Abstract
The draft genome (∼160 Mb) of the urochordate ascidian Ciona intestinalis has been sequenced by the whole-genome shotgun method and should provide important insights into the origin and evolution of chordates as well as vertebrates. However, because this genomic data has not yet been mapped onto chromosomes, important biological questions including regulation of gene expression at the genome-wide level cannot yet be addressed. Here, we report the molecular cytogenetic characterization of all 14 pairs of C. intestinalis chromosomes, as well as initial large-scale mapping of genomic sequences onto chromosomes by fluorescent in situ hybridization (FISH). Two-color FISH using 170 bacterial artificial chromosome (BAC) clones and construction of joined scaffolds using paired BAC end sequences allowed for mapping of up to 65% of the deduced 117-Mb nonrepetitive sequence onto chromosomes. This map lays the foundation for future studies of the protochordate C. intestinalis genome at the chromosomal level.
Ciona intestinalis is one of the most extensively studied ascidian species and is of particular phylogenetic importance for studying the molecular mechanisms underlying the origin and evolution of chordates and vertebrates (Cameron et al. 2000; Satoh 2003). In addition to having a basic chordate body plan and a compact genome, ascidians provide a simple experimental system for investigating gene networks involved in pattern formation and cell-fate specification during chordate development (Satoh 1994, 2003; Jeffery 2001; Nishida 2002).
As a result of these advantages, many efforts have been made in recent years toward developing genomic resources for C. intestinalis. The C. intestinalis genome has been sequenced using the whole-genome shotgun method (Dehal et al. 2002), and eightfold redundant coverage of the genome using random paired-end sequences was generated at the DOE Joint Genome Institute (JGI). Together with sequence data for nearly 480,000 ESTs and 5647 full-length cDNAs from Kyoto University and the National Institute of Genetics (Japan) (Satou et al. 2002), this has permitted the assembly of 2501 scaffolds longer than 3 kb. A total of 60 Mb, or half the assembly, was reconstructed into 177 scaffolds longer than 190 kb, and more than 85% of the assembled sequence was in 905 scaffolds longer than 20 kb. From these data, it is estimated that ∼160 Mb of the C. intestinalis genome is composed of a euchromatic sequence containing 15,852 protein-coding genes (117 Mb), as well as rDNA repeats and other repetitive sequences (Dehal et al. 2002; http://genome.jgi-psf.org/ciona4/ciona4.home.html).
Facilitating the construction of physical maps is the generation of large-insert genomic libraries, which have the advantage of being clone-based. C. intestinalis BAC libraries are readily available (Kobayashi et al. 2002), with 6150 clones already end-sequenced by the National Institute of Genetics (Japan) and reported in the data set of the Ciona draft genome (Dehal et al. 2002). The BAC libraries are available from Kyoto University (http://hoya.zool.kyoto-u.ac.jp/cgi-bin/gbrowse/ci). Together with an additional 148 BAC end sequences (BESs) that were analyzed during the sequencing process, this has generated a set of 12,448 raw sequences (6224 paired BAC end sequences) that are now available in the Ghost database (http://hoya.zool.kyoto-u.ac.jp/cgi-bin/gbrowse/ci). However, the assignment of every BES to scaffolds using a high-stringency assembly program is difficult, because polymorphisms and repeat sequences cause multiple end matches to several different scaffolds (e.g., Vinson et al. 2005). Recently, the genomic sequence for another Ciona species, Ciona savignyi, was made available on the internet (http://www.broad.mit.edu/annotation/ciona/). These data have already made it possible to compare sequences in regulatory regions between the two Ciona species (Johnson et al. 2004; Kusakabe et al. 2004).
While this sequence information represents an important and valuable resource, in the absence of a genetic context, the system remains largely inaccessible to genome-wide approaches to answering various biological questions. For example, the draft genome has not yet been mapped onto chromosomes, and the assembly of whole-genome shotgun sequences is still fragmented. C. intestinalis has 28 (n = 14) chromosomes (Colombera and Lazzaretto-Colombera 1978). We previously established a method of two-color fluorescent in situ hybridization (FISH) in Ciona (Shoguchi et al. 2004) and observed that counterstaining of Ciona chromosome preparations with DAPI made it possible to recognize the centromere, although the other banding landmarks were not observed (Shoguchi et al. 2004). We also identified 20 metacentric chromosomes and eight submetacentric or subtelocentric chromosomes in our karyotype analysis of Ciona chromosomes (Shoguchi et al. 2005). The largest pair of metacentric chromosomes was named chromosome 1, while the largest pair of submetacentric/subtelocentric chromosomes was named chromosome 2. However, the small size of the chromosomes (most pairs measuring <2 μm) and morphological polymorphisms made accurate pairing of the other chromosomes difficult, so that a precise karyotype was not feasible based on morphology alone. In further studies (Shoguchi et al. 2005), FISH with bacterial artificial chromosome (BAC) clones containing rDNA clusters, as well as staining of chromosome preparations with propidium iodide indicated that 18S/28S ribosomal gene repeats are present on the short arms of three pairs of chromosomes, and that these short arms show remarkable size polymorphism. In addition, each chromosome was characterized by FISH with a representative BAC clone, although we could not reach a final determination of the karyotype (Shoguchi et al. 2005).
