Abstract
The mammalian genome depends on patterns of methylated cytosines for normal function, but the relationship between genomic methylation patterns and the underlying sequence is unclear. We have characterized the methylation landscape of the human genome by global analysis of patterns of CpG depletion and by direct sequencing of 3073 unmethylated domains and 2565 methylated domains from human brain DNA. The genome was found to consist of short (<4 kb) unmethylated domains embedded in a matrix of long methylated domains. Unmethylated domains were enriched in promoters, CpG islands, and first exons, while methylated domains comprised interspersed and tandem-repeated sequences, exons other than first exons, and non-annotated single-copy sequences that are depleted in the CpG dinucleotide. The enrichment of regulatory sequences in the relatively small unmethylated compartment suggests that cytosine methylation constrains the effective size of the genome through the selective exposure of regulatory sequences. This buffers regulatory networks against changes in total genome size and provides an explanation for the C value paradox, which concerns the wide variations in genome size that scale independently of gene number. This suggestion is compatible with the finding that cytosine methylation is universal among large-genome eukaryotes, while many eukaryotes with genome sizes <5 × 108 bp do not methylate their DNA.
The methylation of CpG-rich promoter sequences imposes strong transcriptional silencing that can be transmitted by clonal inheritance in somatic cells for >80 cell generations (Wigler et al. 1981; Stein et al. 1982; Schubeler et al. 2000). Genetic studies in mice and humans (for review, see Goll and Bestor 2005) have shown that demethylation causes the reactivation of retrotransposons in both germ and somatic cells and the loss of monoallelic expression at imprinted loci. Global reductions in methylation levels caused by mutations in DNA methyltransferase genes are lethal (Li et al. 1992). Overexpression of Dnmt1 in transgenic mice causes de novo methylation at imprinted loci and is also lethal (Biniszkiewicz et al. 2002), while partial demethylation of the genome in mice that carry hypomorphic alleles of Dnmt1 leads to high rates of lymphoma (Gaudet et al. 2002). Global demethylation (Goelz et al. 1985) with ectopic de novo methylation of certain CpG islands (Greger et al. 1989) has been reported in human cancers. The human chromosome instability and immunodeficiency disorder known as ICF syndrome (OMIM 242860) is caused by mutations in the DNA methyltransferase 3B (DNMT3B) gene (Xu et al. 1999).
The actual methylation landscape of the human genome remains poorly defined, in part because current methods for methylation profiling depend on hybridization of probes or primers, which requires prior selection of regions to be tested. Sequence selection introduces bias, and hybridization methods cannot distinguish between members of repeat families (Weber et al. 2005). This is a major shortcoming, as interspersed repeats are abundant within and between genes, and most 5-methylcytosine lies within tandem and dispersed repeats (Yoder et al. 1997). CpG islands are thought to be normally unmethylated and are associated with 75% of human genes (Ioshikhes and Zhang 2000). CpG islands are defined operationally as sequences that have G+C contents of >0.55, observed versus expected CpG densities of >0.5, and a length of >300 bp (although very few are <500 bp) (Aerts et al. 2004). The CpG island compartment has been held to be constitutively unmethylated. Existing techniques for methylation profiling have not produced a uniform or widely accepted description of genomic methylation patterns.
While many findings confirm that the normal function of the genome depends on methylation patterns and that perturbations of genomic methylation patterns are often lethal, the structure of genomic methylation patterns is not well understood and the biological functions of cytosine methylation have long been controversial. We have developed new analytical tools and new software that allows the mapping of sequence annotation onto large assemblages of genomic sequence that has been fractionated by methylation status in a sequence-independent manner. The result is a coherent image of the structure of genomic methylation that suggests a resolution to the long-standing C-value paradox (Thomas Jr. 1971).
Results
Germ-line methylation patterns inferred from genome-wide patterns of CpG depletion
The availability of an essentially complete sequence of the euchromatic human genome allowed the development of new approaches to the definition of the large-scale structure of genomic methylation patterns. In the first approach, analysis of genome-wide CpG depletion in different sequence compartments was used to deduce germ-line methylation status, and in the second approach, the structure of methylation patterns in somatic cells was deduced from high-resolution methylation data obtained from a large (∼3 × 106 bp) random sample of methylated and unmethylated domains from human brain.
