Abstract
Duplication of chromosomal segments and associated genes is thought to be a primary mechanism for generating evolutionary novelty. By comparative genome hybridization using a full-coverage (tiling) human BAC array with 79-kb resolution, we have identified 63 chromosomal segments, ranging in size from 0.65 to 1.3 Mb, that have inferred copy number increases in human relative to chimpanzee. These segments span 192 Ensembl genes, including 82 gene duplicates (41 reciprocal best BLAST matches). Synonymous and nonsynonymous substitution rates across these pairs provide evidence for general conservation of the amino acid sequence, consistent with the maintenance of function of both copies, and one case of putative positive selection for an uncharacterized gene. Surprisingly, the core histone genes H2A, H2B, H3, and H4 have been duplicated in the human lineage since our split with chimpanzee. The observation of increased copy number of a human cluster of core histone genes suggests that altered dosage, even of highly constrained genes, may be an important evolutionary mechanism.
Gene duplication has long been considered a primary mechanism of adaptive evolution (Ohno 1970). In theory, newly duplicated genes are redundant, and relaxed functional constraints allow acquisition of sequence changes in support of new functions and expression patterns. The importance of gene duplication in human evolution is supported by numerous studies that have documented DNA copy number differences between human and nonhuman primates. These studies have used diverse molecular approaches based on karyotyping (Yunis et al. 1980), physical clone maps (Fujiyama et al. 2002), partial coverage genomic arrays (Locke et al. 2003), cDNA arrays (Fortna et al. 2004), end-sequence profiling (Newman et al. 2005), draft genome sequence (Chimpanzee Sequencing and Analysis Consortium 2005), and high-quality sequence from chimp Chromosome 21 (orthologous to human Chromosome 22) (Watanabe et al. 2004). Careful alignment of accurately finished genome sequences from human and other primates promises to reveal DNA copy number differences at the highest possible resolution. Finished sequence for human is available, but the whole-genome shotgun (WGS) assembly for chimpanzee remains in draft form. The WGS approach is of proven value for rapidly and economically generating a full genome sequence, but it is clear that current approaches to assembling WGS data sets can underrepresent recently duplicated genome segments (She et al. 2004). Thus, to achieve a complete genome survey of DNA copy number in human versus chimpanzee, we have performed comparative genome hybridization (CGH) using the first full-coverage bacterial artificial chromosome (BAC) array of the human genome, which consists of 32,855 overlapping clones providing ∼79 kb average resolution. This is a direct approach and yields the greatest coverage at the highest level of resolution thus far achieved for comparison of these three species.
Results
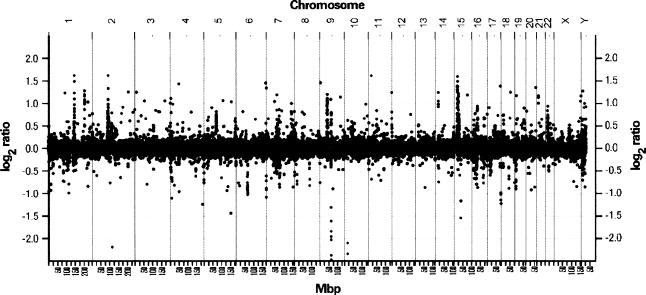
Using full-coverage BAC array CGH, we executed a three-phased approach to identify segments of human genomic DNA that have likely been acquired since divergence from the common ancestor we share with chimpanzee. First, two samples of human genomic DNA (gDNA), one pooled from seven unrelated males and the other pooled from four unrelated females, were cohybridized to identify and exclude nodes on the array that gave aberrant ratios in a human-only comparison. Pooled DNAs were used in order to minimize the number of hybridization experiments, and to favor the detection of fixed rather than polymorphic copy number differences. Of the 31,842 mapped autosomal clones on the array, 212 showed aberrant ratios (>1.5 H-spread; see Methods) in the human-human comparison, and were excluded from further analysis. Next, we hybridized the human test DNA sample pooled from seven human males to a reference DNA sample comprised of DNA pooled from three unrelated male chimpanzees (Coriell Institute, Repository numbers NAO3448, NAO3450, NAO3452) (Fig. 1). These hybridizations were repeated under dye reversal, and a total of 1319 clones (855 increases, 464 decreases) were identified that consistently showed ratios that exceeded threshold in both dye orientations. As an added measure of stringency, we retained clones only if (1) they were confirmed by an equivalent copy number aberration in at least one additional overlapping clone, or (2) their location in the human reference genome sequence (NCBI_34) is supported by both their restriction digest pattern and BAC end sequence placement (Krzywinski et al. 2004). Under these criteria, a total of 585 clones gave elevated ratios in human relative to chimpanzee.
Figure 1.
Human chimpanzee DNA copy number ratio determined by full-coverage BAC array CGH.
We used gorilla as an outgroup to determine the most likely ancestral copy number state. By parsimony, human chromosomal segments showing an increased copy number ratio relative to both chimpanzee and gorilla most likely represent insertions specific to the human lineage. This is true regardless of whether the human genomic region containing the given segment is more similar to chimpanzee or gorilla. Note, however, that there are further caveats to the parsimony approach that must be considered. While the widely accepted species tree of hominoids places human and chimpanzee as a clade, with gorilla as an outgroup, there are regions of the genome that are incongruent with the species tree. For regions of the genome consistent with a human-gorilla clade, the assignment of a copy number increase to human is unaffected, that is, parsimony still favors a single event in the human branch over two independent events in the chimp and gorilla branches. However, a study by Chen and Li (2001) reported that 12 of 53 surveyed loci were more consistent with a chimp-gorilla clade than a chimp-human clade. For these regions only, parsimony is blind to whether there is a copy number increase in the human branch, versus a decrease in the branch leading from the human/chimp/gorilla node to the chimp/gorilla node. Thus, regarding the ancestral copy number state, our hybridization results will be ambiguous for ∼20% of the genome. In some regards, orangutan may be a more suitable outgroup, since the ratio of unresolved ancestral polymorphism to divergence is much lower because of the longer divergence time. However, a potential drawback in using orangutan as an outgroup in these experiments is that the arrays are spotted with human genomic clones, and hybridization becomes less reliable when more distant species are evaluated.
