Abstract
Animal mitochondrial DNA is characterized by a remarkably high level of within-species homoplasy, that is, phylogenetic incongruence between sites of the molecule. Several investigators have invoked recombination to explain it, challenging the dogma of maternal, clonal mitochondrial inheritance in animals. Alternatively, a high level of homoplasy could be explained by the existence of mutation hot spots. By using an exhaustive mammalian data set, we test the hot spot hypothesis by comparing patterns of site-specific polymorphism and divergence in several groups of closely related species, including hominids. We detect significant co-occurrence of synonymous polymorphisms among closely related species in various mammalian groups, and a correlation between the site-specific levels of variability within humans (on one hand) and between Hominoidea species (on the other hand), indicating that mutation hot spots actually exist in mammalian mitochondrial coding regions. The whole data, however, cannot be explained by a simple mutation hot spots model. Rather, we show that the site-specific mutation rate quickly varies in time, so that the same sites are not hypermutable in distinct lineages. This study provides a plausible mutation model that potentially accounts for the peculiar distribution of mitochondrial sequence variation in mammals without the need for invoking recombination. It also gives hints about the proximal causes of mitochondrial site-specific hypermutability in humans.
Mitochondrial DNA sequence variation in animals is notoriously characterized by a high amount of homoplasy, i.e., phylogenetic/genealogic conflict between sites. This is true between species, decreasing the efficiency of mitochondrial markers for phylogenetic analyses (see Springer et al. 2001; Delsuc et al. 2003), and also within species, as reported in many population genetic and phylogeographic surveys (see Vigilant et al. 1991; Vandewoestijne et al. 2004). Given the prevalence of mitochondrial data in molecular biodiversity, it is essential to understand the reasons for such a high amount of discrepancy between sites and its consequences on the interpretation of data sets. Eyre-Walker et al. (1999) took this point of view when analyzing third-codon-position variations between 29 nearly complete human mitochondrial genomes. They argued that the high amount of homoplasy they found was due to recombination between mitochondrial lineages—a strong claim challenging the “dogma” of maternal, clonal mitochondrial inheritance in animals. This report initiated a controversy.
There are two major mechanisms potentially explaining the occurrence of homoplasy within species. The first one is recombination. When partial genetic exchanges occur between distantly related individuals, the various segments of the recombined molecules are phylogenetically incongruent, because they actually have distinct genealogical histories. Alternatively, homoplasy can be generated by convergence due to multiple mutations. If two distantly related individuals independently receive the same mutation at site i, then site i will wrongly support their grouping, in conflict with other sites in the data set. The high amount of homoplasy in mitochondrial DNA could therefore be due to the phylogenetic noise introduced by mutation hot spots. In principle, an obvious difference between the two models is the expected distribution of the number of distinct states taken by polymorphic sites: Mutation hot spots, not recombination, should generate three- or four-state polymorphisms. Mitochondrial DNA, however, undergoes more transitions (C↔T and A↔G changes) than transversions, so that a majority of two-state polymorphisms is expected under both the recombination and the hot spots hypothesis.
A number of studies have attempted to demonstrate the occurrence of recombination in animal mitochondria. Recombination first appeared supported in humans by linkage disequilibrium (Awadalla et al. 1999) and geographic (Hagelberg et al. 1999) analyses, but these studies were criticized (Kivisild and Villems 2000; Hagelberg 2003) and/or could not be reproduced when the data set increased in size (Ingman et al. 2000; Innan and Nordborg 2002). Given the cytological evidence for maternal mitochondrial inheritance in animals (see Birky 1995), several investigators called for prudence before invoking recombination and suggested that the mutational hypothesis should be favored until it is formally rejected (Hey 2000). Recent reports, however, provided indirect evidence for mitochondrial recombination in several animal species (Piganeau et al. 2004; Gantenbein et al. 2005; Tsaousis et al. 2005), as well as direct proof of paternal leakage (Schwartz and Vissing 2002) and subsequent recombination (Kraytsberg et al. 2004) in one human.
Curiously, despite the importance of the debate, the alternative mutation hot spots hypothesis has not been examined in depth. In their seminal article, Eyre-Walker et al. (1999) considered various mutational models potentially explaining homoplasy in humans, i.e., unidirectional and bidirectional mutation hot spots. They found that not one of them was supported by the data. Stoneking (2000) and Pesole and Saccone (2001) showed that very recent somatic and germline mutations in the human mitochondrial control region tend to occur at evolutionarily hypervariable sites, suggesting that these sites are true hot spots. Aside from these reports, the debate has focused on recombination, mutation hot spots being invoked when the recombination hypothesis was not firmly supported. In particular, no response has been provided to Eyre-Walker et al.'s (1999) rejection of various mutation hot spots models in human coding regions.
In this article, we take the point of view of trying to detect mutation hot spots from mitochondrial DNA sequence variation, and asking whether they can explain the high level of homoplasy observed within species. Mutation hot spots, if any, should result in the co-occurrence of polymorphisms between closely related species, assuming that a hot spot in species 1 is still hot in species 2. They should also imply a correlation across sites of within-species and between-species variability—a hot spot should contribute both to polymorphism and divergence and be variable both within and between species. By using mammals as a model taxon, we take a phylogenetic approach to check these predictions of the mutation hot spots hypothesis and to try to elucidate the causes of the high level of homoplasy in mitochondrial DNA.