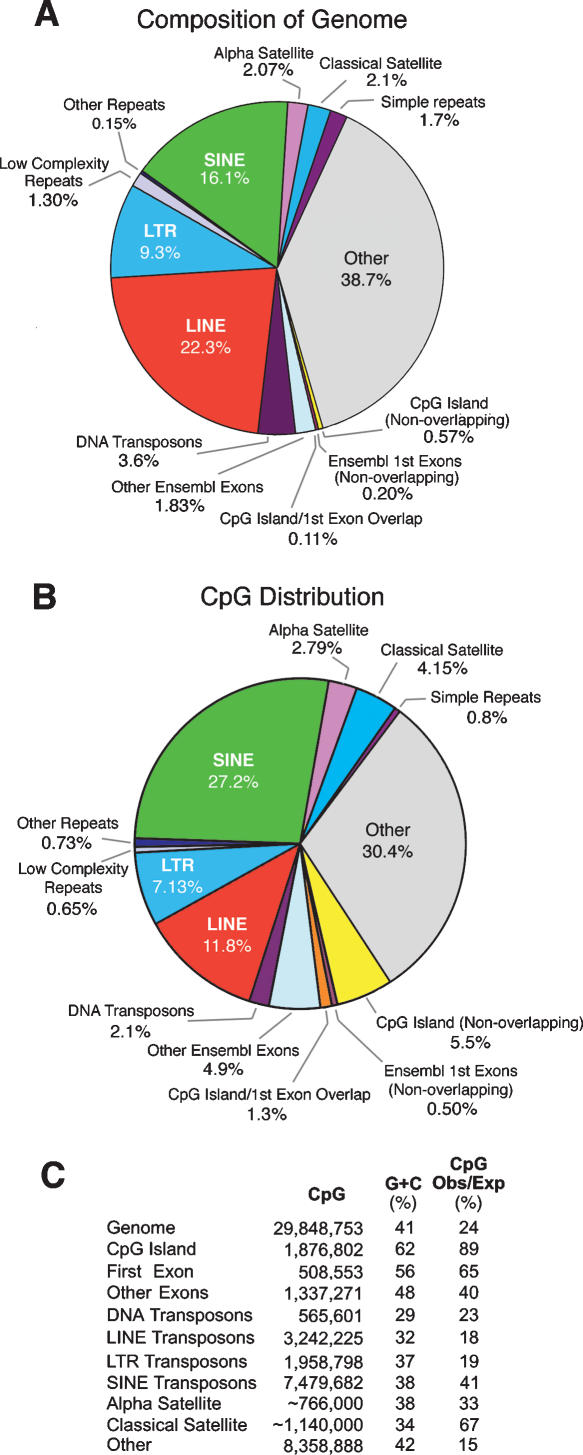
Indirect evidence that a sequence exists in the methylated state in the germ line is provided by an observed density of CpG sites below that predicted by local nucleotide frequencies. Deamination of m5C converts the base directly to thymine, and C→T transition mutations at methylated CpG sites are ∼18-fold more frequent than the mean of other point mutations (Kondrashov 2002). Figure 1, B and C, show that CpG depletion is especially pronounced in LINE (long interspersed nuclear element) transposons and the LTRs (long terminal repeats) of endogenous retroviruses, which contain, respectively, only 18% and 19% of the CpG sites predicted by nucleotide composition. CpG retention in the SINE (short interspersed nuclear element) class is 41% of the expected value; this reflects the relatively recent accumulation of large numbers of primate-specific Alu SINE transposons, most of which have accumulated in the last 60 million years (Lander et al. 2002). The most severe CpG depletion (15% of expected) is seen in unannotated single-copy sequences denoted as “other” in Figure 1. This compartment contains the oldest transposons, pseudogenes, and other sequences that have been evolving under the neutral rate for long periods of time and have been severely eroded by the accumulation of mutations (Lander et al. 2002). Genome-wide analysis showed that CpG islands (which are associated with the promoters of at least 75% of human genes [Ioshikhes and Zhang 2000] and have a high G+C content that is almost entirely the result of a high CpG density [Aerts et al. 2004]) represent only 0.68% of the genome (Fig. 1A) but contain 6.8% of all CpG sites (Fig. 1B) and have a CpG content that is only 11% below that predicted by nucleotide composition (Fig. 1C). CpG islands were found to represent the only major sequence compartment subject to CpG depletion of less than twofold, which suggests that CpG islands represent the only major sequence compartment that is unmethylated in the germ line. This analysis of CpG depletion indicates that transposons are methylated in the germ line, while CpG islands are largely unmethylated.
Figure 1.
Composition of the human genome and distribution of CpG sites. (A) Representation of sequence compartments in the human genome. (B) CpG content of the sequence compartments shown in A. (C) Total CpG numbers, G+C densities, and observed versus expected CpG frequencies for the compartments shown in A and B. The CpG island category included both first exon overlapping and nonoverlapping classes. Estimates of amount and CpG content of α and classical satellite DNA were from Eichler et al. (2004) and E.E. Eichler, pers. commun.
