Abstract
The ie2 gene of Autographa californica multicapsid nuclear polyhedrosis virus is 1 of the 10 baculovirus genes that have been identified as factors involved in viral DNA replication. IE2 is detectable in the nucleus as one of the major early-expressed proteins and exhibits a dynamic localization pattern during the infection cycle (D. Murges, I. Quadt, J. Schröer, and D. Knebel-Mörsdorf, Exp. Cell Res. 264:219-232, 2001). Here, we investigated whether IE2 localized to regions of viral DNA replication. After viral DNA was labeled with bromodeoxyuridine (BrdU), confocal imaging indicated that defined IE2 domains colocalized with viral DNA replication centers as soon as viral DNA replication was detectable. In addition, a subpopulation of IE2 structures colocalized with two further virus-encoded replication factors, late expression factor 3 (LEF-3) and the DNA binding protein (DBP). While DBP and LEF-3 structures always colocalized and enlarged simultaneously with viral DNA replication sites, only those IE2 structures that colocalized with replication sites also colocalized with DBP. Replication and transcription of DNA viruses in association with promyelocytic leukemia protein (PML) oncogenic domains have been observed. By confocal imaging we demonstrated that the human PML colocalized with IE2. Triple staining revealed PML/IE2 domains in the vicinity of viral DNA replication centers, while IE2 alone colocalized with early replication sites, demonstrating that PML structures do not form common domains with viral DNA replication centers. Thus, we conclude that IE2 colocalizes alternately with PML and the sites of viral DNA replication. Small ubiquitin-like modifier SUMO-1 has been implicated in the nuclear distribution of PML. Similar to what was found for mammalian cells, small ubiquitin-like modifiers were recruited to PML domains in infected insect cells, which suggests that IE2 and PML colocalize in conserved cellular domains. In summary, our results support a model for IE2 as part of various functional sites in the nucleus that are connected with viral DNA replication.
The replication of viral DNA genomes within host cell nuclei takes place in discrete regions, which can be visualized as distinctive structures specific to virus-infected cells. Centers of viral DNA replication represent an accumulation of replication proteins and other viral and cellular components in addition to newly synthesized viral DNA. This has been best characterized in herpes simplex virus type 1 (HSV-1)-infected cells (24). Viral replication centers have been proposed to assemble nonrandomly but appear to be determined by preexisting host nuclear structures (3). There is evidence that viral DNA replication sites are located adjacent to promyelocytic leukemia protein (PML) bodies. PML is the defining component of PML bodies, PML oncogenic domains (PODs) or nuclear domain 10, which are thought to be involved in the modulation of cell growth and proliferation (for a review see reference 17).
We demonstrated a dynamic nuclear localization pattern for early baculovirus proteins IE2 and PE38 during the infection cycle of Autographa californica multicapsid nuclear polyhedrosis virus (AcMNPV) (22). The genes encoding both factors belong to a set of 10 viral genes involved in baculovirus DNA replication. Based on transient complementation assays using origin-containing plasmids, IE2 and PE38 are not essential but rather have a stimulatory effect on DNA replication (12, 16). How they contribute to the replication of baculovirus DNA in infected cells is still speculative. It has been suggested that other factors, such as late expression factor 3 (LEF-3), a homotrimeric single-stranded DNA binding protein (SSB) which improves the strand displacement ability of the viral DNA polymerase, are essential in viral DNA replication (5, 8, 12, 19). In addition, LEF-3 may be involved in the transport of putative helicase P143 to the nucleus, which is in line with the observation that LEF-3 interacts directly with P143 (6, 30).
Our study of the functional role of IE2 and PE38 is based on experiments in which their localization within infected cells was determined. We previously demonstrated that IE2 is localized in nuclear domains that changed to threadlike structures late in infection, while PE38 was only partly present in nuclear dots very early in infection (22). After transient expression both proteins were found to be associated with PODs in mammalian cells; however, during the early phase of infection only IE2 and human PML colocalized in insect cells (22). If both PE38 and IE2 were directly involved in viral DNA replication, their localization would be expected to be close to that of replicating DNA. Since PE38 shows mainly a sandy nuclear staining pattern, its spatial distribution hardly contributes to a better understanding of whether PE38 is associated with sites of viral DNA replication. Therefore, we focused on the spatial and temporal distribution of IE2 in the context of viral DNA replication.
Here, we present the first evidence that AcMNPV replication takes place at distinct nuclear domains which form large foci during DNA replication. IE2 domains colocalized with the sites of viral DNA replication and partly colocalized with LEF-3 domains. We characterized a further putative replication factor of AcMNPV, designated DNA binding protein (DBP), which forms common domains with replication centers in cells infected with Bombyx mori nucleopolyhedrovirus (BmNPV), a close homologue of AcMNPV (23). DBP of BmNPV was found to bind preferentially to single-stranded DNA and to unwind partial DNA duplexes in vitro (20). We confirmed the colocalization of AcMNPV DBP and DNA replication sites and found that IE2 structures associated with those of DBP. Although IE2 and human PML localized in common domains, PML was not within viral DNA replication sites but was found in close proximity.
MATERIALS AND METHODS
Cells and viruses.
Spodoptera frugiperda IPLB21 (29) and Trichoplusia ni TN-368 (9) insect cells were grown in TC100 medium (7) supplemented with 10% fetal calf serum. Infections with AcMNPV plaque isolate E (28), a wild-type virus, and with recombinant virus AcMNPV-PML/e (22) were performed at a multiplicity of 10 to 20 PFU per cell. Time zero was defined as the time when the AcMNPV inoculum was added to the cells. DNA synthesis was visualized by adding thymidine analogue 5-bromo-2′-deoxyuridine (BrdU) (Sigma) to the cell medium 1 h prior to fixation with 2% paraformaldehyde. After the cells were permeabilized with Triton X-100, DNA was denatured for 2 min in 0.07 M NaOH. To inhibit AcMNPV DNA replication, TN-368 cells were treated with aphidicolin (Sigma; stock solution 1 mg/ml in ethanol; diluted 1:200 in TC100) at 1 h postinfection (p.i.). The cells were incubated in the presence of aphidicolin until they were fixed at 4, 6, and 8 h p.i.