Results
Data sets
Two data sets were built from public databases. We first extracted from Polymorphix (Bazin et al. 2005), a data set of mammalian species for which the (nearly) complete cytochrome b (MT-CYB) gene has been sequenced in six individuals or more. Cytochrome b was chosen because it is the only mitochondrial protein-coding gene for which polymorphism data are available in several groups of closely related mammals. Sequence alignments were inspected by eye and corrected when required. Dubious sequences were manually removed. These include potential nuclear pseudogenes and sequences with many gaps or undetermined nucleotides. Then sequences from congeneric species were gathered, and maximum-likelihood phylogenetic trees were built. Several species were not monophyletic, because of genetic introgression, incomplete lineage sorting, or taxonomic uncertainties. A small number of sequences were removed or reassigned to ensure species monophyly. One highly polyphyletic species had to be removed, and the two species from genus Pseudois were fused in a single data set because of their intricate genealogies. We ended up with 113 mammalian cytochrome b data sets (see Supplemental material), each with six to 105 sequences; among the hundreds of human cytochrome b sequences available, the 105 ones obtained by Ingman et al. (2000) and Ingman and Gyllensten (2003) were used, to avoid a strong size discrepancy with other data sets. These data sets encompassed 11 orders, 32 families, and 66 genera, among which 27 were represented by more than one species (data set available from http://kimura.univ-montp2.fr/data). Since this study aims at addressing mutational effects, we focused on transitions (C↔T and A↔G changes) at third-codon positions, following the method of Eyre-Walker et al. (1999). All these changes are synonymous and therefore presumably neutral. They correspond to ∼90% of observed polymorphisms. Data sets included 368-380 third-codon positions, and the percentage of polymorphic third-codon positions varied from 1.05% (Lemur catta) to 51.05% (Phyllotis xanthopygus).
In addition to this mammalian cytochrome b data set (see Supplemental material), a Hominoidea full-genome data set was built by gathering 560 human mitochondrial sequences (Herrnstadt et al. 2002), the two sequences available from GenBank for chimpanzee (Pan troglodytes) and gorilla (Gorilla gorilla), and the one sequence available for bonobo (P. paniscus), Borneo orangutan (Pongo pygmaeus), Sumatra orangutan (P. pygmaeus abelii), and one gibbon (Hylobates lar). The human data set includes African, European, and Asiatic samples. The control region was excluded because it has its own specific evolutionary process and cannot be easily aligned between species so divergent; 15,370 gap-free sites were finally analyzed.
Mammalian cytochrome b
Homoplasy analysis
We first checked whether the amount of within-species homoplasy in mammalian cytochrome b was actually higher than expected. Homoplasy is defined as the sum across sites of the parsimony score plus one minus the number of distinct states (Kluge and Farris 1969). The homoplasy is high when sites disagree with respect to the underlying phylogeny. This number was calculated for each species and compared with the expectation under the null hypothesis of equal mutation rates across sites, i.e., no hot spot. The null distribution was obtained by simulation. For each species, a maximum-likelihood analysis was conducted assuming constant rates for sites, and 1000 data sets were generated by using the inferred tree, branch lengths, and rate matrix. We controlled that the proportions of polymorphic sites in simulated data sets were comparable to those of real data sets (not shown). Then homoplasy was calculated for each simulated data set, and the P-value was defined as the proportion of simulated data sets showing more homoplasy than the real data.
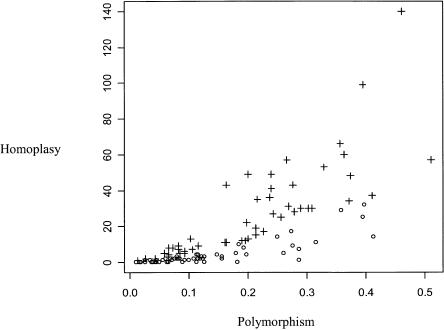
Results are given in Figure 1. Many but not all data sets showed significant homoplasy. The amount of homoplasy is correlated to the proportion of polymorphic sites, as expected—little variation implies little conflict. The human data set showed significant homoplasy (observed: eight, expected: 1.74, P < 10-3), consistent with previous studies (Eyre-Walker et al. 1999). Although not every species showed the same pattern, this analysis overall confirmed the strong level of within-species homoplasy in mammalian cytochrome b, asking for an explanation.
Figure 1.
Cytochrome b polymorphism and homoplasy in 113 mammalian species. Each symbol is for one species: x-axis, proportion of third-codon position polymorphic sites; y-axis, total within-species homoplasy at third-codon position; open circles, nonsignificant homoplasy; and crosses, significant homoplasy.
Co-occurrence analysis
Mutation hot spots, if any, should tend to generate polymorphisms in several species, resulting in the co-occurrence of polymorphic sites among closely related species. We first checked this prediction at the genus level—this concerns only polyspecific genera, i.e., genera represented by more than one species. For a given polyspecific genus, we call co-occurrence a site polymorphic in strictly more than half the number of species represented. This (arbitrary) threshold is two for genera represented by two or three species, three for genera represented by four or five species, etc. Note that such co-occurrences actually correspond to several mutations having appeared independently in distinct species. Shared alleles due to ancestral polymorphism or secondary introgression were removed before the analysis when making each species monophyletic (see Data sets section).
For every genus, the observed number of co-occurrences was compared to the expectation under the hypothesis of independence of mutation events between species. Sites in each species were randomly permuted 1000 times, and the amount of co-occurrence of polymorphisms was recomputed from shuffled data sets. Randomizing the location of polymorphic sites within species removes the effect of potential mutation hot spots on polymorphism co-occurrence. The P-value was defined as the proportion of randomized data sets for which co-occurrence was higher than in the real data. Purines and pyrimidines were randomized separately in this procedure; i.e., a purine site in the real data set was kept a purine in randomized data sets. We did that because the purine transition rate is generally higher than is the pyrimidine one (Tamura and Nei 1993; confirmed from our data), which slightly contributes to the probability of co-occurrence of polymorphisms between species.
An excess of co-occurrence of polymorphisms was detected in 22 polyspecific genera out of 27, and it was significant in 11 cases. These proportions reach 15 out of 18 and 10 out of 18, respectively, if one considers only genera in which at least one species shows significant homoplasy. This analysis supports the existence of mutation hot spots in mammalian cytochrome b third-codon positions. The effect is strong in genera Clethrionomys (Rodentia, Arvicolinae), Neotoma (Rodentia, Sigmodontinae), and Sorex (Insectivora, Soricidae), for instance (Table 1). In other groups, however, no significant co-occurrence was detected, although homoplasy is strong (e.g., Apodemus, Sigmodon). For these groups, and more generally, we asked whether the observed amounts of co-occurrence are compatible with a “pure” hot spots model.