Direct analysis of methylated and unmethylated domains
Human brain DNA was fractionated into methylated and unmethylated compartments to test whether methylation patterns in somatic cells had a structure similar to that inferred from patterns of CpG depletion. New methods for the mass isolation of methylated and unmethylated domains were developed in order to allow determination of methylation status of all sequences, including repeats, and to avoid the biases that arise from prior selection of particular genomic regions, which is required by methods of methylation profiling that involve the use of probes or primers. Unmethylated domains were isolated by limit digestion of DNA from a full-term male human brain with McrBC, an enzyme complex from Escherichia coli that degrades DNA between methylated half sites of the form Rm5C-N40–500-Rm5C. McrBC has been used in other applications to remove methylated sequences from DNA of plants and vertebrates and to map methylated sites (Lee et al. 2002; Palmer et al. 2003). Methylated domains were isolated by means of their resistance to the methylation-sensitive restriction endonucleases TaiI (ACGT), BstUI (CGCG), HhaI (GCGC), HpaII (CCGG), and AciI (CCGC and GCGG).
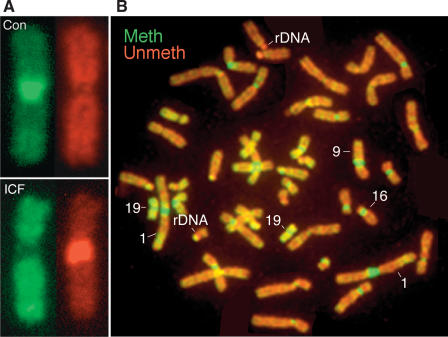
The organization of genomic methylation patterns in brain DNA was first characterized at cytogenetic scale by in situ hybridization of methylated and unmethylated DNA against normal human metaphase chromosomes. DNA from ICF syndrome cells (Xu et al. 1999) was used to confirm that fractionation was on the basis of methylation status only and not other attributes (Fig. 2A; Supplemental Fig. S1). Figure 2A shows that the classical satellite DNA (satellite 3) on chromosome 9 is sensitive to McrBC and resistant to restriction endonucleases in the control sample, and that the pattern is reversed in DNA from ICF syndrome patients; these individuals are unable to methylate classical satellite DNA as a result of mutations in DNMT3B (Xu et al. 1999). As shown in Figure 2B, those sequences that are least methylated and those that are most methylated correspond to tandem-repeated sequences that are not present in current genome assemblies. Classical satellite DNA on the juxtacentromeric long arms of chromosomes 1, 9, and 16 is heavily methylated, as previously reported (Miller et al. 1974), while the rDNA repeats on the acrocentric short arms of group D chromosomes (13, 14, and 15) and group G chromosomes (21 and 22) are largely unmethylated. Cytogenetic analysis of genomic methylation patterns by hybridization analysis is consistent with earlier findings that used antibodies to m5C (Miller et al. 1974). This further confirms that the fractionation method used in this study does separate methylated sequences from unmethylated sequences.
Figure 2.
Analysis of genomic methylation patterns at cytogenetic scale. (A) Methylated and unmethylated DNA was isolated from control and ICF syndrome fibroblasts, labeled with contrasting fluorophores, and hybridized against normal human metaphase chromosomes. Satellite 3 on chromosome 9 can be seen to be methylated in the control (top) and unmethylated in ICF syndrome DNA (bottom). Methylated DNA is labeled in green, unmethylated DNA in red. (B) As in A, except that DNA was from normal human brain and all chromosomes are shown. Dense methylation is seen as green and is present at classical and α satellite DNA; demethylation is most conspicuous at rDNA repeats on acrocentric short arms. The regions of most intense labeling with both methylated and unmethylated probes correspond to tandem repeat sequences that are not part of current genome assemblies.
High-resolution methylation profiling of brain DNA was performed by direct sequencing of ends of inserts from plasmid libraries that contained methylated or unmethylated domains. Alignment of sequence reads to genome sequence via BLAT allowed addition of sequence between the paired end reads. Of the 4252 unmethylated and 3501 methylated domains that were sequenced, 3072 and 2565, respectively, could be oriented with regard to the direction of transcription of an overlapping gene; these sequences were subjected to further analysis via new graphical methods developed to allow display of annotated features on large assemblages of genomic sequences (Fig. 3). Methylated and unmethylated domains were sorted by length and displayed as stacks of sequences, with 5 kb of genomic sequence (of undetermined methylation status) added to either end to provide context. Methylated McrBC cleavage sites that border unmethylated domains, and unmethylated restriction endonuclease cleavage sites that border methylated domains, are shown as short gaps in the sequence stacks. Total unmethylated sequence analyzed was 13,795,647 bp; total methylated sequence was 8,235,466 bp. All alignment was by methylation status rather than by sequence. Annotated features from the University of California, Santa Cruz genome browser were mapped onto the sequence stacks and shown in a contrasting color, with a threefold increase in line weight to aid visualization.