Transcriptional analysis.
AcMNPV-infected S. frugiperda cells were harvested to prepare polyadenylated RNA, which was analyzed by Northern blotting as described previously (13). RNAs were visualized by hybridization to cDNA clone 58 including the DBP open reading frame (ORF) of AcMNPV. 32P-labeled cDNA clone 58 was prepared by nick translation.
Plasmid constructions and transfection experiments.
cDNA synthesis from polyadenylated RNA isolated 1 h p.i. was performed as described previously (13). Plasmid pCMV-DBP carries ORF 25 of AcMNPV (1) under the control of the early cytomegalovirus promoter/enhancer and was generated by insertion of the BamHI/Asp718 fragment of cDNA clone 58 into the corresponding sites of plasmid pSCTEV3S (25) (see Fig. 4). Plasmid pBs-pe38PML-HR1 includes the pml gene under the control of the AcMNPV pe38 promoter (22), and plasmid pPst-N carries the PstN fragment of AcMNPV DNA expressing the IE2 gene (14). TN-368 cells were transfected as described previously (22), and immunofluorescence was performed 45 h after transfection.
FIG. 4.
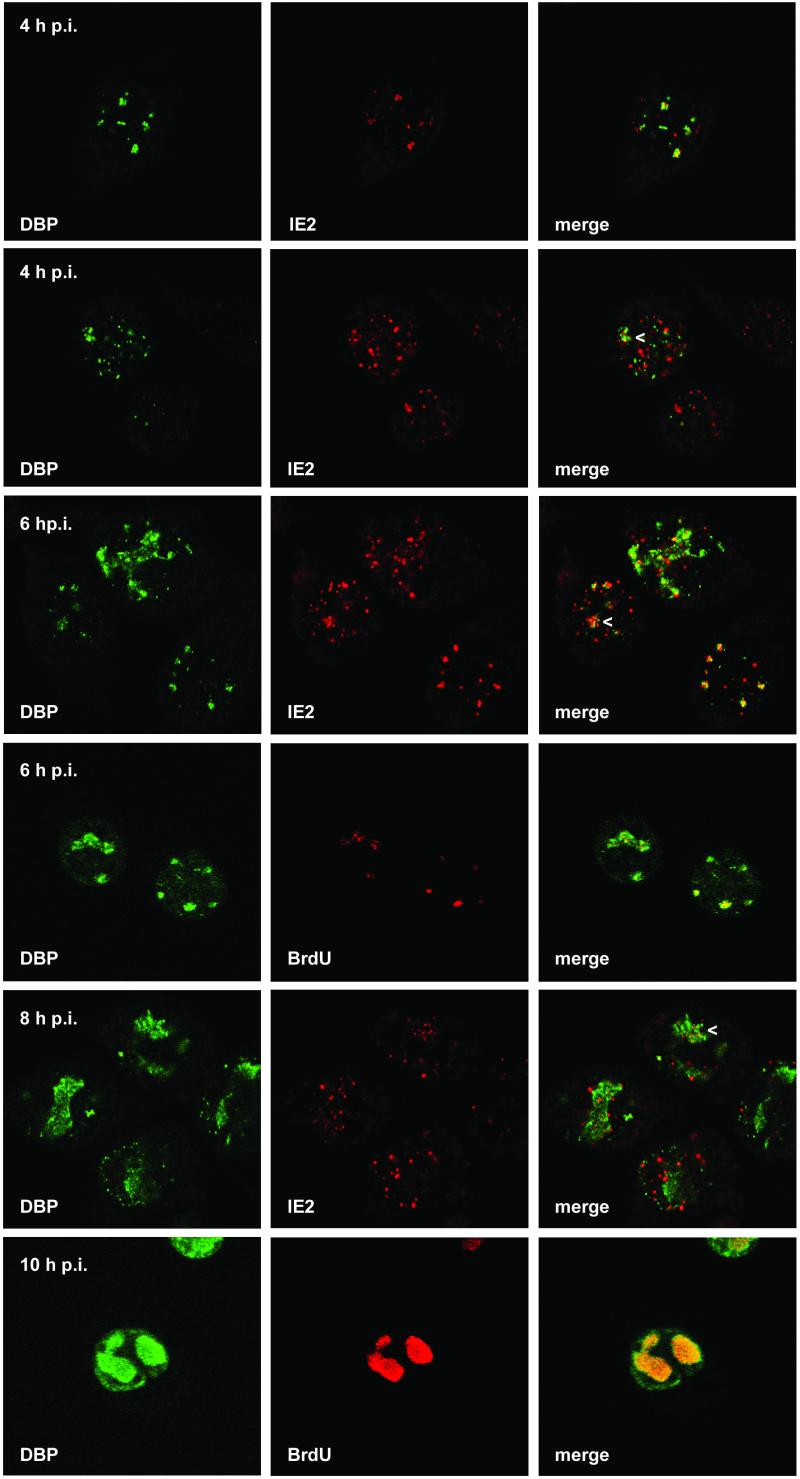
Localization of DBP and IE2 and of DBP and viral DNA replication sites after AcMNPV infection of TN-368 cells. AcMNPV-infected TN-368 cells were fixed at 4, 6, 8, and 10 h p.i. and costained with rabbit anti-DBP antiserum (green) and mouse anti-IE2 antiserum (red) or mouse BrdU MAb (red). Arrowheads indicate IE2 dot clusters. Confocal images and the merges are shown.
Immunocytochemistry and antibodies.