Thus, we proceeded with hybridization of the pooled human male test DNA sample to reference DNA from a single female gorilla (Coriell Institute, Repository number NGO5251). We decided to use human male test DNA rather than female test DNA for consistency with previous experiments. Because we had fewer chimpanzee and gorilla samples than human samples, there is some possibility that sites that are polymorphic in chimpanzee and gorilla have impacted our analysis. The fact that we restrict analysis to genomic segments where chimpanzee and gorilla copy number agree, relative to human, minimizes this impact. Of the 585 clones that had an elevated ratio in human relative to chimpanzee, 235 also gave elevated ratios relative to gorilla and therefore likely represent human-specific copy number increases. Presumably, the subset of clones that did not show elevated ratios relative to gorilla represent copy number decreases in chimpanzee relative to the ancestral state. Again, as an added measure of stringency, clones have been retained in the set of 235 only if they are confirmed by an equivalent copy number aberration in at least one additional overlapping clone, or their location in the human reference genome sequence (NCBI_34) is supported by both their restriction digest pattern and BAC-end sequence placement. These 231 clones collapse into 55 contiguous chromosomal segments (43 with multiple clones, and 12 singletons) with minimum, maximum, and average segment lengths of 65,252 bp, 1,133,633 bp, and 308,959 bp, respectively, and a cumulative genome footprint of 16,992,728 bp.
Separately, we evaluated ratios of clones located on the X and Y chromosomes. We identified a total of eight X chromosome (ChrX) and 28 Y chromosome (ChrY) clones that met the criteria of concordant dye-flip ratios and an equivalent copy number difference in at least one overlapping clone. Sex chromosome ratios from the female gorilla sample are not directly comparable to those from the male chimpanzee and human reference samples, thus for sex chromosome differences we are unable to infer human increase rather than chimp decrease. However, evaluation of duplicate segments within the human reference genome sequence (below) supports the notion that these are copy number increases in the human lineage. These eight ChrX clones and 28 ChrY clones collapse into two ChrX contigs and six ChrY contigs covering 415,787 bp and 1,190,263 bp, respectively, bringing the cumulative genomic footprint of all segments (autosomal plus sex chromosome) to 18,598,778 bp (Table 1). These segments are the basis of further analysis. While loss of genetic material on the human lineage is of considerable interest, here we consider only observed copy number increases. This is because copy number increases, as opposed to losses, can be readily validated through design of quantitative PCR experiments and through evaluation of signatures of duplication events in the reference human genome sequence as we describe below.
Table 1.
Position in the human genome (NCBI build 34) of 63 DNA segments with increased copy number relative to chimpanzee and gorilla, as determined by full-coverage BAC array CGH
| Chromosome | Start | End | Size (kb) | Band |
|---|---|---|---|---|
| 1 | 16,264,647 | 16,504,347 | 239,700 | 1p36.13 |
| 1 | 103,514,958 | 103,782,587 | 267,629 | 1p21.1 |
| 1 | 141,699,150 | 141,904,149 | 204,999 | 1q21.1 |
| 1 | 142,697,868 | 142,832,783 | 134,915 | 1q21.1 |
| 1 | 145,670,162 | 146,179,409 | 509,247 | 1q21.1–1q21.2 |
| 1 | 146,800,368 | 147,090,803 | 290,435 | 1q21.2 |
| 2 | 87,255,531 | 88,008,041 | 752,510 | 2p11.2 |
| 2 | 87,880,005 | 87,967,456 | 87,451 | 2p11.2 |
| 2 | 89,146,792 | 89,285,612 | 138,820 | 2p11.2 |
| 2 | 89,290,779 | 89,526,330 | 235,551 | 2p11.2 |
| 2 | 89,855,739 | 90,009,157 | 153,418 | 2p11.2 |
| 2 | 91,079,860 | 91,278,418 | 198,558 | 2p11.2 |
| 2 | 110,374,075 | 110,628,474 | 254,399 | 2q13 |
| 2 | 112,292,849 | 112,493,396 | 200,547 | 2q13 |
| 2 | 132,579,662 | 132,809,765 | 230,103 | 2q21.1–2q21.2 |
| 4 | 13,423 | 177,154 | 163,731 | 4p16.3 |
| 4 | 70,432,214 | 70,618,319 | 186,105 | 4q13.2 |
| 5 | 26,149,382 | 26,333,080 | 183,698 | 5p14.1 |
| 5 | 69,007,757 | 69,735,382 | 727,625 | 5q13.2 |
| 5 | 69,955,192 | 70,348,029 | 392,837 | 5q13.2 |
| 5 | 112,886,922 | 113,064,958 | 178,036 | 5q22.2 |
| 6 | 170,704,322 | 170,894,763 | 190,441 | 6q27 |
| 7 | 60,835,494 | 61,035,840 | 200,346 | 7q11.1 |
| 7 | 64,392,229 | 64,533,018 | 140,789 | 7q11.21 |
| 7 | 71,931,071 | 72,131,909 | 200,838 | 7q11.23 |
| 7 | 73,503,814 | 73,734,450 | 230,636 | 7q11.23 |
| 7 | 73,914,085 | 74,089,099 | 175,014 | 7q11.23 |
| 7 | 142,658,374 | 142,723,626 | 65,252 | 7q34 |
| 7 | 143,134,261 | 143,465,465 | 331,204 | 7q35 |
| 8 | 47,000,811 | 47,258,017 | 257,206 | 8q11.1 |
| 9 | 38,905,428 | 39,384,898 | 479,470 | 9p13.1 |
| 9 | 39,765,473 | 39,978,303 | 212,830 | 9p12 |
| 9 | 40,174,404 | 40,340,379 | 165,975 | 9p12 |
| 9 | 40,495,504 | 40,759,034 | 263,530 | 9p12 |