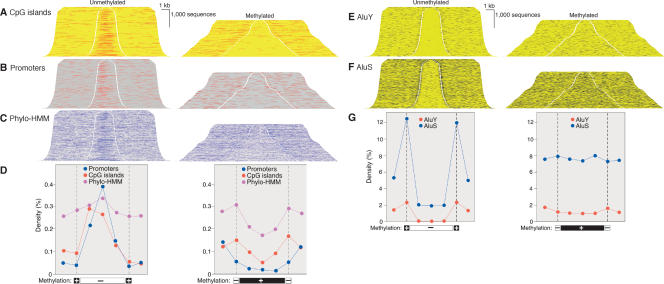
Figure 3.
Regulatory sequences and Alu transposons in unmethylated and methylated domains. DNA was fractionated as shown in Figure 2 and cloned into plasmid vectors. BLAT alignment of paired end sequence reads allowed addition of sequence between end reads and of external sequence. Sequences were sorted by insert length and assembled into stacks; the gap indicates position of cleavage by McrBC (left) or RE (right). Genome features annotated at the UCSC genome browser (http://genome.ucsc.edu) were mapped onto sequence stacks in contrasting colors and with a threefold increase in line weight to improve visibility. All sequences shown were associated with a transcript annotated in Ensembl and transcription was from left to right.(A) Enrichment of CpG islands in unmethylated domains and depletion in methylated domains. (B,C) Enrichment in unmethylated domains and depletion in methylated domains of promoters (defined as in Trinklein et al. 2003) and sequences conserved among human, mouse, and rat by phylogenetic Hidden Markov Model analysis (Jojic et al. 2004). (D) Quantitative analysis of data shown in A,B,C. Methylated and unmethylated domains were normalized for length and divided into thirds. Feature density is plotted against the mean of the 5 kb of flanking sequence, at the cleavage site (indicated by broken vertical line) and for each of the three segments of the methylated and unmethylated domains. There were 142,694 CpG sites in the methylated domains and 75,017 in the unmethylated domains; total sequence lengths were 13,795,647 and 8,235,466 bp, respectively. (E,F) Exclusion of AluS and AluY transposons from unmethylated domains. (G) Quantitation of data of E and F as in Figure 1D.
As shown in Figure 3, unmethylated domains are strongly enriched in features annotated as CpG islands (Tykocinski and Max 1984; Gardiner-Garden and Frommer 1987). The CpG islands in unmethylated domains had a median length of 771 nucleotides (standard deviation of 518 nucleotides) but only 354 nucleotides (standard deviation 334) in methylated domains. This suggests that many of the short sequences annotated as CpG islands in methylated domains may represent false positives in the sequence annotation, which predict CpG islands according to somewhat arbitrary thresholds of length, CpG density, and G+C content. CpG islands associated with first exons of known genes are rarely <500 bp (Aerts et al. 2004). Inspection of each of the annotated CpG islands shown in the methylated domains of Figure 3A showed that only one was associated with the 5′ end of an annotated gene; further analysis of this island showed that it was associated with the Synaptonemal Complex Protein 3 (SYCP3) gene and was methylated in all tested tissues other than sperm (data not shown). The significance of this single methylated promoter is unknown. As is also shown in Figure 3, promoters (defined empirically as the 500 bp 5′ of a known first exon or EST cluster [Trinklein et al. 2003]) are strongly enriched in unmethylated domains and are depleted from methylated domains. Enrichment in unmethylated domains and depletion from methylated domains is also apparent in the density of sequences that are conserved among human, mouse, and rat as established by phylogenetic Hidden Markov Model analysis (Jojic et al. 2004), but the magnitude of the trend was much reduced by comparison to the enrichment of CpG islands in the unmethylated sequences and depletion from methylated sequences (Fig. 3D,E,F). This suggests that methylation status, which is not directly sequence based, will be a useful discriminant in efforts to identify regulatory regions that currently depend on sequence-based metrics. Consideration of methylation status may reduce the false-positive rates in current methods of promoter prediction.
Transposon proliferation is a penalty of sexual reproduction and DNA methylation has been proposed to have evolved to defend the host against the deleterious effects of transposon accumulation (Bestor 2003). In support of this view is the finding that transposons are heavily methylated in all known eukaryotes that have methylated genomes, and in the ascomycete fungus Neurospora crassa all methylated sequences were found to be transposon derived (Selker et al. 2003). Transposons are also specific targets of methylation in plants (Tompa et al. 2002) and mammals (Bourc'his and Bestor 2004). The relationship of methylation and transposons in the human genome was studied by mapping annotated transposons onto sequence stacks arranged by methylation status. As shown in Figure 3, E,F,G, Alu transposons (the most numerous human transposon at 1.2 × 106 copies per haploid genome; Lander et al. 2002) are largely excluded from unmethylated domains. Figure 3, E,F,G, also shows that the boundaries of unmethylated domains tend to be occupied by methylated Alu transposons of the younger AluS and AluY families.