TN-368 cells were grown on coverslips, fixed in 2% paraformaldehyde, and permeabilized by incubation with 0.1% Triton X-100 as described previously (22). Alternatively, cells were fixed in 100% acetone (22). Cells on coverslips were blocked with 2% bovine serum albumin and then incubated for 1 h with either rabbit or mouse anti-IE2 antiserum (1:1,000 or 1:100 dilution) (14, 22), rabbit anti-PE38 antiserum (1:1,000 dilution) (14), mouse monoclonal antibody (MAb) 5E10 (1:10 dilution) to detect PML (27), rabbit polyclonal antibodies directed against BmNPV DBP (1:4,000 dilution) (23), rabbit anti-AcMNPV LEF-3 antiserum (1:100 dilution) (5), and rabbit anti-dSmt3 antiserum (1:200 dilution) (15) in phosphate-buffered saline containing 2% bovine serum albumin. The BrdU-labeled cells were incubated with monoclonal anti-BrdU antibodies (1:50 dilution) derived either from mice (clone B44; Becton Dickinson) or from rats (clone BU1/75; Harlan SeraLab). Primary antibodies were visualized with fluorochrome-conjugated anti-rabbit, anti-mouse, or anti-rat immunoglobulin G (Molecular Probes). Specimens were viewed with a Zeiss Axiovert 135 with an Intas digital camera system (see Fig. 6a to c), and confocal imaging was performed with a Leica DM IRBE microscope linked to Leica TCS-SP. Cells were imaged with the confocal microscope with either a 63× objective and a 1.32 numerical aperture (NA) (see Fig. 1B) or a 100× objective and a 1.25 NA (see Fig. 1A, 2, and 4 to 6). Images were scanned with Umax PowerLookIII and assembled by Adobe Photoshop, version 6.0.
FIG. 6.
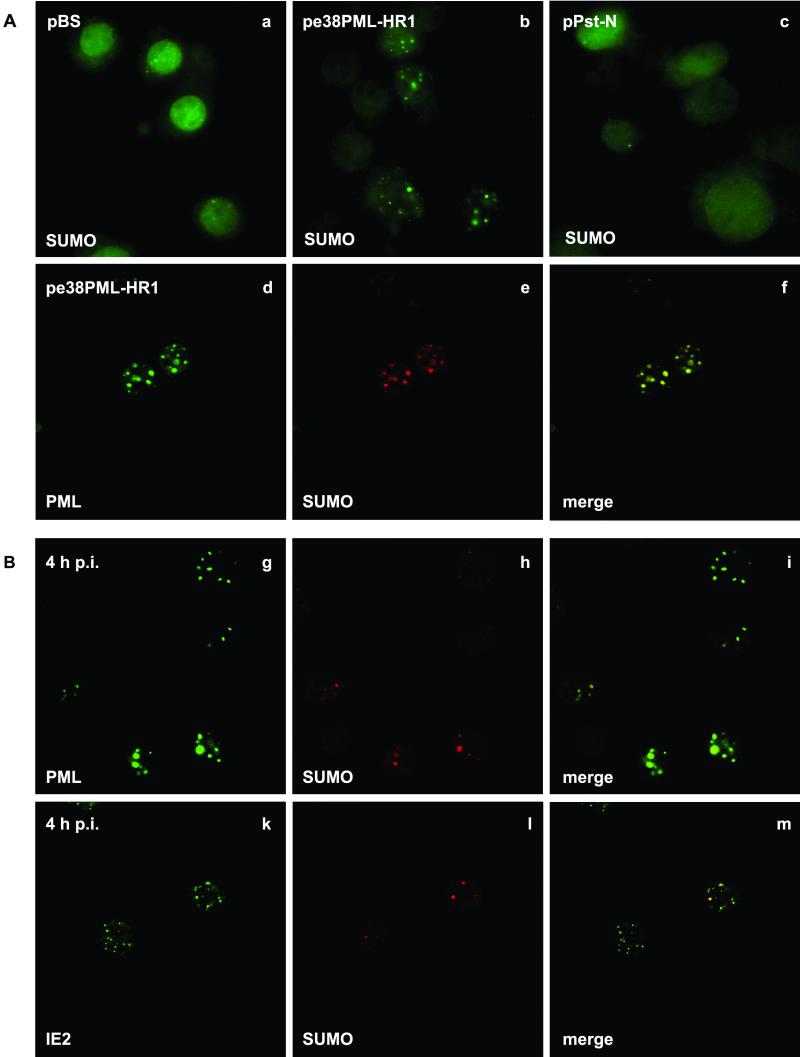
Distribution of SUMO in transfected and AcMNPV-infected TN-368 cells. TN-368 cells were transfected with plasmid pBluescript (pBS), pe38PML-HR1, expressing the pml gene, or pPst-N, expressing the ie2 gene (A) , and were infected with recombinant virus AcMNPV-PML/e (B). After fixation the cells were stained with anti-dSmt3 antiserum (a to c). Costaining was performed with anti-dSmt3 antiserum (red) and MAb 5E10 against PML (green) or with mouse anti-IE2 antiserum (green) at 4 h p.i. Confocal images and the merges are shown (d to m).
FIG. 1.
