Abstract
Alternative splicing is a major mechanism for gene product regulation in many multicellular organisms. By using different exon combinations, some coding regions can encode amino acids in multiple reading frames in different transcripts. Here we performed a systematic search through a set of high-quality human transcripts and show that ∼7% of alternatively spliced genes contain dual (multiple) coding regions. By using a conservative criterion, we found that in these regions most secondary reading frames evolved recently in mammals, and a significant proportion of them may be specific to primates. Based on the presence of in-frame stop codons in orthologous sequences in other animals, we further classified ancestral and derived reading frames in these regions. Our results indicated that ancestral reading frames are usually under stronger selection than are derived reading frames. Ancestral reading frames mainly influence the coding properties of these dual coding regions. Compared with coding regions of the whole genome, ancestral reading frames largely maintain similar nucleotide composition at each codon position and amino acid usage, while derived reading frames are significantly different. Our results also indicated that prior to acquisition of a new reading frame, the suppression of in-frame stop codons in the ancestral state is mainly achieved by one-step transition substitutions at the first or second codon position. Finally, the selective forces imposed on these dual coding regions will also be discussed.
Alternative splicing is a major mechanism for functional regulation in many multicellular organisms, and it plays an important role in increasing the diversity of protein products from an individual gene locus. Estimates suggest that alternative splicing occurs in 40%∼60% of human genes (Mironov et al. 1999; Croft et al. 2000; Kan et al. 2001; Lander et al. 2001; Modrek et al. 2001). By using different exon combinations, alterative splicing can conceivably give birth to some dual coding regions in which the same exon sequence shared between different transcripts can encode amino acids in different reading frames.
Recently several specific examples of dual coding regions have been reported in the human and mouse genomes (Scorilas et al. 2001; Zhao et al. 2004); however, there has been no large-scale study of dual coding regions in alternatively splicing genes. The importance of comprehensively characterizing such genes, especially in the human genome, is at least twofold: (1) dual coding regions generate distinct amino acid sequences in functionally related proteins, increasing genome complexity, and the systematic and precise characterization of existing dual coding regions in alternatively spliced genes can therefore lead to improvements in gene prediction and annotation; and (2) during evolution, dual coding regions must simultaneously maintain two reading frames and are therefore shaped by unusual selective forces. Thus they provide a special opportunity to trace the origin of unique splicing patterns and to understand the evolutionary constraints on coding sequences in double reading frames.
Overlapping genes in mammalian genomes have recently been examined (Veeramachaneni et al. 2004). We note that “dual coding regions” and “overlapping genes” are related but distinct concepts. Dual coding regions do not necessarily involve more than one gene. Most dual coding regions come from alternatively spliced transcripts from the same gene. Overlapping genes, on the other hand, need not generate dual coding regions, since a lot of overlapping regions either do not encode amino acids or maintain the same reading frame.
Here we performed a systematic search through high-quality mRNAs to identify regions in the human genome that encode amino acids in more than one reading frame due to alternative splicing. We then studied the evolution of these unusual coding regions by comparative analysis of orthologous sequences in other animal genomes. Throughout this study, we asked the following questions: (1) how frequently do human alternatively spliced genes contain dual or multiple coding regions, (2) when were the secondary reading frames acquired in these coding regions, and (3) how do these regions evolve after both reading frames are established?
Results
Characterization of dual coding regions in alternatively spliced genes
To estimate the frequency of dual coding regions in alternatively spliced genes, we performed a systematic search through a set of high-quality and well-annotated human transcripts (see Methods). Among 2585 alternatively spliced genes, we found 173 genes that contain coding sequences for two reading frames (one example is shown in Figure 1, with details provided in Supplemental File 1) and six genes that can encode amino acid sequences in all three reading frames. Thus ∼7% of alternatively spliced genes in the human genome contain multiple coding regions. Because our data set does not include all spliced versions of each gene, this number can be considered an underestimate.
