Abstract
Coronavirus nucleoproteins (N proteins) localize to the cytoplasm and the nucleolus, a subnuclear structure, in both virus-infected primary cells and in cells transfected with plasmids that express N protein. The nucleolus is the site of ribosome biogenesis and sequesters cell cycle regulatory complexes. Two of the major components of the nucleolus are fibrillarin and nucleolin. These proteins are involved in nucleolar assembly and ribosome biogenesis and act as chaperones for the import of proteins into the nucleolus. We have found that fibrillarin is reorganized in primary cells infected with the avian coronavirus infectious bronchitis virus (IBV) and in continuous cell lines that express either IBV or mouse hepatitis virus N protein. Both N protein and a fibrillarin-green fluorescent protein fusion protein colocalized to the perinuclear region and the nucleolus. Pull-down assays demonstrated that IBV N protein interacted with nucleolin and therefore provided a possible explanation as to how coronavirus N proteins localize to the nucleolus. Nucleoli, and proteins that localize to the nucleolus, have been implicated in cell growth-cell cycle regulation. Comparison of cells expressing IBV N protein with controls indicated that cells expressing N protein had delayed cellular growth. This result could not to be attributed to apoptosis. Morphological analysis of these cells indicated that cytokinesis was disrupted, an observation subsequently found in primary cells infected with IBV. Coronaviruses might therefore delay the cell cycle in interphase, where maximum translation of viral mRNAs can occur.
Infectious bronchitis virus (IBV), a member of the Coronavirus genus of the Coronaviridae family, order Nidovirales (13), is an enveloped virus with a single-stranded, positive-sense RNA genome of 27,608 nucleotides (9) that is 5′ capped and 3′ polyadenylated which replicates in the cytoplasm of infected cells. The 5′ two-thirds of the IBV genome encodes the replicase-transcription complex, Rep1a and Rep1ab, the latter resulting from a −1 frameshift (10). During IBV replication, a 3′-coterminal nested set of six subgenomic mRNAs are synthesized that encode other viral proteins, including nucleoprotein (N protein). Recently, we have reported that IBV N protein localizes to the cytoplasm and a structure in the nucleus proposed to be the nucleolus both in IBV-infected cells and in cells transfected with a plasmid expressing IBV N protein under the control of a PolII promoter (26), a result subsequently confirmed in species-specific and -nonspecific cells expressing the mouse hepatitis virus (MHV) and porcine transmissible gastroenteritis virus (TGEV) N proteins (64).
The nucleolus is only present during interphase in mammalian cells (1) and is formed around ribosomal DNA repeats, which cluster at chromosomal loci called nucleolar organizer regions. It is the site where 5.8S, 18S, and 28S rRNAs are transcribed, processed, and assembled into ribosome subunits (11, 51). The nucleolus also sequesters regulatory complexes and has been implicated in the regulation of the cell cycle, telomerase activity, signal recognition particle biogenesis, small RNA processing, and mRNA transport (40, 41). The nucleolus is a dynamic structure composed of (or contains) at least 271 proteins (4), including nucleolin, fibrillarin, spectrin, B23, and the ribosomal proteins S5 and L9 (12, 51). Nucleolin (also called C23) represents 10% of the total nucleolar protein content, is highly phosphorylated and methylated, and also can be ADP-ribosylated (21). One of the main functions of nucleolin is processing the first cleavage step of rRNA in the presence of U3 snoRNP (21). Nucleolin may also function as a chaperone for correct folding in pre-rRNA processing (2). Fibrillarin is highly conserved in sequence, structure, and function in eukaryotes (5) and is directly involved in many posttranscriptional processes, including pre-rRNA processing, pre-rRNA methylation, and ribosome assembly (60).
As a consequence of infection, a number of viral proteins interact with the nucleolus and can reorganize nucleolar antigens (25), with examples from retroviruses, DNA viruses, and RNA viruses. These include human immunodeficiency virus type 1 (HIV-1) Rev (16) and tat (56), Newcastle disease virus matrix protein (42), adenovirus IVa2 gene product (37) and V protein (39), Marek's disease virus MEQ protein (36), hepatitis D virus large-delta antigen (54), and porcine reproductive and respiratory syndrome virus nucleocapsid protein (49). The nucleolus is also the site of Borna disease virus replication and transcription (47). Virus infection can also result in the redistribution of nucleolar antigens. For example, adenovirus infection results in the redistribution of nucleolin and B23 (38) and fibrillarin (46), and nucleolin is redistributed in poliovirus-infected cells (62). Many of these nucleolar antigens are involved in ribosome biogenesis (15) and possibly in cell division (65) and, thus, viruses might target these proteins in order to favor transcription or translation of virus mRNAs or possibly to alter the cell cycle machinery.
In this study we investigated whether the coronavirus N proteins associated with fibrillarin and/or nucleolin in the context of primary infected cells and in cells that expressed N protein. The coronavirus model chosen was IBV (Beaudette strain) because of its ability to grow in both primary cells (chicken kidney) and in continuous cells. Expression studies of IBV N protein were carried out in Vero cells, which are permissive for transfection (and infection). Where antibody combinations permitted, specific properties of the N protein were confirmed with MHV (JHM strain). In addition, because the nucleolus, and proteins that associate with it, have been implicated in cell cycle regulation, we sought to determine whether the IBV N protein disrupts host cell division. We show here for the first time that both IBV and MHV N proteins interact with fibrillarin and nucleolin and that IBV N protein delays cell growth, possibly by disrupting cytokinesis.
MATERIALS AND METHODS
Cells and viruses.
Vero (simian) and HeLa cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 5% fetal calf serum (FCS) at 37°C in a humidified atmosphere. Sf9 cells were cultured in Sf-900II SFM medium (Invitrogen) at 28°C. IBV Beaudette, a Vero cell-adapted strain, was grown in 11-day-old embryonated domestic fowl eggs at 37°C and harvested from allantoic fluid at 24 h postinfection (p.i.). The growth of IBV Beaudette in chicken kidney cells was performed as described previously (43). The recombinant baculovirus, BacIBVN, that expressed IBV N protein was grown as described previously (64).
Plasmids.