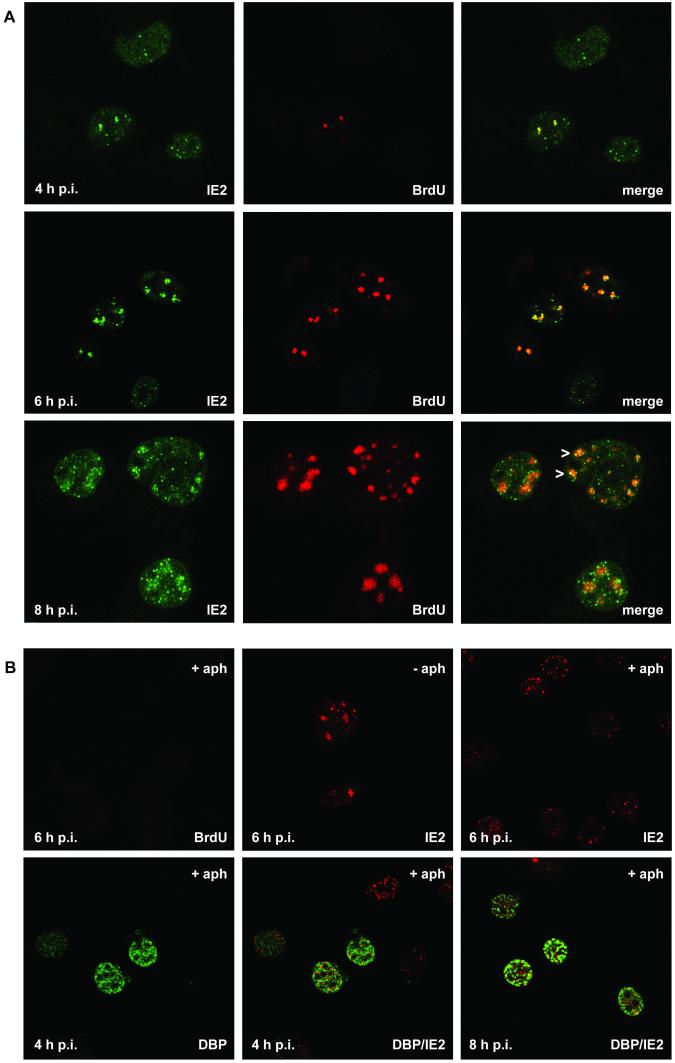
Colocalization of IE2 and viral DNA replication sites after AcMNPV infection of TN-368 cells. (A) AcMNPV-infected TN-368 cells were fixed at 4, 6, and 8 h p.i. and costained with rabbit anti-IE2 antiserum (green) and mouse BrdU MAb (red). Arrowheads indicate IE2 dot clusters. (B) AcMNPV-infected TN-368 cells were treated with aphidicolin (+ aph) or mock treated (− aph) at 1 h p.i. Cells were fixed at 4, 6, and 8 h p.i. and stained with mouse BrdU MAb (green) or mouse anti-IE2 antiserum (red). Costaining was performed with rabbit anti-DBP antiserum (green) and mouse anti-IE2 antiserum (red). Confocal images and the merges are shown.
Cell extracts and immunoblotting.
Detergent-based subcellular fractionation of TN-368 cells was performed as previously described (22). Proteins were resolved by sodium dodecyl sulfate (SDS)-10 or 7.5% polyacrylamide gel electrophoresis and processed as described previously (22). The primary rabbit antibodies were diluted 1:2,000 for anti-PE38 antiserum, 1:10,000 for anti-IE2 antiserum, and 1:10,000 for anti-DBP antiserum. The antigen-antibody complexes were identified by enhanced chemiluminescence (ECL or ECLplus system; Amersham).
RESULTS
Localization of IE2 in viral DNA replication centers during AcMNPV infection.
TN-368 cells were infected with AcMNPV, and viral DNA replication centers were visualized after incorporation of BrdU by staining with an anti-BrdU antibody. In mock-infected cells cellular DNA replication was visible as tiny dots throughout the nucleus (data not shown). In contrast, staining of viral DNA replication centers was intense and visible in distinct domains, which were detectable in about 5% of the infected cells at 4 h p.i (Fig. 1A, first [top] row). By 6 h p.i. viral DNA replication centers had enlarged and formed foci of various sizes (Fig. 1A; see Fig. 4, fourth row, and Fig. 5E). By 8 h p.i. viral DNA replication foci had enlarged further. These foci showed a granular staining and could have been formed by replication dots growing together (Fig. 1A, third row). In some cells the granular staining of the DNA replication sites had already covered most of the nucleus (see Fig. 5F).
FIG. 5.
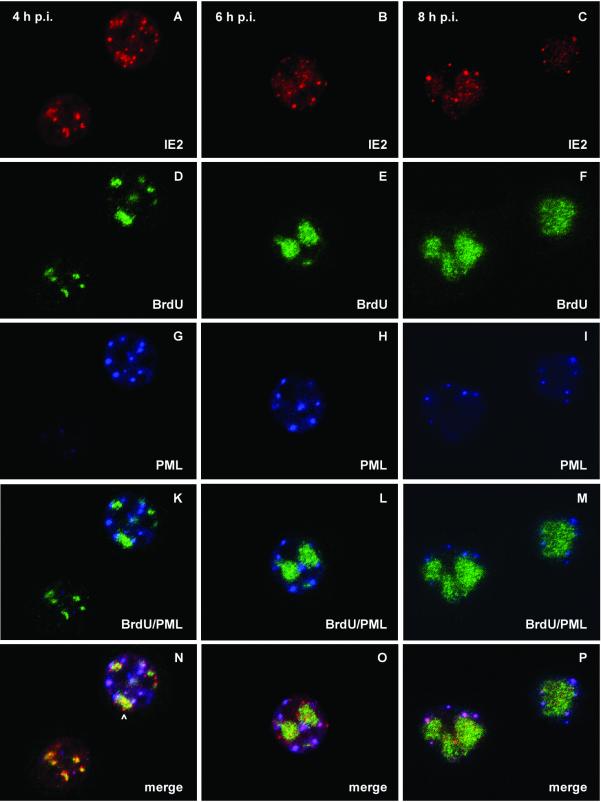
Localization of IE2, PML, and viral DNA replication sites after AcMNPV infection of TN-368 cells. TN-368 cells were infected with recombinant virus AcMNPV-PML/e. Cells were fixed at 4, 6, and 8 h p.i. and triple stained with rabbit anti-IE2 antiserum (red), rat BrdU MAb (green), and mouse MAb 5E10 (blue) to visualize PML. The arrowhead indicates IE2 dot clusters. Confocal images with double (K, L, and M) and triple merges (N, O, and P) are shown.