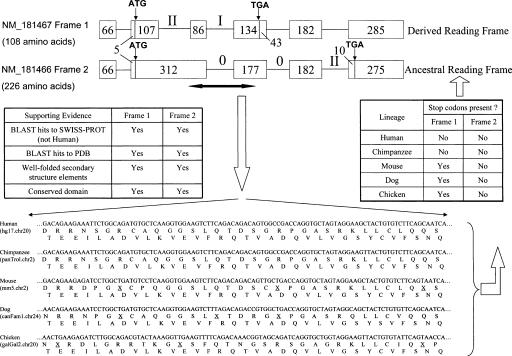
Figure 1.
Schematic representation of a dual coding region in the human ITGB4BP gene. Exons are represented by boxes, and introns are represented by connecting lines. Numbers inside the boxes refer to base pairs. Roman numerals indicate intron phases. The dual coding region is marked by a black horizontal arrow. Orthologous sequences for this region are shown in other species, and in-frame stop codons are marked by an underlined X. Based on this alignment, the table on the right summarizes the presence of stop codons in two reading frames. Bioinformatic supporting evidence for both reading frames is shown in the table on the left. White arrows indicate direction of data flow. NM_181466 and NM_181467 are RefSeq accession numbers.
In general, the multiple coding regions we identified are not very long, with an average length of 133 nucleotides. They are distributed over 22 out of 24 chromosomes (all but chromosomes 13 and Y), indicating that the phenomenon is genome wide (Supplemental File 2). We also examined the distribution of these genes in different biological processes, function categories, and components, respectively (eight GO biological processes, 16 GO functional categories, and seven GO biological components) (Supplemental File 2). No obvious bias was detected, and the distributions appear to be random. Because very few genes contain triple coding regions, in the following analysis we focus on the dual coding regions in the human genome.
In addition, multiple coding regions in alternatively spliced genes are not unique to humans. In a broader survey, we extended this analysis to two other model organisms Caenorhabditis elegans and Drosophila melanogaster, and we found that at least 6.8% and 2.3% of alternatively spliced genes, respectively, contain multiple coding regions (Table 1).
Table 1.
Statistical summary of alternatively spliced genes in three model organisms
| No. of transcripts | No. of genes | Transcripts per gene | Genes with dual coding regions | Frequency | |
|---|---|---|---|---|---|
| H. sapiens | 7146 | 2585 | 2.76 | 179 | 6.9% |
| D. melanogaster | 8691 | 3086 | 2.82 | 69 | 2.2% |
| C. elegans | 2884 | 1246 | 2.31 | 85 | 6.8% |
Evolutionary origin of dual coding regions
For a representative gene with a dual coding region, a second splicing pattern presumably arose later during evolution, leading to creation of an alternate reading frame. Ideally, we would be able to identify the origin of the secondary frame directly, if we knew whether this reading frame can be expressed in orthologous genes in other organisms. However, currently, alternative splicing data in animals other than human are quite incomplete in terms of both scope and annotation. As an alternative, we inferred the presence of dual coding regions by using orthologous genomic sequences. Our method is based on a simple observation: If two reading frames encode amino acids, then neither frame should contain a stop codon. On the contrary, the presence of stop codons in one reading frame is strong negative evidence, indicating that this reading frame is probably not translated. Thus, by identifying whether or not orthologous sequences in other animal species are translatable in both corresponding reading frames, we can trace the evolutionary origin of dual coding regions in the human genome (Fig. 1).