Table 1.
Cytochrome b polymorphism co-occurrence analysis in 27 mammalian genera
| Order | Family | Genus | nb-sa | nb-hb | obs-coocc | exp-coocd | P-value |
|---|---|---|---|---|---|---|---|
| Carnivora | Felidae | Panthera | 2 | 1 | 2 | 0.236 | 0.022* |
| Carnivora | Mustelidae | Martes | 2 | 0 | 1 | 0.901 | 0.621 |
| Carnivora | Viverridae | Genetta | 2 | 2 | 11 | 6.483 | 0.029* |
| Cetartio. | Camelidae | Camelus | 2 | 0 | 1 | 0.648 | 0.499 |
| Chiroptera | Phyllostomidae | Carollia | 4 | 2 | 23 | 12.243 | 0** |
| Chiroptera | Phyllostomidae | Glossophaga | 2 | 1 | 9 | 6.705 | 0.126 |
| Chiroptera | Phyllostomidae | Artibeus | 2 | 0 | 13 | 10.891 | 0.261 |
| Chiroptera | Rinholophidae | Rhinolophus | 2 | 0 | 1 | 0.354 | 0.299 |
| Insectivora | Soricidae | Crocidura | 2 | 0 | 8 | 6.32 | 0.265 |
| Insectivora | Soricidae | Blarina | 3 | 1 | 27 | 20.951 | 0.061 |
| Insectivora | Soricidae | Sorex | 6 | 5 | 15 | 6.906 | 0.005** |
| Lagomorpha | Leporidae | Lepus | 4 | 1 | 0 | 0.358 | 1 |
| Primates | Cheirogaleidae | Microcebus | 2 | 0 | 8 | 8.175 | 0.619 |
| Rodentia | Echimyidae | Proechimys | 2 | 0 | 2 | 2.38 | 0.719 |
| Rodentia | Geomyidae | Cratogeomys | 2 | 0 | 28 | 21.971 | 0.048* |
| Rodentia | Geomyidae | Geomys | 2 | 1 | 40 | 30.494 | 0.011* |
| Rodentia | Muridae | Clethrionomys | 3 | 2 | 47 | 35.431 | 0.003** |
| Rodentia | Muridae | Eothenomys | 2 | 2 | 10 | 5.756 | 0.047* |
| Rodentia | Muridae | Microtus | 3 | 3 | 73 | 64.39 | 0.041* |
| Rodentia | Muridae | Apodemus | 2 | 1 | 5 | 5.901 | 0.748 |
| Rodentia | Muridae | Eliurus | 5 | 1 | 53 | 46.293 | 0.07 |
| Rodentia | Muridae | Baiomys | 2 | 1 | 11 | 10.212 | 0.437 |
| Rodentia | Muridae | Calomys | 3 | 2 | 14 | 8.651 | 0.026* |
| Rodentia | Muridae | Neotoma | 5 | 3 | 51 | 39.536 | 0.005** |
| Rodentia | Muridae | Peromyscus | 2 | 0 | 4 | 3.424 | 0.466 |
| Rodentia | Muridae | Sigmodon | 3 | 2 | 23 | 18.212 | 0.104 |
| Rodentia | Sciuridae | Spermophilus | 3 | 1 | 60 | 64.035 | 0.857 |
Number of represented species in the genus
Number of species showing significant homoplasy
Observed number of polymorphism co-occurrence
Expected number of polymorphism co-occurrence under the no hot spot hypothesis
significant at the 5% level
significant at the 1% level
Hot spots model
To achieve this aim, we simulated data sets at the genus level under the hot spots hypothesis. For each polyspecific genus, sequences from all species were gathered in a single file, and a maximum-likelihood phylogenetic analysis was performed. Then 100 data sets were simulated by using the inferred tree, branch lengths, and rate matrix, and a γ distribution of rates across sites, thus mimicking mutation hot spots and assuming shared hot spots between species. We call this model the constant hot spots model. The shape parameter of the assumed γ distribution was tuned so that simulated data sets resemble the observed one with respect to the number of polymorphic sites and amount of homoplasy within species. Co-occurrence of polymorphisms was then computed for each simulated data set and compared with the actual one. For 20 genera out of 27, the observed level of co-occurrence of polymorphisms between species was lower than expected under the constant hot spots hypothesis, and this trend was significant in eight genera. For these genera, the constant hot spots model cannot explain both the observed level of homoplasy within species and the observed level of polymorphism co-occurrence between species: When more hot spots were introduced in the simulations by increasing the variance of the assumed γ distribution in order to equate the expected and observed amounts of co-occurrence, the simulated data sets showed significantly more homoplasy within species than did the actual ones (data not shown).
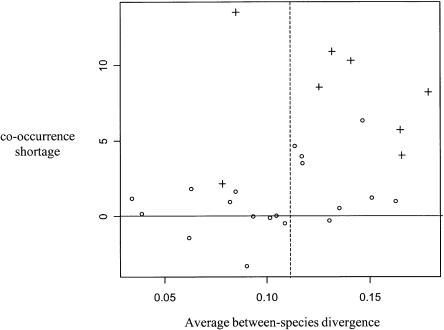
Something in the constant hot spots model must therefore be wrong, at least for eight genera. This model assumes a common genealogy for all the sites, i.e., no recombination. A departure from this assumption could of course explain the observed pattern—recombination generates some homoplasy but no co-occurrence (since it does not affect the location of polymorphic sites). Another assumption of the constant hot spots model, however, is constancy in time of the mutation rate of every site, which implies that hot spots are shared between species. A departure from this assumption, i.e., site-specific mutation rate variation, would also decrease the observed co-occurrence. One prediction of this hypothesis is that polymorphism co-occurrence should decrease as species diverge; closely related species should tend to share more hot spots than do distantly related species. We calculated for each polyspecific genus the difference between the expected amount of co-occurrence under the constant hot spots model (averaged over simulations) and the observed one. This co-occurrence shortage was plotted against the average nucleotide divergence between species. Figure 2 shows that the constant hot spots model correctly fits the data when species are closely related, whereas genera including distantly related species tend to show a lower level of co-occurrence than expected. This pattern is consistent with the hypothesis of site-specific mutation rate variation in time, and between species. Recombination applies independently of species divergence, so that recombination alone cannot generate such a relationship.
