Abstract
Latent infections by herpes simplex virus are characterized by repression of productive-cycle gene expression. Several hypotheses to explain this repression involve inhibition of expression of the immediate-early gene activator ICP0 during latency. To address these hypotheses, we developed quantitative reverse transcriptase-PCR assays that detected spliced and intron-containing ICP0 transcripts in mouse ganglia latently infected with wild-type virus. In these ganglia, the numbers of spliced ICP0 transcripts correlated better with the numbers of transcripts from the immediate-early gene encoding ICP4 than with those from the early gene encoding thymidine kinase. There were fewer spliced than intron-containing ICP0 transcripts on average, with considerable ganglion-to-ganglion variation. We then investigated whether ICP0 expression in latently infected ganglia is reduced by the latency-associated transcripts (LATs) and whether splicing of ICP0 transcripts is inhibited by the product of open reading frame (ORF) P. A LAT deletion mutation which essentially eliminates expression of the major LATs did not appreciably increase levels of ICP0 transcripts. LAT deletion mutants did, however, appear to express reduced levels of intron-containing ICP0 transcripts. ORF P mutations did not alter levels of ICP0 transcripts in a manner consistent with inhibition of ICP0 splicing by ORF P. Although these results argue against antisense inhibition of ICP0 expression by LATs or inhibition of ICP0 splicing by ORF P, they are consistent with the possibilities of a block between immediate-early and early gene expression and regulation of spliced versus intron-containing ICP0 transcripts in latently infected ganglia.
Herpes simplex virus (HSV) has two distinct lifestyles, productive infection and latent infection. During productive infection (reviewed in reference 37), three major kinetic classes of viral genes, immediate-early (IE) (also called α), early (E) (also called β) and late (L) (also called γ), are abundantly expressed in an ordered cascade. Four of the five IE genes (infected-cell proteins [ICP] 0, 4, 22, and 27) encode regulators of viral gene expression during productive infection. Of these IE proteins, ICP4 and ICP27 are essential for viral replication. Although ICP0 is not essential, it is critical for efficient viral replication and for the full expression of all three gene classes, especially at low multiplicities of infection (MOIs) (4, 5, 7, 13, 38, 42).
During latent infection of sensory ganglia of mammalian hosts (reviewed in references 34, 37, and 40), the only viral gene products that are abundantly expressed are the latency-associated transcripts (LATs) (39). These arise from the RL (b) repeat segments of the HSV genome (Fig. 1). The major LAT species are ≈1.4- to ≈2.2-kb nuclear RNAs (12, 15, 48, 50). These are thought to be stable introns that arise from less abundant ≈8.5-kb primary transcripts. There is evidence that these latter minor LAT species also exist in latently infected ganglia (32, 51). The high levels of expression of IE, E, and L genes observed during productive infection are dramatically repressed during latency. The mechanisms responsible for this repression, which is the hallmark of latent infection, are poorly understood.
FIG. 1.
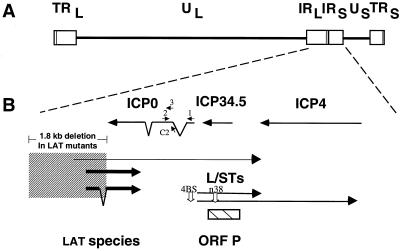
Locations of relevant HSV transcription units. (A) Diagram of the prototype arrangement of the HSV genome, with the unique long (UL) and unique short (US) sequences (lines) bracketed by the terminal repeat (TR) and internal repeat (IR) regions (boxes). (B) Expanded diagram of the internal repeat region showing relative sizes, locations, and orientations of transcripts encoded in this region, with bent lines indicating sequences removed by splicing and arrowheads indicating the 3′ ends of the transcripts. The locations and orientations of the 0-1, 0-2, and 0-C2 oligonucleotides used as primers (indicated as 1, 2, and C2, respectively) and the 0-3 oligonucleotide used as a probe (indicated as 3) are shown. Major and a presumed minor LAT species are indicated with thick and thin lines, respectively, and the locations of the two most abundant L/ST species are shown (49). The stippled box encompassing the 5′ ends of the LATs and extending upstream represents the location of the ≈1.8-kb deletion in LAT mutants dlLAT1.8 and KdlLAT1.8, which is restored in KFSLAT+. This deletion removes the promoter, transcriptional start site, and 1,015 bp of transcribed sequences of LATs. The location of ORF P is shown as a hatched box. The positions of the 4BS mutation upstream of the L/STs and the n38 mutation in ORF P are shown by open arrows.
To investigate mechanisms regulating HSV gene expression during latency, we have developed sensitive, quantitative reverse transcriptase-PCR (RT-PCR) assays. Using these assays, we have detected and quantified levels of transcripts derived from certain productive-cycle genes, notably those encoding ICP4 and the E protein thymidine kinase (TK) in ganglia latently infected with wild-type strain KOS and certain KOS-derived mutants (9, 19, 20). Despite the presence of these productive-cycle viral transcripts, no infectious virus has ever been detected in scores of such ganglia (26; unpublished results; D. Leib, personal communication). Moreover, there is no apparent correlation between amounts of ICP4 and tk transcripts in ganglia latently infected with wild-type virus (9). One possible explanation for the lack of infectious virus and the lack of correlation between ICP4 and tk transcripts is that certain virally encoded factors are not expressed. Specifically, because of the importance of ICP0 for productive gene expression and replication at low MOI (4, 5, 7, 13, 38, 42), for escape from quiescent infection in cell culture (reviewed in reference 32), and for reactivation of virus from latency (3, 18), much interest has focused on the hypothesis that the absence of ICP0 could be crucial for the establishment and maintenance of latency (reviewed in reference 34).
There is also evidence for the existence of viral factors that contribute to repression of productive-cycle gene expression during latency. In particular, we have shown that a LAT deletion mutation results in increased and better-correlated expression of transcripts from the ICP4 and tk genes in latently infected mouse ganglia (9). The same LAT mutation also results in increased expression of productive-cycle transcripts in acutely infected ganglia, as detected by in situ hybridization (17). Thus, a viral function associated with the LAT locus represses productive-cycle gene expression in ganglia.
There are several potential mechanisms for LAT-mediated repression of viral gene expression in latently infected ganglia. Several investigators have hypothesized that LAT-mediated repression entails inhibition of ICP0 expression. One long-standing and attractive hypothesis is that the LATs, the most abundant of which are partially complementary to ICP0 mRNA (Fig. 1), repress ICP0 expression via an antisense mechanism (15, 29, 41). In cultured cells, repression appears to occur by reduction of ICP0 mRNA levels (29), as expected for nuclear antisense repression (22) and as observed with other HSV antisense arrangements (11, 25, 36). Thus, if the LATs repress ICP0 expression during latency by an antisense mechanism, one would predict that drastic reductions in LAT expression should lead to marked increases in ICP0 transcripts in latently infected ganglia.