Procedures for recombinant DNA techniques were either standard (6, 50) or performed according to the manufacturer's instructions. The construction of pCi-IBV-N, pTriExIBVN, and pCi-MHV-N have been described previously (26, 64), in which the expression of either the IBV or MHV N protein is under the control of a PolII promoter. pCi-Neo was obtained from Promega. In the case of pTriExIBVN, the plasmid also contains a T7 promoter for expression of N protein in Escherichia coli; the N protein is expressed with a C-terminal His tag. The IBV Beaudette N gene was cloned directly into pcDNA4/HisMax-TOPO/lacZ to make pHis-IBV-N, by using the forward primer, ATGGCAAGCGGTAAAGCAGC, and reverse primer, TCAAAGTTCATTCTCTCCTA, according to manufacturer's instructions (Invitrogen) with pCi-IBV-N as a template and under cycling conditions described previously (26). pFibrillarin-GFP (green fluorescent protein [GFP]), a plasmid that expresses human fibrillarin under the control of a PolII promoter, was generously provided by Angus Lamond (University of Dundee). pTarget-CAT was constructed by PCR cloning of the chloramphenicol acetyltransferase (CAT) gene into pTarget (Promega), such that the expression of CAT was under the control of a PolII promoter.
Transfection and fixation.
Mammalian cells (105 cells per 9.6-cm2 dish) were grown on glass coverslips and transfected with 2 μg of plasmid DNA (in the case of two plasmid transfections, 1 μg of each plasmid was used) and 16 μg of Lipofectamine in Opti-MEM (Gibco) for 5 h and replaced with normal growth medium (DMEM-5% FCS) for 24 h prior to fixing with 50% methanol-50% acetone at −20°C.
Antibodies and immunofluorescence.
Coverslips were incubated for 1 h at 37°C with the appropriate primary antibody, washed three times for 10 min each in excess phosphate-buffered saline (PBS), incubated with the appropriate secondary antibody, and then washed three times for 10 min each in excess PBS. The following antibodies and dilutions (in PBS) were used. For primary antibodies, we used fibrillarin (human) mouse monoclonal antibody (diluted 1:100; Cytoskeleton Research), nucleolin (human) mouse monoclonal antibody (Leinco Technologies; diluted 1:160 and preblocked with 0.2% dry skimmed milk [Cadbury] in PBS for 15 min) (primary and secondary antibodies were incubated in blocking solution), rabbit anti-MHV polyclonal sera (diluted 1:100; generously provided by Peter Rottier, Utrecht University), and rabbit anti-IBV polyclonal sera (diluted 1:100). For secondary antibodies, we used fluorescein isothiocyanate (FITC)-labeled goat anti-mouse antibody (diluted 1:100; Harlan Sera-Lab), Alexa Fluor 564 goat anti-rabbit antibody (diluted 1:100; Molecular Probes), and Texas Red goat anti-mouse (diluted 1:100; Harlan Sera-Lab). Actin was visualized with TRITC (tetramethyl rhodamine isocyanate) conjugated to phalloidin (Sigma), and cells were incubated at 1 μg/ml in PBS for 30 min at room temperature. Annexin V-FITC for FACS was obtained from Clontech. Cells were either mounted in mounting media for fluorescence (Vectashield) or stained with propidium iodide (PI) to visualize nuclear DNA (Vectashield). Fluorescence microscopy was carried out with a Leica confocal microscope (TCS NT software) equipped with appropriate filter sets.
Cell proliferation assay.
Vero cells (60% confluence in a 10-cm dish) were transfected as described above, except that quantities were linearly scaled for the increased dish size, and incubated for a further 72 h in normal growth medium at 37°C. Cells were then washed twice with PBS, trypsinized with 2 ml of PBS-EDTA-trypsin, and centrifuged at 1,800 × g for 5 min; the pellet was then resuspended in 7 ml of Osmocell, and the number of cells was counted in a Coulter Counter according to manufacturer's instructions (Coulter Electronics).
Apoptosis assay.
Vero cells were transfected, as described above, at 60% confluence in a 10-cm dish and incubated in normal growth medium for a further 24 h. Cells were washed in PBS and removed from the dish by treatment with PBS-EDTA-trypsin, followed by inactivation with growth medium containing serum, and washed twice with binding buffer (Clontech). The proportion of cells that were apoptotic was determined by staining with Annexin V-FITC and PI according to the manufacturer's instructions (Clontech). To determine the proportion of cells expressing N protein, cells were resuspended in 875 μl of ice-cold PBS with 0.1% sodium azide (solution A) and fixed by the addition of 175 μl of ice-cold PBS with 2% paraformaldehyde (pH 7.4) for 1 h at 4°C (52). Fixative was removed by centrifugation for 5 min (250 × g, 4°C) and aspiration of the supernatant. Cells were permeabilized by resuspending them in 1 ml of PBS with 0.05% Tween 20 for 15 min at 37°C, and then they were washed with 1 ml of solution A and centrifuged for 8 min at 250 × g at 4°C. The presence of IBV N protein was detected by using rabbit polyclonal antibody against IBV (1:100) and goat anti-rabbit phycoerythrin (PE)-conjugated antibody (1:100; Sigma). A total of 10,000 cells were counted by using a Becton Dickson FACScan flow cytometer, and the results were analyzed by using the CellQuest software.
Purification of IBV N protein from either E. coli or Sf9 cells.
pTriExIBVN was transformed into the expression host strain, Tuner(DE3)pLacI (Novagen). The protein was expressed after induction with IPTG (isopropyl-β-d-thiogalactopyranoside). Sf9 cells were infected with BacIBVN (64) at a multiplicity of infection of 10 and incubated for 72 h. In both cases the His-tagged protein was purified by metal chelation chromatography with a His-Bind Kit (Novagen). Briefly, cells were harvested by centrifugation, resuspended in 20 mM Tris-HCl (pH 7.9)-0.5 M NaCl-5 mM imidazole, and sonicated. Unbound proteins were removed by washing, and N protein was recovered with elution buffer (20 mM Tris-HCl, pH 7.9; 0.5 M NaCl; 200 mM imidazole). The eluate was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Western blotting and was shown to contain a protein that corresponded in size to the predicted relative molecular mass of the IBV N protein. Treatment with calf intestinal alkaline phosphatase and mass spectroscopy revealed that N protein prepared from Sf9 was phosphorylated (termed Nphos protein), whereas N protein prepared from E. coli was not (termed Nnonphos protein) (data not shown).
Preparation of nuclear extracts.