Figure 2.
Polymorphism co-occurrence vs. species divergence in 27 mammalian genera. Each symbol is for one polyspecific genus: x-axis, average cytochrome b sequence divergence between species (all three codon positions) in the genus; y-axis, difference between the observed number of synonymous polymorphism co-occurrences between species and the expected number under the constant mutation hot spots model; open circles, nonsignificant co-occurrence shortage; and crosses, significant co-occurrence shortage.
Higher taxonomic levels
Analyses performed at higher taxonomic levels essentially confirmed these results. We built subfamily and family data sets by gathering data from distinct genera. To ensure independence between the analyses at various levels, polyspecific genera were represented by a single species in subfamily data sets. Several combinations of species were tried. When the number of possible combinations did not exceed 10, they were all examined. Otherwise, 10 randomly chosen combinations were used. A similar strategy was used at the family level. Significant co-occurrence was detected in three subfamilies out of nine: Sigmodontinae (eight significant combinations of species out of 10), Arvicolinae (five out of 10), and Soricinae (five out of 10). At the family level, just two combinations of species in Muridae (out of 10) and one in Geomyidae (out of four) showed significant co-occurrence of polymorphisms, and no signal of co-occurrence was detected at all in the other four families. Again, some co-occurrence of polymorphism was found, indicating the existence of mutation hot spots, but the strength of the co-occurrence signal appears to decrease at higher taxonomic levels.
Primate full genome
Human is by far the most thoroughly studied species as far as mitochondrial diversity is concerned. H. sapiens, however, had to be excluded from the co-occurrence analysis because no complete cytochrome b polymorphism data set is available from its close relatives. What we have, however, are hundreds of complete human mitochondrial genomes, plus eight complete sequences from other Hominoidea species. Given the extensive sampling available, we tried to detect potential mutational hot spots through a phylogenetic analysis, first within H. sapiens and then between species. Under the constant hot spots model, sites showing a strong level of variability within species should also be variable between species—note that this rationale requires the additional assumption of selective neutrality of mutations, as we discuss below.
Human hypervariable sites
We analyzed the complete coding region of 560 human mitochondrial genomes, including individuals from Africa, Europe, and Asia (Herrnstadt et al. 2002). Seven polymorphic sites documented as sequencing errors in Herrnstadt et al. (2003) were removed. A maximum-likelihood tree was reconstructed by using the TN93 + γ model. Then the site-specific mutation rate was measured by using the maximum-parsimony method: For each site, the minimum number of changes required given the tree topology was recorded. A maximum-likelihood estimation of site-specific rates was also conducted by using the BASEML program (empirical Bayesian, posterior rate) (Yang 1997) and gave similar results. The parsimony score varied from zero (91.2% of the 15,425 sites) to 11. Twenty-six sites reached a parsimony score of five or more (Table 2), and three sites reached 11, consistent with the many instances of multiple mutations reported in the detailed analysis of Herrnstadt et al. (2002).
Table 2.
Hypervariable sites in human mtDNA
| rCRSa | MP bb | MP wc | MP w2d | Ae | Te | Ge | Ce | Gene | Statusf |
|---|---|---|---|---|---|---|---|---|---|
| 709 | 0 | 11 | 8 | 75 | 485 | 12S rRNA | nc | ||
| 1719 | 0 | 5 | 4 | 29 | 531 | 16S rRNA | nc | ||
| 1888 | 2 | 5 | 1 | 54 | 506 | 16S rRNA | nc | ||
| 3010 | 1 | 7 | 6 | 113 | 447 | 16S rRNA | nc | ||
| 3316 | 1 | 5 | 2 | 6 | 554 | MT-ND1 | ns | ||
| 3705 | 2 | 5 | 1 | 5 | 555 | MT-ND1 | s | ||
| 5237 | 0 | 5 | 2 | 5 | 555 | MT-ND2 | s | ||
| 5460 | 0 | 9 | 6 | 18 | 542 | MT-ND2 | ns | ||
| 6221 | 0 | 5 | 3 | 536 | 24 | MT-CO1 | s | ||
| 6260 | 1 | 6 | 1 | 12 | 548 | MT-CO1 | s | ||
| 6261 | 0 | 5 | 0 | 5 | 555 | MT-CO1 | ns | ||
| 10398 | 3 | 7 | 5 | 401 | 1 | 158 | MT-ND3 | ns | |
| 11800 | 2 | 5 | 0 | 555 | 4 | 1 | MT-ND4 | s | |
| 11914 | 1 | 11 | 6 | 52 | 508 | MT-ND4 | s | ||
| 12007 | 0 | 5 | 4 | 27 | 533 | MT-ND4 | s | ||
| 13106 | 2 | 6 | 3 | 534 | 26 | MT-ND5 | ns | ||
| 13435 | 1 | 5 | 0 | 555 | 5 | MT-ND5 | s | ||
| 13709 | 0 | 11 | 5 | 50 | 510 | MT-ND5 | ns | ||
| 14471 | 0 | 6 | 1 | 4 | 541 | 15 | MT-ND6 | s | |
| 14570 | 0 | 5 | 1 | 7 | 553 | MT-ND6 | s | ||
| 14767 | 1 | 8 | 2 | 322 | 238 | MT-CYB | ns | ||
| 15302 | 0 | 6 | 3 | 73 | 487 | MT-CYB | s | ||
| 15327 | 1 | 6 | 0 | 11 | 549 | MT-CYB | ns | ||
| 15785 | 1 | 7 | 3 | 522 | 30 | MT-CYB | s | ||
| 15885 | 1 | 5 | 1 | 8 | 542 | 8 | MT-CYB | ns | |
| 15925 | 2 | 9 | 4 | 531 | 29 | MT-TT | nc |
Site number (revised Cambridge Reference Sequence, http://www.mitomap.org)
Parsimony score between primate species
Parsimony score within humans
Parsimony score within humans (Ingman control data set)
Absolute nucleotide frequencies (when higher than zero)
nc, non-protein-coding; s, synonymous; ns, nonsynonymous
These sites are obvious outliers of the site-specific rate distribution, i.e., hypervariable sites. The total tree length for the human data set is 0.13 substitution per site. This number becomes 0.53 per site if one conservatively assumes that observable mutations occur only at third-codon positions of protein coding genes, the other sites being invariant due to strong selection. Even with this unrealistic assumption, the probability that one site or more undergoes ≥10 changes is of the order of 10-6 under the hypothesis of evenly distributed mutations and no recombination, while three such sites are observed in the real data set. Similarly, the expected number of sites showing five mutations or more is 1.13 under the hypothesis of equal mutation rate for every site, while we observe 26 such sites. We checked that these results were robust to uncertainties in the phylogenetic reconstruction. We generated 100 alternative tree topologies by using the bootstrap procedure, and reperformed the analysis. The 26 putative hot spots listed in Table 2 essentially remained hot when the tree topology varied: The minimal parsimony score over the 26 × 100 trials was three, and the minimal average (over trees) parsimony score was 4.8.