| 9 | 41,111,334 | 41,356,670 | 245,336 | 9p12 |
| 9 | 41,431,201 | 41,545,828 | 114,627 | 9p11.2 |
| 9 | 41,557,881 | 42,021,347 | 463,466 | 9p11.2 |
| 9 | 43,787,741 | 44,127,010 | 339,269 | 9p11.2 |
| 9 | 63,574,163 | 64,135,077 | 560,914 | 9q13 |
| 9 | 65,216,018 | 65,523,621 | 307,603 | 9q13 |
| 9 | 65,617,653 | 65,814,846 | 197,193 | 9q13–9q21.11 |
| 10 | 46,138,281 | 46,380,466 | 242,185 | 10q11.22 |
| 10 | 57,969,110 | 58,079,974 | 110,864 | 10q21.1 |
| 14 | 18,070,001 | 18,405,573 | 335,572 | 14q11.2 |
| 15 | 18,898,763 | 19,129,849 | 231,086 | 15q11.2 |
| 15 | 19,301,328 | 19,916,381 | 615,053 | 15q11.2 |
| 15 | 20,813,690 | 21,041,101 | 227,411 | 15q11.2 |
| 15 | 22,004,834 | 22,248,614 | 243,780 | 15q11.2 |
| 15 | 26,055,804 | 26,752,132 | 696,328 | 15q13.1 |
| 16 | 21,002,359 | 21,198,510 | 196,151 | 16p12.3 |
| 16 | 32,089,294 | 32,319,125 | 229,831 | 16p11.2 |
| 16 | 70,583,784 | 71,175,111 | 591,327 | 16q22.1–16q22.2 |
| 18 | 42,695,217 | 42,966,859 | 271,642 | 18q21.1 |
| 20 | 58,697,375 | 58,867,659 | 170,284 | 20q13.32–20q13.33 |
| 21 | 21,603,697 | 21,743,689 | 139,992 | 21q21.1 |
| 22 | 14,440,103 | 15,014,058 | 573,955 | 22q11.1 |
| X | 87,762,347 | 87,994,542 | 232,195 | Xq21.31 |
| X | 88,109,770 | 88,293,362 | 183,592 | Xq21.31 |
| Y | 8,909,830 | 9,017,798 | 107,968 | Y |
| Y | 9,598,790 | 9,717,440 | 118,650 | Y |
| Y | 19,570,010 | 19,871,468 | 301,458 | Y |
| Y | 21,103,620 | 21,264,863 | 161,243 | Y |
| Y | 25,095,852 | 25,388,816 | 292,964 | Y |
| Y | 27,429,589 | 27,637,569 | 207,980 | Y |
Since it is expected that genomic segments recently gained in the human lineage have originated through duplication of existing sequence, we evaluated the degree of overlap between segments identified by CGH and segments identified by in silico analysis (BLAST matches >1 kb long with >90% identity, as described in Krzywinski et al. 2004) of the reference human genome sequence (NCBI_34). Whereas coverage of the genome by in silico predicted segmental duplications is 5.2%, in silico predicted duplications covered 73.4% of the sequence represented in the 63 CGH-identified chromosomal segments, more than an order of magnitude enrichment. This is consistent with the notion that copy number gains as identified by CGH have arisen through segmental duplication. While the sequence identity between in silico defined duplicates can suggest an approximate duplication date, only phylogenetic analysis allows a time interval to be directly estimated for the duplication event. Of the total 8499 in silico defined duplicated segments, 176 (2.1%) intersect with the portion of the genome that appears from CGH to be have been gained specifically in the human lineage, suggesting that these duplications occurred in the past 4 to 6 million years (the divergence time of human and chimpanzee). It is important to note, however, that this is a minimal estimate for the proportion of recent duplicates, since segments smaller than the resolution of the BAC array (∼79 kb) will not have been detected by CGH.
A total of 192 non-pseudogene Ensembl genes were detected on the 63 duplicated segments. If these segments arose through segmental duplication, we would expect representation from paralogous genes within this set. The coding sequences of these genes were compared by reciprocal BLAST analysis (expect-value cutoff = 10-10), which identified 41 strict paralogous gene pairs (82 genes total) (Table 2). For these genes, pairwise synonymous and nonsynonymous substitution rates were estimated for the aligned sequence using the codon substitution models of Yang and Nielsen (2000) as implemented in PAML (phylogenetic analysis by maximum likelihood) (Yang 1997). The mean dN/dS ratio of the gene set is 0.433, the median ratio is 0.388, and the highest observed ratio is 1.56. Likelihood ratio tests were applied to test the null hypothesis of equal rates of substitution (H0: dN = dS) versus the alternative hypothesis (HA: dN ≠ dS). For 15 of the 41 genes, the null hypothesis had a nominal P-value <5%, and in all but one of these significant cases, dN/dS < 1, implying that purifying selection had been removing deleterious amino acid variants, and that both copies of the gene must have retained function for at least a fraction of the time subsequent to the gene duplication. An extreme case of protein conservation since the time of gene duplication would be if dN/dS = 0 (i.e., no amino acid changes were tolerated), and in this case it might be appropriate to compare the null hypothesis to the alternative with dN/dS < 0.5. This null hypothesis arises because we allow for one homolog to be strictly conserved (dN/dS = 0) and the other to be strictly neutral (dN/dS = 1), thus a somewhat more liberal test of one of the pair showing excess divergence considers as a null hypothesis the average of dN/dS = 0.5 (Thornton and Long 2002). This test identified as significant three genes that had dN/dS > 0.5 (Table 2), but in no case have these genes been functionally characterized.
Table 2.