An alternative hypothesis for inhibition of ICP0 expression during latency involves a protein encoded by an open reading frame (ORF) called ORF P. The ORF P protein is encoded either by transcripts called L/STs that span the L/S junction of HSV DNA (49) or, conceivably, by certain minor LATs (23-25). Based on several lines of evidence, it was hypothesized that the ORF P protein might repress ICP0 expression during latency by inhibiting splicing of ICP0 transcripts (2). If ORF P is encoded by LATs and if repression by LATs during latency operates via ORF P, then one would predict that drastic reductions in LAT expression would lead to increased levels of spliced ICP0 transcripts and decreased levels of unspliced ICP0 transcripts. Regardless of the involvement of LATs, this hypothesis predicts that inactivation of ORF P should lead to increased levels of spliced ICP0 transcripts and decreased levels of unspliced ICP0 transcripts, while increasing ORF P expression should result in decreased levels of spliced and increased levels of unspliced ICP0 transcripts.
To address these multiple hypotheses, we developed RT-PCR assays to quantify (i) ICP0 transcripts in which the first intron is spliced out and (ii) ICP0 transcripts that retain at least a portion of these intron sequences. We then measured the levels of these ICP0 transcripts in ganglia latently infected with wild-type virus or with viral mutants with lesions that either essentially eliminate LAT expression, inactivate ORF P protein, or lead to overexpression of intact ORF P protein. Although our studies provide little support for the various hypotheses tested, they suggest the existence of interesting control points for gene expression in latent infections.
MATERIALS AND METHODS
Viruses and cells.
Wild-type HSV-1 strain KOS; the KOS-derived LAT deletion mutants dlLAT1.8 and K dlLAT1.8, which were independently engineered to contain a 1.8-kbp deletion in the LAT transcription unit; the marker-rescued derivative of KdlLAT1.8, KFSLAT+ (17, 26); the KOS-derived ORF P nonsense mutant L/ST-n38 and its marker-rescued derivative L/ST-n38R; and the KOS-derived L/ST promoter mutant L/ST-4BS, which contains mutations in the ICP4 binding site, and its marker-rescued derivative L/ST-4BSR (25) were propagated and assayed on Vero cell monolayers as described previously (10). The locations of the mutations are shown in Fig. 1.
RNA standards.
Plasmids used for in vitro transcription to generate RNA quantification standards from mouse β-actin cDNA (pSPMβA) and from the LAT (pKS+5′LAT) and ICP4 (pKS+5′ICP4) and tk (pSVtk1) genes were described previously (21). To generate synthetic, spliced ICP0 transcripts, pKS+ICP0-cDNA was constructed by cloning a 2.7-kb BglII fragment of wild-type HSV-1 strain 17 syn+ ICP0 cDNA from pDS18 (generously provided by S. Silverstein) into the BamHI site of BlueScriptII KS+ (Stratagene), placing it under the control of the bacteriophage T7 promoter. To generate synthetic, unspliced ICP0 transcripts, pBKS+ICP0-DNA was constructed by cloning a 4.6-kb SacI-HindIII fragment of wild-type strain KOS ICP0 DNA from pSH (4) into the SacI and HindIII sites of BlueScriptII KS+, placing genomic (intron-containing) ICP0 sequences under the control of the bacteriophage T7 promoter. RNA was transcribed in the presence of labeled GTP and quantified as previously described (21).
Infection of mice and tissue collection.
Eight-week-old male CD-1 (Charles River Laboratories) or CD-1-derived Hsd:ICR (Harlan Sprague Dawley) mice (similar results were obtained with mice from both suppliers) were anesthetized and inoculated with 2 × 106 PFU of each virus per eye via corneal scarification as previously described (27). At 30 days postinoculation, mice were sacrificed, and individual trigeminal ganglia were removed, rapidly frozen in liquid nitrogen as described previously (21), and stored at −80°C.
Quantitative PCR and RT-PCR analysis.
Frozen ganglia were homogenized in guanidine thiocyanate, and 1/10 of each ganglion homogenate was assayed by quantitative PCR for viral DNA and cellular DNA as previously described (21). Total RNA purified from the remainder of the homogenate as well as serial dilutions of synthetic transcript standards reconstituted with mouse brain RNA were treated with DNase to eliminate cell and viral DNA and reverse transcribed in the presence of mouse β-actin-specific (Act-2), ICP0-specific (0-2; CAGGTCTCGGTCGCAGGGAAAC), and other virus-specific (LAT-2, 4-1 or 4-2 and tk-2) primers (9, 21). The cDNAs were then amplified by PCR. For the spliced ICP0 transcripts, the 0-1 (AGCGAGTACCCGCCGGCCTG) and 0-2 primer pair was used. For the intron-containing ICP0 transcripts, the 0-C2 (CTTTGGTTGCAGACCCCTTTCTC) and 0-2 primer pair was used (Fig. 1). The various ICP0 primers used are complementary to both strain 17 and strain KOS sequences (30; S.-H. Chen and D. M. Coen, unpublished results) (GenBank accession no. AF431736).
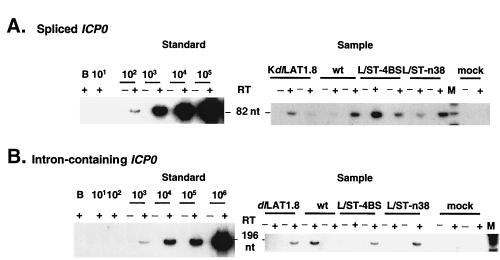
PCR was performed as previously described (9, 21), with the following modifications. The spliced and intron-containing ICP0 reaction mixture contained 1.5 mM Mg2+ and the annealing temperatures were 60 and 55°C. The cDNAs of spliced ICP0 transcripts were amplified for 30 cycles, and those of intron-containing ICP0 transcripts were amplified for 32 cycles. The RT-PCR product is 82 nucleotides for spliced and 196 nucleotides for intron-containing ICP0 transcripts (Fig. 2). Product specificities of ICP0 sequences amplified from tissue were verified by predicted sizes, hybridization with the internal oligonucleotide probe 0-3 (Fig. 2), and restriction endonuclease analysis (not shown). PCR components for the other gene-specific assays have been described previously (9, 21). Positive and negative controls were included in each amplification.
FIG. 2.