The detected hypervariable sites are located in eight distinct protein-coding genes, the two ribosomal RNA genes, and one transfer RNA gene (Table 2). The density of hypervariable sites appears higher around the D-loop and lower opposite to the D-loop (no hot spots were detected between positions 6300 and 10300). Three of the detected sites showed three distinct nucleotide states. Most mutations in hypervariable sites are transitions (Table 2), which is typical of mammalian mitochondrial DNA evolution (Reyes et al. 1998; Raina et al. 2005), suggesting that the highly variable sites detected are not in general due to sequencing errors. The high level of quality control and corrections performed by the investigators is another argument with this respect (see Bandelt et al. 2002; Herrnstadt et al. 2002, 2003). We further checked the sequencing error issue by using an independent human mitochondrial data set (105 sequences) (Ingman et al. 2000; Ingman and Gyllensten 2003). Among the 26 candidate hot spots, 23 were variable in the Ingman data set and 13 had a parsimony score of three or higher, supporting that the hypervariable sites listed in Table 2 correspond to actual sequence variation in human populations.
Among the 26 hypervariable sites listed in Table 2, 22 involve an A↔G polymorphism. This is significantly higher (P < 10-4) than the proportion of A↔G versus C↔T polymorphisms in the whole data set (48.8%). This result is incompatible with a recombinational origin of highly homoplasic sites: Recombination should lead to an increase of the parsimony score of a random set of sites, irrespective of their state. A prevalence of A↔G hypervariable sites is consistent, however, with specificities of the mitochondrial mutation process, guanines from the heavy strand being highly mutable (Raina et al. 2005). That guanines are more likely than other bases to be a mutation hot spot appears a plausible hypothesis.
We asked whether apparent mutation hot spots in H. sapiens were also divergence hot spots between Hominoidea. We built a data set including six nonhuman Hominoidea species and reproduced the above described analysis by using the well-supported (H. lar, ((P. pygmaeus, P. abelii), (G. gorilla, (P. paniscus, P. troglodytes)))) model tree. The site-specific parsimony score varied from zero to four (total tree length: 0.723 substitution per site). Among the 26 sites showing five or more mutations in humans, 11 showed no substitution between nonhuman Hominoidea species. One site showed 11 mutations in humans but no change between species. These results appear in contradiction with the constant hot spots hypothesis. The discrepancy, however, could be caused by natural selection. Slightly deleterious mutations can segregate as polymorphic but have a low fixation probability, so that they rarely contribute to divergence between species. Sites detected as hot spots in humans but invariant between species might be so because they involve deleterious mutations. To approach mutational effects only, we focused on the third-codon positions of protein-coding sequences.
Third-codon positions
Three thousand seven hundred fifty-nine third-codon positions were extracted from the alignment, and the above analyses were reperformed. The maximal site-specific parsimony score was 11 in the human data set and four in the Hominoidea data set. The correlation between the within-human and between-species parsimony scores was very low but significant (r = 0.05, P < 10-3). The average within-human parsimony scores were 0.23, 0.27, 0.29, and 0.44, respectively, for sites showing zero, one, two, and three minimal substitutions between species, suggesting the existence of mutation hot spots. The relatively large divergence between species probably reduces the power of this analysis by saturating the between-species parsimony score: Some sites in the data set have probably underwent dozens of substitutions since the divergence between Hylobatidae and Hominidae, but the parsimony score is bounded by the small number of species compared.
We performed simulations to further check the hot spots hypothesis. Data sets were simulated under the TN93 + γ model by using the maximum-likelihood estimates of tree topology, branch lengths, and rate matrix, as well as four candidate values for the shape of the γ distribution, with the aim of approaching what the data set should look like if sequences had evolved under the constant hot spots model. The parsimony analyses presented above were reperformed on simulated data sets and compared with real data (Table 3). A γ shape of 0.5 or one appeared to plausibly fit the real data: The within-humans and between-species diversity and homoplasy were correctly reproduced, as was the number of human hypervariable sites. The simulated data sets differed from the real one in showing a stronger correlation between the within-humans and between-species parsimony scores, a result illustrated by the excess in real data of sites detected as hot within humans, but invariant between species. This indicates that the constant hot spots model cannot account for every aspect of the Hominoidea data set, in agreement with the analysis of mammalian cytochrome b.
Table 3.