Vero cell nuclear proteins were extracted with Nu-CLEAR Extraction Kit (Sigma) according to the manufacturer's instructions. Vero cells were cultured in DMEM with 10% FCS to reach 70 to 90% confluence. The cells were washed twice with PBS, scraped into a centrifuge tube, and collected by centrifugation for 5 min at 450 × g. The cell pellet was resuspended in a ×5 packed cell volume of lysis buffer (10 mM HEPES, pH 7.9; 1.5 mM MgCl2; 10 mM KCl; 1 mM dithiothreitol; protease inhibitor cocktail) and incubated on ice for 15 min. A 10% Igepal CA-630 solution was mixed with the cell suspension to a final concentration of 0.6%. The crude nuclei pellet was collected by centrifugation for 30 s at 10,000 × g, resuspended in ×2/3 packed cell volume of extraction buffer (20 mM HEPES, pH 7.9; 1.5 mM MgCl2; 0.42 M NaCl; 0.2 mM EDTA; 25% glycerol; 1 mM dithiothreitol; protease inhibitor cocktail), incubated on ice with regular stirring for 30 min, and centrifuged for 5 min at 20,000 × g. The supernatant was dialyzed against PBST (80 mM disodium hydrogen orthophosphate anhydrous; 20 mM sodium dihydrogen orthophosphate; 100 mM sodium chloride; 0.01% Tween 20, pH 7.5) for 6 h at 4°C. The precipitated proteins were removed by centrifugation for 5 min at 20,000 × g. Nuclear extracts were used in pull-down assays with IBV N protein (described below). These assays were conducted with nuclear extracts that either contained RNA or were RNA-free. In this case, extracts were treated with 7 U of RNase A (Sigma) per μl of extract.
Immobilization of N protein and pull-down assay.
Either C-terminal His-tagged IBV N protein, His-tagged HIV core protein, or His-tagged E. coli DucR (the latter two proteins generously provided by Ian Jones and Simon Andrews, respectively, University of Reading) were immobilized by adding 50 μl of a 5% Ni-nitrilotriacetic acid (NTA) magnetic agarose beads (Qiagen) suspension and 20 μg of purified protein into 500 μl of PBST, followed by shaking on an end-over-end shaker for 1 h at room temperature. The beads were collected on a magnetic separator and washed once with 500 μl of PBST. Dialyzed nuclear extract was added to the beads, incubated on a shaker for 1 h at room temperature. The bound proteins were washed once with 500 μl of PBST and then eluted with 50 μl of elution buffer (20 mM sodium phosphate, pH 7.4; 500 mM sodium chloride; 500 mM imidazole).
Western blotting.
Western blotting was performed by using the ECL Detection Kit (Amersham/Pharmacia) according to the manufacturer's instructions. Briefly, the protein sample to be analyzed was separated on a 10% Tris-HCl precast polyacrylamide gel (Bio-Rad), and SDS was present in the loading and running buffer. The proteins were transferred onto an Immobilon-P Transfer Membrane (Millipore) in 39 mM glycine-48 mM Tris base-0.01% SDS-20% methanol for 1 h at room temperature. The membrane was blocked with 5% milk-PBS plus 0.1% Tween 20 for 1 h at room temperature. Primary antibody was diluted in blocking solution and incubated with the membrane for 2 h at room temperature. Secondary antibody incubation was performed with a 1:2,000 dilution of the appropriate antibody conjugated to horseradish peroxidase in PBS for 1 h at room temperature. The membrane was then washed three times in PBST for 10 min. The membrane was then treated with ECL reagents (Amersham/Pharmacia) and subjected to autoradiography. Guinea pig anti-IBV polyclonal sera (generously provided by D. Cavanagh, IAH Compton) was used as the primary antibody to detect IBV N protein (diluted 1:1,000) and C23 (human) goat polyclonal antibody (Santa Cruz Laboratories) was used as the primary antibody to detect nucleolin (diluted 1:500).