Staining of IE2 revealed the viral product in all cells as nuclear dots which were variable in size but which never formed foci during the infection cycle (Fig. 1A, first [left] column). As shown by confocal imaging IE2 colocalized with viral DNA replication foci, which was rather evident early in infection (Fig. 1A, third column). However, there were always more IE2 dots than initiating replication centers (Fig. 1A, third column). When DNA replication foci enlarged, clusters of distinct IE2 signals colocalized with BrdU staining, indicating that IE2 forms subdomains within the replication sites (Fig. 1A, third row). In addition, single IE2 dots were observed in close proximity to DNA replication foci (Fig. 1). Since the presence of IE2 dot clusters coincided with the enlargement of the DNA replication sites, we studied the effect of AcMNPV DNA replication inhibitor aphidicolin on the distribution of IE2 dots. In aphidicolin-treated cells, no BrdU staining was detectable (Fig. 1B). In contrast to what was found for untreated cells, a rather uniform distribution of nuclear IE2 domains, instead of clusters of IE2 dots, was observed, suggesting that the redistribution of IE2 to clusters of distinct dots is coupled to the formation of DNA replication foci (Fig. 1B).
We conclude that IE2 is present at the site of AcMNPV DNA replication from the beginning. Later in infection, IE2 forms distinct domains in the DNA replication centers, in addition to IE2 nuclear domains, that are in the vicinity of replication sites.
Localization of IE2, LEF-3, and DBP in viral DNA replication centers.
Virus-encoded factors LEF-3 and DBP have been shown to colocalize with viral DNA replication factories in cells infected with BmNPV (23). Therefore, we analyzed whether IE2 is present together with LEF-3 and DBP in common nuclear structures during AcMNPV infection.
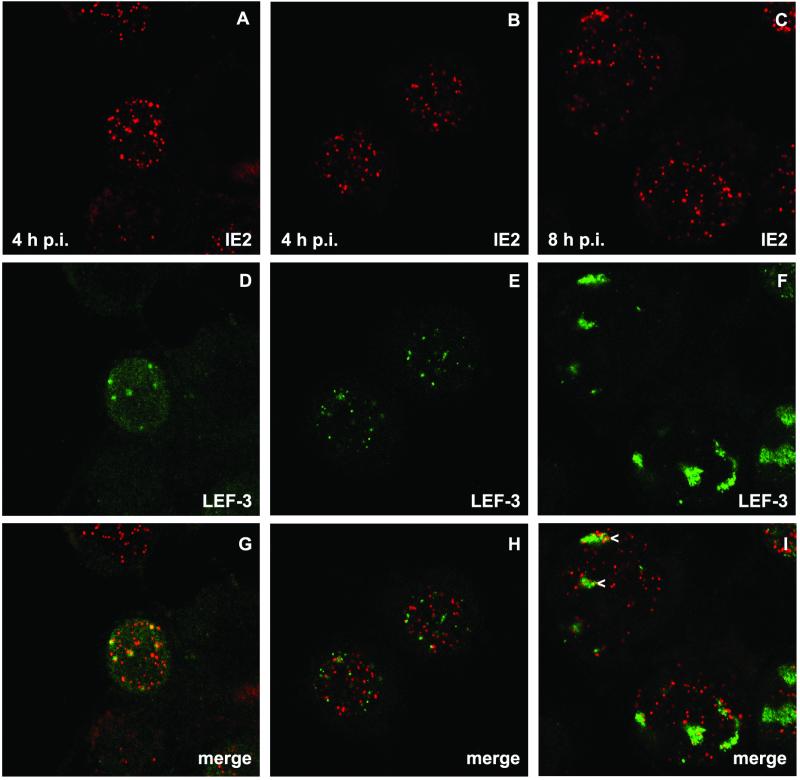
LEF-3 staining revealed nuclear dots, which were detectable in a few cells at 4 h p.i., when all cells contained distinct IE2 structures (Fig. 2D and E). By 8 h p.i. LEF-3 dots enlarged to foci that showed a granular staining reminiscent of DNA replication centers (Fig. 2F). Costaining demonstrated the colocalization of IE2 and LEF-3, at least in some nuclei, at 4 h p.i. (Fig. 2G); however, other nuclei showed LEF-3 dots that rarely had IE2 counterparts (Fig. 2H). Once LEF-3 dots enlarged, they could be found in common structures with IE2 dot clusters, a colocalization which resembles the colocalization of IE2 and viral DNA replication centers (Fig. 2I). In all costaining experiments, LEF-3 colocalized with a subpopulation of IE2 structures.
FIG. 2.
Localization of IE2 and LEF-3 after AcMNPV infection of TN-368 cells. AcMNPV-infected TN-368 cells were fixed at 4 and 8 h p.i. and costained with rabbit anti-LEF-3 antiserum (green) and mouse anti-IE2 antiserum (red). Arrowheads indicate IE2 dot clusters. Confocal images and the merges are shown.
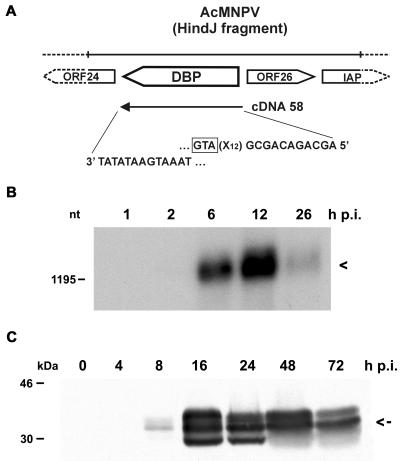
So far replication factor DBP has only been described for BmNPV (20). Therefore, one prerequisite for the colocalization experiments with IE2 and DBP was evidence for DBP of AcMNPV. The ORF encoding DBP in AcMNPV (ORF 25) has a 96% nucleotide sequence identity with the corresponding BmNPV gene (1). We isolated a DBP-specific cDNA clone from a library prepared from RNA at 1 h p.i. and analyzed the transcriptional activity during AcMNPV infection (Fig. 3A and B). Although the cDNA clone indicated the presence of the dbp transcript at 1 h p.i., it was only very weakly detectable at 2 h p.i., even when polyadenylated RNA on Northern blots was investigated (Fig. 3B). After hybridization to DBP-specific cDNA clone 58, a transcript of about 1,300 nucleotides (nt) was observed mainly at 6 and 12 h p.i., with a decrease at 26 h p.i. (Fig. 3B). Its size is consistent with the predicted ORF of 948 nt and a poly(A) tail of about 300 nt.
FIG. 3.