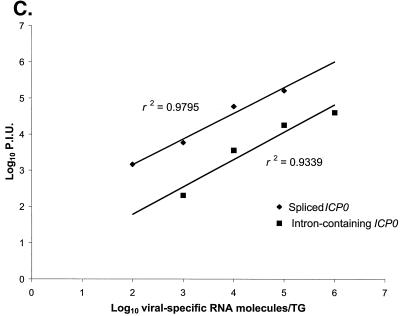
Quantitative RT-PCR assays of ICP0 transcripts. Phosphorimages of assays of spliced ICP0 transcripts (A) and intron-containing ICP0 transcripts (B). RT-PCR products generated using the 0-2 and 0-1 primer pair (A) or 0-2 and C2 primer pair (B) were separated on polyacrylamide gels, blotted, and probed with radiolabeled 0-3 oligonucleotide. Equivalent aliquots of either synthetic transcript mixes derived by in vitro transcription of pKS+ICP0-cDNA (A) or pBKS+ICP0-DNA (B), prepared with 5 μg of mouse brain RNA (A and B, left, under Standard) or individual ganglion RNAs (A and B, right, under Sample) were reacted with (+) or without (−) RT, and the products are displayed in adjacent lanes. In each of the left panels, the number of synthetic transcript molecules added to mouse brain RNA is indicated above the lanes. The B lane indicates where water was added instead of transcripts. The sizes of the RT-PCR products are indicated between the left and right panels. In each of the right panels, samples were derived from individual ganglia harvested at 30 days postinoculation from mock-infected mice (mock) or from mice infected with each of the indicated viruses. Lane M, molecular size markers (φX174 DNA digested with HinfI and end-labeled with 32P). (C) Linearity of RT-PCR assays. The phosphorimages shown on the left of panels A and B were quantified, and the phosphorimager units (P.I.U.) were plotted versus the synthetic RNA molecules added to mouse brain RNA. The best-fit lines were generated by linear regression analysis, and the correlation coefficients (r2) are displayed adjacent to the lines. Solid diamonds, spliced ICP0 transcripts; solid squares, intron-containing ICP0 transcripts.
Products of PCR were separated by electrophoresis on nondenaturing polyacrylamide gels (21). Amounts of adipsin PCR products and β-actin RT-PCR products were quantified by analysis of gels stained with SYBR Green I (Molecular Probes, Eugene, Oreg.) using a FluorImager (Molecular Dynamics, Sunnyvale, Calif.). Other products were transferred to nylon membranes and probed with labeled oligonucleotides as described previously (21), except that oligonucleotide 0-3 (AGCCCGCCCCGGATGTCTGGG) was used as a probe for both spliced and intron-containing ICP0 PCR products. Blots were exposed and quantified using a PhosphorImager. For each sample, the amount of viral DNA was normalized to cellular DNA encoding adipsin, and the amounts of viral mRNAs were normalized to cellular mRNA encoding β-actin and calculated on a per ganglion basis, as described previously (9, 21).
RESULTS
Spliced ICP0 transcripts can be detected in ganglia latently infected with wild-type HSV.
The expression of ICP0 has been hypothesized to be an important control point for latency (34). To investigate whether latency is due to an absence of ICP0 expression, we developed sensitive, quantitative RT-PCR assays for transcripts derived from the ICP0 gene. Unlike most HSV-encoded proteins, ICP0 is translated from spliced mRNA, which arises from a gene that contains two introns. We therefore developed an assay to detect ICP0 transcripts from which the 5′ intron (intron 1) has been spliced. As a source of synthetic spliced ICP0 transcripts with which to standardize the assay, we cloned a 2.7-kb fragment of ICP0 cDNA into a transcription vector, which permitted generation of known amounts of intronless ICP0 transcripts. These synthetic transcripts were reverse transcribed with primer 0-2, which anneals just downstream of the splice junction. The resulting cDNA was then subjected to amplification by PCR using 0-2 and primer 0-1, which anneals upstream of the splice junction (Fig. 1; primers 0-2 and 0-1 are labeled 2 and 1, respectively). This produced an 82-bp PCR product (Fig. 2A). The assay was highly sensitive, able to detect fewer than 10 transcripts. Consequently, we could detect ≈100 transcripts per ganglion in our standard experimental protocol, which assays ≈1/10 of the ganglionic RNA for each transcript (Fig. 2A). The assay was also linear through several orders of magnitude (Fig. 2C).
We then applied this assay to 28 ganglia that were latently infected with wild-type strain KOS. For normalization, we assayed amounts of β-actin transcripts and viral and cellular DNAs. On average, there were 104 viral DNA molecules/ganglion (Table 1), which is very consistent with what we have observed previously (8, 9, 20, 21). It has been estimated that there are ≈2 × 104 neurons per ganglion, of which only a minority (1 to 30%, i.e., 200 to 6,000 neurons, depending on virus, inoculating dose, and assay method) are latently infected (reference 39 and references therein). Thus, on average, we expect 2 to 50 viral genomes per latently infected neuron, but values for individual neurons can be expected to vary widely (37). Similarly, we have previously detected a mean of ≈1 ICP4 transcript per viral genome in latently infected ganglia (9), which results in an expected value of 2 to 50 transcripts per latently infected neuron. This range of values is lower than the 600 ICP4 transcripts/cell we detected at 2 h postinoculation in Vero cells infected at an MOI of 20 (M. F. Kramer, S.-H. Chen, and D. M. Coen, unpublished results), but again, values for individual neurons can be expected to vary widely.
TABLE 1.
Viral genomes and transcripts per viral genome in latently infected ganglia
| Virus (no. of ganglia assayed) | No. of viral genomes/gangliona | No. of LATs/ viral genomeb |
ICPO transcripts/viral DNAb
|
|
|---|---|---|---|---|
| Spliced (no. of positive ganglia/total) | Intron-containingc (no. of positive ganglia/total) | |||
| KOS (28) | 1 × 104 ± 0.3 × 104 | 1 × 105 ± 0.3 × 105 | 0.3 ± 0.2 (27/28) | 2 ± 1 (13/28) |
| dlLAT1.8 (10) | 2 × 104 ± 0.4 × 104 | 0.04 ± 0.03 | 0.5 ± 0.4 (10/10) | 0.3 ± 0.2 (1/10) |
| K dlLAT1.8 (10) | 6 × 103 ± 3 × 103 | 0.08 ± 0.08 | 0.08 ± 0.04 (6/10) | <0.7 (0/10) |
| KFSLAT+ (10) | 6 × 103 ± 2 × 103 | 4 × 105 ± 2 × 105 | 0.2 ± 0.1 (9/10) | 2 ± 0.8 (2/10) |
| L/ST-n38 (14) | 5 × 103 ± 1 × 103 | 2 × 105 ± 0.4 × 105 | 0.6 ± 0.2 (14/14) | 6 ± 4 (4/14) |
| L/ST-n38R (6) | 3 × 103 ± 0.6 × 103 | 2 × 105 ± 0.6 × 105 | 0.4 ± 0.2 (6/6) | 10 ± 10 (3/6) |
| L/ST-4BS (13) | 4 × 103 ± 2 × 103 | 1 × 105 ± 0.2 × 105 | 0.2 ± 0.1 (13/13) | 4 ± 2 (4/13) |
| L/ST-4BSR (8) | 5 × 103 ± 1 × 103 | 1 × 105 ± 0.2 × 105 | 0.2 ± 0.1 (8/8) | 0.7 ± 0.3 (2/8) |
Amounts of viral genomes per ganglion are expressed as the mean number of DNA molecules per ganglion ± standard error of the mean for each group.