RESULTS
As a consequence of virus infection, a number of viruses reorganize nucleolar antigens and use these antigens to enter the nucleolus. We sought to investigate whether the same events occurred in cells infected with the coronavirus IBV and, because N protein localized to the nucleolus, whether these antigens played a role in this process. We investigated the potential interaction of IBV with two nucleolar antigens: fibrillarin and nucleolin. Because proteins that associate with the nucleolus have been implicated in the control of cell growth and/or regulation of the cell cycle, we examined the potential effect of N protein on cell division.
Distribution of fibrillarin in cells infected with IBV and in cells that express IBV N protein.
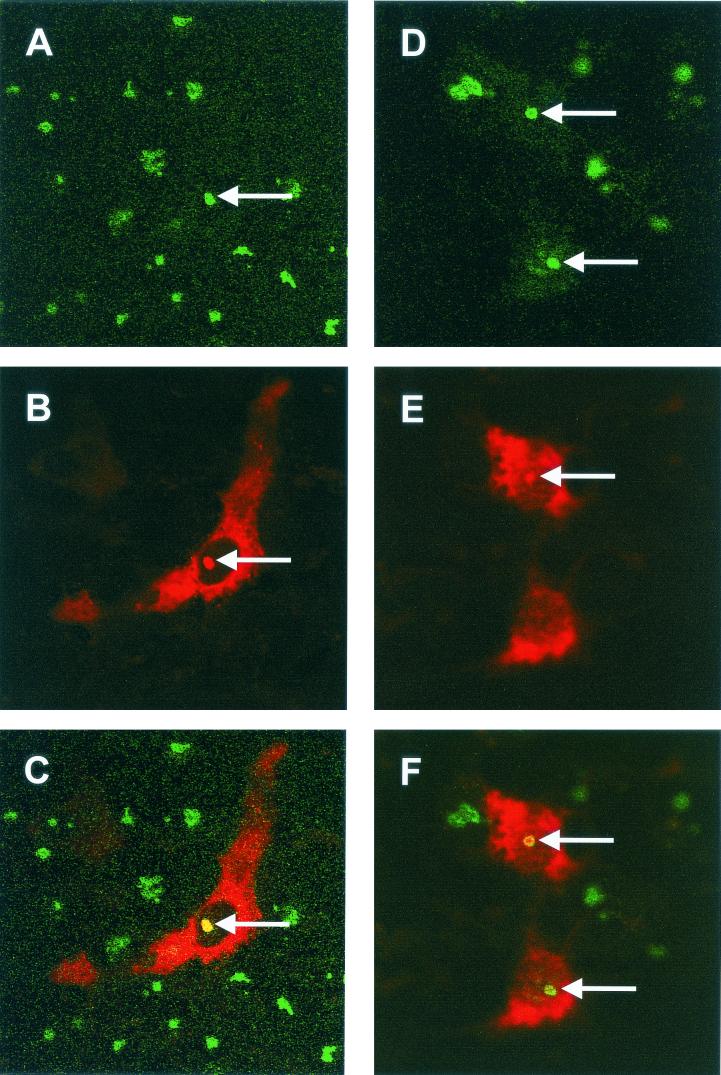
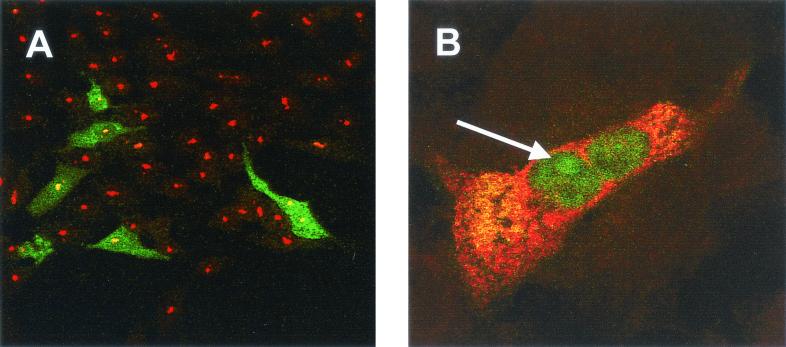
Primary chicken kidney cells were infected with IBV Beaudette as described previously (43), fixed 8 h p.i., and analyzed by indirect immunofluorescence by staining for fibrillarin (green) (Fig. 1A and D) and IBV proteins (red) (Fig. 1B and E) with colocalization of both proteins, if it occurred, appearing in yellow (Fig. 1C and F). Nucleoli were identified as distinct regions within the nucleus in which localization of IBV proteins to the nucleolus was confirmed, as indicated by an arrow (Fig. 1A to F). Based upon the expression profile of the IBV N protein from expression plasmids the most likely IBV protein, in the absence of other IBV proteins, observed to be localized in the nucleoli of infected cells is the N protein (26, 64). Interestingly, the pattern of fibrillarin staining in infected cells (Fig. 1A and D) appeared to be generally uniform in appearance, with perhaps higher concentrations around the periphery of the nucleolus, compared to adjacent uninfected or mock-infected cells (Fig. 2F), where fibrillarin was present as a characteristic globular (Christmas tree-like) structure (5). Analysis of 20 fields of view in duplicate experiments indicated that the Christmas tree structure of fibrillarin was never observed in infected cells.
FIG. 1.
Primary chicken kidney cells were infected with IBV, fixed, and analyzed for indirect immunofluorescence with rabbit anti-IBV polyclonal sera (red), and fibrillarin was detected by using anti-fibrillarin (human) mouse monoclonal antibody (green). Differentially fluorescing images were gathered separately from the same 0.5-μm-thick optical section by using a confocal microscope and the appropriate filter. Two pairs of images (A+B and D+E) were digitally superimposed to depict the distribution of IBV and fibrillarin proteins (C and F). Colocalization when it occurs is shown in yellow. The arrows indicate the position of the same nucleolus in the respective optical sections. Magnification, ×60.
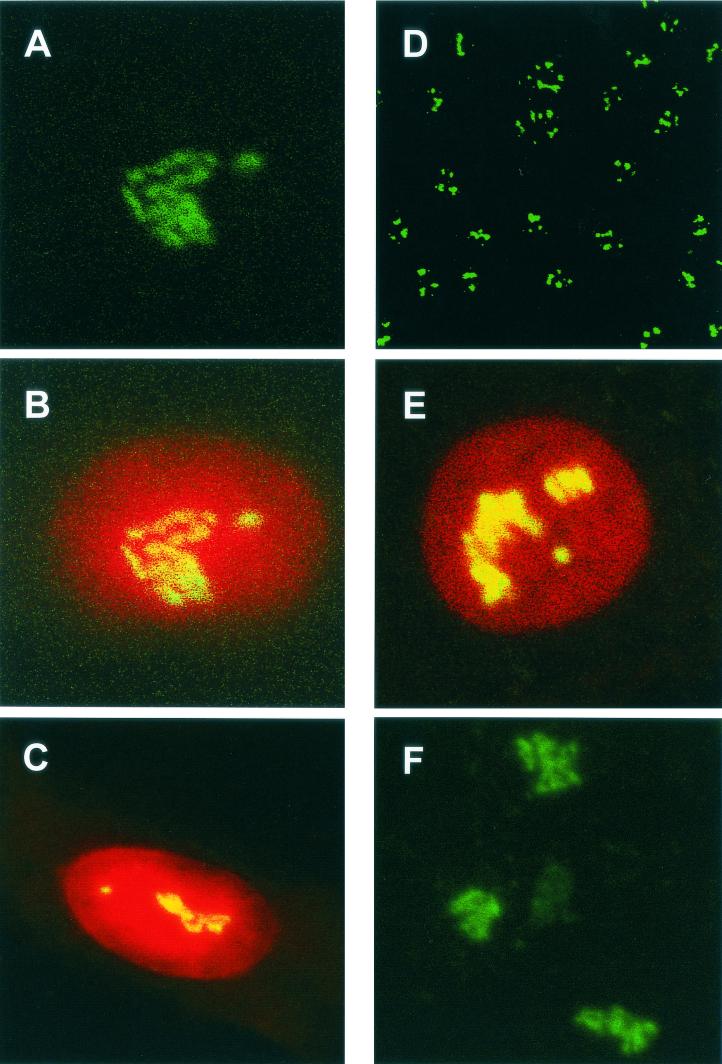
FIG. 2.
Detection of fibrillarin by indirect immunofluorescence in mock-transfected Vero cells (A to C) and HeLa cells (D to E) and in mock-infected chicken kidney cells (F). Fibrillarin was detected with anti-fibrillarin (human) mouse monoclonal antibody (green). The nucleus was visualized by staining with PI (B, C, and E). Colocalization when it occurs is shown in yellow. Magnifications: A to C, E, and F, ×60; D, ×15.