Here, we report further molecular cytogenetic characterization of the 14 pairs of C. intestinalis chromosomes and initial large-scale mapping of genomic sequences onto chromosomes by FISH. Using scaffolds that were manually constructed with 6224 paired BAC end sequences, we carried out two-color FISH for a total of 170 BAC clones, which represent larger scaffolds released from JGI as C. intestinalis genome ver. 1. The publicized C. savignyi genome sequences on the Web site were also referenced to select the clones and to make efficient mapping. As a result, we were able to map up to 65% of the nonrepetitive genome sequence onto chromosomes. This map provides useful resources for sequencing entire genomes and is available online at http://hoya.zool.kyoto-u.ac.jp/chromosomeall.html.
Results and Discussion
Efficient mapping of selected BAC clones by two-color FISH
A total of 170 BAC clones were used to map C. intestinalis genomic sequence onto chromosomes (for selected BAC clones, see Supplemental Table S1). We selected 139 BAC clones to cover 100 of the largest scaffolds assembled in the Ciona draft genome (v. 1.0) (Scaffold_1 to Scaffold_100), and another set of 23 BAC clones was selected to increase the accuracy of the scaffolds joined by BAC end sequencing (BES). The remaining eight BAC clones were selected because they were expected to map to chromosome regions that were not targeted by the other 162 clones.
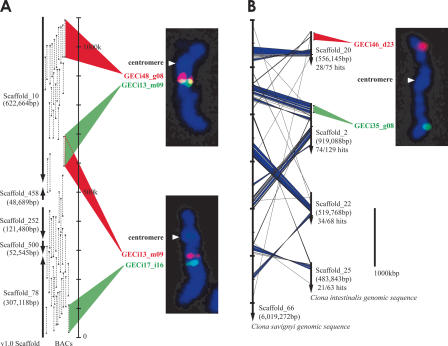
First, to increase the accuracy of mapping and to avoid confusion arising from potential cross-well contamination of clones and other artifacts, we selected two clones for each of the major scaffolds (for example, GECi48_g08 and GECi13_m09 for Scaffold_10, Fig. 1A). In a previous study, each chromosome was characterized by FISH with a representative BAC clone (Shoguchi et al. 2005). To use these data efficiently and to collect two-color FISH data for all 14 chromosomes, we made use of two informational resources, i.e., a database compiling the end sequences of C. intestinalis BACs (http://hoya.zool.kyoto-u.ac.jp/cgi-bin/gbrowse/ci.) and the sequenced genome of C. savignyi (http://www.broad.mit.edu/annotation/ciona/). The paired end sequences of 6224 BAC clones were used to join scaffolds from the JGI database (http://genome.jgi-psf.org/ciona4/ciona4.home.html). Because the strict parameters used in computational assembly of genomic sequences with a high degree of polymorphism would be expected to leave unjoined those scaffolds assembled by the paired BESs, we constructed manually joined scaffolds by using paired BESs, starting from the larger scaffolds. For example, Scaffold_10 and Scaffold_78 were joined by linking Scaffolds_10, _458, _252, _500, and _78 with the aid of BESs (Fig. 1A). By performing FISH with the representative clones, we could confirm that the joined scaffolds mapped to the same chromosome. Moreover, we could determine the orientation of the BES-joined scaffolds on the chromosome based on the relative locations of the constituent scaffolds; for example, we found that Scaffold_10 was closer to the centromere than Scaffold_78 (Fig. 1A).
Figure 1.