Values are expressed as number of RNA molecules normalized to cellular mRNA encoding β-actin per number of viral genomes normalized to cellular (adipsin) DNA, expressed as the mean ± standard error of the mean for each group.
For any ganglia where intron-containing transcripts were not detected, values equivalent to the limit of detection for that transcript were assigned and divided by the number of viral DNA molecules in that ganglion.
Thus, in ganglia expressing average or greater numbers of ICP4 transcripts, if most of those transcripts were in a few neurons, then the numbers of transcripts in those neurons would be similar to or greater than that seen in productively infected Vero cells. Indeed, recent in situ hybridization analyses have detected single neurons in ≈10% of latently infected ganglia that contain relatively high levels of certain productive-cycle transcripts (16). These rare cells, however, are likely to account for only a small portion of the expression of productive-cycle transcripts that we observed (see Discussion).
We detected spliced ICP0 transcripts in almost all (27 of 28) of the KOS-infected ganglia (two examples in Fig. 2A, lane wt). When normalized, the amounts of these transcripts were highly variable, ranging from 0.001 to 5 spliced ICP0 transcripts per viral genome. (Such variability has previously been observed with transcripts from another IE gene, ICP4 [21]. Despite this variability, it is possible to detect differences in expression of this transcript in ganglia infected with LAT− versus LAT+ viruses [9].) The average number of spliced ICP0 transcripts per viral genome (i.e., the arithmetic mean, which is akin to the value that would be obtained if the ganglia were pooled) in these wild-type-infected ganglia was 0.3 molecules per genome (Table 1). This was only slightly lower than the average number of ICP4 transcripts per viral genome detected in these ganglia (0.6), which is similar to the value (≈1) observed previously (9). Thus, transcripts from the ICP0 gene were not absent in latently infected ganglia.
Correlations with ICP4 and tk transcripts.
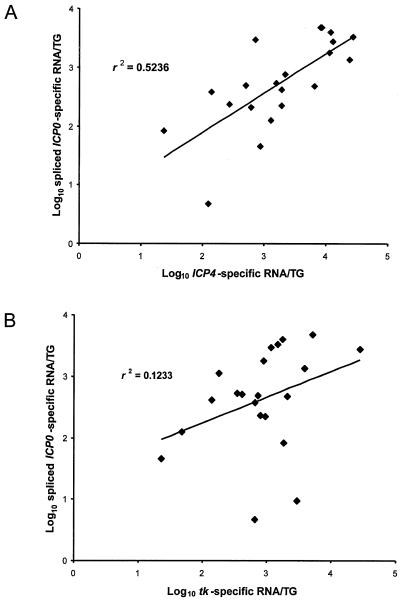
Previously, we reported no apparent correlation between amounts of ICP4 and tk transcripts in a large series of KOS-infected ganglia in which both transcripts could be detected in the same ganglion (9). A similar analysis of KOS-infected ganglia in the present study revealed a modest correlation (r2 = 0.524) between the amounts of ICP0 and ICP4 transcripts (Fig. 3A). This correlation was statistically significant (P < 0.01, using the t test of a correlation coefficient). However, no significant correlation was detected between ICP0 and tk transcripts (Fig. 3B; r2 = 0.123; P > 0.1), nor, as previously reported for a different set of KOS-infected ganglia (9), between ICP4 and tk transcripts (r2 = 0.104; P > 0.1). Thus, the numbers of the two IE transcripts in ganglia correlated better with each other than did the numbers of either IE transcript with the E transcript tk.
FIG. 3.
Correlation of expression of spliced ICP0 transcripts and ICP4 transcripts (A) and spliced ICP0 and tk transcripts (B) in ganglia latently infected with KOS. The numbers of each transcript in each individual ganglion (TG) were plotted against the amount of the other transcript in the same ganglion. The best-fit lines were generated by linear regression, and the correlation coefficients (r2) are displayed in each panel.
Detection of intron-containing ICP0 transcripts in latently infected ganglia.
Because certain hypotheses regarding regulation of gene expression during latency invoke regulation of splicing of ICP0 transcripts (2, 47) we also developed an assay for ICP0 transcripts that fail to accurately splice out sequences from intron 1. We cloned a fragment of genomic KOS DNA into a transcription vector and generated synthetic unspliced transcripts. The RT-PCR primers that we used to assay spliced transcripts failed to detect such synthetic unspliced transcripts, perhaps due to inefficient reverse transcription of the relatively long GC-rich intron 1 sequence. We therefore developed an alternative assay in which we used the 0-2 primer for reverse transcription and as the downstream primer for PCR and primer 0-C2, which hybridizes to intron 1 sequences in cDNA, as the upstream PCR primer (Fig. 1; 0-C2 is labeled C2). These primers yielded a 196-bp RT-PCR product from unspliced synthetic transcripts (Fig. 2B). The assay was linear through several orders of magnitude and was sensitive, but less so than the assay for spliced ICP0 transcripts (Fig. 2C). The limit of detection of this assay was ≈100 transcripts, which translates to a limit of detection of ≈103 transcripts per ganglion in our protocol.
When this assay was applied to the 28 ganglia latently infected with strain KOS, we detected intron-containing ICP0 transcripts in 13 of the ganglia (examples in Fig. 2B). (As it is possible that these transcripts are spliced elsewhere, for example, removing intron 2, we refer to them as intron-containing rather than as unspliced.) The inability to detect these transcripts in as many ganglia as we detected spliced transcripts is most likely a consequence of the different sensitivities of the assays. Levels of intron-containing ICP0 transcripts per viral genome were variable, ranging from 0.1 to 26 transcripts per genome. The average number of these transcripts per viral genome (2 transcripts/viral genome) was higher than that of the spliced transcripts (Table 1). However, on a ganglion-by-ganglion basis, this was not always the case. The two ganglia with the highest levels of spliced ICP0 transcripts contained very low or undetectable levels of intron-containing transcripts and vice versa. Regardless, intron-containing ICP0 transcripts could be detected, sometimes at levels higher than those of spliced transcripts, in ganglia latently infected with wild-type virus.
