE, Hsp15, Hsp33, and HtpG). The retention of these products may reflect Sodalis' adaptation to the inhospitable physiological environment of the midgut.
Sodalis genes associated with virulence in related pathogenic organisms
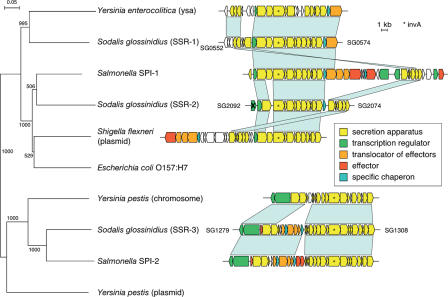
The Sodalis chromosome encodes three putative Type-III secretion systems (TTSS) that are organized into distinct clusters referred to as Sodalis symbiosis regions (SSRs) SSR-1, SSR-2, and SSR-3 (Fig. 5). The symbiosis islands have a similar GC content to the chromosomal average and show no anomalies in GC skew plot, suggesting that the acquisition of these TTSSs may be due to multiple ancient horizontal transfer events that have subsequently been modified through divergent evolution, yielding the systems evident today. Two of these regions were previously described as SSR-1 and SSR-2 (Dale et al. 2005), while SSR-3 was discovered in this study. SSR-1 (20.3 kb in length, SG0552–SG0574) is most similar in gene content and order to ysa from Y. enterolitica. Our analysis of SSR-1 noted two differences compared to a previous description (Dale et al. 2005), namely, the lack of a yspD homolog and the presence of a functional yspA gene (Foultier et al. 2002). SSR-2 (17.5 kb in length, SG2074-SG2092) is related to SPI-1 of Salmonella (Lostroh and Lee 2001) and Mxi-Spa of the Shigella virulence plasmid (Buchrieser et al. 2000). SSR-3 (23.3 kb in length, SG1279–SG1308), the newly recognized region in Sodalis, is most similar to SPI-2 of Salmonella (Hensel et al. 1997) and the chromosomally encoded island of Y. pestis (Deng et al. 2002). SSR-3 encodes secretion system apparatus components, secretion system effectors and chaperons, and an operon for the two-component regulatory system SseAB (secretion system regulator). SSR-3 has also retained all of the virulence-associated genes of Salmonella SPI-2, including the effector SsaB and chaperonin SscB.
Figure 5.
Phylogenetic relationship shared between Sodalis' three TTSS structures and those characterized from pathogenic microbes. The tree was based on amino acid sequence alignments of the invA gene product. Scale bars representing branch lengths and bootstrap values are displayed at each internal node. Genes associated with each cluster are depicted as arrows that indicate the direction of transcription. The arrows with a cross denote pseudogenes. The light blue bars between loci indicate the regions of sequence similarity and gene order conservation. The different colors depict the functional roles of their putative products.
Preliminary analysis of the TTSSs indicate that the envelope-associated structural components, encoded within tightly linked operons, have been conserved, while some of the secreted effectors and transcriptional regulators found at positions adjacent to the core components appear absent, nonfunctional, or under relaxed selection. The nonfunctional proteins include sipACD (from SSR-1) and invE and hilA (from SSR-2), while those that display relaxed selection include prgIJ, spaN, and invB from SSR-2 (Dale et al. 2005). Interestingly, homologs of several virulence factors—including the Salmonella SPI-1 effector SipD; systemic infection regulators phoPQ, RcsC/YojN/RcsB, EnvZ/OmpR, and SsrAB; as well as hemolysin (SG1998), invasion proteins (SG0550 and SG0551), and phospholipases (SG2218 and SG2338)—are present on Sodalis' chromosome. The function of these genes in this symbiont remains to be elucidated.