Strategy for mapping BAC clones to chromosomes. (A) Correspondence between the distribution of BAC end sequences (BESs) on joined scaffolds and the cytogenetic location of FISH signals. The construction of joined scaffolds was performed using paired BAC end sequences. The left arrows indicate joined scaffolds from version 1.0 of the C. intestinalis draft genome. The regions of paired BAC end sequences corresponding to each scaffold are indicated by black triangles and are connected by dashed lines. For example, Scaffold_10 and Scaffold_458 are joined by two paired BAC end sequences in a tail-to-tail arrangement. Two-color FISH was performed using GECi48_g08 (red) and GECi13_m09 (green) BACs. The distribution of BESs on joined scaffolds indicates a gap of about 300 kb between the two clones, which gives an indication of the resolution achieved by FISH. GECi13_m09 (red) and GECi17_i16 (green) were also found to hybridize to the same chromosome. The cytogenetic locations of FISH signals correlate well to a first approximation with the order of joined scaffolds (Scaffold_10, Scaffold_458, Scaffold_252, Scaffold_500, and Scaffold_78). (B) A sample result from comparison of C. intestinalis and C. savignyi sequences. The vertical arrows represent scaffolds of the draft genomes (left) C. savignyi; (right) C. intestinalis. Diagonal lines connect best hit regions as determined by a FASTA comparison of C. intestinalis and C. savignyi sequences, with blue areas indicating regions with five sequential hits (within a distance of less than or equal to one gene), indicating strong similarity. Twenty-eight of 75 predicted genes in Scaffold_20 of C. intestinalis found best hit matches in Scaffold_66 of C. savignyi. Scaffold_2, Scaffold_22, and Scaffold_25 of C. intestinalis showed similar results, suggesting that Scaffold_20, Scaffold_25, Scaffold_22, and Scaffold_2 of C. intestinalis map to the same chromosome. These data corroborate results from comparative genomics studies that suggest a low rate of chromosome translocation between these two species. GECi46_d23 (red), which corresponds to Scaffold_20 and GECi35_g08 (green), which corresponds to Scaffold_2, map to different arms of the same chromosome.
Second, to increase the accuracy of the scaffolds joined by BESs, we selected 23 BAC clones corresponding to these scaffolds (for example, Scaffold_151) to obtain BAC–FISH data. Third, the other eight BAC clones were selected at random to map onto chromosomal regions that were poorly covered by the previously used 162 BAC clones. For example, Scaffold_107 had no linkage with the large scaffolds. The construction of the joined scaffolds began with Scaffold_1, and the construction was checked by carefully selecting and examining many mapped BAC clones, as described below.
As shown in Figure 1A, mapping of BACs onto Ciona chromosomes by two-color FISH was effective, but the paucity of cytogenetic markers makes more detailed mapping difficult. To overcome this problem, we compared the syntenic relationships between genomic sequences of C. intestinalis and C. savignyi in order to further refine our map. This analysis was based on a FASTA algorithm comparison of the 15,852 C. intestinalis gene models to C. savignyi draft genome sequences (Pearson 2000). The rough syntenic relationships were determined by counting the best hits manually. For example, as shown in Figure 1B, 74 of 129 gene models (57%) in C. intestinalis (Ci) Scaffold_2 showed a best hit relationship to those found in the largest C. savignyi (Cs) scaffold, Scaffold_66 (∼6 Mb in length) (Data version 4/25/2003), while 34 of 68 gene models (50%) in Ci Scaffold_22 had a best-hit relationship to those found in Cs Scaffold_66. Sequential hits were also found, indicating stronger syntenic relationships (Fig. 1B, blue color regions). Similarly, we could also predict that Ci Scaffold_20 and Ci Scaffold_25 mapped to the same chromosome. This method can be applied when comparative genomics show a low rate of chromosome translocation (Stein et al. 2003; Richards et al. 2005). We hypothesize that the majority of Ciona genes have remained on the same chromosome arm during the evolution of the two Ciona species. Thus, Scaffold_20 and Scaffold_2 were mapped to the same chromosome by FISH, even though the red and green signals were not close (Fig. 1B).
Molecular cytogenetic characterization and karyotype of C. intestinalis chromosomes
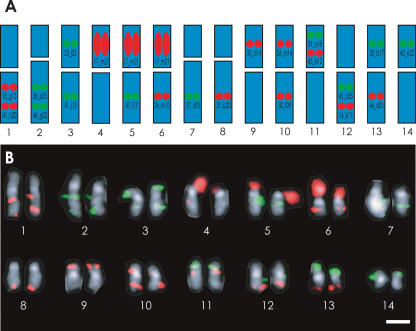
It was impossible to identify the 14 pairs of C. intestinalis chromosome based on morphology alone, although we were previously able to define two large chromosomes (Shoguchi et al. 2005). As illustrated in Figure 2A, in order to identify all of the 14 pairs of chromosomes simultaneously, we prepared a set of 21 BAC-derived probes for two-color FISH. Based on independent FISH data using two different clones, we could predict the localization of these BACs. For example, GECi17_m21 is a BAC probe that should broadly label rDNA clusters with red signal (also see Supplemental Fig. S1). GECi18_g12 and GECi41_h20, both labeled red, are expected to hybridize with the long arm of chromosome 1, while GECi38_p05 and GECi48_g02, both labeled green, are expected to hybridize with the long arm of chromosome 2. GECi23_j02 (green) is expected to hybridize with the short arm of chromosome 3, while GECi41_j13 (green) should hybridize with the long arm, and so on. Together, these 21 BACs should permit one to distinguish between all 14 chromosome pairs simultaneously by two-color FISH.