Strict paralogous gene pairs (reciprocal best BLAST matches) present on human DNA segments with increased copy number relative to chimpanzee and gorilla
| Gene 1 | Chr | Start | End | Description | Gene 2 | Chr | Start | End | Description | dN | dS | dN/dS | P(1.0) | P(0.5) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSG00000186301 | 1 | 16,353,203 | 16,360,133 | Similar to macrophage stimulating 1 (hepatocyte growth factor-like) | ENSG00000186715 | 1 | 16,462,486 | 16,468,660 | No description | 0.233 | 0.843 | 0.276 | 0.000 | 0.000 |
| ENSG00000051415 | 1 | 103,423,056 | 103,655,958 | α-Amylase, salivary precursor | ENSG00000174876 | 1 | 103,584,694 | 103,592,978 | α-Amylase salivary precursor | 0.015 | 0.061 | 0.245 | 0.000 | 0.044 |
| ENSG00000132043 | 1 | 146,102,223 | 146,214,718 | No description | ENSG00000168614 | 1 | 145,923,370 | 145,958,684 | No description | 0.268 | 0.260 | 1.032 | 0.694 | 0.000 |
| ENSG00000182639 | 1 | 147,000,080 | 147,000,460 | Histone H2B.J | ENSG00000184678 | 1 | 147,074,392 | 147,074,772 | Histone H2B.Q | 0.004 | 0.188 | 0.021 | 0.001 | 0.004 |
| ENSG00000183598 | 1 | 147,001,408 | 147,001,818 | H3 histone family 2; H3 histone family, member M | ENSG00000183702 | 1 | 147,040,763 | 147,042,418 | H3 histone, family 2; H3 histone family, member M | 0.000 | 0.418 | 0.001 | 0.000 | 0.000 |
| ENSG00000183558 | 1 | 147,030,455 | 147,030,847 | Histone H2A.O | ENSG00000183717 | 1 | 147,039,210 | 147,039,773 | Histone H2A.O | 0.000 | 0.001 | 0.387 | 0.758 | 0.811 |
| ENSG00000182217 | 1 | 147,047,334 | 147,049,286 | Histone H4 | ENSG00000183941 | 1 | 147,020,824 | 147,022,776 | Histone H4 | 0.001 | 0.000 | 0.797 | 0.743 | |
| ENSG00000169769 | 2 | 89,344,195 | 89,344,735 | IG κ chain V-III region NG9 precursor | ENSG00000184817 | 2 | 87,540,174 | 87,540,735 | No description | 0.051 | 0.061 | 0.839 | 0.749 | 0.324 |
| ENSG00000173756 | 2 | 89,423,308 | 89,423,634 | IG κ chain V-II region GM607 precursor | ENSG00000182563 | 2 | 89,330,327 | 89,330,632 | No description | 0.092 | 0.237 | 0.388 | 0.014 | 0.500 |
| ENSG00000131426 | 2 | 89,897,439 | 89,897,914 | IG κ chain V-I region HK 101 precursor | ENSG00000163245 | 2 | 89,301,505 | 89,301,971 | IG κ chain V-I region | 0.027 | 0.029 | 0.903 | 0.888 | 0.403 |
| ENSG00000172038 | 2 | 89,912,079 | 89,912,595 | IG κ chain V-III region CLL precursor (rheumatoid factor) | ENSG00000186192 | 2 | 89,058,824 | 89,248,127 | IG κ chain C region | 0.045 | 0.098 | 0.459 | 0.116 | 0.858 |
| ENSG00000163184 | 2 | 89,957,240 | 89,957,539 | IG κ chain V-I region | ENSG00000186856 | 2 | 89,062,541 | 89,242,332 | No description | 0.000 | 0.021 | 0.001 | 0.040 | 0.105 |
| ENSG00000169606 | 2 | 132,594,457 | 132,595,857 | No description | ENSG00000172981 | 22 | 14,635,352 | 14,636,482 | No description | 0.031 | 0.165 | 0.191 | 0.000 | 0.001 |
| ENSG00000154927 | 2 | 132,690,833 | 132,735,739 | No description | ENSG00000186825 | 2 | 132,763,505 | 132,764,092 | No description | 0.000 | 0.006 | 0.001 | 0.118 | 0.210 |
| ENSG00000177631 | 4 | 49,320 | 78,099 | No description | ENSG00000182141 | 22 | 15,009,448 | 15,026,562 | No description | 0.327 | 0.582 | 0.562 | 0.000 | 0.003 |
| ENSG00000184671 | 5 | 69,006,181 | 69,062,049 | Baculoviral IAP repeat-containing protein 1 (neuronal apoptosis inhibitory protein) | ENSG00000185284 | 5 | 69,659,709 | 69,678,715 | No description | 0.036 | 0.113 | 0.318 | 0.000 | 0.051 |
| ENSG00000182078 | 5 | 69,234,613 | 69,234,750 | No description | ENSG00000186932 | 5 | 70,293,834 | 70,293,971 | No description | 0.000 | 0.001 | 0.001 | 0.766 | 0.874 |
| ENSG00000183666 | 5 | 69,498,165 | 69,537,080 | No description | ENSG00000183761 | 5 | 69,239,339 | 69,301,070 | No description | 0.000 | 0.001 | 0.000 | 0.682 | 0.832 |
| ENSG00000180027 | 5 | 70,249,240 | 70,289,288 | SMA3 protein | ENSG00000185759 | 5 | 69,997,497 | 70,038,832 | SMA3 protein | 0.000 | 0.001 | 0.410 | 0.730 | 0.813 |
| ENSG00000174133 | 5 | 70,271,857 | 70,276,207 | No description | ENSG00000186361 | 5 | 70,010,592 | 70,014,942 | No description | 0.039 | 0.026 | 1.514 | 0.001 | 0.000 |
| ENSG00000170135 | 7 | 73,622,573 | 73,679,914 | Transcription factor GTF2IRD2 | ENSG00000174428 | 7 | 73,962,893 | 73,977,192 | Transcription factor GTF2IRD2 | 0.052 | 0.099 | 0.524 | 0.002 | 0.820 |
| ENSG00000123965 | 7 | 73,718,985 | 73,733,868 | Postmeiotic segregation increased 2-like 5 | ENSG00000135165 | 7 | 71,762,053 | 75,683,468 | Zona pellucida sperm-binding protein 3 precursor | 0.007 | 0.009 | 0.793 | 0.802 | 0.611 |
| ENSG00000165178 | 7 | 73,984,535 | 73,999,836 | No description | ENSG00000182487 | 7 | 72,046,778 | 72,062,076 | No description | 0.003 | 0.023 | 0.137 | 0.007 | 0.072 |