Despite an apparent lack of motility associated with Sodalis either in vivo or under in vitro cultivation conditions, 90 flagellar-related CDSs, contained on two distinct clusters, were identified on Sodalis' chromosome (Supplemental Fig. 4). The first of these (35.3 kb in length, SG0021–SG0059) is predicted to encode a complete flagellar apparatus with motility proteins MotA and MotB, transcriptional regulators FlhCD, and the sigma factor FliA. In addition, the putative chemotaxis-transducing proteins CheW and CheZ are also present. The second locus (SG2052–SG2068, 27.1 kb) contains 15 pseudogenes and thus encodes an incomplete flagellar apparatus.
Expression of functions in Sodalis typically associated with virulence in related pathogenic microbes
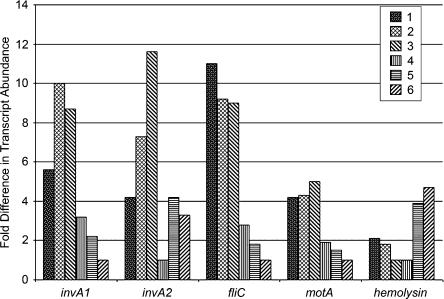
Given their mutualistic association, the expression of TTSSs might mediate different symbiotic functions in the tsetse-Sodalis system. Previously, an in vitro assay using an insect cell line demonstrated a role for SSR-1 in host cell entry and for SSR-2 in post-invasion processes (Dale et al. 2002, 2005). We evaluated the transcript abundance of SSR-specific invA products from adult, larval, and pupal developmental stages of tsetse in vivo (Fig. 6). Our results indicate that invA1 transcripts are produced constitutively, with peak expression levels corresponding to the late larval and early pupal stages. A similar expression profile is detected for invA2, with a peak in the early pupal stage. In contrast, no invA3-specific transcripts could be observed in vivo in the developmental stages tested, although abundant expression is detected under in vitro cultivation conditions on a C6/36 cell line (data not shown). In Salmonella, the SSR-3 homolog SPI-2 plays a central role in virulence and its expression is dependent on the regulatory system ssrAB, although the nature of the signals is unknown (Kuhle and Hensel 2004). Many Salmonella effector proteins translocated by SPI-2 are encoded outside the island and are associated with prophages. Analysis of the putative virulence-related product hemolysin indicates that it is expressed most prominently in adult midgut (Fig. 6). Identification of such potential effectors from Sodalis can now be investigated. A similar analysis of flagella-associated fliC and motA expression indicates peak levels for both products during the larval and early pupal stages, while no expression is observed in adult midgut (Fig. 6). This is in accordance with the previous descriptions of this organism as nonmotile in adult midgut (Dale and Maudlin 1999). Little is known about the mechanism of symbiont transmission into the intrauterine progeny from the mother. TTSSs and flagella may be important during host metamorphosis for establishment and maintenance of symbiosis in progeny.
Figure 6.
Analysis of gene expression representative of SSR, flagella, and putative effector functions during host development. Transcript abundance levels were determined for invA1, invA2, fliC, motA, and hemolysin expressed at the following host developmental stages: 1, early larvae; 2, late larvae; 3, early pupae; 4, late pupae; 5, 6-d-old adults; and 6, 40-d-old fertile females. Results for each gene were normalized against the host developmental stage, during which their expression was least abundant. The data are presented as bar graphs, which represent fold difference in transcript abundance. No invA3 transcripts could be amplified from these cDNA pools tested (data not shown).
Conclusions
Eubacterial genomes range in size from 0.45 Mb to over 10 Mb (Wernegreen 2002). Typically, microbes with free-living lifestyles have larger genomes coding for a wide range of functional capabilities, while symbiotic and pathogenic intracellular organisms have smaller genomes with more limited capabilities. Extreme genome reductions and loss of function are observed in the insect intracellular obligate mutualists (Wernegreen 2002), including Wigglesworthia, which has a dramatically streamlined 700-kb genome (Akman and Aksoy 2001; Akman et al. 2002). These small chromosomes are presumably derived from larger ancestral genomes through a massive loss of genes that are either no longer needed in the restricted host environment or that have products that can be provided by their hosts (Moran 2002). These associations represent long coevolutionary histories with host insects (Chen et al. 1999). In contrast, the size of Sodalis' chromosome is closer to those from free-living microbes, in accordance with its recent symbiotic history. However, the genome contents provide a preview of a streamlining process where pathways no longer required in the restricted host environment have begun to be inactivated through slow erosion at individual loci.