A LAT deletion mutation does not result in increased levels of ICP0 transcripts in latently infected ganglia.
It has long been hypothesized that LATs repress ICP0 expression by an antisense mechanism (15, 29, 41). If this were so, as noted above, one would expect that drastic reductions in LAT expression should lead to an increase in ICP0 transcripts in latently infected ganglia. To test this prediction, we compared the levels of ICP0 transcripts in ganglia latently infected with (i) wild-type virus (KOS), (ii) a LAT deletion mutant, dlLAT1.8 (26), in which the LAT promoter and 5′ transcribed sequences are removed (Fig. 1), (iii) a second, independent LAT deletion mutant, KdlLAT1.8, which contains the same deletion as dlLAT1.8 (17), and (iv) a rescued virus, KFSLAT+, in which the sequences deleted in KdlLAT1.8 are restored (17). The two independent LAT deletion mutants were used to ensure that any phenotypes observed were not due to adventitious mutations. As published previously (8, 9), average numbers of viral DNA molecules in ganglia infected with the two LAT mutants were within ≈2-fold of those in ganglia infected with the wild type or the rescued virus (Table 1). Quantification of major LATs revealed ≈105 molecules per viral genome in KOS- or KFSLAT+-infected ganglia (Table 1), consistent with our previous results (9, 21). As expected and as reported previously (8, 9), the LAT deletion in mutants dlLAT1.8 and KdlLAT1.8 essentially eliminated expression of the major LATs (more than 106-fold reduction; Table 1).
Neither of the two independent LAT deletion mutants exhibited major differences in expression of spliced ICP0 transcripts relative to wild-type or the rescued virus. In dlLAT1.8-infected ganglia, the mean number of spliced ICP0 transcripts per genome was <2-fold higher than that in KOS-infected ganglia (Table 1). In KdlLAT1.8-infected ganglia, the mean number of spliced ICP0 transcripts per genome was just 2- to 4-fold lower than that in ganglia infected with KOS or the rescued virus KFSLAT+.
Intron-containing ICP0 transcripts were detected in only 1 of 10 dlLAT1.8-infected ganglia and in none of 10 KdlLAT1.8-infected ganglia, which was considerably less frequent than our detection of these transcripts in KOS-infected ganglia (13 of 28) (Table 1). These differences in detection between mutant and wild-type virus were statistically significant (using Fisher's exact test, P = 0.02 for KOS versus dlLAT1.8; P = 0.01 for KOS versus KdlLAT1.8). Intron-containing transcripts were detected in 2 of 10 ganglia infected with the rescued virus KFSLAT+. This was more frequent than seen with the corresponding mutant, KdlLAT1.8 (0 of 10), but this difference did not attain statistical significance. It is possible that analyses of more KFSLAT+-infected ganglia would result in a statistically significant difference. The intron-containing ICP0 transcripts were generally detected in 25 to 50% of ganglia infected with other LAT+ viruses (Table 1; see below). The mean numbers of intron-containing ICP0 transcripts per genome detected in the LAT− mutant-infected ganglia were ≥3- to 8-fold lower than those in ganglia infected with the wild-type or the rescued virus (Table 1). (Although intron-containing transcripts were detected in a minority of ganglia, differences in mean numbers per genome were calculated using the conservative assumption that the numbers of intron-containing transcripts in ganglia where none were detected correspond to the limit of detection. Thus, the actual differences may be even greater.) In summary, there was no evident increase in the accumulation of ICP0 transcripts due to the LAT deletion, and the LAT− mutants expressed reduced amounts of intron-containing ICP0 transcripts relative to LAT+ viruses.
Mutations affecting ORF P expression do not significantly alter levels of ICP0 transcripts.
The lower levels of intron-containing ICP0 transcripts in ganglia infected with LAT− mutants could be interpreted as favoring the hypothesis that splicing of ICPO transcripts is inhibited in latently infected ganglia by a LAT or a LAT-encoded product. ORF P, which conceivably could be encoded by a minor LAT species as well as by L/STs, has been hypothesized to repress ICP0 expression during latency by inhibition of ICP0 splicing (2). To address this hypothesis, we inoculated mice with (i) wild-type virus, (ii) an ORF P nonsense mutant, L/ST-n38 (the mutation does not alter the amino acid sequence of the overlapping gene encoding ICP34.5), (iii) a marker-rescued derivative of L/ST-n38, L/ST-n38R, (iv) a mutant, L/ST-4BS, in which the ICP4 binding site in the L/ST promoter was mutated to relieve ICP4 repression of ORF P expression driven by this promoter, and (v) a marker-rescued derivative of L/ST-4BS, L/ST-4BSR (25). Latently infected ganglia were assayed for levels of viral DNA, the major LATs, and ICP0 transcripts. Average amounts of viral DNA in ganglia latently infected with the various viruses are tabulated in Table 1. The numbers of viral DNA molecules in ganglia infected with L/ST-n38 and L/ST-4BS were within ≈2-fold of those in ganglia infected with KOS.
The levels of the major LATs per viral genome in ganglia infected with the various L/ST viruses were similar to those in wild-type-infected ganglia (Table 1). The average numbers of spliced ICP0 transcripts per genome were ≤2-fold higher in ganglia latently infected with the ORF P nonsense mutant (L/ST-n38) than in those infected with KOS or the rescued virus (L/ST-n38R), and the ranges of the standard errors of the means of these values overlapped (Table 2). In ganglia infected with L/ST-4BS, whose mutation relieves ICP4 repression of L/STs, the average numbers of spliced ICP0 transcripts per genome were very similar to those in ganglia infected with either the corresponding rescued virus or KOS (Table 2).
The frequency of detection of intron-containing ICP0 transcripts was roughly similar in ganglia infected with L/ST-n38, L/ST-4BS, the rescued viruses, or KOS (Table 2). Although there were ≈3-fold higher than the numbers of these transcripts per viral genome in ganglia infected with L/ST-n38, this did not correlate with the presence of the mutation, as the numbers were even higher in the rescued virus. (Nor would it conform to the prediction that ORF P inhibits ICP0 splicing.) There were only 2-fold-higher levels of these transcripts in L/ST-4BS-infected ganglia than in wild-type-infected ganglia. For all of the L/ST mutants and rescued viruses, the ranges of the standard errors of the means of these values overlapped those of the wild type (Table 2). Thus, our data suggest that mutations affecting ORF P or its expression do not significantly affect levels of spliced or intron-containing ICP0 transcripts in ganglia.