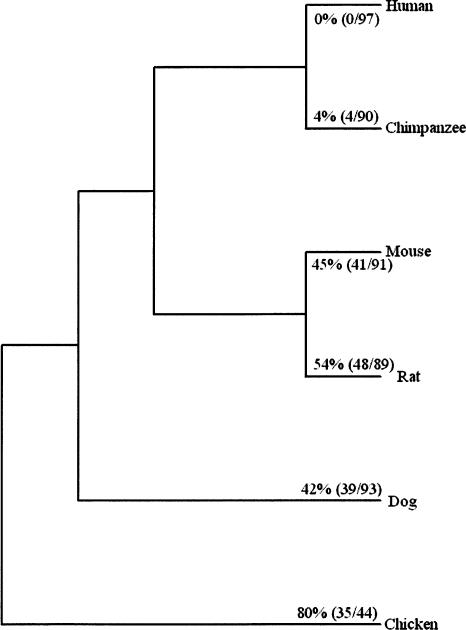
For the dual coding regions in humans, we performed a comparative analysis of orthologous sequences in the chimpanzee, mouse, rat, dog, and chicken genomes. Except in very few cases, in these orthologous regions, stop codons appear consistently in only one of two reading frames. As shown in Figure 2, one of two reading frames contains one or more stop codons in 80% of the dual coding regions in chickens, 45% in mice, and 54% in rats, respectively. These results indicate that most secondary reading frames in the dual coding regions examined evolved recently in mammals, and that a significant proportion of them evolved after the divergence of primates and rodents, possibly just in the primate lineage.
Figure 2.
Evolutionary origin of dual coding regions in the human genome. The frequency that one of two reading frames contains stop codons in orthologous sequences in each species is shown on each lineage. Two numbers are shown in parentheses: One is the number of the sequences in which one reading frame contains stop codons, and the other is the total sequence number in comparison. Only dual coding regions >100 nucleotides are used in this analysis.
This method is intrinsically conservative for inferring nondual coding regions, because the absence of in-frame stop codons does not necessarily imply that the reading frame encodes a protein. Here we used only those relatively long coding sequences in the analysis, to reduce the possibility that one reading frame is free of stop codons by chance.
Moreover, we emphasize that every lineage has surely acquired some unique dual coding regions that are not present in humans. Therefore it remains an open question whether the frequency of dual coding regions increases in specific lineages during evolution.
Selection on two reading frames in dual coding regions
Based on the presence of stop codons in two reading frames in orthologous sequences, we classified all 204 reading frames of 102 dual coding regions as either an “ancestral reading frame” (ARF) or a “derived reading frame” (DRF) with confidence (see Methods). We started our analysis with a set of high-quality RefSeq mRNAs. All these sequences have been manually curated by National Center for Biotechnology Information (NCBI) staff and have solid experimental support for their existence. However, it was still possible that some dual coding regions may not encode functional protein products, or that some mRNAs containing DRFs could be transcriptional noise from incorrect splicing. To address this possibility, we carried out a set of thorough bioinformatic analyses on the peptide sequences translated from DRFs within dual coding regions: We tested (1) whether the peptide sequences show significant sequence similarity to those proteins in the SWISS-PROT database, especially to nonhuman proteins; (2) whether these peptide sequences show significant sequence similarity to proteins in the Protein Data Bank (PDB), since such proteins with known structure are generally considered functional; (3) whether these peptide sequences contain some specific functional motifs; (4) whether these peptide sequences are predicted to contain well-folded secondary structural elements (since, given the vast sequence space, only a tiny fraction can be folded); and (5) whether these peptide sequences can be recognized as distant homologs by sequence-structure comparison onto known proteins (threading). All the above features provide strong supporting evidence for a protein's functionality, since randomly translated sequences from transcriptional noise would not be expected to have such features. As a result, we found that almost all the cases we report are supported by some evidence, and the majority of them are associated with more than one line of evidence (Supplemental File 3). Furthermore, in these peptide sequences, the proportion of hits with sequence similarity to known proteins and with potential structural similarity to known structures is statistically significantly higher than random expectation (Supplemental File 2). These data suggest that most, if not all, of these transcripts are functional.