The Sodalis genome still retains a robust DNA-repair system and extensive synteny with genomes of closely related enterics, possibly again indicative of its recent symbiotic affiliation. Comparative analysis of the putative proteomes of related Yersinia and Photorhabdus indicates that almost a third of Sodalis' capabilities are unique. About two-thirds of the unique gene set corresponds to phage-like sequences and symbiosis region genes, while the remainder comprises putative proteins with no known homologs in the databases. Their future functional characterization can provide insights into the exceptional aspects of Sodalis' symbiotic biology. Interestingly, Sodalis shares all of Wigglesworthia's gene contents except those encoding thiamine, cobalamine, and molybdopterin biosynthesis pathways. One of Wigglesworthia's presumed functions is the supplementation of tsetse's restricted diet with a plethora of vitamins known to be important for fertility (Nogge 1981; Akman et al. 2002). Hence, thiamine may play a key role for the fecundity of tsetse females.
The genome of entomopathogenic Photorhabdus encodes many adhesins, toxins, hemolysins, proteases, and lipases and contains a wide array of antibiotic synthesizing genes. These products are likely to play a role in the elimination of competitors, as well as in colonization, invasion, and degradation of the host insect cadaver (Duchaud et al. 2003). Because maternally transmitted Sodalis is a mutualist microbe with no known adverse impact on host biology, its chromosomally encoded TTSSs, hemolysin, lipases, and adhesions may fulfill functions different from those reported in pathogenic microbes. In a mutant line of Sodalis, where the invC locus of SSR-2 was inactivated by transposon mutagenesis, establishment of Sodalis infections in tsetse progeny was compromised, thus indicating its role in transmission to progeny (Dale et al. 2001). In another study with the related symbiont from Sitophilus zeamais, SSR-2 expression peaked in the pupal stage, again corresponding to metamorphosis and establishment of symbiotic infections (Dale et al. 2002). Our detection of TTSS and flagella transcripts during early larval development further confirms the importance of protein translocation systems for maternal transmission and the establishment of symbiosis. Density analysis of tsetse's three distinct symbionts during host development indicates that Sodalis is maintained at orders of magnitude lower than either parasitic Wolbachia or mutualistic Wiggelsworthia (S. Aksoy, pers. comm.). Furthermore, Sodalis density remains well regulated during development, with the exception of 24–48 h pre- and post-eclosion, during which a window of opportunity for Sodalis replication is apparently available. Host and/or symbiont factors, which closely monitor integrative molecular communication necessary to maintain Sodalis densities for homeostasis, can now be investigated in vivo.
Future Sodalis functional studies will be aided by the availability of an in vitro culture system, episomal and chromosomal transformation systems, and a well-established protocol for reconstituting flies with genetically modified symbionts. Finally, since Sodalis lives in close proximity to pathogenic trypanosomes, a paratransgenic strategy (where trypanocidal products are expressed in tsetse midgut by recombinant Sodalis) is entertained as a means to control parasite infections in the fly. The eventual replacement of natural parasite-susceptible vector populations with refractory flies harboring modified Sodalis could provide an additional strategy to reduce disease transmission (Rio et al. 2004). In this regard, the highly restricted metabolic capabilities of Sodalis would limit its survival in nontarget organisms and enhance the efficacy of this paratransgenic approach.
Methods
Insect and bacterial cultivation
Glossina morsitans morsitans flies were maintained at 24°C with 55% relative humidity and received defibrinated bovine blood through an artificial membrane system every 48 h. The symbiont, S. glossinidius, was isolated from surface-sterilized pupae of G. m. morsitans and cultured on a feeder layer of Aedes albopictus C6/36 cells as described previously (Dale and Maudlin 1999). Pure Sodalis cultures were maintained in vitro at 25°C in Mitsuhashi-Maramorosch (M&M) medium (1 mM CaCl2, 0.2 mM MgCl2, 2.7 mM KCl, 120 mM NaCl, 1.4 mM NaHCO3, 1.3 mM NaH2PO4, 22 mM D (+) glucose, 6.5 g/L lactalbumin hydrolysate, and 5.0 g/L yeast extract) supplemented with 5% heat-inactivated fetal bovine serum (Beard et al. 1993).