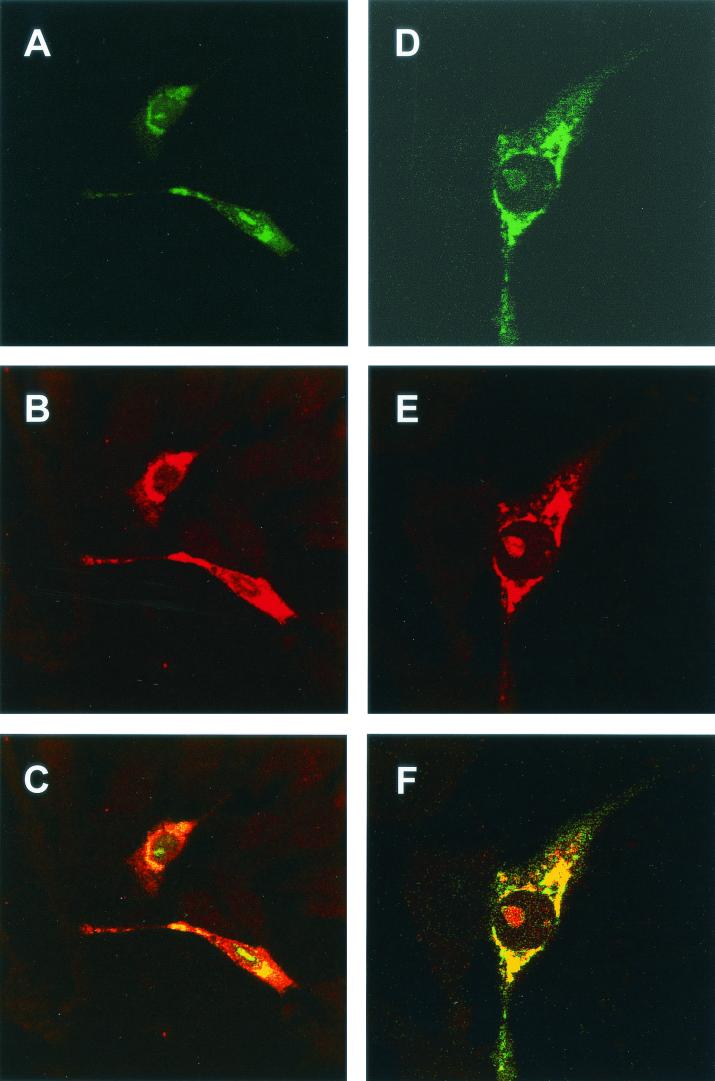
To investigate the pattern of fibrillarin in the presence of IBV N protein, HeLa cells (105 cells per 9.6-cm2 dish) were transfected with pCi-IBV-N, fixed, and analyzed by indirect immunofluorescence to detect IBV N protein (red) and fibrillarin (green). IBV N protein localized to the cytoplasm and colocalized with fibrillarin in the nucleolus (Fig. 3A). Similar to the result observed in primary cells infected with IBV, the pattern of fibrillarin was different compared to adjacent cells (Fig. 3A, arrowed) or mock-transfected cells, in which fibrillarin is shown in green and the nucleus is shown in red (Fig. 2E; also see Fig. 2D for the low-power image [×15.2] of fibrillarin in HeLa cells). These results indicated that fibrillarin does not form the characteristic Christmas tree-like structure (5) in the presence of N protein.
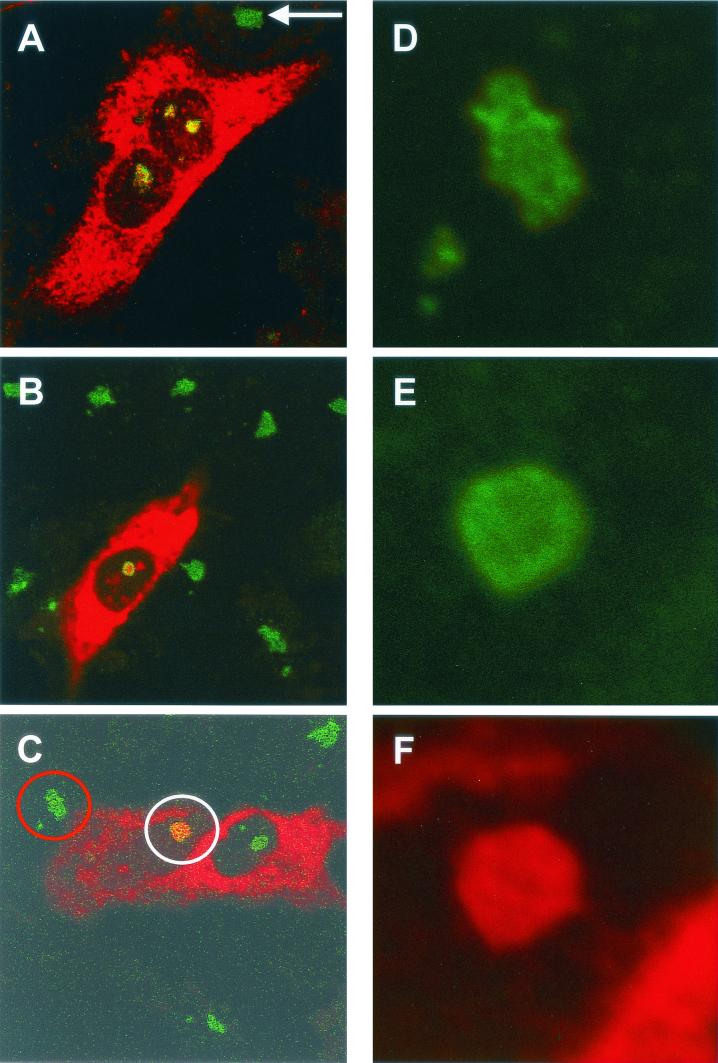
FIG. 3.
HeLa (A and C to F) or Vero (B) cells were transfected with either pCi-MHV-N (B to F) or pCi-IBV-N (A) and then fixed and analyzed by indirect immunofluorescence with rabbit anti-MHV or anti-IBV polyclonal sera (red). The nucleolus was detected with anti-fibrillarin (human) mouse monoclonal antibody (green). The structures identified by the red and white circles in panel C were resolved an additional 12 times (panel D and panels E and F, respectively). Colocalization when it occurs is shown in yellow. The arrow indicates a nucleolus in an adjacent untransfected cell. Magnifications: A, B, and C, ×60 (zoom, ×2).
Interaction of MHV N protein with fibrillarin and nucleolin.
To investigate whether the nonglobular pattern of fibrillarin was restricted to IBV N protein or whether it was was a more common feature of coronaviruses, Vero and HeLa cells (105 cells per 9.6-cm2 dish) were transfected with pCi-MHV-N for 24 h and fixed for analysis by indirect immunofluorescence. MHV N is shown in red, and fibrillarin is shown in green (Fig. 3B [Vero] and C [HeLa]). Analysis of image Fig. 3C at a higher resolution indicated that the distribution of fibrillarin was uniform in the nucleolus of cells expressing N protein, with perhaps a slightly higher concentration around the periphery, (Fig. 3E, denoted by a white circle in Fig. 3C) compared to adjacent cells, which did not display evidence of N protein expression (Fig. 3D, denoted by a red circle in Fig. 3C), or mock-transfected HeLa cells (Fig. 2E). Similar to our previously published results (26, 64), the distribution of MHV N protein appeared to be uniform in the nucleolus (Fig. 3F). Again, in mock-transfected Vero cells (Fig. 2A, B, and C) the distribution of fibrillarin in the nucleolus was Christmas tree-like (5). However, the distribution of fibrillarin in the nucleolus in cells expressing MHV N protein in either Vero or HeLa cells was similar to that observed in primary cells infected with IBV or HeLa cells expressing IBV N protein, indicating that this modified distribution of fibrillarin is a potential common feature of the two coronaviruses and not unique to any particular cell line.