| ENSG00000181881 | 7 | 143,339,071 | 143,443,973 | No description | ENSG00000181926 | 7 | 143,383,314 | 143,389,573 | No description | 0.000 | 0.000 | 0.858 | 0.991 | 0.832 |
| ENSG00000170341 | 7 | 143,406,977 | 143,407,891 | Seven transmembrane helix receptor | ENSG00000170356 | 7 | 143,320,745 | 143,321,671 | Seven transmembrane helix receptor | 0.000 | 0.004 | 0.001 | 0.104 | 0.189 |
| ENSG00000106714 | 9 | 39,063,817 | 39,278,466 | Contactin associated protein-like 3 precursor (cell recognition molecule CASPR3) | ENSG00000154529 | 9 | 39,704,516 | 39,839,734 | No description | 0.009 | 0.020 | 0.422 | 0.229 | 0.809 |
| ENSG00000147926 | 9 | 39,908,370 | 39,912,422 | No description | ENSG00000182355 | 9 | 39,345,699 | 39,351,955 | No description | 0.009 | 0.006 | 1.560 | 0.295 | 0.005 |
| ENSG00000179828 | 9 | 40,483,601 | 40,501,134 | No description | ENSG00000182153 | 9 | 41,776,397 | 41,793,931 | No description | 0.000 | 0.003 | 0.001 | 0.154 | 0.261 |
| ENSG00000170165 | 9 | 43,977,955 | 43,988,544 | No description | ENSG00000176057 | 9 | 43,907,570 | 43,918,159 | No description | 0.000 | 0.003 | 0.001 | 0.147 | 0.251 |
| ENSG00000170161 | 9 | 63,660,398 | 63,661,000 | No description | ENSG00000186383 | 9 | 41,506,025 | 41,506,582 | No description | 0.015 | 0.055 | 0.269 | 0.091 | 0.420 |
| ENSG00000176299 | 14 | 18,238,610 | 18,239,551 | Seven transmembrane helix receptor | ENSG00000182974 | 15 | 19,866,208 | 19,867,149 | Seven transmembrane helix receptor | 0.029 | 0.028 | 1.034 | 0.942 | 0.097 |
| ENSG00000176294 | 14 | 18,285,736 | 18,286,659 | Seven transmembrane helix receptor | ENSG00000183706 | 15 | 19,880,105 | 19,881,055 | Seven transmembrane helix receptor | 0.077 | 0.191 | 0.402 | 0.000 | 0.356 |
| ENSG00000175733 | 15 | 21,016,715 | 21,019,583 | No description | ENSG00000184095 | 15 | 19,057,902 | 19,060,769 | No description | 0.083 | 0.091 | 0.913 | 0.756 | 0.025 |
| ENSG00000128731 | 15 | 25,958,549 | 26,169,609 | HECT domain and RLD 2 | ENSG00000140181 | 15 | 20,829,990 | 20,883,368 | No description | 0.020 | 0.034 | 0.599 | 0.017 | 0.391 |
| ENSG00000153684 | 15 | 26,233,151 | 26,236,568 | No description | ENSG00000183629 | 15 | 26,495,524 | 26,498,941 | No description | 0.000 | 0.001 | 0.001 | 0.590 | 0.671 |
| ENSG00000169861 | 16 | 32,106,704 | 32,113,996 | No description | ENSG00000182414 | 16 | 32,009,310 | 32,099,878 | IG heavy chain V-III region | 0.305 | 0.533 | 0.572 | 0.036 | 0.611 |
| ENSG00000183677 | 18 | 42,811,361 | 42,813,622 | RNA polymerase II transcription factor SIII subunit A2 (elongin A2) | ENSG00000183791 | 18 | 42,806,560 | 42,808,200 | RNA polymerase II transcription factor SIII subunit A3 (elongin A3) | 0.070 | 0.085 | 0.825 | 0.381 | 0.018 |
| ENSG00000130538 | 22 | 14,823,378 | 14,824,325 | No description | ENSG00000186445 | 14 | 18,171,223 | 18,172,203 | Seven transmembrane helix receptor | 0.008 | 0.000 | 0.066 | 0.016 | |
| ENSG00000185912 | Y | 8,898,452 | 8,960,986 | Testis-specific Y-encoded protein | ENSG00000187194 | Y | 9,000,204 | 9,001,613 | No description | 0.052 | 0.126 | 0.412 | 0.089 | 0.699 |
| ENSG00000131007 | Y | 19,638,276 | 19,645,782 | Transcript Y 9 protein | ENSG00000131009 | Y | 19,788,761 | 19,796,267 | Transcript Y 9 protein | 0.000 | 0.002 | 0.001 | 0.821 | 0.941 |
| ENSG00000169953 | Y | 19,788,522 | 19,830,805 | Heat shock transcription factor, Y-linked | ENSG00000172468 | Y | 19,603,741 | 19,646,021 | Heat shock transcription factor, Y-linked | 0.000 | 0.002 | 0.001 | 0.601 | 0.633 |
Gene identifiers, coordinates, and descriptions are from the Ensembl database (v. 27.35a) and are based on the NCBI_34 genome build. P-values for likelihood ratios are given for observed dN/dS compared to a null dN/dS of 1.0 or 0.5, respectively.
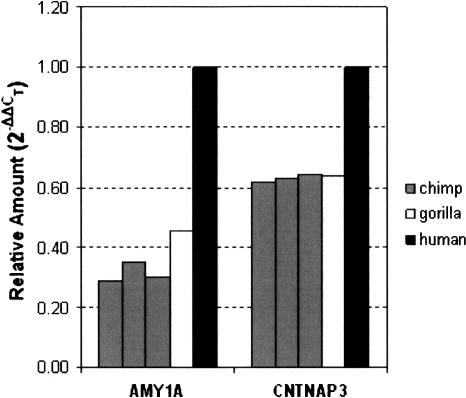
We selected two duplicated loci (AMY1A and CNTNAP3) for evaluation by an independent method (real-time quantitative PCR; Taqman). These loci were sequenced in our human, chimpanzee, and gorilla samples, and then primer/probe sets were designed to regions of sequence that were perfectly conserved between duplicates and among species. Of note, a third human amylase family member (AMY2B) was present on the duplicated segment that contained the two copies of AMY1A; thus the amylase primer probe sets were designed to a region of exact sequence identity among all three amylases. Results from these PCR assays (Fig. 2) verify increased copy number of these loci in human versus chimpanzee and gorilla.
Figure 2.
Relative copy number of the AMY1A and CNTNAP3 loci in three unrelated chimpanzees and a single gorilla compared to a single pooled sample of human gDNA.