Genome sequence analysis
The nucleotide sequence of the Sodalis genome was determined by a whole-genome shotgun strategy, as described previously (Akman et al. 2002). We constructed small-insert (2-kb) and large-insert (10-kb) genomic libraries and generated 68,664 sequences, giving ninefold coverage from both ends of the genomic clones. Sequence assembly was carried out by using the PHRED-PHRAP-CONSED package (Gordon et al. 2001). Remaining gaps were closed by sequencing of clones that spanned the gaps or by polymerase chain reaction (PCR) direct sequencing with oligonucleotide primers designed to anneal to each end of neighboring contigs. To exclude the possibility of sequence error for the assignment of the pseudogenes, we assessed and confirmed the quality of these regions with the Q-value of the finished data, read redundancy in the assembly, and curation of the trace data. Overall accuracy of the finished sequence was estimated to have an error rate of less than one per 10,000 bases (PHRAP score of ≥40). Statistical data regarding the accuracy of this sequencing project are available from our Web site (http://genome.ls.kitasato-u.ac.jp/SG_qual/). The finished sequence was also confirmed by pulsed-field gel electrophoresis by using appropriate restriction enzymes.
Bioinformatics
Putative protein-coding sequences were first identified by using both GenomeGambler 1.51 (Sakiyama et al. 2000) and Glimmer 2.0 (Delcher et al. 1999) programs and then were manually confirmed and corrected by using BLASTP (Altschul et al. 1997). tRNA genes were predicted by tRNAscan-SE (Lowe and Eddy 1997). Functional classification of ORFs was made by homology search against clusters of orthologous groups of proteins (Tatusov et al. 2001) using BLASTP. The SSR phylogenetic tree (Fig. 5) was made by using Njplot, on the basis of a ClustalW sequence alignment (with a bootstrap trial of 1000) (Supplemental Fig. 1B).
Specific loci were designated as pseudogenes when, compared with their functional homologs in related microbes, they encoded small fragmented ORFs or were interrupted by the loss or addition of a large portion of DNA.
Gene expression analysis
Total RNA was extracted by using Trizol reagent (Invitrogen, catalog no. 15596–026) from flies at specified developmental stages: early larva (1- to 3-d-old), late larva (4- to 9-d-old), early pupa (24-h post-deposition), late pupae (24–48 h prior to eclosion), early adult (6-d-old), and late adult (40-d-old). Total RNA was also purified from Sodalis cultures maintained for 12 h on a C6/36 cell line. Adults were processed 48 h after their last blood-meal, and larva were microsurgically dissected from pregnant mothers. Negative reverse transcription (RT)–PCR was used to confirm that all contaminating DNA was removed from RNA samples following treatment with DNase I (Ambion, catalog no. 1906). First-strand cDNA synthesis was primed with random nanomers (New England Biolabs) by using a SuperScript First-Strand Synthesis Kit (Invitrogen, catalog no. 11904–018) according to the manufacturer's protocol. Transcription levels of different genes were measured by performing semiquantitative RT-PCR. The cDNA pools were normalized by determining the linear range of amplification in serially diluted samples using primers specific for GroEL (F, 5′-TAGGTACCATCTCCGCCAAC-3′; R, 5′-GTGGCTTTTTCCAGCTCAAG-3′). The cDNA dilution to be used was empirically chosen where comparable GroEL amplification intensities were observed, while avoiding saturation at 28 cycles of amplification. The number of PCR cycles was constant for a particular sequence in the multiple