It was possible to investigate, by using the MHV system in HeLa cells, whether N protein redistributed nucleolin in cells transfected with MHV N protein. We have previously shown that human anti-nucleolin monoclonal antibody (Leinco) can be used as a marker for nucleoli in HeLa cells transfected with pCi-MHV-N (64). Unfortunately, this was not possible in Vero cells expressing IBV N protein because the monoclonal antibody to human nucleolin did not recognize the simian form. Similar to our previous findings (64), MHV N protein did not localize to the nucleolus in all cells that showed evidence of N protein expression (e.g., Fig. 4A and B). Comparison of nucleolin in these cells with mock-transfected cells (Fig. 4B, inset panel) indicated that the distribution of nucleolin was unchanged. Interestingly, MHV N protein also formed a speckled pattern in the nucleus of two transfected cells (Fig. 4B), and these may correspond to Cajal bodies or ND-10 domains (31). We also observed such speckles in Vero cells that express IBV N protein (25).
FIG. 4.
HeLa cells were transfected with pCi-MHV-N (A and B [except inset panel]), fixed, and analyzed by indirect immunofluorescence with rabbit anti-MHV polyclonal sera (red). The nucleolus was detected with anti-nucleolin (human) mouse monoclonal antibody (green). (A) Colocalization when it occurs is shown in yellow and is denoted by an arrow. (B) Inset panel is from mock-transfected HeLa cells. Magnifications: A and B, ×15 (resolved an additional four times) and ×60, respectively.
MHV and IBV N proteins associate with fibrillarin in the perinuclear region (and nucleolus).
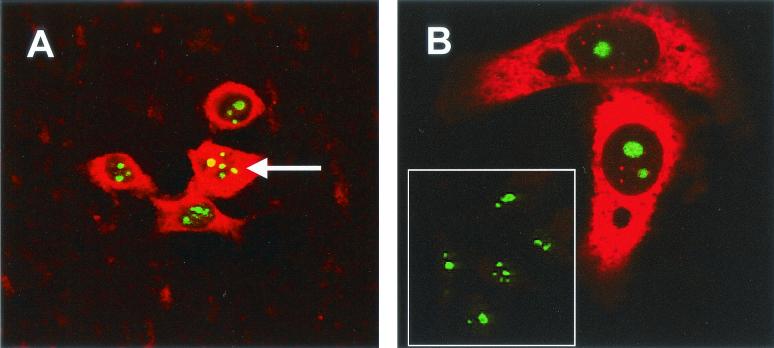
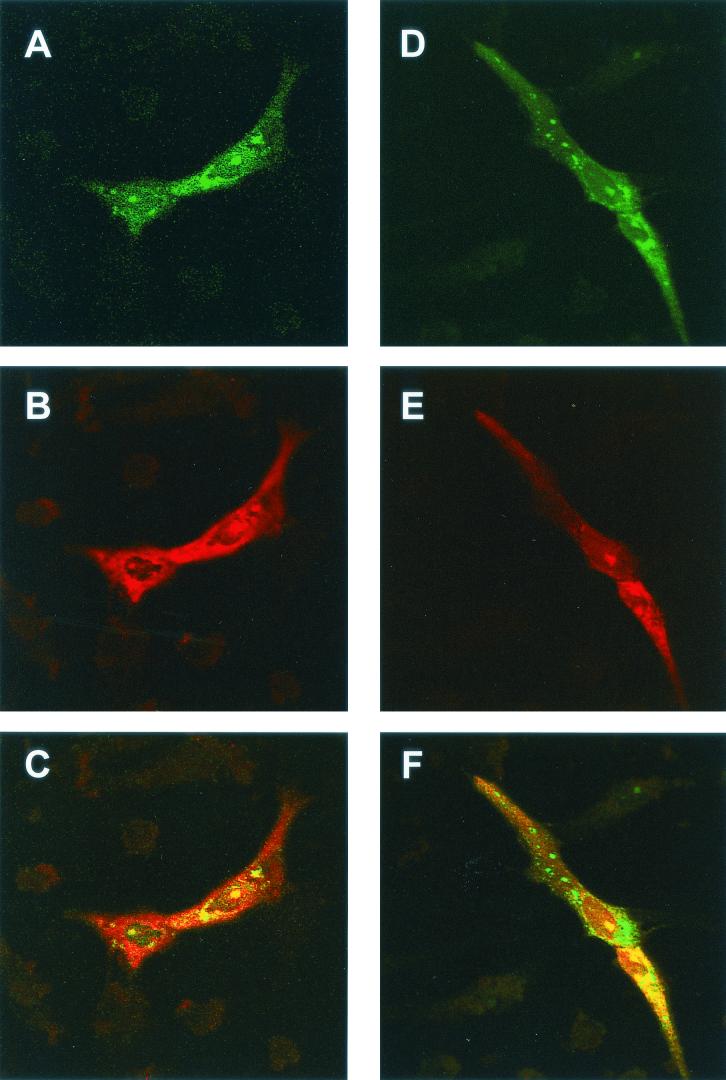
As a result of fibrillarin containing nucleolar localization signals and localizing to the nucleolus, fibrillarin within the cytoplasm is difficult to detect with specific antibodies. However, to investigate whether N protein and fibrillarin colocalized in the cytoplasm, we made use of the plasmid pFibrillarin-GFP, which expressed a fibrillarin-GFP fusion protein under the control of a PolII promoter. Confocal microscopy showed that the fibrillarin-GFP fusion protein localized both to the nucleolus and to the perinuclear region. However, anti-human fibrillarin antibody (Fig. 5A) detected fibrillarin in the nucleolus but not the cytoplasm. Fibrillarin-GFP may have accumulated in the cytoplasm because it was expressed in excess, because the presence of the GFP moiety reduced the efficiency of nucleolar localization, or because the presence of GFP altered the structure of fibrillarin. However, GFP-tagged nucleolar antigens, including fibrillarin, have been used successfully to investigate their functions and distribution (15, 45, 58). With these potential caveats in mind, Vero cells were transfected with pFibrillarin-GFP and either pCi-IBV-N or pCi-MHV-N. Confocal microscopy showed that the fibrillarin-GFP fusion protein localized both to the nucleolus and the perinuclear region (Fig. 6A and D and 7A and D). Both IBV and MHV N proteins localized to the cytoplasm and the nucleolus (Fig. 6B and E and 7B and E, respectively). Both N proteins and the GFP-fibrillarin fusion protein colocalized in the perinuclear region and nucleolus (Fig. 6C and F and 7C and F). Interestingly, the GFP-fibrillarin fusion protein localized preferentially to the nucleus and nucleolus (arrowed) in cells identified as undergoing aberrant cell division (Fig. 5B) (64). Unfortunately, the antibody available to detect fibrillarin by using immunofluorescence, when used in Western blots is known to cross-react with arginine-rich proteins (Cytoskeleton, Inc., and our unpublished data), and therefore we could not investigate whether N protein interacted directly with fibrillarin rather than just undergoing colocalization.