Real and simulated third-codon position variation within humans and between primates
| Dataa | Pwb | Hwc | Pbb | Hbc | σd | max_we | w > 5f | w > 10f | w > 5, b = 0g |
|---|---|---|---|---|---|---|---|---|---|
| Real | 0.190 | 270 | 0.624 | 583 | 0.058 | 11 | 9 | 1 | 5 |
| α = 0.1 | 0.069 | 187 | 0.257 | 243 | 0.404 | 9.8 | 14.2 | 1 | 0.6 |
| α = 0.5 | 0.161 | 254 | 0.543 | 487 | 0.247 | 9.1 | 10.7 | 0.5 | 1.4 |
| α = 1 | 0.180 | 230 | 0.611 | 529 | 0.148 | 7.7 | 8.5 | 0 | 1.9 |
| α = 10 | 0.218 | 170 | 0.651 | 555 | 0.055 | 5 | 1.4 | 0 | 0.8 |
Data set analyzed: real, or simulated by using four distinct γ shape parameters (10 simulations each, average results are given)
Proportion of variable sites within humans (Pw) or between primates (Pb)
Total homoplasy within humans (Hw) or between primates (Hb)
Correlation coefficient between the site specific parsimony scores within humans vs. between primates
Maximal site-specific parsimony score within humans
Number of sites showing a within-humans parsimony score higher than five, respectively 10
Number of sites showing a within-humans parsimony score higher than five and a between-primates parsimony score equal to zero
Discussion
We made use of two mammalian polymorphism DNA sequence data sets to try and detect the existence of mutation hot spots in the mitochondrial genome. The cytochrome b analysis revealed a significant amount of third-codon-position polymorphism co-occurrence among related species, rejecting the hypothesis of equal mutation rates across synonymous sites. Simulations showed that a pure hot spots model can account for the observed within-species homoplasy and between-species polymorphism co-occurrence in genera including little-divergent species. When species reach 10%-12% sequence divergence or more, the constant hot spots model predicts too much co-occurrence (Fig. 2), suggesting that site-specific mutation rates vary in time. The Hominoidea full genome analysis confirmed these findings and generalized it to noncytochrome b data: An A↔G-biased set of hypervariable sites was detected in humans, and the within-human and between-primates site-specific parsimony scores were weakly but significantly correlated, implying the existence of mutation hot spots. This correlation, however, was lower than expected under a constant hot spots model.
Eyre-Walker et al. (1999), applying a similar reasoning, did not detect any correlation between the within-human and between-primates site-specific variability and therefore rejected the mutation hot spots model. The differences between the two studies include the size of the data sets (560 human sequences in this study, 29 in the previous one) and the methods used: We took a phylogenetic approach to estimate the site-specific mutation rate, while Eyre-Walker et al. (1999) could only qualify a site as constant or variable (or at best homoplasic), further decreasing the power of the analysis.
One surprising finding of this study is the high apparent rate of evolution of site-specific mutation rates. As little as 10%-12% sequence divergence between congeneric species is enough to generate a detectable shortage of polymorphism co-occurrence. At the family level, virtually all the co-occurrence signal vanishes: Knowing that site i is polymorphic in species 1 does not increase the probability that it is found polymorphic in species 2.
The process of site-specific variation of evolutionary rate, known as covarion or heterotachy (Fitch 1971; Galtier 2001; Lopez et al. 2002), is usually considered as the consequence of changes in the selective constraints applying to specific sites (see Gu 1999; Pupko and Galtier 2002). Our results suggest that, at least for mammalian mitochondrial DNA, such patterns can also occur neutrally as the consequence of mutational effects. Heterotachy might partly explain the discrepancy between pedigree and evolutionary estimates of the mitochondrial mutation rate (Ho et al. 2005): A site can be very fast at the population/pedigree level but slower in the long run. From an empirical point of view, this “mutational covarion” means that one can hardly learn from one species which mitochondrial sites are going to be variable in another one. It would be worth knowing whether this statement also applies at the level of the gene or genome fragment. It is tempting, when starting a molecular biodiversity project in a new species, to target markers known to be polymorphic in related species. The current study suggests that this practice might be of little relevance in many cases. Crochet and Desmarais (2000) reached a similar conclusion when they compared the level of polymorphism in the control region versus cytochrome b in several gull species, and found that the two were essentially unrelated across species.
These results also ask the question of the causes of a variable in time site-specific mutation rate: What makes the mutation rate of a site increase or decrease? We examined the 11 human hypervariable sites in Table 2 showing a common state in all nonhuman primates. Nine of these sites are G↔A polymorphisms with a higher frequency of allele G. Among these, seven show state A in all six nonhuman Hominoidea species. Given the high prevalence of G→A mutations in mammalian mitochondria, this pattern strongly suggests that these sites correspond to unidirectional G→A hot spots. Such sites would most often be in state A and not hypermutable. They would occasionally substitute to G, as they did in the human lineage, and transiently become G→A hypermutable until state A fixes again. This simple process might partly explain the decrease of polymorphism co-occurrence with species divergence: The mutation rate of a site simply varies when it substitutes from a mutable nucleotide state to a little-mutable one, or reciprocally. The high mutability of guanines, however, cannot explain all the hypervariability observed in humans: Some of the detected hot spots involve numerous T↔C or A→G changes within H. sapiens and several changes between species. These sites might be unconditional, bidirectional hot spots.
Three of the 26 human hypervariable sites listed in Table 2 are located in the 16S ribosomal RNA, a gene whose within-species diversity has been investigated in five additional Hominoidea species, i.e., P. paniscus, P. troglodytes, G. gorilla, P. pygmaeus, and Hylobates syndactylus (between 10 and 35 individuals surveyed per species) (Noda et al. 2001). All three sites are G↔A polymorphisms in humans, with a high (>80%) frequency of allele G. Site 1888 is highly variable between species as well and is polymorphic in P. pygmaeus. It might be a constant hot spot, shared by all Hominoidea species. Site 1719 shows state A in all Hominoidea species excepting human and is monomorphic in the five species investigated by Noda et al. (2001). This site might correspond to a unidirectional G→A hot spot, hypermutable only when in state G. Site 3010, finally, shows state G in four of the five species surveyed by Noda et al. (2001) but appears monomorphic in these species. More data would be required to decide whether this site is actually a human-specific hot spot. The current paucity of mitochondrial polymorphism data sets in nonhuman Hominoidea species reduces the power of such analyses, which is regret-table.