samples analyzed in a given experiment. The specific primers used were as follows: FliC (F, 5′-GCAGTTTCAGGATACCC-3′; R, 5′-GGCGGAAAATGGTATAG-3′); MotA (F, 5′-ATCCCGGTAAGTAGCC-3′; R, 5′-CAGCCGATTCCATCAGA-3′); Hemolysin (F, 5′-ATGGGAAACAAACCATTAGCCA-3′; R, 5′-TCAAGTGACAAACAGATAAATC-3′); InvA1 (F, 5′-CAGTTTACACCCACCTCTCG-3′; R, 5′-CAATAGCAGAGCGGGTAACG-3′); InvA2 (F, 5′-GCTGATCTCAATCAGCGCA-3′; R, 5′-TACCGTCGCGTGGCGCAACAC-3′), and InvA3 (F, 5′-CGGGGTCATTGATGCGGTCCA-3′; R, 5′-GTTATCTGTCCCATGCCGT-3′). Amplification conditions consisted of a 3-min hotstart at 95°C followed by a specified number of cycles of 30 sec at 95°C, 40 sec at 58°C, and 60 sec at 72°C and a final 10-min elongation at 72°C in an MJ Research PTC-200 thermal cycler. Following electrophoresis on 1% agarose gels, bands were visualized with a Kodak 2000R imager, and mean intensities were compared from three independent experiments.
Acknowledgments
We thank K. Furuya, C. Yoshino, A. Nakazawa, Y. Yamashita, and N. Itoh for technical assistance. This work was supported in part by Grant-In-Aid of the Ministry of Education, Culture, Sports, Science and Technology, Japan; Grant of the 21st Century COE Program, Ministry of Education, Culture, Sports, Science and Technology, Japan; and Research for the Future Program from the Japan Society for the Promotion of Science and NIH/NIAID grant AI-34033 and NSF/MCB 0237305.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4106106.
Footnotes
References
- Akman, L. and Aksoy, S. 2001. A novel application of gene arrays: Escherichia coli array provides insight into the biology of the obligate endosymbiont of tsetse flies. Proc. Natl. Acad. Sci. 98 7546-7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akman, L., Rio, R.V.M., Beard, C.B., and Aksoy, S. 2001. Genome size determination and coding capacity of Sodalis glossinidius, an enteric symbiont of tsetse flies, as revealed by hybridization to Escherichia coli gene arrays. J. Bacteriol. 183 4517-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akman, L., Yamashita, A., Watanabe, H., Oshima, K., Shiba, T., Hattori, M., and Aksoy, S. 2002. Genome sequence of the endocellular obligate symbiont of tsetse, Wigglesworthia glossinidia. Nat. Genet. 32 402-407. [DOI] [PubMed] [Google Scholar]
- Aksoy, S. 1995. Molecular analysis of the endosymbionts of tsetse flies: 16S rDNA locus and over-expression of a chaperonin. Insect Mol. Biol. 4 23-29. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Tsetse: A haven for microorganisms. Parasitol. Today 16 114-118. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann, R.I., Ludwig, W., and Schleifer, K.H. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59 143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard, C.B., O'Neill, S.L., Mason, P., Mandelco, L., Woese, C.R., Tesh, R.B., Richards, F.F., and Aksoy, S. 1993. Genetic transformation and phylogeny of bacterial symbionts from tsetse. Insect Mol. Biol. 1 123-131. [DOI] [PubMed] [Google Scholar]
- Buchrieser, C., Glaser, P., Rusniok, C., Nedjari, H., D'Hauteville, H., Kunst, F., Sansonetti, P., and Parsot, C. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38 760-771. [DOI] [PubMed] [Google Scholar]
- Charles, H., Heddi, A., and Rahbe, Y. 2001. A putative insect intracellular endosymbiont stem clade, within the Enterobacteriaceae, infered from phylogenetic analysis based on a heterogeneous model of DNA evolution. C.R. Acad. Sci. III 324 489-494. [DOI] [PubMed] [Google Scholar]