FIG. 5.
Vero cells were transfected with pFibrillarin-GFP (A and B) and pCi-IBV-N (B), fixed, and analyzed by direct fluorescence to visualize GFP and by indirect immunofluorescence with rabbit anti-IBV polyclonal sera (red) (B), and fibrillarin was detected with anti-human antibody (red) (A). Colocalization when it occurs is shown in yellow. The arrow indicates the position of a nucleolus. Magnifications: A, ×15; B, ×60 (zoom, ×4).
FIG. 6.
Vero cells were transfected with pFibrillarin-GFP and pCi-IBV-N, fixed, and analyzed by indirect immunofluorescence with rabbit anti-IBV (red) and direct fluorescence to visualize GFP. Differentially fluorescing images were gathered separately from the same 0.5-μm-thick optical section by using a confocal microscope and the appropriate filter. Two pairs of images (A+B and D+E) were digitally superimposed to depict the distribution of IBV N protein and fibrillarin-GFP fusion protein (C and F, respectively). Magnifications: ×60 (all panels) (zoom, ×2).
FIG. 7.
Vero cells were transfected with pFibrillarin-GFP and pCi-MHV-N, fixed, and analyzed by indirect immunofluorescence with rabbit anti-MHV (red) and direct fluorescence to visualize GFP. Differentially fluorescing images were gathered separately from the same 0.5-μm-thick optical section by using a confocal microscope and the appropriate filter. Two pairs of images (A+B and D+E) were digitally superimposed to depict the distribution of MHV N protein and fibrillarin-GFP fusion protein (C and F, respectively). Magnification, ×60 (all panels).
IBV N protein interacts with nucleolin.
The observations that both IBV and MHV N proteins colocalized with fibrillarin (e.g., Fig. 3, 6, and 7) and that MHV N protein colocalized with nucleolin (Fig. 4A) were consistent with data presented previously in which coronavirus N protein localized to the nucleolus (64) and suggested that the N protein interacted with these nucleolar antigens. Therefore, we investigated whether phosphorylated (Nphos) or nonphosphorylated (Nnonphos) protein could interact directly with nucleolin. We used a recombinant baculovirus, BacIBVN, which expresses IBV N protein with a C-terminal His tag (64), to infect insect (Sf9) cells. If N protein is expressed in insect cells it should be phosphorylated. The vector used in construction of BacIBVN, pTriExIBVN, was also used to express N protein in E. coli, which presumably would be nonphosphorylated. In the present study we used recombinant IBV N protein purified from either E. coli or insect cells to study potential interactions between immobilized Nphos and Nnonphos protein and nucleolin derived from nuclear extracts of Vero cells, a model cell system for IBV infection.
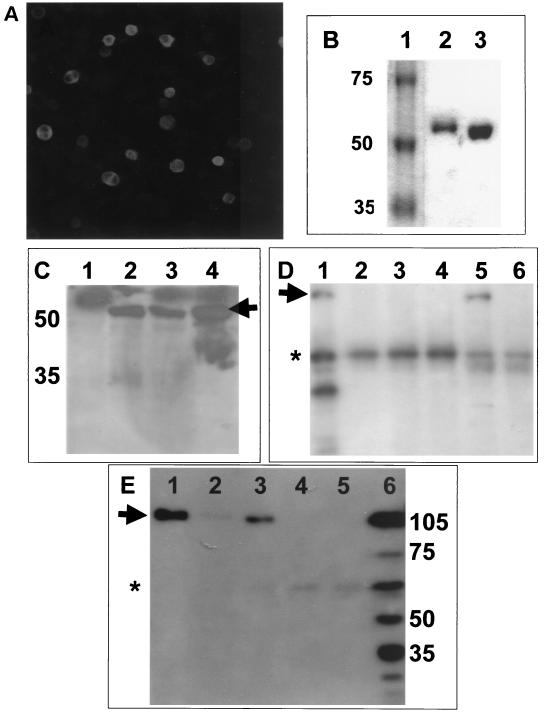
Insect cells were infected with BacIBVN, and expression of N protein was confirmed by using immunofluorescence with rabbit anti-IBV polyclonal sera (Fig. 8A). Tuner(DE3)pLacI (Novagen) was transformed with pTriExIBVN. N protein was purified from these cells by His-tag affinity binding chromatography. Comparison of the electrophoretic mobilities of N protein purified from Sf9 cells (Fig. 8B, lane 2) versus that of N protein purified from E. coli (Fig. 8B, lane 3) indicated that N protein purified from insect cells was slower migrating and therefore had a larger apparent molecular weight, which was possibly due to phosphorylation. The electrophoretic mobility of purified N protein from insect cells (Fig. 8C, lane 4) was compared to N protein isolated from either Vero cells that had been transfected with pTriExIBVN (the construct used to create the recombinant baculovirus (64) (Fig. 8C, lane 3) or Vero cells transduced with BacIBVN (Fig. 8C, lane 2). As a control Vero cells were transfected with pTri-Ex, the backbone vector used to express N protein; no protein corresponding in mobility to N protein was detected (Fig. 8C, lane 1). There were no apparent differences in mobility of the N proteins, indicating that full-length recombinant N protein was produced in insect cells. The mobility of these proteins corresponded in mobility to N protein from infected cells (data not shown). Mass spectroscopic analysis confirmed that N protein purified from Sf9 cells was phosphorylated, whereas N protein purified from E. coli was not (data not shown).
FIG. 8.