Figure 2.
Molecular cytogenetic characterization of all 14 pairs of C. intestinalis chromosomes. (A) Strategy for identifying all 14 pairs of C. intestinalis chromosomes simultaneously. BAC probes used are shown on chromosomes they are predicted to recognize; chromosome 1: GECi18_g12 (red) and GECi41_h20 (red), both on the long arm (the GECi designation was omitted in the figure); chromosome 2: 38_p05 (green) and 48_g02 (green), both on the long arm; chromosome 3: 23_j02 (green) on the short arm and 41_j13 (green) on the long arm; chromosome 4: 17_m21 (rDNA probe; red) on the short arm; chromosome 5: 17_m21 (rDNA probe; red) on the short arm and 47_l17 (green) on the long arm; chromosome 6: 17_m21 (rDNA probe; red) on the short arm and 26_m10 (red) on the long arm; chromosome 7: 37_e03 (green) on the long arm; chromosome 8: 31_b20 (red) on the long arm; chromosome 9: 30_d14 (red) on the short arm; chromosome 10: 26_m14 (red) on the short arm and 30_f09 (red) on the long arm; chromosome 11: 39_o18 (green) and 45_b12 (red), both on the short arm; chromosome 12: 45_o05 (green) and 16_k11 (red), both on the long arm; chromosome 13: 20_b17 (green) on the short arm and 46_d01 (red) on the long arm; and chromosome 14: 40_e22 (green) on the short arm. (B) Karyotyping of all 14 pairs of C. intestinalis chromosomes by FISH. Metaphase chromosomes were counterstained with DAPI and imaged monochromatically. The centromere regions were brightly stained. Scale bar, 2 μm.
As shown in Figure 2B, 14 pairs of C. intestinalis chromosomes with distinctive staining patterns could be aligned according to size. Chromosomes 1, 3, 4, 5, 6, 9, 10, 11, 12, 13, and 14 were metacentric, while chromosomes 2, 7, and 8 were submetacentric. rDNA clusters were detected on the short arms of chromosomes 4, 5, and 6. As pointed out in a previous study (Shoguchi et al. 2005), the short arms of chromosome 2 and 4–6, which contain rDNA clusters, showed size polymorphism. Thus, we were able to fully characterize and karyotype all 14 pairs of chromosomes.
The size of the haploid genome of C. intestinalis is ∼160 Mb. When chromosomes were giemsa stained, they appeared larger than when counterstained with DAPI, which often did not appear to label telomeric regions (data not shown). As a result, portions of the merged signal from broadly labeling BACs, such as GECi38_p05 of chromsome 2, may appear on neighboring chromosomes. Alternatively, these areas of overlap may represent less condensed chromosomal regions. Nevertheless, measurements of the length of giemsa-stained metaphase chromosomes and correlation with the total genome size suggested that the Ciona chromosomes range in size between about 8 and 16 Mb, as shown in Table 1. The largest chromosome, 1, was estimated to be 16.5 Mb, which is approximately half the size of the smallest human chromosome, 21 (Ichikawa et al. 1993). Chromosome 2 was estimated to be 15.3 Mb, while chromosomes 3–12 ranged in size between about 10 and 13 Mb. The smallest chromosomes, 13 and 14, were very similar in size and were estimated to be about 8 Mb. Each chromosome arm was estimated to be between 3 and 11 Mb in size, which is about the same as the major band length of human metaphase chromosomes, as defined by classical cytogenetics (Paris Conference 1971). These data provide basic information needed to map each BAC probe to the identified chromosomes, as described below.
Table 1.