Characterization of DBP from AcMNPV. (A) Schematic representation of the dbp ORF on the AcMNPV genome. The sequences at the 5′ and 3′ ends of dbp cDNA clone 58 are indicated. IAP, inhibitor of apoptosis genes. The translational start codon of the DBP ORF (GTA) is boxed. (B) Polyadenylated RNA (10 μg) isolated from S. frugiperda cells at 1, 2, 6, 12, and 26 h p.i. was analyzed on a 1.2% agarose gel containing 2.2 M formaldehyde. The Northern blot was hybridized to DBP-specific cDNA clone 58. Arrowhead, transcript of about 1,300 nt. The position of the DNA size marker is given on the left. (C) Nuclear protein extracts were prepared from uninfected cells (lane 0) and from cells at 4, 8, 16, 24, 48, and 72 h p.i. Proteins were resolved on SDS-10% polyacrylamide gels and stained with anti-DBP antiserum. Arrow, middle-sized protein band of about 34 kDa. Protein size markers are given on the left.
In nuclear protein fractions of AcMNPV-infected cells the antibody directed against BmNPV DBP recognized a protein of about 34 kDa seen as multiple bands in SDS-polyacrylamide gel electrophoresis; this might indicate posttranslational modifications (Fig. 3C). The middle-sized DBP band was also found in cytoplasmic protein fractions (data not shown). DBP expression was weakly observed at 8 h p.i.; there was a significant increase until 24 h p.i. and a decrease at 48 h p.i. (Fig. 3C). While the onset of DBP expression coincides roughly with its transcriptional activity, the presence of DBP during the late phases of infection suggests its high stability.
Indirect immunofluorescence studies of AcMNPV-infected cells demonstrated DBP in nuclear dots that rapidly expanded to foci (Fig. 4, first column). As with LEF-3 the first DBP staining was detected in the nucleus at 4 h p.i. When viral DNA replication centers and DBP were costained, the merging of the signals indicated their colocalization in nearly all DBP foci and even in the smaller domains (Fig. 4, fourth row). By 10 h p.i. the enlarged DBP foci corresponded to the DNA replication centers although DBP staining was also observed at the rim of the nuclei (Fig. 4, sixth row). The staining pattern suggests that formation of DBP foci is coupled to the initiation of viral DNA replication. When DBP was transiently expressed in TN-368 cells, distinct DBP structures were evenly distributed in the nucleus and never formed foci, even in cells with high DBP expression (data not shown). Similar results were obtained when viral DNA replication was blocked with aphidicolin, which abolished the formation of DBP foci (Fig. 1B). Our observations are in agreement with previous results that demonstrate a similar staining pattern of BmNPV DBP in B. mori cells and suggest the association of enlarged DBP foci and viral DNA replication (23).
By 4 h p.i. some cells expressed both DBP and IE2, mainly with more IE2 dots than DBP dots. In a subpopulation of these cells colocalization of IE2 and DBP was rather evident, while in other cells common domains of IE2 and DBP were rarely detectable (Fig. 4, first and second rows). Similar observations were made at 6 h p.i., when more cells showed DBP dots and DBP foci were already detectable in a few cells (Fig. 4, third row). As soon as IE2 formed dot clusters, colocalization with DBP was observed, indicating that IE2 colocalized with DBP in DNA replication sites (Fig. 4, second, third and fifth rows). In addition, IE2 structures were present in close proximity to DBP foci (Fig. 4, third column). After inhibition of viral DNA replication with aphidicolin, colocalization of IE2 and DBP became less evident, suggesting that the formation of common IE2/DBP domains presupposes the establishment of viral DNA replication sites (Fig. 1B).
In summary, the costaining results for IE2/DBP and IE2/LEF-3 demonstrate comparable extents of colocalization, which means that only a subpopulation of IE2 structures colocalized with LEF-3 and DBP, respectively. In contrast to those of IE2, DBP and LEF-3 structures always colocalized and enlarged simultaneously with the sites of viral DNA replication, except when DBP was present at the rim of the nuclear membrane. Therefore, we assume that LEF-3 and DBP structures represent viral DNA replication sites with which certain IE2 structures become associated. Since DBP and LEF-3 appear later in the infection cycle than IE2, one might envision that DBP and LEF-3 structures assemble at preexisting IE2 nuclear domains. However, our analysis demonstrated that, even when very few DBP and LEF-3 dots were present early in infection, there were always some dots that had no IE2 counterpart, suggesting that DBP and LEF-3 structures may assemble independently from IE2. We assume that the assembly of LEF-3 and DBP structures depends on the initiation of viral DNA replication followed by colocalization with IE2.
Localization of IE2 and PML with respect to viral replication centers.
Previous infection studies with recombinant virus AcMNPV-PML/e, which expresses the human pml gene, demonstrate the colocalization of IE2 and PML during the early phase of infection, followed by a redistribution of both proteins (22). Since we observed that a subpopulation of IE2 structures colocalized with viral DNA replication sites, we asked whether PML also forms common domains with viral DNA replication centers or whether PML colocalizes with IE2 adjacent to replication sites. After infection with recombinant virus AcMNPV-PML/e, PML staining was rarely detectable at 4 h p.i., even in those cells that already showed staining of DNA replication sites. Only at 4 h p.i., when viral DNA replication sites had enlarged, was PML staining detectable (Fig. 5D and G). Costaining of PML and viral DNA replication sites indicated no significant colocalization (Fig. 5K to M). This observation is in line with the finding that colocalization of PML and DBP was not detectable (data not shown). However, from 4 to 8 h p.i., PML domains were detectable in the vicinity of DNA replication centers rather than randomly distributed throughout the nucleus (Fig. 5K to M). Triple staining of IE2, PML, and viral DNA replication centers revealed that IE2 colocalized with PML adjacent to DNA replication sites while few IE2/PML domains seemed to be unrelated to DNA replication sites (Fig. 5N to P). When IE2 dot clusters colocalized with DNA replication sites, no colocalization with PML was observed (Fig. 5N). As indicated by inhibitor studies the formation of IE2/PML domains was independent of viral DNA replication (data not shown).