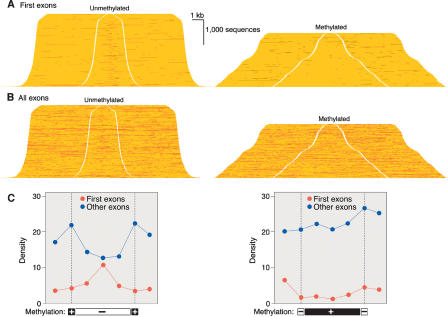
The data of Figure 3 indicate that promoters and first exons are located within unmethylated domains, while exons other than first exons are more prone to occur within methylated domains. Exons have retained a relatively high CpG density (40% of that expected on the basis of nucleotide frequencies) due to negative selection constraints imposed by codon usage and by a marked CpG enrichment in exonic sequences near splice sites due to a postulated role of CpG-rich sequences in RNA splicing (Majewski and Ott 2002). The methylated state observed here for non-first exons is in agreement with the finding that CpG sites in protein-coding exons are hotspots for mutation, and ∼30% of both somatic and germ-line mutations are C→T transition mutations at CpG sites (Kondrashov 2002).
A Monte Carlo simulation was used to estimate the effects of inhomogeneity in the distribution of cleavage sites for restriction endonucleases and McrBC on the data of Figures 3 and 4. Each of the 217,711 CpG sites in the sequences depicted in Figures 3 and 4 were assigned a random methylation status with a probability of methylation of 0.6 per CpG dinucleotide, given that 60% of CpG sites are normally methylated in somatic cell DNA (Goll and Bestor 2005). Virtual McrBC and restriction endonuclease cleavage patterns were generated computationally on the randomly methylated sequences. The virtual patterns were compared with the experimentally determined patterns, and the process was iterated 107×. The relative frequency with which CpG sites were located in methylated or unmethylated domains was calculated by comparison of the virtual cleavage data to the actual cleavage data. By this method, a CpG site located in an AluY transposon was calculated to be 342-fold (and an AluS 80-fold) more likely to be located in a methylated domain than was a CpG dinucleotide in a CpG island, while CpG sites in exons other than first exons were 10.3-fold more likely to reside in methylated domains than were CpG sites in first exons. This latter finding indicates that unmethylated first exons and methylated other exons are a general property of the genome, as the mean exon content per gene is ∼10 (Lander et al. 2002). These findings indicate that recognition-site distributions did not skew the results and that the depiction of methylation profiles in Figures 3 and 4 is valid.
Figure 4.
Frequency of first exons and all exons in methylated and unmethylated domains. (A) First exons displayed as in Figure 3. (B) All exons. (C) Quantitation of A and B as in Figure 3. Exons other than first exons are largely methylated; the peak in unmethylated exons at left is due almost entirely to first exons.
Discussion
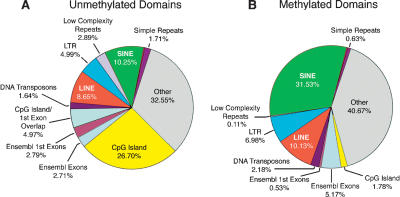
The computational analysis of genome-wide CpG depletion in germ-line DNA and direct methylation profiling of brain DNA yielded very similar views of the large-scale structure of genomic methylation patterns, as is summarized in Figure 5. The most prominent difference between unmethylated and methylated domains is the high content of CpG islands in the former; 31.67% of all CpG sites in unmethylated domains are in CpG islands, while the corresponding figure for methylated domains is 1.78%, an enrichment of 17.8-fold. This is a minimum estimate, as many of the annotated CpG islands in the methylated domains are much shorter than the mean CpG island length (Aerts et al. 2004) and are likely to reflect false positives in the annotation. The enrichment of CpG islands in unmethylated domains is the largest difference between the unmethylated and methylated compartments, and the sequence composition of methylated domains closely resembles that of the genome as a whole (Fig 1A). These data indicate that the only large and constitutive unmethylated compartment in the human genome is the CpG island compartment, which contains ∼75% of all promoters and 6.8% of all CpG sites. The most prominent component of methylated domains was transposons, especially Alu transposons. These data explain the intense staining of chromosome 19 with both methylated and unmethylated probes in the in situ hybridization data of Figure 2B. Chromosome 19 has more unmethylated CpG dinucleotides in CpG islands than any chromosome other than chromosome 1 (147,530 vs. 170,395) and has the highest density of Alu transposons (Lander et al. 2002).
Figure 5.
CpG distributions in unmethylated and methylated domains. All percentages shown were corrected for G+C content and are displayed as observed versus expected values. There is a marked enrichment of CpG island sequences in unmethylated domains (A,B), while the sequence composition of methylated domains resembles that of the genome as a whole, as shown in Figure 1B.