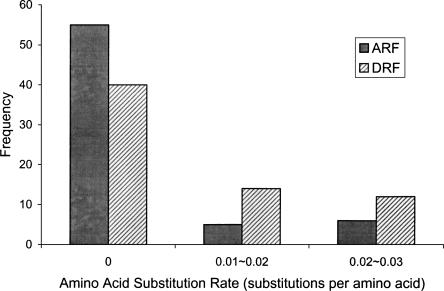
From an evolutionary point of view, it is meaningful to compare the selection intensity imposed on both reading frames in the same coding region. We performed an analysis of 66 pairs of human and chimpanzee orthologs, where both reading frames lack internal stop codons in both species and amino acid sequences in both reading frames are well conserved. Assuming that these chimpanzee sequences encode amino acids in two reading frames, we compared the amino acid substitution rates in ARFs and DRFs. (In dual coding regions, the ratio of nonsynonymous substitution rate to synonymous substitution rate, Ka/Ks, is no longer a valid index for selection intensity, since synonymous sites in one reading frame can be nonsynonymous sites in the other frame.) Our results show that amino acid sequences in ARFs are usually more conserved than are those in DRFs, and only 9% of the regions have a lower amino acid substitution rate in DRFs than in corresponding ARFs. As shown in Figure 3, the distributions of amino acid substitution rates between ARFs and DRFs are significantly different (P < 0.003). These results suggest that ARFs are usually under stronger selective constraint than DRFs.
Figure 3.
Selection on ARFs and DRFs in dual coding regions. The black bars represent the frequency of amino acid substitution rate in ARFs; the striped bars represent the frequency of amino acid substitution rate in DRFs in dual coding regions. For all regions included in this analysis, the amino acid substitution rates between human and chimpanzee sequences are <0.03 substitutions per amino acid in both reading frames.
Coding properties of dual coding regions
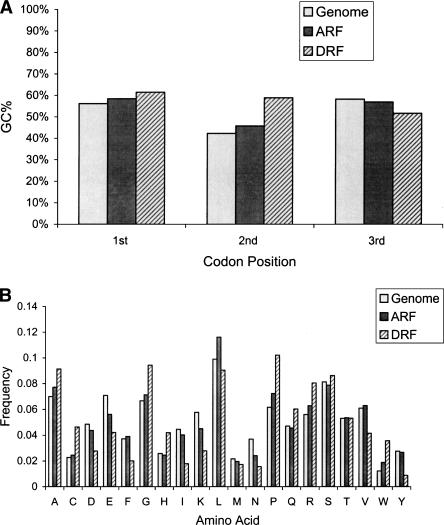
We calculated nucleotide composition at each codon position and amino acid usage in ARFs and DRFs, respectively, and then compared these values to the whole-genome usage. Similar to coding regions of the whole genome, nucleotide composition in ARFs is GC-rich at the first and third codon positions (Fig. 4A). As a direct result of frameshifts from ARFs, GC% at the second codon position in DRFs strikingly increases, because the second codon position in DRFs is the first or third codon position in ARFs. Moreover, ARFs maintain similar amino acid usage to the whole genome, and most amino acids (15 out of 20) show a similar frequency. In contrast to ARFs, most amino acids (16 out of 20) in DRFs are significantly over- or underrepresented (P < 10-3) (Table 2; Fig. 4B). The great variation in amino acid usage in DRFs can be explained largely by the increase of GC% at the second codon position. Mapping the amino acid variation in DRFs onto the genetic code table revealed a strong correlation between change in amino acid usage and the GC usage at the second codon position (Fig. 4C).
Figure 4.
(A) GC% at each codon position in dual coding regions. The gray bars represent GC% at each codon position in the whole-genome coding regions, the black bars represent GC% in ARFs, and the striped bars represent GC% in DRFs in dual coding regions. (B) Amino acid usage in dual coding regions. The gray bars represent the frequency of amino acids in the whole-genome coding regions, the black bars represent the frequency of amino acids in ARFs, and the striped bars represent the frequency of amino acids in DRFs in dual coding regions, (C) The correlation between amino acid usage and nucleotides at codon positions in DRFs in dual coding regions. The overrepresented amino acids are colored in blue, the underrepresented amino acids are colored in green, and the stop codons are in red. The statistically significantly overrepresented amino acids (Phe, Ile, Val, Tyr, Asn, Lys, Asp, and Glu) are colored dark blue (P <10-3); the statistically significantly underrepresented amino acids (Pro, Ala, His, Gln, Cys, Trp, Arg, and Gly) are colored dark green (P <10-3).