Chromosomal mapping of BAC clones in Ciona intestinalis
| Chromosome | Chromosome arm length (Mb) estimated by cytogenetic analysis | Number of BAC clones mapped by FISH | Total size (Mb) of joined and mapped scaffolds | % coverage | Number of BAC clones**corresponding to mapped scaffolds |
|---|---|---|---|---|---|
| 1p | 6.15 | 5 | 4.427703 | 71.9 | 141 |
| 1q | 10.34 | 10 | 5.059860 | 48.9 | 179 |
| 2p | 4.03 | 0 | 0 | 0 | 0 |
| 2q | 11.31 | 14 | 7.652363 | 67.6 | 333 |
| 3p | 5.34 | 6 | 1.807897 | 33.8 | 63 |
| 3q | 7.69 | 11 | 4.971595 | 64.6 | 194 |
| 4p | 4.56 | * | * | * | * |
| 4q | 8.47 | 10 | 5.372748 | 63.4 | 220 |
| 5p | 4.25 | * | * | * | * |
| 5q | 7.95 | 11 | 5.268640 | 66.3 | 179 |
| 6p | 4.85 | * | * | * | * |
| 6q | 6.2 | 7 | 2.267187 | 36.6 | 71 |
| 7p | 3.21 | 0 | 0 | 0 | 0 |
| 7q | 7.84 | 11 | 6.392846 | 81.5 | 217 |
| 8p | 2.78 | 0 | 0 | 0 | 0 |
| 8q | 7.94 | 23 | 6.421300 | 80.8 | 269 |
| 9p | 4.85 | 4 | 2.887692 | 59.5 | 110 |
| 9q | 5.87 | 8 | 3.683591 | 62.7 | 155 |
| 10p | 4.27 | 4 | 1.809801 | 42.3 | 70 |
| 10q | 6.45 | 8 | 3.222832 | 49.9 | 125 |
| 11p | 4.27 | 7 | 2.430748 | 56.9 | 75 |
| 11q | 5.63 | 5 | 1.579050 | 28.0 | 47 |
| 12p | 4.23 | 3 | 1.465373 | 34.6 | 42 |
| 12q | 5.67 | 7 | 3.741953 | 65.9 | 146 |
| 13p | 3.67 | 1 | 0.192510 | 5.2 | 4 |
| 13q | 4.25 | 3 | 1.436447 | 33.7 | 45 |
| 14p | 3.62 | 4 | 1.851839 | 51.1 | 61 |
| 14q | 4.3 | 8 | 2.191802 | 50.9 | 70 |
| Total | 159.99 | 170 | 76.135777 | 47.6 | 2816 |
18S/28S rDNA repeat-containing regions, as also indicated in Figure 2
BAC clones are counted only when both paired-BAC end sequences corresponding to the scaffolds map to the same chromosome arm (BLASTN hit with >90% identity, alignment length >100, probability <e-80) and 45.2% of the BAC clones are annotated
Locations of 170 mapped BAC clones
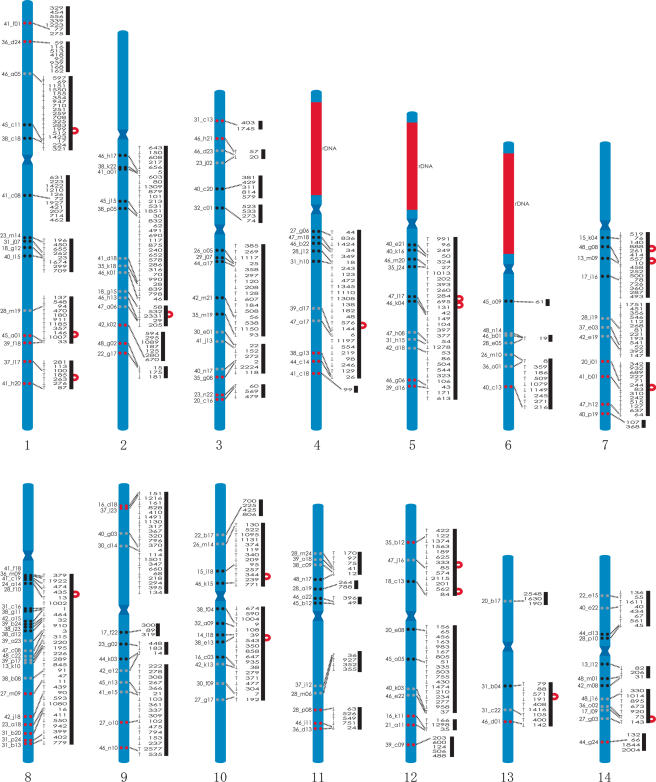
As noted in our previous two-color FISH study (Shoguchi et al. 2004), DAPI staining marks the centromere, which can be used as a landmark for orienting the two-color FISH data (see also Supplemental Fig. S1). As mentioned above, we carried out two-color FISH of a total of 170 BAC probes, and obtained 172 data sets that showed signals on the same chromosome of 14 linkage groups (Supplemental Table S2). The scaffolds constructed using paired BESs were used to select two cohybridizing BAC clones, and BAC–FISH was used to confirm these assemblies. Thus, 17 regions were joined by using a pair of BESs (Fig. 3). The relative order of clones that mapped very close to one another, such as GECi23_m14 and GECi31_j07 on chromosome 1, was also determined by the mapped sequence data for joined scaffolds. FISH with two BAC clones (e.g., GECi23_m14 and GECi40_l15) was thus able to determine the orientation of a joined scaffold (Fig. 3).