Cytosine methylation is proposed to have two major roles in genome evolution. First, cytosine methylation accelerates the rate of accumulation of C→T mutations at methylated CpG sites, which leads to irreversible inactivation of promoters that are methylated in the germ line; most promoters that are methylated reside in transposons. Second, DNA methylation might act to constrain the effective size of the genome by selective exposure of unmethylated CpG island promoters and the masking of the remainder of the genome. Other data have shown that it is the unmethylated compartment of the genome that is accessible to diffusible factors (Antequera et al. 1989), and that methylation controls accessibility (Keshet et al. 1986). This masking function of cytosine methylation is suggested to underlie the C value paradox (Thomas Jr. 1971), which refers to the finding that related taxa with similar gene numbers can have genomes that differ in size by several orders of magnitude; genome size within free-living eukaryotes varies by a factor of >80,000; within the chordates by a factor of 1800; and within the vertebrates by a factor of 330 (Gregory 2005). Many mammalian transcription factors have short and degenerate recognition sequences with small (<100-fold) preferences for specific over nonspecific binding sequences (Oda et al. 1998), and the coordinated function of networks of DNA-binding regulatory factors will therefore be sensitive to the amount of DNA available for interaction. It is suggested that cytosine methylation provides a masking function that buffers against the increasing delays required for diffusion-limited regulatory factors to acquire their recognition sequences in expanding genomes; this masking function also prevents the diversion of transcription factors to cryptic and nonfunctional binding sites or to transposons and their remnants, many of which contain functional Pol II or Pol III promoters. It can be noted that >45% of the human genome is derived from transposons, while ∼2% encodes functional RNA (Lander et al. 2002). It is suggested that cytosine methylation acts to maintain the accessible compartment of the genome at a constant level and thereby buffers the genome against large changes in size that result from sequence duplications and the accumulation of transposons. This suggestion in consistent with the finding that all known large-genome (>5 × 108 bp) organisms have methylated DNA (Kidwell 2002) and contain genes for members of the DNA methyltransferase 1 and 3 families (Goll and Bestor 2005), while many small genome eukaryotes lack recognizable DNA methyltransferase genes altogether.
Methods
DNA preparation
ICF Syndrome lymphoblastoid cell line GM08714 and the parental control cell line GM08728 were obtained from the Coriell Institute for Medical Research, NIGMS Human Genetic Mutant Cell Repository. ICF cells were grown at 37°C in RPMI plus 15% FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin. Cerebral cortex and cerebellum from a full-term human brain was obtained from the tissue bank of the Herbert Irving Comprehensive Cancer Center under an IRB-approved protocol. Genomic DNA from brain was obtained by rapidly freezing the tissue in liquid nitrogen, grinding the frozen tissue into a fine powder with a mortar and pestle, and incubating at 50°C for ∼12–18 h in digestion buffer (100 mM NaCl, 10 mM Tris-HCl at pH 8, 25 mM EDTA at pH 8, 0.5% SDS, and 0.1 mg/mL proteinase K). Following three phenol/chloroform extractions, genomic DNA was precipitated by adding 0.5 vol of 7.5 M NH4Ac and 2 vol of 100% ethanol. Genomic DNA was recovered by centrifugation and resuspended in TE buffer at 37°C. Genomic DNA from cultured cells was isolated using the DNeasy DNA extraction protocol (Qiagen). Pools of unmethylated genomic DNA were obtained by digestion with the methylation-dependent endonuclease McrBC (New England Biolabs). A total of 5–10 μg of genomic DNA was digested with a 10-fold excess of enzyme at 37°C in the recommended reaction buffer (50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT supplemented with 100 μg/mL BSA, and 5 mM GTP). Following completion of the digestion, samples were phenol/chloroform extracted and precipitated with 0.1 vol of 3 M sodium acetate (pH 5.2) and 2 vol of 100% ethanol, and resuspended in TE buffer. Size fractionation was performed as described in Supplemental Figure S1.
To prepare unmethylated genomic probes for the Methylation-Sensitive Comparative Genomic Hybridization procedure, McrBC and restriction endonuclease digestions were size fractionated on 1% low melting-point agarose gels, and appropriate size ranges (see Supplemental Fig. S1) were gel purified according to standard protocols. Genomic fragments between 2 and 9 kb were purified using a QIAquick gel-extraction kit (Qiagen), while genomic fragments ≥9 kb were isolated using a QIAEX II DNA extraction protocol (Qiagen).