As a control, cells infected with wild-type AcMNPV were compared to those infected with recombinant virus AcMNPV-PML/e to exclude the possibility that PML expression influences the localization of IE2 in viral replication centers. Infection studies confirmed that the IE2 localization in wild-type virus-infected cells did not differ from that in recombinant virus-infected cells (data not shown).
In summary, IE2/PML domains represent nuclear structures that are juxtaposed to, but that do not resemble, viral DNA replication sites. In addition, IE2 forms subdomains in DNA replication sites that are different from PML domains. Thus, it is conceivable that IE2 is part of various functional sites that are related to viral DNA replication.
Localization of a small ubiquitin-like modifier in insect cells.
Nuclear PML in mammalian cells was described as one of the proteins to which small ubiquitin-like modifier SUMO-1 can be covalently conjugated (2, 26). It has been proposed that SUMO-1 modification regulates the nuclear localization of PML and can modulate its cellular interactions (4, 21). Recently, the Drosophila melanogaster homologue of SUMO-1, designated dSmt3, was characterized (10, 15). We used a polyclonal anti-dSmt3 antiserum to address the question of whether PML or IE2 or both are modified by SUMO in insect cell line TN-368; the answer might have implications for their nuclear localization patterns. Thus, we examined the subcellular distributions of dSmt3 homologues in uninfected TN-368 cells and cells infected with the recombinant virus AcMNPV-PML/e. While uninfected cells exhibited a diffuse nuclear staining pattern, punctate structures in the nuclei of infected cells were observed, which indicates that a SUMO homologue is detectable with the anti-dSmt3 antiserum in TN-368 cells (data not shown). To understand whether PML or IE2 or both cause the distinct change in SUMO staining, we performed transient-expression studies. After transfection of a plasmid that expressed PML, anti-dSmt3 antibodies stained nuclear structures, which was in contrast to the diffuse staining pattern after transient expression of IE2 and in mock-transfected cells (Fig. 6a to c). Colocalization of SUMO and PML signals both after transient expression and in AcMNPV-PML/e-infected cells was confirmed by confocal imaging (Fig. 6f and i). When IE2 and SUMO were costained, the merging of the signals in part of the IE2 domains was observed; this suggests that conjugation with SUMO is only detectable in common IE2/PML domains (Fig. 6m). Taken together, our results support the notion that a SUMO homologue of TN-368 cells is recruited to PML dots.
DISCUSSION
By investigating the spatial distribution of IE2 in the nuclei of AcMNPV-infected cells with respect to the sites of viral DNA synthesis, we demonstrated that IE2 structures localized to DNA replication centers and were associated with nuclear structures formed by viral DNA replication factors LEF-3 and DBP. Thus, we conclude that IE2 is part of the viral DNA replication compartments. The dynamic pattern of colocalization of IE2 with viral and cellular factors during the infection cycle gave further insights into how IE2 may contribute to viral DNA replication. Early in infection the sites of initiation of viral DNA synthesis localized always to IE2 structures, although only to some of the IE2 domains, which argues that viral DNA replication initiated at distinct IE2 structures. Since LEF-3 and DBP are SSBs, one might expect their colocalization with IE2/early viral DNA replication sites. Furthermore, LEF-3 and DBP expression is delayed in comparison with that of IE2; hence, distinct IE2 structures might target the viral SSBs to the sites of viral DNA replication initiation. DBP formed common domains with early replication centers and was found to colocalize with IE2; however, DBP and LEF-3 were also present in structures distinct from IE2 domains. These observations suggest that DBP and LEF-3 may assemble independently from IE2 structures and that both factors can colocalize with IE2 in replication sites as soon as viral DNA replication is detectable.
While the LEF-3 and DBP structures enlarged simultaneously with viral DNA replication centers, IE2 formed clusters of distinct domains within the replication compartments. Blockage of viral DNA synthesis by aphidicolin indicates that both the enlargement of the foci and the formation of IE2 subcompartments within DNA replication centers depend on viral DNA synthesis.
During infection with nuclear DNA viruses such as simian virus 40, adenovirus 5, and HSV-1 the initiation of DNA replication adjacent to PML domains has been frequently observed (for a review see reference 17). Interestingly, parental viral genomes show the tendency, initially described for HSV-1 genomes, to associate with the periphery of PML domains (18). In earlier studies we demonstrated that human PML and AcMNPV protein IE2 colocalize at the early phase of infection (22). Although it was expressed in a heterologous cellular environment, PML formed domains in AcMNPV-infected insect cells that localized in the vicinity of initiating viral DNA replication centers. These findings suggest that POD-like structures are conserved in nonvertebrates, thereby strengthening the assumption that incoming viral genomes are targeted in a nonrandom manner. Since the common domains of IE2 and PML were adjacent to the viral DNA replication sites, it is possible that IE2 has to be released from PML domains to colocalize with the sites of viral DNA replication initiation. It will be of great interest to identify further cellular and viral interaction partners that accumulate both at the IE2/PML domains and the IE2 subdomains in the DNA replication sites to shed light on the functional role of IE2 in various nuclear structures during viral DNA replication.
We present the first evidence that a SUMO homologue which can be recruited to PML domains exists in TN-368 cells. SUMO modification is thought to affect protein targeting, and SUMO is thought to be an antagonist of its structural counterpart ubiquitin (for a review see reference 11). Although the SUMO-modified form of PML in mammalian cells is thought to be restricted to PODs, the molecular role of SUMO-1 modification of various PML isoforms is still unclear. For TN-368 insect cells, whether colocalization of PML and SUMO indicates a SUMO-conjugated PML form and whether IE2 is conjugated to SUMO once it is recruited to PML domains remain to be shown.
In summary, our results lead us to assume that during AcMNPV infection IE2 structures are formed in association with cellular and viral factors in a nonrandom manner and that these structures resemble various functional domains in the nucleus that are related to viral DNA replication.