Table 2.
Statistical significance of amino acid frequency in ancient reading frames and derived reading frames
|
P (being over- or underrepresented)a
|
||
|---|---|---|
| Amino acid | Ancient reading frames | Derived reading frames |
| A | 0.088 | <0.001 ↑ |
| C | 0.432 | <0.001 ↑ |
| D | 0.119 | <0.001 ↓ |
| E | 0.003 ↓ | <0.001 ↓ |
| F | 0.504 | <0.001 ↓ |
| G | 0.228 | <0.001 ↑ |
| H | 0.170 | <0.001 ↑ |
| I | 0.077 | <0.001 ↓ |
| K | 0.002 ↓ | <0.001 ↓ |
| L | 0.002 ↑ | 0.063 |
| M | 0.419 | 0.081 |
| N | <0.001 ↓ | <0.001 ↓ |
| P | 0.026 | <0.001 ↑ |
| Q | 0.457 | <0.001 ↑ |
| R | 0.081 | <0.001 ↑ |
| D | 0.468 | 0.079 |
| T | 0.370 | 0.395 |
| V | 0.431 | <0.001 ↓ |
| W | 0.001 ↑ | <0.001 ↑ |
| Y | 0.196 | <0.001 ↓ |
↑ indicates overrepresented; ↓ underrepresented. P-value cut-off = 0.01
Lastly, to infer the substitutions that suppressed in-frame stop codons prior to acquisition of DRFs, we calculated the distribution of the amino acids in human DRFs that correspond to stop codons in dog DRFs. The most frequently used amino acids are Q, R, and W, and statistical analysis shows that these three amino acids are significantly enriched at these locations (P = 10-32) (see Table 3; Supplemental File 2).
Table 3.
The association between Q, R, and W amino acids in human DRFs and the amino acids corresponding to stop codons in dog DRFs
| Amino acids corresponding to stop codons in dog DRF | Amino acids not corresponding to stop codons in dog DRF | Total | |
|---|---|---|---|
| Q, R, and W in human DRF | 53 | 451 | 504 |
| Other amino acids in human DRF | 26 | 2359 | 2385 |
| Total | 79 | 2810 | 2889 |
χ2 = 139, P = 10-32
Discussion
Our results show that a small but significant fraction of human genes contain dual or multiple coding regions due to recently acquired differences in splicing patterns. In a sense, such alternative splicing can be viewed as a mechanism to generate a frameshift at the RNA level. This mechanism differs from ribosomal frameshifting, where a slippery signal in mRNA triggers a programmed reading-frame change during translation (Baranov et al. 2004). Thus two totally distinct approaches seem to have evolved to achieve the same goal—that is, to generate different functional products from the same genetic materials. The alternative processing of dual coding protein-coding regions may provide more combinatorial options.
During the process of acquiring a new reading frame, the effect of various exon combinations would be relentlessly evaluated by selection. Only those splicing patterns without major deleterious fitness effects would survive. Thus, many potential splicing patterns would be blocked by in-frame stop codons at undesirable locations, either through the process of nonsense-mediated mRNA decay (NMD) (Maquat 2004) or through the expression of rogue proteins that produce a selective disadvantage. Suppression of stop codons in the DRF would therefore be required to establish this reading frame. For example, at the locations corresponding to in-frame stop codons in dog DRFs, we find the three most frequently used amino acids are Q, R, and W. These three amino acids share the common feature that some or all of their codons can be converted to stop codons by one-step transition substitutions at the first or second codon position (Q: CAA and CAG; R: CGA; W: TGG. For R, CGA is the most frequent Arg codon at these sites.) This observation should not be surprising, because (1) transition substitutions occur at a much higher frequency than do transversion substitutions (Topal and Fresco 1976); and (2) the first or second codon position in DRFs often corresponds to the third codon position, the most degenerate, in ARFs.