- Chen, X.A., Li, S., and Aksoy, S. 1999. Concordant evolution of a symbiont with its host insect species: Molecular phylogeny of genus Glossina and its bacteriome-associated endosymbiont, Wigglesworthia glossinidia. J. Mol. Evol. 48 49-58. [DOI] [PubMed] [Google Scholar]
- Cheng, Q. and Aksoy, S. 1999. Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect Mol. Biol. 8 125-132. [DOI] [PubMed] [Google Scholar]
- Cheng, Q., Ruel, T.D., Zhou, W., Moloo, S.K., Majiwa, P., O'Neill, S.L., and Aksoy, S. 2000. Tissue distribution and prevalence of Wolbachia infections in tsetse flies, Glossina spp. Med. Vet. Entomol. 14 44-50. [DOI] [PubMed] [Google Scholar]
- Cole, S.T., Eiglmeier, K., Parkhill, J., James, K.D., Thomson, N.R., Wheeler, P.R., Honore, N., Garnier, T., Churcher, C., Harris, D., et al. 2001. Massive gene decay in the leprosy bacillus. Nature 409 1007-1011. [DOI] [PubMed] [Google Scholar]
- Dale, C. and Maudlin, I. 1999. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int. J. Systematic Bacteriol. 49 267-275. [DOI] [PubMed] [Google Scholar]
- Dale, C. and Welburn, S.C. 2001. The endosymbionts of tsetse flies: Manipulating host-parasite interactions. Int. J. Parasitol. 31 628-631. [DOI] [PubMed] [Google Scholar]
- Dale, C., Young, S.A., Haydon, D.T., and Welburn, S.C. 2001. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl. Acad. Sci. 98 1883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale, C., Plague, G.R., Wang, B., Ochman, H., and Moran, N.A. 2002. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc. Natl. Acad. Sci 99 12397-12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale, C., Jones, T., and Pontes, M. 2005. Degenerative evolution and functional diversification of type-III secretion systems in the insect endosymbiont Sodalis glossinidius. Mol. Biol. Evol. 22 758-766. [DOI] [PubMed] [Google Scholar]
- Delcher, A.L., Harmon, D., Kasif, S., White, O., and Salzberg, S.L. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, W., Burland, V., Plunkett, G., Boutin, A., Mayhew, G.F., Liss, P., Perna, N.T., Rose, D.J., Mau, B., Zhou, S., et al. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184 4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaud, E., Rusniok, C., Frangeul, L., Buchrieser, C., Givaudan, A., Taourit, S., Bocs, S., Boursaux-Eude, C., Chandler, M., Charles, J.F., et al. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21 1307-1313. [DOI] [PubMed] [Google Scholar]
- Foultier, B., Troisfontaines, P., Muller, S., Opperdoes, F.R., and Cornelis, G.R. 2002. Characterization of the ysa pathogenicity locus in the chromosome of Yersinia enterocolitica and phylogeny analysis of type III secretion systems. J. Mol. Evol. 55 37-51. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Desmarais, C., and Green, P. 2001. Automated finishing with autofinish. Genome Res. 11 614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker, C.S. and Kilama, W.L. 1974. The relationship between Plasmodium gallinaceum density and the fecundity of Aedes aegypti. J. Invert. Pathol. 23 101-105. [DOI] [PubMed] [Google Scholar]
- Hao, Z., Kasumba, I., Lehane, M.J., Gibson, W.C., Kwon, J., and Aksoy, S. 2001. Tsetse immune responses and trypanosome transmission: Implications for the development of tsetse-based strategies to reduce trypanosomiasis. Proc. Natl. Acad. Sci. 98 12648-12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel, M., Shea, J.E., Raupach, B., Monack, D., Falkow, S., Gleeson, C., Kubo, T., and Holden, D.W. 1997. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella Pathogenicity Island 2. Mol. Microbiol. 24 155-167. [DOI] [PubMed] [Google Scholar]
- Hu, Y. and Aksoy, S. 2005. An antimicrobial peptide with trypanocidal activity characterized from Glossina morsitans morsitans. Insect Biochem. Mol. Biol. 35 105-115. [DOI] [PubMed] [Google Scholar]