Discussion
Using full-coverage BAC array CGH, we have identified 63 genomic segments with an increased hybridization ratio in human versus chimpanzee. Because these segments also show an increased hybridization ratio in human versus gorilla, the most parsimonious explanation is that these CGH-defined segments have been duplicated very recently in human evolutionary history, subsequent to our divergence from chimpanzee. This interpretation is supported by the high representation within these segments of in silico defined human segmental duplications, and the verification by real-time quantitative PCR of copy number differences at selected loci. However, the formal possibility remains that some subset of these CGH-defined segments has been independently lost in both chimpanzee and gorilla, rather than gained in humans. Owing to high sequence similarity among these three closely related primates and the substantial length of the CGH BAC probes (∼200 kb), it is exceedingly unlikely that sequence divergence is responsible for any observed differences. It must be considered that a portion of the genome does not represent the species tree but, rather, supports a chimp-gorilla clade over a chimp-human clade. For copy number differences in this portion of the genome, which remains to be accurately mapped, parsimony is not effective in assigning the ancestral copy number state. However, the fact that we have relied on gorilla as an outgroup should not have a significant impact on the results of the present study because we evoke parsimony only in the first data-filtering step of our analysis. Subsequent analysis is strictly focused on paralogous gene pairs within genomic segments with copy number alteration. Tandem duplication is a signature of DNA copy number increase, and provides a level of internal validation to our analysis. Furthermore, where we have done quantitative gene dosage analysis for further verification of human copy number gains (Fig. 2), the data have supported this interpretation.
Interestingly, we observe a substantial DNA copy number increase at chromosome 2q13 in human. This is the site of the telomeric fusion between chimpanzee chromosomes 12 and 13 in the human/chimp common ancestor that resulted in human chromosome 2 (Yunis et al. 1980). Furthermore, the expansions of human chromatin adjacent to the centromeres of chromosomes 1, 9, and 16 that were noted by Yunis et al. (1980) were also observed in the present study (Table 1). Fortna et al. (2004) previously used cDNA arrays for comparative analysis of hominoid genomes, and found a total of 25 relatively large DNA segments (11.1 Mb average length) that appeared to be duplicated in the human lineage. Of the 63 human copy number gains we measured in the present study, 30 segments have coordinates that agree with this previous study. The remaining 33 segments appear to be novel findings from our complete coverage array.
We analyzed the gene content of the 63 chromosomal segments with increased copy number in human. Genes within these regions were subjected to reciprocal BLAST analysis to find duplicated copies. Among the 41 high-confidence paralogous gene pairs we detected, the most highly represented gene family is immunoglobulin (IGK) genes, with five paralogous pairs. This is consistent with earlier whole-genome comparative analysis of, for example, fly and mosquito (Christophides et al. 2002; Hill et al. 2002; Zdobnov et al. 2002) or rat and mouse (Gibbs et al. 2004), where immune-related gene families have been found to be prone to expansion. The second most highly represented gene family is histones, with four paralogous pairs. In particular, the present study highlights human-specific duplications of members of the core histone minor cluster at chromosome 1q21 (Table 2). Histone octamers (comprised of two proteins each from core histone multimember families H2A, H2B, H3, and H4) form the core of the nucleosome, and are among the most evolutionarily conserved of all proteins, with substantial sequence conservation between humans and organisms as divergent as sea urchin (Grunstein et al. 1976). It is possible, given the relatively strict structural requirements of the nucleosome and exceptional sequence conservation across species, that variation in gene dosage becomes a viable alternative to sequence variation for adapting histone expression and function in accordance with selective pressures. While it is not clear what selective advantage might be conferred by many of the human duplicated genes highlighted in this study, of primary interest are genes with a potential role in nervous system development. An interesting candidate in this regard is ENSG00000106714 (Contactin-associated protein, CNTNAP3) and its uncharacterized paralog ENSG00000154529. CNTNAP3 is a member of the neurexin family of cell recognition molecules. Neurexins and their membrane-bound ligands (neuroligins) are thought to mediate interactions between neurons, including synapse formation. CNTNAP3, a member of the NCP subgroup of neurexins, is expressed throughout the human brain and is important in ion channel localization and neuron-glial interactions (Spiegel et al. 2002). Should further investigation of CNTNAP3 and its paralog verify non-pseudogene status, it will be important to evaluate what role these genes might have in synaptic function.
Under neutral evolution, coding mutations will be fixed at the same rate as silent mutations, giving a dN/dS ratio of 1. The median dN/dS ratio observed in our gene set was 0.388, which is consistent with net purifying selection acting on these recently duplicated genes. This observation is consistent with previous reports of reduced dN/dS ratios between paralogous genes in Drosophila (Thornton and Long 2002) and Arabidopsis (Zhang et al. 2002), and is indicative of continued function of both gene copies subsequent to the duplication event. While several genes in our set had dN/dS ratios >1, and a strict application of the PAML test of H0: dN/dS = 1 versus HA: dN/dS > 1 identified only one case of positive selection, a more liberal test that uses H0: dN/dS = 1/2 (and an alternative hypothesis with dN/dS as a free parameter) identifies an additional two unannotated genes with weak support for significant positive selection. This latter test might, instead, be considered a test for constraint, however, as it assumes that the original functional gene copy tolerates zero nonsynonymous changes.
The dN/dS ratios reported here are average ratios for the aligned length of each protein pair. Identification and sequencing of the strict orthologs of these genes in chimpanzee and additional primates will allow evaluation of synonymous and nonsynonymous substitution rates in a site-specific and lineage-specific manner and will likely yield further insight into human adaptive evolution. Further exploration of genes and noncoding functional sequences within the boundaries of these variable segments will be helpful for elucidating the genetic basis of human-specific traits.