Figure 3.
Mapping of BAC clones and draft genome sequence (version 1) onto C. intestinalis chromosomes. The locations of 170 FISH-mapped BACs are shown relative to DAPI-counterstained chromosomes with centromere bands (at an approximately relative scale). Clones that mapped proximal to the centromere are represented by a pair of black circles. Clones that mapped distal to the centromere are represented by a pair of red circles, and those that mapped to the middle portion of the arms are shown by a pair of gray circles. Red areas indicate three regions with rDNA clusters on the short arms of chromosomes 4, 5, and 6. The paired-BAC end sequences assembled the 468 scaffolds into 48 joined scaffolds and three scaffolds that were not joined to any of the others, as shown by bold lines. Seventeen scaffolds assembled using one pair of BAC end sequences for each are shown by red curved lines (for example, Scaffold_199 and Scaffold_512) and map to regions in between FISH-mapped BACs. Arrows on the left side of the scaffold number indicate the orientation of the mapped sequences.
Results of the two-color FISH analysis are summarized in Figure 3. We found that BAC clones GECi38_g11 and GECi42_a15 on Scaffold_1 mapped close to one another on the long arm of chromosome 8, near the centromere (8q). In addition, clones GECi47_c08 and GECi39_a23 on Scaffold_3 also mapped to chromosome 8, but were more distant from the centromere. Therefore, Scaffolds_1 and _3 were mapped to the long arm of chromosome 8. As mentioned above, we also manually constructed joined scaffolds using paired BAC end sequences. The largest joined scaffold was about 6.4 Mb, and hybridized to 23 BAC clones mapped to the long arm of chromosome 8.
In a similar manner, a total of 166 other BAC clones were mapped onto the 14 pairs of C. intestinalis chromosomes. The short arm of chromosome 1 was marked by five BAC clones and the long arm by 10 clones (Table 1). Similarly, each of the other chromosomes hybridized to multiple BAC probes, as summarized in Table 1. The FISH data corresponded precisely to the joined scaffolds generated by the paired BES data. The only exception was GECi31_c16, which mapped to chromosome 8 by FISH, but whose end sequences corresponded to both Scaffold_1 and Scaffold_9. While Scaffold_1 was mapped to chromosome 8, Scaffold_9 was mapped to chromosome 10 using two other BAC clones (see Supplemental Table S1). The small partial sequence of Scaffold_9 is likely to map to chromosome 8, because no secondary signal was observed on chromosome 10. One BAC clone, GECi17_p11, only produced a secondary signal (Supplemental Fig. S1), because nonspecific signals caused by the hybridization of repetitive sequences, which were often present in large DNA fragments, were suppressed with the genomic DNA (Shoguchi et al. 2004).
Genes for ribosomal RNA seem to be clustered within the C. intestinalis genome, as appears to be the case for other animals (Shoguchi et al. 2005). A probe made from a BAC clone containing rDNA gene clusters stained a broad region of the short arm of chromosomes 4, 5, and 6, suggesting that these regions are likely to be occupied by clusters of ribosomal genes (Fig. 2B; Supplemental Fig. S1). Chromosomes 2, 7, and 8 are submetacentric, and therefore also have short arms, but genomic sequence information has not yet been mapped to these arms.
We estimate that nearly 65% of the nonrepetitive genome sequence information has been mapped to chromosomes by FISH, although some highly polymorphic regions of the genome made accurate assembly of joined scaffolds difficult. In addition, cytogenetic analysis was performed for an estimated 47.6% of the known 160-Mb sequence, which includes repetitive sequences and corresponds to 45.2% of the BAC clones (2816/6224 clones).
Although we were able to map about 65% of C. intestinalis genomic information onto chromosomes, further mapping efforts will be required in the future. Future studies to fill in gaps between scaffolds as well as within a scaffold will also be helpful for further mapping of the genome information onto the chromosomes.