None of the reported hot spots correspond to known disease-associated mutations, as we checked from the MITOMAP database (Brandon et al. 2005). Some of these hot spots might, however, have some functional relevance. Site 15925 is in close proximity to tRNA Thr anticodon (1592-15923) and to two known polymorphisms (15923 and 15924) that were associated with the lethal infantile mitochondrial myopathy (Yoon et al. 1991). Sites 709 (12S rRNA) and 6261 (cox1, Ala→Thr nonsynonymous change), finally, are G→A hypervariable in humans and invariably show state G in all six nonhuman Hominoidea species. We argue that these mutations are probably slightly deleterious, so that negative selection prevents their fixation in natural populations—note, however, that site 709 is found in state A in many nonprimate mammals. Alternatively, they might be very recent human-specific hot spots, but in this case the putative increase of mutation rate in humans would have occurred without a change in nucleotide state. Again, polymorphism data from nonhuman primates would enlighten this question. Further understanding of the specificities of the mutation process in mammalian and primate mitochondrial genomes is obviously of primary interest given the many diseases associated with mitochondrial mutations in humans.
Now back to the mutation hot spots versus recombination debate. This study aimed at testing whether mutation hot spots actually occurred in the coding region of the mitochondrial genome. The answer is obviously positive: Significant co-occurrence among species of synonymous cytochrome b polymorphism was detected in many mammalian genera, and a correlation between within-species and between-species synonymous site-specific variability, together with an A↔G-biased set of hypervariable sites, were found in Hominoidea. We also asked whether these hot spots could explain the relatively high level of mitochondrial homoplasy within species. A pure hot spots model appeared to fit many of the cytochrome b data sets. The model was rejected only for genera including relatively divergent species, which we interpret as evidence for site-specific mutation rate variation in time. A link between nucleotide state and mutation rate is proposed to explain, at least partly, this peculiarity. This study therefore provides a plausible mutational model to mitochondrial within-species homoplasy and shows that recombination alone cannot explain these patterns.
It should be noted that our results do not exclude the occurrence of recombination. Mutation hot spots might actually make the detection of recombination more difficult by generating patterns of linkage disequilibrium independent of physical distance, so that this work could paradoxically content supporters of the recombination hypothesis as well. What we have, however, is a mutation model potentially accounting for the distribution of mitochondrial DNA sequence variation within and between species. Whether recombination also plays a significant evolutionary role is still an open question, but we are now entitled to demand strong evidence to believe in it.
Methods
Cytochrome b alignments were performed by using MABIOS (Abdeddaim 1997), and full-genome sequences were aligned with MUSCLE (Edgar 2004). Maximum-likelihood phylogenetic analyses were conducted with program PHYML (Guindon and Gascuel 2003) using the TN93 model (Tamura and Nei 1993), and a constant or γ (five classes) distribution of rates across sites. The TN93 model accounts for unbalanced base composition and allows three distinct rates for transversions, A↔G transitions, and C↔T transitions. Constant rates across sites mean no hot spots, while a γ distribution is intended to reflect the existence of hypervariable sites. The other analyses are described in the Results section. They were achieved by using homemade C, PERL, and R programs.
Acknowledgments
This work was supported by French Ministère de la Recherche ACI IMPBio and CNRS-INRA Equipe Projet Multi Laboratoire “Méthodes informatiques pour la phylogénie moléculaire.”
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4305906.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Abdeddaim, S. 1997. Fast and sound two-step algorithms for multiple alignment of nucleic sequences. Int. J. Artif. Intell. Tools 6 179-192. [Google Scholar]
- Awadalla, P., Eyre-Walker, A., and Maynard-Smith, J. 1999. Linkage disequilibrium and recombination in hominid mitochondrial DNA. Science 286 2524-2525. [DOI] [PubMed] [Google Scholar]
- Bandelt, H.J., Quintana-Murci, L., Salas, A., and Macaulay, V. 2002. The fingerprint of phantom mutations in mitochondrial DNA data. Am. J. Hum. Genet. 71 1150-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin, E., Duret, L., Penel, S., and Galtier, N. 2005. Polymorphix, a polymorphism sequence database. Nucleic Acids Res. 33 481-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky, C.W. 1995. Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution. Proc. Natl. Acad. Sci. 92 11331-11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon, M.C., Lott, M.T., Nguyen, K.C., Spolim, S., Navathe, S.B., Baldi, P., and Wallace, D.C. 2005. MITOMAP, a human mitochondrial genome database: 2004 update. Nucleic Acids Res. 33 611-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crochet, P.A. and Desmarais, E. 2000. Slow rate of evolution in the mitochondrial control region of gulls (Aves: Laridae). Mol. Biol. Evol. 17 1797-1806. [DOI] [PubMed] [Google Scholar]
- Delsuc, F., Stanhope, M.J., and Douzery, E.J. 2003. Molecular systematics of armadillos (Xenarthra, Dasypodidae): Contribution of maximum likelihood and Bayesian analyses of mitochondrial and nuclear genes. Mol. Phyl. Evol. 28 261-275. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker, A., Smith, N.G.C., and Maynard-Smith, J. 1999. How clonal are human mitochondria? Proc. Biol. Sci. 266 477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch, W.M. 1971. Rate of change of concomitantly variable codons. J. Mol. Evol. 1 84-96. [DOI] [PubMed] [Google Scholar]