Methods
Comparative genomic hybridization
Hybridizations were done using the whole-genome SMRT array (Ishkanian et al. 2004), which consists of amplified MseI fragments from 32,433 tiled Human BACs (Krzywinski et al. 2004) spotted in triplicate on two aldehyde-coated slides, and gives an effective resolving power of 79 kb. Test and reference DNAs were digested with MseI, labeled by random priming with the fluorescent nucleotide analogs Cy5-dCTP and Cy3-dCTP, and purified using Sephadex G50. For each hybridization experiment, the test and reference DNAs were combined and denatured, and repetitive sequences were blocked by coincubation with denatured human Cot-1 DNA (Invitrogen). Repeat-blocked DNA was then hybridized at 45°C for 48 to 72 h. After hybridization, the slides were washed at 45°C for 15 min in 80% DIGEasy hybridization solution (Roche Scientific), 2× SSC (pH 7), followed by three washes of 5 min at room temperature with 0.1× SSC (pH 7), 0.1% SDS, four rinses of 30 sec each in 0.1× SSC (pH 7) at room temperature, a brief rinse with deionized, distilled water, and then dried. Imaging was done using a Packard Biosciences Scan Array Express instrument.
Microarray analysis
An open-source software package, called MIA, was developed and implemented to extract intensity ratios from the raw images. The analysis of CGH array images is divided into several broad steps: (1) addressing, which consists of finding the location of subarrays and individual tiles containing one and only one spot; (2) segmentation, which consists of identifying the pixels belonging to the spot within each tile; (3) extraction of spot and background intensities; and (4) normalization of data. These steps are not specific to CGH experiments, and the software can therefore be useful for any two-color microarray work such as the analysis of gene expression with spotted arrays. Similar to the approach of Yang et al. (2001), the addressing and segmentation steps are performed on a combined 8-bit image obtained by a square-root transformation, but the intensities are extracted from the original raw 16-bit images. Several mathematical techniques were applied in order to obtain a completely automated addressing procedure. The average spacing between spots is deduced by analyzing the Fourier transform of one-dimensional spectra projected in both the horizontal and vertical directions. The average spot size is obtained by granulometry, more precisely, by studying the effect of successive morphological openings with structural elements of increasing size (Soille 2003). Once the subarrays are located, individual tiles are first positioned on a regular two-dimensional square lattice, then an optional optimization can be performed for each tile. The Seeded Region Growing algorithm (Adams and Bischof 1994) was used in the segmentation process of each tile. A square seed (by default of size two by two pixels) associated with the spot is positioned at the center of intensity of the tile. The pixels on the edge of the tile serve as background seeds unless their intensity falls within the top 10% of that tile, and the pixels with intensities above that threshold form the seeds for artifacts. Therefore, following the application of the Seeded Region Growing algorithm, each pixel within the tile is assigned as being part of the spot, part of the background, or part of an image artifact. There are no geometrical constraints applied to the shape of the spot except that it is obviously constrained to the limits of the tile as determined at the addressing stage. Spot intensities are extracted from the raw 16-bit images. The spot pixels as determined in the segmentation phase are averaged to extract the spot intensity. Similar to the work of Yang et al. (2002), the background intensity is obtained by probing a morphological opening obtained with a large structural element (default of 2.5 times the average spot spacing). Intensity ratios were normalized with the help of the robust LOESS regression on the so-called M-A plot, where M = log2(I1/I2) and A = log2[(I1 × I2)1/2], with I1 and I2 being the background subtracted intensities of the spot in the two images.
We selected individual thresholds for each array using the same type of calculations used to compute box-and-whisker plots (Tukey 1977). In a box-and-whisker plot, the difference between the upper (H2) and lower (H1) hinges is called the H-spread. The definition of hinge is similar to that of quartile and therefore H-spread is similar to the interquartile range. The upper (lower) whisker is located 1.5 × the H-spread above (below) the upper (lower) hinge, and values outside the whiskers are considered extreme. For each individual array, the thresholds for DNA copy number aberrations were set at the whisker levels. A MySQL relational database called CGHdb was built and populated with all ratio and ancillary data. Results from a human female (test) versus human male (reference) hybridization allowed estimation of true-positive, false-negative, and false-positive rates based on sex chromosome copy number. A total of 1134 of the 1430 X-chromosome clones had increased copy number, and 157 of the 196 Y chromosome clones had decreased copy number in the female test DNA sample. These values give a true-positive rate of 79.2% for increases and 80.1% for decreases, with corresponding false-negative rates of 20.8% for increases and 19.9% for decreases. Of the 30,216 autosomal clones, 665 (2.2%) showed increased copy number, and 592 (2.0%) showed decreased copy number. It is not possible to determine if these autosomal copy number differences reflect true positives or false positives.
Quantitative PCR
Primer/probe sets for two separate test genes (AMY1A and CNTNAP3) were designed using the repeat masked reference human genome sequence (NCBI_34; April 2003 release; http://genome.ucsc.edu/) (Table 1). Primers and probes were selected in regions of exact match between the test gene and its top paralog, as defined below (Identification of Paralogs). The SNAP25 gene, previously established to be single copy in the human genome (Bailey et al. 2002), was selected as a reference gene. The target sequences for test and reference primers and probes were verified in our human, chimp, and gorilla samples by direct sequencing. To determine relative copy number, 10 ng of genomic DNA was assayed in triplicate in 20-μL reactions containing 1× final concentration TaqMan Universal Master Mix (ABI part number 4304437), 200 nM each primer and probe, and 10 ng of template DNA. Each experiment was performed using a 384-well optical PCR plate and the ABI 7900HT machine with default cycling conditions. Copy number of the test locus in chimp or gorilla versus human was defined as 2-ΔΔCT, where ΔCT is the difference in threshold cycles for the test and reference loci.
Identification of paralogs
We sought to identify paralogous genes within the 55 recently duplicated human genomic regions. cDNA sequences for 206 Ensembl genes from these regions were collected with the Ensembl API v.27_35a. Using BioPerl v1.4 (Stajich et al. 2002), all sequences were transformed into peptides. Each gene's best partner within this set was determined by using the BLAST algorithm (Altschul et al. 1997) (E-value 10-10, ungapped alignments) against the peptide library. Once a gene's best partner was identified, the BLAST algorithm was rerun with the best hit as the query sequence. If, in the second iteration, the best BLAST hit was the original query sequence, then this gene pair was deemed paralogous. High-scoring segment pairs (HSPs) from the aligned regions were extracted in-frame in nucleotide form for subsequent analysis of substitution rates.