- Kuhle, V. and Hensel, M. 2004. Cellular microbiology of intracellular Salmonella enterica: Functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell Mol. Life Sci. 61 2812-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lostroh, C.P. and Lee, C.A. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3 1281-1291. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W.C. and Denlinger, D.L. 1974. Secretory discharge and microflora of milk gland in tsetse flies. Nature 247 301-303. [Google Scholar]
- Maudlin, I., Welburn, S.C., and Mehlitz, D. 1990. The relationship between rickettsia-like organisms and trypanosome infections in natural populations of tsetse in Liberia. Trop. Med. Parasitol. 41 265-267. [PubMed] [Google Scholar]
- Moran, N.A. 2002. Microbial minimalism: Genome reduction in bacterial pathogens. Cell 108 583-586. [DOI] [PubMed] [Google Scholar]
- Nogge, G. 1981. Significance of symbionts for the maintenance of an optimal nutritional state for successful reproduction in haematophagous arthropods. Parasitology 82 101-104. [Google Scholar]
- Parkhill, J., Dougan, G., James, K.D., Thomson, N.R., Pickard, D., Wain, J., Churcher, C., Mungall, K.L., Bentley, S.D., Holden, M.T., et al. 2001a. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413 848-852. [DOI] [PubMed] [Google Scholar]
- Parkhill, J., Wren, B.W., Thomson, N.R., Titball, R.W., Holden, M.T., Prentice, M.B., Sebaihia, M., James, K.D., Churcher, C., Mungall, K.L., et al. 2001b. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413 523-527. [DOI] [PubMed] [Google Scholar]
- Rio, R.V., Hu, Y., and Aksoy, S. 2004. Strategies of the home-team: Symbioses exploited for vector-borne disease control. Trends Microbiol. 12 325-336. [DOI] [PubMed] [Google Scholar]
- Sahu, S.N., Acharya, S., Tuminaro, H., Patel, I., Dudley, K., LeClerc, J.E., Cebula, T.A., and Mukhopadhyay, S. 2003. The bacterial adaptive response gene, barA, encodes a novel conserved histidine kinase regulatory switch for adaptation and modulation of metabolism in Escherichia coli. Mol. Cell. Biochem. 253 167-177. [DOI] [PubMed] [Google Scholar]
- Sakiyama, T., Takami, H., Ogasawara, N., Kuhara, S., Kozuki, T., Doga, K., Ohyama, A., and Horikoshi, K. 2000. An automated system for genome analysis to support microbial whole-genome shotgun sequencing. Biosci. Biotechnol. Biochem. 64 670-673. [DOI] [PubMed] [Google Scholar]
- Tatusov, R.L., Natale, D.A., Garkavtsev, I.V., Tatusova, T.A., Shankavaram, U.T., Rao, B.S., Kiryutin, B., Galperin, M.Y., Fedorova, N.D., and Koonin, E.V. 2001. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29 22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn, S.C. and Maudlin, I. 1999. Tsetse-typanosome interactions: Rites of passage. Parasitol. Today 15 399-403. [DOI] [PubMed] [Google Scholar]
- Welburn, S.C., Maudlin, I., and Ellis, D.S. 1987. In vitro cultivation of rickettsia-like organisms from Glossina spp. Ann. Trop. Med. Parasitol. 81 331-335. [DOI] [PubMed] [Google Scholar]
- Wernegreen, J.J. 2002. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 3 850-861. [DOI] [PubMed] [Google Scholar]
- Xu, J., Bjursell, M.K., Himrod, J., Deng, S., Carmichael, L.K., Chiang, H.C., Hooper, L.V., and Gordon, J.I. 2003. A genomic view of the human Bacteroides thetaiotaomicron symbiosis. Science 299 2074-2076. [DOI] [PubMed] [Google Scholar]
- Zhou, D., Hardt, W.D., and Galan, J.E. 1999. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect. Immun. 67 1974-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]