Preparation of probes for in situ hybridization
Restriction enzyme and McrBC genomic pools were labeled by nick translation using SpectrumRed (McrBC) and SpectrumGreen (RE) dUTP (Vysis). Nick translation reactions were carried out according to the Vysis protocol. Approximately 1 μg of genomic DNA was nick translated in the presence of 0.01 mM SpectrumRed or SpectrumGreen dUTP, 0.01 mM dTTP, 0.02 mM dATP, dCTP, and dGTP, nick translation buffer, and nick translation enzyme in a final volume of 50 μL. Reactions were incubated at 15°C for ∼4 h and stopped by heating at 70°C for 10 min. To prepare the probe mix, 10 μL (200 ng) of SpectrumRed McrBC DNA and 10 μL (200 ng) of SpectrumGreen McrBC DNA (Control) were mixed together with 3 M sodium acetate (pH 5.2) and 100% ethanol, vortexed briefly, and precipitated on dry ice for 15 min or overnight at -30°C. Following centrifugation at 12,000 RPM for 30 min at 4°C, DNA pellets were dried for 10–15 min under reduced pressure. Pellets were resuspended in 3 μL of nuclease-free H20 and 7 μL of CGH Hybridization Buffer (Vysis).
Target metaphases were from phytohemagglutinin-stimulated lymphocytes of normal male donors. Chromosomes were denatured in 70% formamide/2× SSC (pH 7.0–7.5) at 73°C for 5 min. Chromosomes were then dehydrated in graded ethanols and dried in air for 5–10 min. Probes were denatured at 73°C for 5 min and 10 μL of denatured probe was added to each hybridization area. After applying plastic coverslips, slides were incubated at 37°C for 24–48 h, then washed in several changes of 0.4× SSC/0.3% NP40 at 74°C prior to mounting.
Preparation of methylated and unmethylated DNA libraries
Unmethylated and methylated domains were obtained by limit digestion with McrBC or restriction endonucleases, respectively. DNA fragment ends were rendered flush with T4 DNA polymerase in the presence of all four dNTPs and cloned into pZero vectors (Invitrogen). After transfection into E. coli, plasmid DNA from individual colonies was released by heat at 100°C and amplified with Templify (Amersham) and paired end reads were obtained on Amersham Megabase or ABI 3730 capillary sequencers. The end reads were mapped to the genome via BLAT (Kent 2002) and sequences between the reads extracted by a custom PERL script. More than 90% of the paired ends reads were on opposite strands and within 10 kb of each other. Additional PERL scripts were compiled to provide graphical display of sequence annotation superimposed on the large assemblages of methylated and unmethylated domains, as shown in Figures 3, 4, 5. All PERL scripts are available by request to F.G.H. (fgh3@columbia.edu). The sequence coordinates for all methylated and unmethylated sequences shown here are available by request.
Acknowledgments
This work was supported by grants from the NIH to J.J., M.Q.Z., and T.H.B. and by a Fellowship from the Leukemia and Lymphoma Society to R.A.R. We thank K. Anderson, D. Bourc'his, M. Damelin, E.E. Eichler, and M. Goll for discussions and T.-J. Nguyen and J. Lee for DNA sequencing.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4362006.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Aerts, S., Thijs, G., Dabrowski, M., Moreau, Y., and De Moor, B. 2004. Comprehensive analysis of the base composition around the transcription start site in Metazoa. BMC Genomics 5 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera, F., Macleod, D., and Bird, A.P. 1989. Specific protection of methylated CpGs in mammalian nuclei. Cell 58 509-517. [DOI] [PubMed] [Google Scholar]
- Bestor, T.H. 2003. Cytosine methylation mediates sexual conflict. Trends Genet. 19 185-190. [DOI] [PubMed] [Google Scholar]
- Biniszkiewicz, D., Gribnau, J., Ramsahoye, B., Gaudet, F., Eggan, K., Humpherys, D., Mastrangelom, M.A., Jun, Z., Walter, J., and Jaenisch, R. 2002. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol. Cell. Biol. 22 2124-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his, D. and Bestor, T.H. 2004. Meiotic catastrophe and retrotransposons in male germ cells lacking Dnmt3L. Nature 431 96-99. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E., Clark, R., and She, X. 2004. An assessment of the sequence gaps: Unfinished business in a finished human genome. Nat. Rev. Genet. 5 345-354. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden, M. and Frommer, M. 1987. CpG islands in vertebrate genomes. J. Mol. Biol. 196 261-282. [DOI] [PubMed] [Google Scholar]
- Gaudet, F., Hodgson, J.G., Eden, A., Jackson-Grusby, L., Dausman, J., Gray, J.W., Leonhardt, H., and Jaenisch, R. 2002. Induction of tumors in mice by genomic hypomethylation. Science 300 489-492. [DOI] [PubMed] [Google Scholar]
- Goelz, S.E., Vogelstein, B., Hamilton, S.R., and Feinberg, A.P. 1985. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science 228 187-190. [DOI] [PubMed] [Google Scholar]