Acknowledgments
We thank Wan Lam for fabrication of arrays, and Evan Eichler for in silico predictions of segmental duplications in the human reference genome sequence. We thank the Canadian Institutes of Health Research and the BC Cancer Agency for funding this study. R.A.H. is a Michael Smith Foundation for Health Research scholar. A.G.C. is supported by NIH grant HG003229.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4456006.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Adams, R. and Bischof, L. 1994. Seeded region growing. IEEE Trans. Pattern Anal. Machine Intell. 16 641-647. [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297 1003-1007. [DOI] [PubMed] [Google Scholar]
- Chen, F.-C. and Li, W.-H. 2001. Genomic differences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68 444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437 69-87. [DOI] [PubMed] [Google Scholar]
- Christophides, G.K., Zdobnov, E., Barillas-Mury, C., Birney, E., Blandin, S., Blass, C., Brey, P.T., Collins, F.H., Danielli, A., Dimopoulos, G., et al. 2002. Immunity-related genes and gene families in Anopheles gambiae. Science 298 159-165. [DOI] [PubMed] [Google Scholar]
- Fortna, A., Kim, Y., MacLaren, E., Marshall, K., Hahn, G., Meltesen, L., Brenton, M., Hink, R., Burgers, S., Hernandez-Boussard, T., et al. 2004. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2 e207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama, A., Watanabe, H., Toyoda, A., Taylor, T.D., Itoh, T., Tsai, S.F., Park, H.S., Yaspo, M.L., Lehrach, H., Chen, Z., et al. 2002. Construction and analysis of a human-chimpanzee comparative clone map. Science 295 131-134. [DOI] [PubMed] [Google Scholar]
- Gibbs, R.A., Weinstock, G.M., Metzker, M.L., Muzny, D.M., Sodergren, E.J., Scherer, S., Scott, G., Steffen, D., Worley, K.C., Burch, P.E., et al. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428 493-521. [DOI] [PubMed] [Google Scholar]
- Grunstein, M., Schedl, P., and Kedes, L. 1976. Isolation and sequence analysis of sea urchin (Lytechinus pictus) histone H4 messenger RNA. J. Mol. Biol. 104 351-369. [DOI] [PubMed] [Google Scholar]
- Hill, C.A., Fox, A.N., Pitts, R.J., Kent, L.B., Tan, P.L., Chrystal, M.A., Cravchik, A., Collins, F.H., Robertson, H.M., and Zwiebel, L.J. 2002. G protein-coupled receptors in Anopheles gambiae. Science 298 176-178. [DOI] [PubMed] [Google Scholar]
- Ishkanian, A.S., Malloff, C.A., Watson, S.K., DeLeeuw, R.J., Chi, B., Coe, B.P., Snijders, A., Albertson, D.G., Pinkel, D., Marra, M.A., et al. 2004. A tiling resolution DNA microarray with complete coverage of the human genome. Nat. Genet. 36 299-303. [DOI] [PubMed] [Google Scholar]
- Krzywinski, M., Bosdet, I., Smailus, D., Chiu, R., Mathewson, C., Wye, N., Barber, S., Brown-John, M., Chan, S., Chand, S., et al. 2004. A set of BAC clones spanning the human genome. Nucleic Acids Res. 32 3651-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke D.P., Segraves, R., Carbone, L., Archidiacono, N., Albertson, D.G., Pinkel, D., and Eichler, E.E. 2003. Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res. 13 347-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, T.L., Tuzun, E., Morrison, V.A., Hayden, K.E., Ventura, M., McGrath, S.D., Rocchi, M., and Eichler, E.E. 2005. A genome-wide survey of structural variation between human and chimpanzee. Genome Res. 15 1344-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene duplication. Springer-Verlag, Berlin.
- She, X., Jiang, Z., Clark, R.A., Liu, G., Cheng, Z., Tuzun, E., Church, D.M., Sutton, G., Halpern, A.L., and Eichler, E.E. 2004. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature 431 927-930. [DOI] [PubMed] [Google Scholar]
- Soille, P. 2003. Morphological image analysis: Principles and applications, 2d ed. Springer-Verlag, Berlin, Heidelberg.
- Spiegel, I., Salomon, D., Erne, B., Schaeren-Wiemers, N., and Peles, E. 2002. Caspr3 and caspr4, two novel members of the caspr family are expressed in the nervous system and interact with PDZ domains. Mol. Cell. Neurosci. 20 283-297. [DOI] [PubMed] [Google Scholar]
- Stajich, J.E., Block, D., Boulez, K., Brenner, S.E., Chervitz, S.A., Dagdigian, C., Fuellen, G., Gilbert, J.G., Korf, I., Lapp, H., et al. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12 1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, K. and Long, M. 2002. Rapid divergence of gene duplicates on the Drosophila melanogaster X chromosome. Mol. Biol. Evol. 19 918-925. [DOI] [PubMed] [Google Scholar]
- Tukey, J.W. 1977. Explanatory data analysis. Addison-Wesley, Reading, MA.
- Watanabe, H., Fujiyama, A., Hattori, M., Taylor, T.D., Toyoda, A., Kuroki, Y., Noguchi, H., BenKahla, A., Lehrach, H., Sudbrak, R., et al. 2004. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature 429 382-388. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13 555-556. [DOI] [PubMed] [Google Scholar]
- Yang, Z. and Nielsen, R. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17 32-43. [DOI] [PubMed] [Google Scholar]
- Yang, Y.H., Buckley, M.J., and Speed, T.P. 2001. Brief. Bioinformatics 2 341-349. [DOI] [PubMed] [Google Scholar]
- Yang, Y.H., Dudoit, S., Luu, P., Li, D.M., Peng, V., Ngai, J., and Speed, T.P. 2002. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis, J.J., Sawyer, J.R., and Dunham, K. 1980. The origin of man: A chromosomal pictorial legacy. Science 208 1145-1148. [DOI] [PubMed] [Google Scholar]
- Zdobnov, E.M., von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R.R., Christophides, G.K., Thomasova, D., Holt, R.A., Subramanian, G.M., et al. 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298 149-159. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Vision, T.J., and Gaut, B.S. 2002. Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol. Biol. Evol. 19 1464-1473. [DOI] [PubMed] [Google Scholar]