Due to the small size and high degree of morphological polymorphism in chromosomes of invertebrate animals, it is sometimes difficult to perform precise karyotyping for these organisms using conventional methods. As shown in this study, two-color FISH is very effective for analyzing the genomes of organisms without extensive prior genetic characterization. Moreover, we show that comparison between genome sequences of closely related species is a very useful tool for mapping. In this era of comparative genomics, in which whole-genome shotgun technology has made it possible to sequence the genome of a variety of different organisms, the approach we have developed will be very useful. Indeed, there are currently several ongoing genome sequencing projects that have targeted such organisms as sea urchins and amphioxus, which will likely face similar challenges as have been encountered with ascidian genome analysis. Our data suggest that analysis by two-color FISH will make it possible to perform chromosomal mapping of their draft sequences.
Methods
Biological materials
C. intestinalis adults were cultured at the Maizuru Fisheries Research Station of Kyoto University and the International Coastal Research Center, Ocean Research Institute of the University of Tokyo. In this study, we used embryos at the neurula and early tailbud stages. Handling of gametes and embryos was carried out as described by Shoguchi et al. (2004).
Fluorescent in situ hybridization with BAC clones
A library of BAC clones with an average length of 135 kb was constructed from C. intestinalis sperm DNA (Kobayashi et al. 2002). Chromosome preparation was performed as described in Shoguchi et al. (2005). FISH was carried out as described in Shoguchi et al. (2004) and modified as follows: To distinguish between the 14 pairs of chromosomes simultaneously using FISH, 21 BAC clones were selected from among those tested by FISH analyses, in which two BAC clones were hybridized at the same time. The 21 BAC-derived probes (as shown in Fig. 2A) were hybridized simultaneously. The hybridization solution consisted of 50% formamide, 2× SSC, 10% dextran sulfate, 10 μg sheared C. intestinalis sperm DNA, 0.15% SDS, and a total of 20 μL of this solution was used per slide. At first, an aliquot containing about one-eighth of each labeled probe (derived from 1 μg of BAC DNA) was added to the hybridization mix to make the karyotyping mixture. Hybridizaton was performed for 55 h at 42°C. Signal from biotin-labeled probes was amplified and detected with sequential applications of avidin FITC DCS (Vector Laboratories), biotinylated anti-avidin D (Vector Laboratories), and then avidin FITC DCS again. For detection of DIG-labeled probes, anti-DIG-rhodamine Fab fragments (ROCHE) was applied, followed by Texas Red anti-sheep secondary antibody (Vector Laboratories). After the first experiment, the concentration of each probe in the karyotyping mixture was adjusted by increasing the concentration of probes that showed weaker signals, or by decreasing the concentration of probes that showed stronger signals in order to obtain signals of similar strength. Image processing was done with Adobe Photoshop 6.0. Computerized background processing and image-feature intensification were conducted using Photoshop 6.0 to make the weak signals recognizable in aligned- and paired-chromosome figures (Fig. 2B). This process caused strong signals to appear enlarged, resulting with some signals appearing outside of the chromosomes. The counterstained blue image was changed to gray scale in Photoshop 6.0 for the sake of image clarity. Following the collection of FISH images and pairing, the preparation was stained in Giemsa solution, as described by Shoguchi et al. (2005). Images were collected with a Zeiss epi-fluorescence microscope equipped with an Axiocam camera. Measurements were done with Adobe Photoshop 6.0. Five preparations were analyzed and the average chromosome arm lengths (Mb) were estimated.
Construction of joined-scaffolds by using BAC end sequences
Of a total of 12,448 BAC end sequences (6224 clones), 11,420 corresponded to scaffolds (Assembly v1.0), as shown by using the BLASTN algorithm (probability <e-30). Manual assembly was performed on those gaps covered by more than two pairs of BAC end sequences. By comparison with FISH data, we also joined some scaffolds using one pair of BAC end sequences. The information is available at http://hoya.zool.kyoto-u.ac.jp/cgi-bin/gbrowse/ci. Each set of joined scaffolds was constructed independently by two people and was verified by comparison of the two data sets.
Acknowledgments
We thank the members of the Joint Genome Institute for helpful advice on bioinformatics. We also thank Drs. Yoichi Matsuda and Chizuko Umehara for useful discussions concerning karyotype analysis and FISH methods, Dr. Shungo Kano for incisive comments, and Chikako Imaizumi for her support in the maintenance of sequence information in our laboratory. We thank the Ciona savignyi genome consortium for the draft genome information on their Web site. This manuscript benefited from the helpful comments of three reviewers. Kazuko Hirayama, Maizuru Fisheries Research Station of Kyoto University, International Coastal Research Center of the University of Tokyo, and Sugashima Marine Biological Laboratory of Nagoya University areacknowledged for their help in culturing of Ciona intestinalis. The present study was supported by Grants-in-Aid from MEXT, Japan to N.S. (12202001, 17018018). Work at JGI was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48, Lawrence Berkeley National Laboratory under Contract No. DE-AC02-05CH11231 and Los Alamos National Laboratory under Contract No. W-7405-ENG-36. The Center for Integrative Genomics is supported by a grant from the Gordon and Betty Moore Foundation.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4156606.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Cameron, C.B., Garey, J.R., and Swalla, B.J. 2000. Evolution of the chordate body plan: New insights from phylogenetic analyses of deuterostome phyla. Proc. Natl. Acad. Sci. 97 4469-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombera, D. and Lazzaretto-Colombera, I. 1978. Chromosome evolution in some marine invertebrates. In Marine organisms (eds. B. Battaglia et al.), pp. 487-525. Plenum, New York.