- Goll, M.G. and Bestor, T.H. 2005. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74 481-514. [DOI] [PubMed] [Google Scholar]
- Greger, V., Passarge, E., Hopping, W., Messmer, E., and Horsthemke, B. 1989. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum. Genet. 83 155-158. [DOI] [PubMed] [Google Scholar]
- Gregory, T.R. 2005. The C-value enigma in plants and animals: A review of parallels and an appeal for partnership. Ann. Bot. (Lond). 95 133-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshikhes, I.P. and Zhang, M.Q. 2000. Large-scale human promoter mapping using CpG islands. Nat. Genet. 26 61-63. [DOI] [PubMed] [Google Scholar]
- Jojic, V., Jojic, N., Meek, C., Geiger, D., Siepel, A., Haussler, D., and Heckerman, D. 2004. Efficient approximations for learning phylogenetic HMM models from data. Bioinformatics 20 I161-I168. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT-the BLAST-like alignment tool. Genome Res. 12 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet, I., Lieman-Hurwitz, J., and Cedar, H. 1986. DNA methylation affects the formation of active chromatin. Cell 44 535-543. [DOI] [PubMed] [Google Scholar]
- Kidwell, M.G. 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115 49-63. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A.S. 2002. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Human Mutat. 21 12-27. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2002. Initial sequencing and analysis of the human genome. Nature 409 860-921. [DOI] [PubMed] [Google Scholar]
- Lee, D.U., Agarwal, S., and Rao, A. 2002. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity 16 649-660. [DOI] [PubMed] [Google Scholar]
- Li, E., Bestor, T.H., and Jaenisch, R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69 915-926. [DOI] [PubMed] [Google Scholar]
- Majewski, J. and Ott, J. 2002. Distribution and characterization of regulatory elements in the human genome. Genome Res. 12 1827-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, O.J., Schnedl, W., Allen, J., and Erlanger, B.F. 1974. 5-Methylcytosine localised in mammalian constitutive heterochromatin.Nature 251 636-637. [DOI] [PubMed] [Google Scholar]
- Oda, M., Furukawa, K., Ogata, K., Sarai, A., and Nakamura, H. 1998. Thermodynamics of specific and non-specific DNA binding by the c-Myb DNA-binding domain. J. Mol. Biol. 276 571-590. [DOI] [PubMed] [Google Scholar]
- Palmer, L.E., Rabinowicz, P.D., O'Shaughnessy, A.L., Balija, V.S., Nascimento, L.U., Dike, S., de la Bastide, M., Martienssen, R.A., and McCombie, W.R. 2003. Maize genome sequencing by methylation filtration. Science 302 135-138. [DOI] [PubMed] [Google Scholar]
- Schubeler, D., Lorincz, M.C., Cimbora, D.M., Telling, A., Feng, Y.Q., Bouhassira, E.E., and Groudine, M. 2000. Genomic targeting of methylated DNA: Influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol. Cell. Biol. 20 9103-9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker, E.U., Tountas, N.A., Cross, S.H., Margolin, B.S., Murphy, J.G., Bird, A.P., and Freitag, M. 2003. The methylated component of the Neurospora crassa genome. Nature 422 893-897. [DOI] [PubMed] [Google Scholar]
- Stein, R., Razin, A., and Cedar, H. 1982. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc. Natl. Acad. Sci. 79 3418-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas Jr., C.A. 1971. The genetic organization of chromosomes. Annu. Rev. Genet. 5 237-256. [DOI] [PubMed] [Google Scholar]
- Tompa, R., McCallum, C.M., Delrow, J., Henikoff, J.G., van Steensel, B., and Henikoff, S. 2002. Genome-wide profiling of DNA methylation reveals transposon targets of CHROMOMETHYLASE3. Curr. Biol. 12 65-68. [DOI] [PubMed] [Google Scholar]
- Trinklein, N.D., Aldred, S.J., Saldanha, A.J., and Myers, R.M. 2003. Identification and functional analysis of human transcriptional promoters. Genome Res. 13 308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tykocinski, M.L. and Max, E.E. 1984. CG dinucleotide clusters in MHC genes and in 5′ demethylated genes. Nucleic Acids Res. 12 4385-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, M., Davies, J.J., Wittig, D., Oakeley, E.J., Haase, M., Lam, W.L., and Schubeler, D. 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37 853-862. [DOI] [PubMed] [Google Scholar]
- Wigler, M., Levy, D., and Perucho, M. 1981. The somatic replication of DNA methylation. Cell 24 33-40. [DOI] [PubMed] [Google Scholar]
- Xu, G.L., Bestor, T.H., Bourc'his, D., Hsieh, C.L., Tommerup, N., Bugge, M., Hulten, M., Qu, X., Russo, J.J., and Viegas-Pequignot, E. 1999. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402 187-189. [DOI] [PubMed] [Google Scholar]
- Yoder, J.A., Walsh, C.P., and Bestor, T.H. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13 335-340. [DOI] [PubMed] [Google Scholar]