After establishing a second reading frame, natural selection would constrain both reading frames. Our comparative analysis between human and chimpanzee revealed that the reading frames from the original splicing patterns undergo many fewer substitutions than do later derived ones, suggesting that ARFs are maintained by stronger selection. This result is consistent with a recent study that concluded that the products from splicing patterns shared by humans and mice tend to be expressed as the major form over human-specific exons and are therefore likely to be under stronger selection (Modrek and Lee 2003). Furthermore, in terms of amino acid usage and GC content at each codon position, the coding properties of these dual coding sequences appear to be mainly influenced by ARFs.
Because two overlapping reading frames cannot explore sequence space independently, some evolutionary cost arises for the superposition of coding regions. A simulation of such an evolutionary search process showed a reduction of allowable point mutations and the possibility of small-scale adaptation (Huynen et al. 1993). Given this cost, what are possible selective advantages of dual coding regions? Their most obvious benefit is to increase the amount of information per unit length, as discussed elsewhere (Eigen and Schuster 1979; Huynen et al. 1993). Dual coding often occurs in viral and prokaryotic genomes and in prokaryote-derived organelles such as mitochondria (Normark et al. 1983), where reduced genome sizes may be too small to store all the necessary information in a sequential manner. The space of dual or multiple coding regions in the human genome is <0.5% of exons. However, considering the vast space of noncoding DNA in the human genome, information storage efficiency is unlikely to be the driving force for evolving dual coding regions. A second explanation comes from a recent theoretical study, which suggests that when the overlapping region involves common crucial residues, the direct evolutionary advantage results from reducing the number of vulnerable points in the DNA sequence (Peleg et al. 2004). A third possible benefit is to enhance partial functions of one gene without reproducing a full-length product. Many dual coding regions occur at transcript termini, and alternative splicing usually yields a truncated version. Nevertheless, such products often perform biological functions and thus may offer a novel way to differentiate protein function.
Methods
Identification and characterization of dual coding regions
To detect multiple coding regions with confidence, we used high-quality and fully annotated mRNAs from the NCBI Reference Sequence (RefSeq) Database in our analysis. The RefSeq Database reflects our current knowledge of biology and is generally accepted as a standard for genome annotation (Pruitt et al. 2005). To further decrease background noise in the analysis, only the RefSeq mRNAs marked as reviewed, validated, or provisional were included, because they are all thought to represent a valid transcript and protein. Sequences marked predicted, inferred, unknown, and model were excluded from our data set. Redundant entries (with the same accession number) were removed from the data set. According to the annotation in the UCSC Genome Browser, the genome locus for each RefSeq transcript was identified. Together, 18,082 transcripts were included in our data set, and they represent 13,584 distinct genes. Among these genes, 2585 are alternatively spliced.
Coding regions were then translated in different reading frames. Amino acid sequences in the annotated reading frames are biologically meaningful and used as the protein database for further BLAST searches. Each amino acid sequence in other reading frames (in-frame stop codons were treated as one distinct amino acid in these frames) was used as a query to blast the protein database using the BLASTP program (downloaded from the NCBI Web site) with a conservative E-value threshold of 0.001 to retain all the potential dual coding cases, including very short regions. After this initial screening, the significant hits were parsed by PERL scripts for further automated and manual analysis. Then only when we found a significant hit to the same gene locus as the query and also when the aligned regions between the hit and the query were exactly the same, did we consider the gene further as a potential candidate containing multiple coding regions. To be more cautious, by using the UCSC Genome Browser, we analyzed gene structures one-by-one to confirm the occurrence of alternative splicing and to identify the exact boundaries of multiple coding regions. Occasionally, a few genes cannot be explained by alternative splicing (presumably due to sequence error or other biological processes), and they were excluded from our analysis. Finally, we identified 179 genes containing multiple coding regions (173 genes for dual coding and six genes for triple coding). GO identifications for these genes were parsed from the GenBank files, and their distributions in different biological process, function categories and components were then analyzed. Among the dual coding genes, five genes contain more than one dual coding region, due to unrelated splicing patterns. These dual coding regions were treated as distinct cases in the following analysis.