- Dehal, P., Satou, Y., Campbell, R.K., Chapman, J., Degnan, B., DeTomaso, A., Davidson, B., DiGregorio, A., Gelpke, M., Goodstein, D.M., et al. 2002. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 298 2157-2167. [DOI] [PubMed] [Google Scholar]
- Ichikawa, H., Hosoda, F., Arai, Y., Shimizu, K., Ohira, M., and Ohki, M. 1993. A NotI restriction map of the entire long arm of human chromosome 21. Nat. Genet. 4 361-366. [DOI] [PubMed] [Google Scholar]
- Jeffery, W.R. 2001. Determinants of cell and positional fate in ascidian embryos. Int. Rev. Cytol. 203 3-62. [DOI] [PubMed] [Google Scholar]
- Johnson, D.S., Davidson, B., Brown, C.D., Smith, W.C., and Sidow, A. 2004. Noncoding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Res. 14 2448-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, M., Matsuda, M., Asakawa, S., Shimizu, N., Nagahama, Y., Satou, Y., and Satoh, N. 2002. Construction of BAC libraries derived from the ascidian Ciona intestinalis. Genes Genet. Syst. 77 283-285. [DOI] [PubMed] [Google Scholar]
- Kusakabe, T., Yoshida, R., Ikeda, Y., and Tsuda, M. 2004. Computational discovery of DNA motifs associated with cell type-specific gene expression in Ciona. Dev. Biol. 276 563-580. [DOI] [PubMed] [Google Scholar]
- Nishida, H. 2002. Specification of developmental fates in ascidian embryos: Molecular approach to maternal determinants and signaling molecules. Int. Rev. Cytol. 217 227-276. [DOI] [PubMed] [Google Scholar]
- Paris Conference. 1971. Standardization in human cytogenetics. Cytogenetics 11 313-362. [PubMed] [Google Scholar]
- Pearson, W.R. 2000. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 132 185-219. [DOI] [PubMed] [Google Scholar]
- Richards, S., Liu, Y., Bettencourt, B.R., Hradecky, P., Letovsky, S., Nielsen, R., Thornton, K., Hubisz, M.J., Chen, R., Meisel, R.P., et al. 2005. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 15 1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, N. 1994. Developmental biology of ascidians. Cambridge University Press, New York.
- ———. 2003. The ascidian tadpole larva: Comparative molecular development and genomics. Nat. Rev. Genet. 4 285-295. [DOI] [PubMed] [Google Scholar]
- Satou, Y., Yamada, L., Mochizuki, Y., Takatori, N., Kawashima, T., Sasaki, A., Hamaguchi, M., Awazu, S., Yagi, K., Sasakua, Y., et al. 2002. A cDNA resource from the basal chordate Ciona intestinalis. Genesis 33 153-154. [DOI] [PubMed] [Google Scholar]
- Stein, L.D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M.R., Chen, N., Chinwalla, A., Clarke, L., Clee, C., Coghlan, A., et al. 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PloS Biol. 1 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoguchi, E., Ikuta, T., Yoshizaki, F., Satou, Y., Satoh, N., Asano, K., Saiga, H., and Nishikata, T. 2004. Fluorescent in situ hybridization to ascidian chromosomes. Zool. Sci. 21 153-157. [DOI] [PubMed] [Google Scholar]
- Shoguchi, E., Kawashima, T., Nishida-Umehara, C., Matsuda, Y., and Satoh, N. 2005. Molecular cytogenetic characterization of Ciona inestinalis chromosomes. Zool. Sci. 22 517-524.15930824 [Google Scholar]
- Vinson, J.P., Jaffe, D.B., O'Neill, K., Karlsson, E.K., Stange-Thomann, N., Anderson, S., Mesirov, J.P., Satoh, N., Satou, Y., Nusbaum, C., et al. 2005. Assembly of polymorphic genomes: Algorithms and application to Ciona savignyi. Genome Res. 15 1127-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]