DISCUSSION
We initiated these studies to test specific hypotheses regarding the regulation of ICP0 expression during latency. Our results provide little if any experimental support for these hypotheses. However, in the course of performing these experiments, we uncovered evidence for the possible existence of certain levels of regulation of viral gene expression in latently infected ganglia, especially in terms of a block between IE and E gene expression and spliced versus intron-containing ICP0 transcripts. Our results are discussed below.
ICP0 transcripts can be detected in latently infected ganglia.
Previous failures to detect ICP0 transcripts in latently infected ganglia by RT-PCR (28, 31, 43, 44) have contributed to the hypothesis that a lack of ICP0 could be crucial for the establishment and maintenance of latency. Our ability to detect both spliced and intron-containing ICP0 transcripts was due largely to the development of highly sensitive assays. Keys to the success of these assays include the specific primers used (several primer sets were tried before we settled on those used) and the amplification conditions (e.g., relatively low magnesium). Our results add ICP0 to the list of productive-cycle transcripts from which RT-PCR products can be detected in ganglia latently infected with wild-type HSV (9, 21, 43).
As we have not yet characterized full-length ICP0 transcripts, we cannot be certain that the transcripts that we have detected can encode authentic ICP0 protein. However, the apparently accurate removal of intron 1 in the spliced transcripts argues for at least some degree of authenticity. Further exploration of the coding potential of these ICP0 transcripts seems warranted, especially as portions of ICP0 proteins and ICP0 proteins from mRNAs containing intron 2 are reported to exhibit distinct biological and biochemical activities (1, 14, 45-47). Regardless, latency cannot be explained simply by the complete absence of ICP0 expression.
Is ICP0 expression associated with reactivating virus?
Using the mouse and viral strains employed in this study, no infectious virus has ever been detected in scores of trigeminal ganglia harvested 30 days postinoculation (24; unpublished results; D. Leib, personal communication). Based on this criterion, the ganglia we studied were latently infected. Recently, thorough in situ hybridization analyses have detected single neurons that contain relatively high levels of both IE and E transcripts in ≈10% of latently infected ganglia (16). It is possible that these rare neurons are undergoing reactivation of virus that is not detectable by infectivity assays, and this may account for some of the ICP0 expression that we detected. Nevertheless, these rare neurons would not explain the relatively high expression of productive-cycle transcripts that we observed in ≥20% of ganglia (9, 21; this study). They also would not account for the many wild-type-infected ganglia that express relatively high numbers of IE transcripts but do not express relatively high numbers of an E transcript and vice versa (9) (Fig. 3). Furthermore, ganglia infected with TK− mutants, which do not reactivate even upon explant, can express relatively high numbers of productive-cycle transcripts per viral genome (21) and exhibit persistently elevated expression of cytokine transcripts (8). Thus, there is accumulating evidence for expression of productive-cycle genes in latently infected ganglia in the absence of reactivation of virus. Regardless, whether the ICP0 expression that we have detected reflects rare reactivation events that result in undetectable infectious virus or a stage of latency in which a block to ICP0 expression has been overcome, the mechanisms that repress or promote such expression in latently infected ganglia are relevant to the maintenance of latency.
No evidence for antisense repression of ICP0 by LATs.
As reviewed in the introduction, a long-standing and attractive hypothesis for repression of productive-cycle gene expression during latency has been that the LATs repress ICP0 expression via an antisense mechanism (15, 29, 41), and this hypothesis predicts that drastically reducing LAT expression would lead to an increase in ICP0 transcripts in latently infected ganglia. Notably, we did not observe a meaningful increase in either spliced or intron-containing ICP0 transcripts resulting from a LAT deletion mutation that essentially eliminates LAT expression. It is possible that individual neurons in wild-type-infected ganglia that contain high levels of LAT have the lowest levels of ICP0 transcripts; however, if that were a mechanism for repressing ICP0, one would still expect that the LAT deletion would lead to increases in ICP0 expression. Indeed, in this study, we did observe increases in ICP4 and tk transcripts in LAT− mutant-infected ganglia (not shown), as reported previously (9). Therefore, LAT-mediated repression of productive-cycle gene expression is not due to reductions in ICP0 transcript levels by LATs acting via an antisense mechanism.
ORF P mutants.
Similarly, as reviewed in the introduction, the hypothesis that ORF P, which is encoded by the L/STs but also conceivably by minor LATs, may repress ICP0 expression by inhibition of splicing during latency (2) predicts that drastically reducing expression of LATs or inactivation of ORF P by a nonsense mutation would lead to increases in spliced ICP0 transcripts. These predictions were not met. The hypothesis also predicts that if ORF P expression in ganglia arises from L/ST expression, then mutant L/ST-4BS should synthesize reduced levels of spliced ICP0 transcripts and increased levels of intron-containing transcripts. We found little or no evidence for either of these effects. Our results with the ORF P mutants in ganglia were consistent with the results obtained in cultured cells (25). Thus, we do not favor the hypothesis that ORF P represses gene expression during latency via effects on ICP0 splicing.
We did not observe any apparent effects of the L/ST mutations on expression of the LATs (Table 1). The absence of an effect on LATs is in apparent contrast to the effect of the L/ST-4BS mutation on LAT expression in neurally derived mouse NB41A3 and other cells (25). This apparent discrepancy may be explained by L/STs' being expressed in insufficient amounts in latently infected ganglia or in fewer or different neurons than LATs or by other differences between ganglia and cultured neurally derived cells.
Correlation with ICP4 transcripts: evidence for a block between IE and E gene expression.
Our finding that the numbers of ICP0 and ICP4 transcripts in wild-type-infected ganglia correlated better than the numbers of either of these IE transcripts with tk transcripts suggests that wild-type-infected ganglia can express two different IE genes in a coordinated fashion without coordinate expression of the E gene tk (Fig. 3). This, in turn, suggests the possibility that, at least in a subset of ganglia, a block exists between IE and E gene expression. We have previously presented evidence that a viral function associated with the LAT locus could be responsible for this block, in that the dlLAT1.8 deletion increases the correlation between ICP4 and tk transcript levels (9). Consistent with this evidence, in the present study, stronger correlations were found between ICP0 and tk transcripts in LAT− mutant-infected ganglia than in wild-type-infected ganglia (not shown). Additional studies are needed to examine further the hypothesis of a block between IE and E gene expression.
Intron-containing ICP0 transcripts.