Comparative analysis of orthologous sequences in other animal genomes
For each dual coding region, the orthologous sequences were extracted from Multiz genome alignments (human May 2004 [hg17], chimp November 2003 [panTro1], mouse May 2004 [mm5], rat June 2003 [rn3], dog July 2004 [canFam1], and chicken February 2004 [galGal2]) using UCSC Genome Browser (Schwartz et al. 2003; Blanchette et al. 2004). To reduce background noise, only well-aligned parts were included in the analysis. Orthologous sequences were translated in both reading frames by a PERL script. For the vast majority of genes, stop codons consistently appear in only one of two reading frames. However, there are only six cases where stop codons appear in both reading frames in the orthologous sequences, yielding confusing biological signals. These cases were excluded from our analysis. Then, for dual coding regions >100 nucleotides in each species, we scored the frequency that one of two reading frames contains stop codons.
Based on the presence of stop codons in orthologous sequences in other animals, we classified the ARF and DRF for each dual coding region, whenever possible. Together, there are 102 cases in which we can infer the order of acquisition of the two reading frames in evolution (ARF vs. DRF). Among these, 89 have orthologs in chimpanzees that also contain no stop codons in either reading frame. For each case of the 89, the number and rate of amino acid substitutions in both reading frames were calculated from the human and chimpanzee sequence alignment. To understand the relative selection intensity on two reading frames, 66 cases were used for further comparison. Here we excluded the cases with high amino acid substitution rates (>0.03 substitutions per amino acid) to ensure that the chimpanzee DRFs encode amino acids. A Wilcoxon matched-pairs signed-ranks test was used to determine whether the amino acid substitution rates in ARFs and DRFs are significantly different. We also carried out a set of bioinformatic analyses on peptide sequences translated from DRFs within these 102 dual coding regions. Sequence similarity searches were performed by using BLASTP. The SWISS-PROT database and PDB database were downloaded from the NCBI Web site. Compared with the randomly shuffled control group, the statistical significance of the frequency of hits (PDB) was determined by a χ2 independence test. Motif searching was performed by using PS_SCAN (Gattiker et al. 2002). To be strict, we excluded motifs with a high probability of occurrence from our analysis. Secondary structure prediction was performed by using NNPREDICT (Kneller et al. 1990). Again, to have a strict threshold, we only counted a sequence as containing well-folded elements when more than three amino acid positions in a row were predicted to be in the same type of secondary structure (α helix/β sheet). Protein sequence-structure comparison (threading) was performed by using FUGUE (Shi et al. 2001). This program scans a database of structural profiles, calculates the sequence-structure compatibility scores, and produces a list of potential homologs. Finally, the statistical significance of the difference between observed prediction in these sequences and random expectation was determined by a χ2 independence test.
The amino acid usage and nucleotide composition at each codon position were calculated for both ARFs and DRFs in dual coding regions and for the whole genome, respectively. Here the coding regions in all 13,584 genes (only one transcript was chosen randomly if there were more than one transcript for a gene) represent the whole-genome coding regions. To determine statistically whether a specific amino acid is overrepresented or underrepresented in dual coding regions, the same length of coding regions was randomly sampled from the whole genome 1000 times and the amino acid usage was calculated for each sample. The statistical significance of an observed amino acid frequency was calculated by the probability of not being smaller (larger) in the simulation. To understand the substitutions related to in-frame stop codon suppression, we calculated the distribution of amino acids in human DRFs that aligned with stop codons in orthologous dog DRFs. A χ2 test was used to determine whether the three most frequently used amino acids Q, R, and W are significantly enriched in these locations.