Acknowledgments
We thank Keiju Okano for the gift of the antiserum against BmNPV DBP, George F. Rohrmann for the gift of the anti-LEF-3 antiserum, Anne Dejean for the gift of anti-dSmt3 antibodies, Roel van Driel for kindly providing MAb 5E10, and Neil Smyth and Mario Schelhaas for critical comments on the manuscript.
This research was supported by grant SFB-A3 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 2.Boddy, M. N., K. Howe, L. D. Etkin, E. Solomon, and P. S. Freemont. 1996. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukemia. Oncogene 13:971-982. [PubMed] [Google Scholar]
- 3.de Bruyn Kops, A., and D. M. Knipe. 1994. Preexisting nuclear architecture defines the intranuclear location of herpesvirus DNA replication structures. J. Virol. 68:3512-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duprez, E., A. J. Saurin, J. M. Desterro, V. Lallemand-Breitenbach, K. Howe, M. N. Boddy, E. Solomon, H. de The, R. T. Hay, and P. S. Freemont. 1999. SUMO-1 modification of the acute promyelocytic leukemia protein PML: implications for nuclear localization. J. Cell Sci. 112:381-393. [DOI] [PubMed] [Google Scholar]
- 5.Evans, J. T., and G. F. Rohrmann. 1997. The baculovirus single-stranded DNA binding protein, LEF-3, forms a homotrimer in solution. J. Virol. 71:3574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans, J. T., G. S. Rosenblatt, D. J. Leisy, and G. F. Rohrmann. 1999. Characterization of the interaction between the baculovirus ssDNA-binding protein (LEF-3) and putative helicase (P143). J. Gen. Virol. 80:493-500. [DOI] [PubMed] [Google Scholar]
- 7.Gardiner, G. R., and H. Stockdale. 1975. Two tissue culture media for production of lepidopteran cells and nuclear polyhedrosis viruses. J. Invertebr. Pathol. 25:363-370. [Google Scholar]
- 8.Hang, X., W. Dong, and L. A. Guarino. 1995. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein. J. Virol. 69:3924-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hink, W. F. 1970. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature 226:466-467. [DOI] [PubMed] [Google Scholar]
- 10.Huang, H. W., S. C. Tsoi, Y. H. Sun, and S. S. Li. 1998. Identification and characterization of the SMT3 cDNA and gene encoding ubiquitin-like protein from Drosophila melanogaster. Biochem. Mol. Biol. Int. 46:775-785. [DOI] [PubMed] [Google Scholar]
- 11.Jentsch, S., and G. Pyrowolakis. 2000. Ubiquitin and its kin: how close are the family ties. Trends Cell Biol. 10:335-342. [DOI] [PubMed] [Google Scholar]
- 12.Kool, M., C. Ahrens, R. W. Goldbach, G. F. Rohrmann, and J. M. Vlak. 1994. Identification of genes involved in DNA replication of Autographa californica baculovirus. Proc. Natl. Acad. Sci. USA 91:11212-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krappa, R., and D. Knebel-Mörsdorf. 1991. Identification of the very early transcribed baculovirus gene PE-38. J. Virol. 65:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krappa, R., R. Roncarati, and D. Knebel-Mörsdorf. 1995. Expression of PE38 and IE2, viral members of the C3HC4 finger family, during baculovirus infection: PE38 and IE2 localize to distinct nuclear regions. J. Virol. 69:5287-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehembre, F., P. Badenhorst, S. Müller, A. Travers, F. Schweisguth, and A. Dejean. 2000. Covalent modification of the transcriptional repressor tramtrack by the ubiquitin-related protein Smt3 in Drosophila flies. Mol. Cell. Biol. 20:1072-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu, A., and L. K. Miller. 1995. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J. Virol. 69:975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660-667. [DOI] [PubMed] [Google Scholar]
- 18.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67-75. [DOI] [PubMed] [Google Scholar]
- 19.McDougal, V. V., and L. A. Guarino. 1999. Autographa californica nuclear polyhedrosis virus DNA polymerase: measurements of processivity and strand displacement. J. Virol. 73:4908-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikhailov, V. S., A. L. Mikhailova, M. Iwanago, S. Gomi, and S. Maeda. 1998. Bombyx mori nucleopolyhedrovirus encodes a DNA-binding protein capable of destabilizing duplex DNA. J. Virol. 72:3107-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murges, D., I. Quadt, J. Schröer, and D. Knebel-Mörsdorf. 2001. Dynamic nuclear localization of the baculovirus proteins IE2 and PE38 during the infection cycle: the promyelocytic leukemia protein colocalizes with IE2. Exp. Cell Res. 264:219-232. [DOI] [PubMed] [Google Scholar]
- 23.Okano, K., V. S. Mikhailov, and S. Maeda. 1999. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J. Virol. 73:110-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 25.Seipel, K., O. Georgiev, and W. Schaffner. 1992. Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions. EMBO J. 11:4961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sternsdorf, T., K. Jensen, and H. Will. 1997. Evidence for covalent modification of the nuclear dot-associated proteins PML and SP100 by PIC1/SUMO-1. J. Cell Biol. 139:1621-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuurman, N., A. de Graaf, A. Floore, A. Josso, B. Humbel, L. de Jong, and R. van Driel. 1992. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101:773-784. [DOI] [PubMed] [Google Scholar]
- 28.Tjia, S. T., E. B. Carstens, and W. Doerfler. 1979. Infection of Spodoptera frugiperda cells with Autographa californica nuclear polyhedrosis virus. II. The viral DNA and the kinetics of its replication. Virology 99:399-409. [DOI] [PubMed] [Google Scholar]
- 29.Vaughn, J. L., R. H. Goodwin, G. J. Tompkins, and P. McCawley. 1977. The establishment of two cell lines from the insect Spodoptera frugiperda. In Vitro 13:213-217. [DOI] [PubMed] [Google Scholar]
- 30.Wu, Y., and E. B. Carstens. 1998. A baculovirus single-stranded DNA binding protein, LEF-3, mediates the nuclear localization of the putative helicase P143. Virology 247:32-40. [DOI] [PubMed] [Google Scholar]