A second possible control point for viral gene expression in latently infected ganglia is suggested by our findings regarding intron-containing ICP0 transcripts. The origin and precise structure of these transcripts are not known. They may be related to intron-containing transcripts detected in cultured cells (6), which, even though their 3′ ends lie upstream of the site where primer 0-2 hybridizes, suggests the existence of ICP0 mRNAs containing various amounts of intron 1. Interestingly, in contrast to what is seen in HSV-infected cells in culture (6, 33), the average level of expression of intron-containing transcripts in ganglia latently infected with wild-type virus was higher than that of spliced transcripts. Also, the ratio of intron-containing to spliced transcripts varied substantially from ganglion to ganglion. These observations, together with results with LAT mutants (see below), are consistent with the possibility of latency-specific regulation of splicing of ICP0 transcripts or differential stabilities of spliced and intron-containing transcripts. In this regard, the finding that an ICP22/US1.5 deletion mutant expresses a greater ratio of intron-containing to spliced ICP0 transcripts (6) may be relevant.
A surprising result was that ganglia latently infected with the two independent LAT− mutants exhibited detectable expression of intron-containing ICP0 transcripts significantly less frequently than did wild-type virus. Two other observations are consistent with the possibility that the LAT mutation results in decreased accumulation of the intron-containing transcripts. (i) The average level of expression of these transcripts was lower in ganglia infected with the LAT− mutants than it was in ganglia infected with wild-type virus, a virus in which the LAT deletion was rescued, or the L/ST mutants. (ii) The transcripts were more frequently detected in ganglia infected with the L/ST mutants and their rescued derivatives, which express wild-type levels of LATs. However, because the virus in which the LAT deletion was rescued did not exhibit a significantly greater frequency of detectable intron-containing transcripts than did the mutant virus (which may be due simply to the small number of ganglia tested), further work is warranted to establish if the reduced expression of the intron-containing transcripts is indeed due to the LAT mutation. If so, there may be a role for a LAT-associated gene product in promoting the accumulation of intron-containing ICP0 transcripts, perhaps by regulating splicing or by regulating the stabilities of spliced versus intron-containing species. Such a function might contribute to LAT-mediated repression of productive-cycle gene expression or other biological effects.
How is latency maintained?
We have shown previously that a viral function associated with the LAT locus, most probably one or more of the LATs, represses productive-cycle gene expression and thus promotes maintenance of latency in HSV-infected ganglia (9, 17). Interestingly, the LAT deletion studied here results in both higher expression of both ICP4 and tk transcripts and stronger correlations between IE and tk transcripts (9) (data not shown). We have previously identified transcripts that are antisense to ICP4 transcripts, whose levels are decreased by the LAT deletion, that could explain a block between IE and E gene expression (9).
Our inability to find evidence for repression of ICP0 expression by LATs provides impetus for investigating this and other possible mechanisms for LAT-mediated repression, which might include regulated accumulation of spliced versus intron-containing ICP0 transcripts during latency. Regardless, LAT− and ORF P mutants, including those studied here, efficiently establish and maintain latent infections and exhibit repression of productive-cycle gene expression (9, 17, 25, 26, 35; this report). Thus, aside from understanding the mechanism of LAT-mediated repression, a major task is to identify other viral and host factors involved in the maintenance of latency.
Acknowledgments
We thank S. Silverstein for generously providing pDS18; D. Leib, L. Feldman, and T. Margolis for communicating results ahead of publication; M. Chen for confirmatory studies; M. Chen and A. Griffiths for help with figure preparation; F. Roth for advice on statistics; and J. Pesola, M. Kramer, and members of the Schaffer lab for helpful discussions.
This research was supported by NIH program project grant PO1 NS35138 (D.M.C., D.M.K, and P.A.S) and by grant NSC 89-2320-B-006-028 from the National Science Council in Taiwan (S.-H.C).
REFERENCES
- 1.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated ring finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruni, R., and B. Roizman. 1996. Open reading frame P—a herpes simplex virus gene repressed during productive infection encodes a protein that binds a splicing factor and reduces synthesis of viral proteins made from spliced mRNA. Proc. Natl. Acad. Sci. USA 93:10423-10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai, W., T. L. Astor, L. M. Liptak, C. Cho, D. M. Coen, and P. A. Schaffer. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J. Virol. 67:7501-7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, W., and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63:4579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, W., and P. A. Schaffer. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66:2904-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter, K. L., and B. Roizman. 1996. Alternatively spliced mRNAs predicted to yield frame-shift proteins and stable intron 1 RNAs of the herpes simplex virus 1 regulatory gene α0 accumulation in the cytoplasm of infected cells. Proc. Natl. Acad. Sci. USA 93:12535-12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J., and S. Silverstein. 1992. Herpes simplex viruses with mutations in the gene encoding ICP0 are defective in gene expression. J. Virol. 66:2916-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, S.-H., D. A. Garber, P. A. Schaffer, D. M. Knipe, and D. M. Coen. 2000. Persistent elevated expression of cytokine transcripts in ganglia latently infected with herpes simplex virus in the absence of ganglionic replication or reactivation. Virology 278:207-216. [DOI] [PubMed] [Google Scholar]
- 9.Chen, S.-H., M. F. Kramer, P. A. Schaffer, and D. M. Coen. 1997. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 71:5878-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coen, D. M., H. E. Fleming, Jr., L. K. Leslie, and M. J. Retondo. 1985. Sensitivity of arabinosyladenine-resistant mutants of herpes simplex virus to other antiviral drugs and mapping of drug hypersensitivity mutations to the DNA polymerase locus. J. Virol. 53:477-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook, W. J., K. K. Wobbe, J. Böni, and D. M. Coen. 1996. Regulation of neighboring gene expression by the herpes simplex virus type 1 thymidine kinase gene. Virology 218:193-203. [DOI] [PubMed] [Google Scholar]
- 12.Dobson, A. T., F. Sederati, G. Devi-Rao, W. M. Flanagan, M. J. Farrell, J. G. Stevens, E. K. Wagner, and L. T. Feldman. 1989. Identification of the latency-associated transcript promoter by expression of rabbit beta-globin mRNA in mouse sensory nerve ganglia latently infected with a recombinant herpes simplex virus. J. Virol. 63:3844-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D. 1989. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate-early gene 1. J. Gen. Virol. 70:1185-1202. [DOI] [PubMed] [Google Scholar]
- 14.Everett, R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74:9994-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farrell, M. J., A. T. Dobson, and L. T. Feldman. 1991. Herpes simplex virus latency-associated transcript is a stable intron. Proc. Natl. Acad. Sci. USA 88:790-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman, L. T., A. R. Ellsion, C. C. Voytek, L. Yang, P. Krause, and T. P. Margolis. 2002. Spontaneous molecular reactivation of herpes simplex virus type 1 latency in mice. Proc. Natl. Acad. Sci. USA 99:978-983. [DOI] [PMC free article] [PubMed]
- 17.Garber, D. A., P. A. Schaffer, and D. M. Knipe. 1997. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J. Virol. 71:5885-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halford, W. P., and P. A. Schaffer. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 75:3240-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kramer, M. F. 1995. Ph.D. thesis. Harvard University, Cambridge, Mass.