Acknowledgments
We thank laboratory members Dr. Andre Cavalcanti for critical manuscript reading and Dr. Tom Doak for helpful discussion. We thank Drs. Judy Swan and Moshe Pritsker for critical manuscript reading, Dong Li, Peking University for kind help in GO analysis, and Dr. Steven Pechous of NCBI for helpful discussion. We also thank three anonymous reviewers for valuable suggestions. This work was supported by National Institute of General Medical Sciences grant GM59708 and National Science Foundation grant DBI-9875184 to L.F.L.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4246506.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Baranov, P.V., Gesteland, R.F., and Atkins, J.F. 2004. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA 10 221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette, M., Kent, W.J., Riemer, C., Elnitski, L., Smit, A.F., Roskin, K.M., Baertsch, R., Rosenbloom, K., Clawson, H., Green, E.D., et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14 708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft, L., Schandorff, S., Clark, F., Burrage, K., Arctander, P., and Mattick, J.S. 2000. ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nat. Genet. 24 340-341. [DOI] [PubMed] [Google Scholar]
- Eigen, M. and Schuster, P. 1979. The hypercycle: A principle of natural self-organzation. Springer-Verlag, Berlin.
- Gattiker, A., Gasteiger, E., and Bairoch, A. 2002. ScanProsite: A reference implementation of a PROSITE scanning tool. Appl. Bioinformatics 1 107-108. [PubMed] [Google Scholar]
- Huynen, M.A., Konings, D.A., and Hogeweg, P. 1993. Multiple coding and the evolutionary properties of RNA secondary structure. J. Theor. Biol. 165 251-267. [DOI] [PubMed] [Google Scholar]
- Kan, Z., Rouchka, E.C., Gish, W.R., and States, D.J. 2001. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 11 889-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneller, D.G., Cohen, F.E., and Langridge, R. 1990. Improvements in protein secondary structure prediction by an enhanced neural network. J. Mol. Biol. 214 171-182. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W. et al. 2001. Initial sequencing and analysis of the human genome. Nature 409 860-921. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. 2004. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell. Biol. 5 89-99. [DOI] [PubMed] [Google Scholar]
- Mironov, A.A., Fickett, J.W., and Gelfand, M.S. 1999. Frequent alternative splicing of human genes. Genome Res. 9 1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C.J. 2003. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 34 177-180. [DOI] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., and Lee, C. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29 2850-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark, S., Bergstrom, S., Edlund, T., Grundstrom, T., Jaurin, B., Lindberg, F.P., and Olsson, O. 1983. Overlapping genes. Annu. Rev. Genet. 17 499-525. [DOI] [PubMed] [Google Scholar]
- Peleg, O., Kirzhner, V., Trifonov, E., and Bolshoy, A. 2004. Overlapping messages and survivability. J. Mol. Evol. 59 520-527. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D., Tatusova, T., and Maglott, D.R. 2005. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33 D501-D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R.C., Haussler, D., and Miller, W. 2003. Human-mouse alignments with BLASTZ. Genome Res. 13 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorilas, A., Kyriakopoulou, L., Yousef, G.M., Ashworth, L.K., Kwamie, A., and Diamandis, E.P. 2001. Molecular cloning, physical mapping, and expression analysis of a novel gene, BCL2L12, encoding a proline-rich protein with a highly conserved BH2 domain of the Bcl-2 family. Genomics 72 217-221. [DOI] [PubMed] [Google Scholar]
- Shi, J., Blundell, T.L., and Mizuguchi, K. 2001. FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310 243-257. [DOI] [PubMed] [Google Scholar]
- Topal, M.D. and Fresco, J.R. 1976. Complementary base pairing and the origin of substitution mutations. Nature 263 285-289. [DOI] [PubMed] [Google Scholar]
- Veeramachaneni, V., Makalowski, W., Galdzicki, M., Sood, R., and Makalowska, I. 2004. Mammalian overlapping genes: The comparative perspective. Genome Res. 14 280-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, F.Q., Zheng, Y., Dong, B., and Oka, T. 2004. Cloning, genomic organization, expression, and effect on β-casein promoter activity of a novel isoform of the mouse Oct-1 transcription factor. Gene 326 175-187. [DOI] [PubMed] [Google Scholar]