- Galtier, N. 2001. Maximum likelihood phylogenetic analysis under a covarion-like model. Mol. Biol. Evol. 18 866-873. [DOI] [PubMed] [Google Scholar]
- Gantenbein, B., Fet, V., Gantenbein-Ritter, I.A., and Balloux, F. 2005. Evidence for recombination in scorpion mitochondrial DNA (Scorpiones: Buthidae). Proc. Biol. Sci. 272 697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X. 1999. Statistical methods for testing functional divergence after gene duplication. Mol. Biol. Evol. 16 1664-1674. [DOI] [PubMed] [Google Scholar]
- Guindon, S. and Gascuel, O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52 696-704. [DOI] [PubMed] [Google Scholar]
- Hagelberg, E. 2003. Recombination or mutation rate heterogeneity? Implications for Mitochondrial Eve. Trends Genet. 19 84-90. [DOI] [PubMed] [Google Scholar]
- Hagelberg, E., Goldman, N., Lio, P., Whelan, S., Schiefenhovel, W., Clegg, J.B., and Bowden, D.K. 1999. Evidence for mitochondrial DNA recombination in a human population of island Melanesia. Proc. Biol. Sci. 266 485-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt, C., Elson, J.L., Fahy, E., Preston, G., Turnbull, D.M., Anderson, C., Ghosh, S.S., Olefsky, J.M., Beal, M.F., Davis, R.E. et al. 2002. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am. J. Hum. Genet. 70 1152-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt, C., Preston, G., and Howell, N. 2003. Errors, phantoms and otherwise, in human mtDNA sequences. Am. J. Hum. Genet. 72 1585-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey, J. 2000. Human mitochondrial DNA recombination: Can it be true? Trends Ecol. Evol. 15 181-182. [DOI] [PubMed] [Google Scholar]
- Ho, S.Y., Philips, M.J., Cooper, A., and Drummond, A.J. 2005. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 22 1561-1568. [DOI] [PubMed] [Google Scholar]
- Ingman, M. and Gyllensten, U. 2003. Mitochondrial genome variation and evolutionary history of Australian and New Guinean aborigines. Genome Res. 13 1600-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman, M., Kaessmann, H., Pääbo, S., and Gyllensten, U. 2000. Mitochondrial genome variation and the origin of modern humans. Nature 408 708-713. [DOI] [PubMed] [Google Scholar]
- Innan, H. and Nordborg, M. 2002. Recombination or mutational hot spots in human mtDNA? Mol. Biol. Evol. 19 1122-1127. [DOI] [PubMed] [Google Scholar]
- Kivisild, T. and Villems, R. 2000. Questioning evidence for recombination in human mitochondrial DNA. Science 288 1931. [PubMed] [Google Scholar]
- Kluge, A.G. and Farris, J.S. 1969. Quantitative phyletics and the evolution of anurans. Syst. Zool. 18 1-32. [Google Scholar]
- Kraytsberg, Y., Schwartz, M., Brown, T.A., Ebralidse, K., Kunz, W.S., Clayton, D.A., Vissing, J., and Khrapko, K. 2004. Recombination of human mitochondrial DNA. Science 304 981. [DOI] [PubMed] [Google Scholar]
- Lopez, P., Casane, D., and Philippe, H. 2002. Heterotachy, an important process of protein evolution. Mol. Biol. Evol. 19 1-7. [DOI] [PubMed] [Google Scholar]
- Noda, R., Kim, C.G., Takenaka, O., Ferrell, R.E., Tanoue, T., Hayasaka, I., Ueda, S., Ishida, T., and Saitou, N. 2001. Mitochondrial 16S rRNA sequence diversity of hominoids. J. Hered. 92 490-496. [DOI] [PubMed] [Google Scholar]
- Pesole, G. and Saccone, C. 2001. A novel method for estimating substitution rate variation among sites in a large data set of homologous sequences. Genetics 157 859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piganeau, G., Gardner, M., and Eyre-Walker, A. 2004. A broad survey of recombination in animal mitochondria. Mol. Biol. Evol. 21 2319-2325. [DOI] [PubMed] [Google Scholar]
- Pupko, T. and Galtier, N. 2002. A covarion-based method for detecting molecular adaptation: Application to the evolution of primate mitochondrial genomes. Proc. Biol. Sci. 269 1313-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina, S.Z., Faith, J.J., Disotell, T.R., Seligmann, H., Stewart, G.B., and Pollock, D.D. 2005. Evolution of base-substitution gradient in primate mitochondrial genomes. Genome Res. 15 665-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, A., Gissi, C., Pesole, G., and Saccone, C. 1998. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol. Biol. Evol. 15 957-966. [DOI] [PubMed] [Google Scholar]
- Schwartz, M. and Vissing, J. 2002. Paternal inheritance of mitochondrial DNA. N. Engl. J. Med. 347 576-580. [DOI] [PubMed] [Google Scholar]
- Springer, M.S., DeBry, R.W., Douady, C., Amrine, H.M., Madsen, O., de Jong, W.W., and Stanhope, M.J. 2001. Mitochondrial versus nuclear gene sequences in deep-level mammalian phylogeny reconstruction. Mol. Biol. Evol. 18 132-143. [DOI] [PubMed] [Google Scholar]
- Stoneking, M. 2000. Hypervariable sites in the mtDNA control region are mutational hotspots. Am. J. Hum. Genet. 67 1029-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. and Nei, M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10 512-526. [DOI] [PubMed] [Google Scholar]
- Tsaousis, A.D., Martin, D.P., Ladoukakis, E.D., Posada, D., and Zouros, E. 2005. Widespread recombination in published animal mtDNA sequences. Mol. Biol. Evol. 22 925-933. [DOI] [PubMed] [Google Scholar]
- Vandewoestijne, S., Baguette, M., Brakefield, P.M., and Saccheri, I.J. 2004. Phylogeography of Aglais urticae (Lepidoptera) based on DNA sequences of the mitochondrial COI gene and control region. Mol. Phyl. Evol. 31 630-646. [DOI] [PubMed] [Google Scholar]
- Vigilant, L., Stoneking, M., Harpending, H., Hawkes, K., and Wilson, A.C. 1991. African populations and the evolution of human mitochondrial DNA. Science 253 1503-1507. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13 555-556. [DOI] [PubMed] [Google Scholar]
- Yoon, K.L., Aprille, J.R., and Ernst, S.G. 1991. Mitochondrial tRNA Thr mutation in fatal infantile respiratory enzyme deficiency. Biochem. Biophys. Res. Commun. 176 1112-1115. [DOI] [PubMed] [Google Scholar]