- 20.Kramer, M. F., S.-H. Chen, D. M. Knipe, and D. M. Coen. 1998. Accumulation of viral transcripts and DNA during establishment of latency by herpes simplex virus. J. Virol. 72:1177-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer, M. F., and D. M. Coen. 1995. Quantification of transcripts from the ICP4 and thymidine kinase genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 69:1389-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar, M., and G. G. Carmichael. 1998. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 62:1415-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagunoff, M., and B. Roizman. 1994. Expression of a herpes simplex virus 1 open reading frame antisense to the gamma 1 34.5 gene and transcribed by an RNA 3′ coterminal with the unspliced latency-associated transcript. J. Virol. 68:6021-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagunoff, M., and B. Roizman. 1995. The regulation of synthesis and properties of protein product of open reading frame P of the herpes simplex virus 1 genome. J. Virol. 69:3615-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, L. Y., and P. A. Schaffer. 1998. A virus with a mutation in the ICP4-binding site in the L/ST promoter of herpes simplex virus type 1, but not a virus with a mutation in open reading frame P, exhibits cell type-specific expression of γ134.5 transcripts and latency-associated transcripts. J. Virol. 72:4250-4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leib, D. A., C. L. Bogard, M. Kosz-Vnenchak, K. A. Hicks, D. M. Coen, D. M. Knipe, and P. A. Schaffer. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J. Virol. 63:2893-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leib, D. A., D. M. Coen, C. L. Bogard, K. A. Hicks, D. R. Yager, D. M. Knipe, K. L. Tyler, and P. A. Schaffer. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63:759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynas, C., K. A. Laycock, S. D. Cook, T. J. Hill, W. A. Blyth, and N. J. Maitland. 1989. Detection of herpes simplex virus type 1 gene expression in latently and productively infected mouse ganglia using the polymerase chain reaction. J. Gen. Virol. 70:2345-2355. [DOI] [PubMed] [Google Scholar]
- 29.Mador, N., D. Goldenberg, O. Cohen, A. Panet, and I. Steiner. 1998. Herpes simplex virus type 1 latency-associated transcripts suppress viral replication and reduce immediate-early gene mRNA levels in a neuronal cell line. J. Virol. 72:5067-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 31.Minagawa, H., S. Tanaka, Y. Toh, and R. Mori. 1994. Detection of herpes simplex virus type 1-encoded RNA by polymerase chain reaction: different pattern of viral RNA detection in latently infected murine trigeminal ganglia following in vitro or in vivo reactivation. J. Gen. Virol. 75:647-650. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell, W. J., R. P. Lirette, and N. W. Fraser. 1990. Mapping of low abundance latency-associated RNA in the trigeminal ganglia of mice latently infected with herpes simplex virus type 1. J. Gen. Virol. 71:125-132. [DOI] [PubMed] [Google Scholar]
- 33.Perry, L. J., F. J. Rixon, R. D. Everett, M. C. Frame, and D. J. McGeoch. 1986. Characterization of the IE110 gene of herpes simplex virus type 1. J. Gen. Virol. 67:2365-2380. [DOI] [PubMed] [Google Scholar]
- 34.Preston, C. M. 2000. Repression of viral transcription during herpes simplex virus latency. J. Gen. Virol. 81:1-19. [DOI] [PubMed] [Google Scholar]
- 35.Randall, G., M. Lagunoff, and B. Roizman. 2000. Herpes simplex virus 1 open reading frames O and P are not necessary for establishment of latent infection in mice. J. Virol. 74:9019-9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Randall, G., and B. Roizman. 1997. Transcription of the derepressed open reading frame P of herpes simplex virus 1 precludes the expression of the antisense γ134.5 gene and may account for the attenuation of the mutant virus. J. Virol. 71:7750-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 38.Sacks, W. R., and P. A. Schaffer. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61:829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawtell, N. M. 1997. Comprehensive quantification of herpes simplex virus latency at the single-cell level. J. Virol. 71:5423-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevens, J. G. 1989. Human herpesviruses: a consideration of the latent state. Microbiol. Rev. 53:318-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens, J. G., E. K. Wagner, G. B. Devi-Rao, M. L. Cook, and L. T. Feldman. 1987. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235:1056-1059. [DOI] [PubMed] [Google Scholar]
- 42.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate-early polypeptide Vmw110. J. Gen. Virol. 67:2571-2585. [DOI] [PubMed] [Google Scholar]
- 43.Tal-Singer, R., T. M. Lasner, W. Podrzucki, A. Skokotas, J. J. Leary, S. L. Berger, and N. W. Fraser. 1997. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue cultures. J. Virol. 71:5268-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka, S., H. Minagawa, Y. Toh, Y. Liu, and R. Mori. 1994. Analysis by RNA-PCR of latency and reactivation of herpes simplex virus in multiple neuronal tissues. J. Gen. Virol. 75:2691-2698. [DOI] [PubMed] [Google Scholar]
- 45.Van Sant, C., R. Hagglund, P. Lopez and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber, P. C., J. J. Kenny, and B. Wigdahl. 1992. Antiviral properties of a dominant negative mutant of the herpes simplex virus type-1 regulatory protein ICP0. J. Gen. Virol. 73:2955-2961. [DOI] [PubMed] [Google Scholar]
- 47.Weber, P. C., and B. Wigdahl. 1992. Identification of dominant-negative mutants of the herpes simplex virus type 1 immediate-early protein ICP0. J. Virol. 66:2261-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, T.-T., Y.-H. Su, T. M. Block, and J. M. Taylor. 1998. Atypical splicing of the latency-associated transcripts of herpes simplex type 1. Virology 243:140-149. [DOI] [PubMed] [Google Scholar]
- 49.Yeh, L., and P. A. Schaffer. 1993. A novel class of transcripts expressed with late kinetics in the absence of ICP4 spans the junction between the long and short segments of the herpes simplex virus type 1 genome. J. Virol. 67:7373-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zabolotny, J. M., C. Krummenacher, and N. W. Fraser. 1997. The herpes simplex virus type 1 2.0-kilobase latency-associated transcript is a stable intron which branches at a guanosine. J. Virol. 71:4199-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zwaagstra, J. C., H. Ghiasi, S. M. Slanina, A. B. Nesburn, S. C. Wheatley, K. Lillycrop, J. Wood, D. S. Latchman, K. Patel, and S. L. Wechsler. 1990. Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: evidence for neuron specificity and for a large LAT transcript. J. Virol. 64:5019-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]