(A) Insect cells were infected with the recombinant baculovirus, BacIBVN, which expressed IBV N protein. Magnification, ×9.6. (B) Comparison of the electrophoretic mobility of recombinant His-tagged N protein purified from either Sf9 cells (lane 2) or E. coli (lane 3) by SDS-polyacrylamide gel electrophoresis. The molecular weight marker is shown lane 1, and the corresponding mass is shown on the left. (C) Western blot analysis with polyclonal guinea pig anti-IBV sera of Vero cells transfected with pTRiEx1.1 (lane 1), Vero cells transduced with BacIBVN (lane 2), Vero cells transfected with pTriExIBVN (lane 3), or N protein purified from insect cells infected with BACIBVN (lane 4). The positions of the molecular mass markers (in kilodaltons) are shown to the left, and the arrow indicates the position of the IBV N protein. (D) Western blot analysis of nucleolin from RNase-treated nuclear extract (lane 1); RNase-treated nuclear extract passed over NTA beads (lane 2), immobilized DcuR protein (lane 3), immobilized HIV core protein (lane 4), and immobilized phosphorylated IBV N protein (lane 5); and untreated extract passed over immobilized phosphorylated IBV N protein (lane 6). The position of mature nucleolin (∼105 kDa) is indicated by an arrow. The asterisk indicates nonspecific binding of an unidentified protein of ∼60 kDA. (E) Western blot analysis of nucleolin from RNase-treated nuclear extracts passed over immobilized phosphorylated (lane 1) and nonphosphorylated (lane 3) N protein and NTA beads (lane 5); nucleolin from untreated extracts passed over immobilized phosphorylated (lane 2); and nonphosphorylated (lane 4) N protein. Nucleolin from whole-cell lysate is shown in lane 6. The positions of molecular mass markers (in kilodaltons) are shown to the right, and the arrow indicates the position of mature nucleolin. The asterisk indicates nonspecific binding of an unidentified protein of ∼60 kDA.
Nuclear extracts were prepared from Vero cells and, in some cases, treated after extraction and purification with RNase A, which was reported to induce the release of nucleolin from nucleoli (50). As a control, nuclear extract was passed over the NTA beads in the absence of any immobilized protein. Recombinant His-tagged N protein was immobilized on NTA beads (Qiagen), mixed with nuclear extracts prepared from Vero cells, and then washed. Recombinant His-tagged HIV core protein and E. coli DcuR (22) were used as controls to test for specificity of nucleolin binding. Bound protein was eluted and analyzed by Western blotting with goat polyclonal anti-human C23 (nucleolin; Santa Cruz Laboratories).
The results indicated that both mature nucleolin (∼105 kDa) and the nonphosphorylated form (∼75 kDa) were present in the RNase-treated nuclear extracts (Fig. 8E, lane 6) (20, 21). In addition, several faster-migrating protein species were detected with the anti-nucleolin antibody, and these were probably autocatalytic breakdown products of nucleolin (14, 21). No mature nucleolin bound to the NTA beads (Fig. 8D, lane 2), immobilized E. coli DcuR protein (Fig. 8D, lane 3), or HIV core protein (Fig. 8D, lane 4). Nonspecific binding of an unidentified protein of ∼60 kDa was apparent in all three controls (Fig. 8D, lanes 2, 3, and 4 [indicated by an asterisk]). However, this protein did not correspond in mobility to either mature or full-length nucleolin. In addition, the amount of this protein or the degree of nonspecific binding varied between experiments. For example, compare Fig. 8D and E (the position of the nonspecific binding protein marked by an asterisk). In contrast, nucleolin was detected when Nphos protein was immobilized to the NTA beads (Fig. 8D, lane 5), indicating that nucleolin formed a specific interaction with the N protein. Next, we passed untreated nuclear extract over immobilized Nphos protein, and the results indicated that less or no nucleolin bound (Fig. 8D, lane 6) compared to the RNase-treated extract (Fig. 8D, lane 5). This is probably attributable to less nucleolin being accessible from nontreated extracts (53).
Next we compared the binding of immobilized Nphos and Nnonphos protein with nucleolin from RNase-treated and untreated extracts. The data indicated that more nucleolin bound to both types of immobilized N protein from the RNase-treated extracts than the nontreated extracts (compare Fig. 8E, lane 1 with lane 2 and lane 3 with lane 4). In addition, the data indicated that Nphos protein bound more nucleolin than Nnonphos protein (compare Fig. 8E, lane 1 with lane 3, respectively). As a control, untreated nuclear extract was passed over the NTA beads in the absence of any immobilized protein; no nucleolin binding was detectable (Fig. 8E, lane 5). No nucleolin was observed to bind to either immobilized DcuR or HIV core protein (Fig. 8D, lane 3 and 4, respectively), indicating that nucleolin specifically associated with either form of N protein.
IBV N protein delays cell growth.
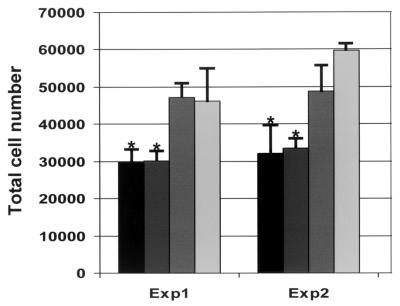
The nucleolus and proteins that associate with it have been implicated in the regulation of the cell cycle (11). We have previously shown that TGEV N protein caused aberrant cell division by disrupting cytokinesis and we hypothesized that this might affect cell growth (64). We wanted to investigate whether IBV N protein affected cell division, either by causing increased or decreased cellular proliferation. To investigate these possibilities, Vero cells were transfected separately with two plasmids that expressed IBV N protein but differed in vector backbone; pCi-IBV-N, which contains the neomycin phosphotransferase gene (which confers resistance to G418), and pHis-IBV-N, which contains the Streptoalloteichus hindustanus bleomycin gene (which confers resistance to Zeocin). pHis-IBV-N was constructed by insertion of a PCR product corresponding to the IBV N gene into pcDNA4/HisMax-TOPO/lacZ. As a control, cells were transfected with vector alone (pCi-Neo; Promega) or mock transfected with Lipofectamine reagent alone. Total cell numbers are shown from triplicate wells, from two separate experiments (Fig. 9). Analysis of variance and the Bonferonni t test indicated that the numbers of cells with the plasmids expressing IBV N protein are significantly less than the numbers of cells transfected with vector alone (P < 0.05).
FIG. 9.
Vero cells were transfected for each experiment in triplicate with pCi-IBV-N (black), pHis-IBV-N (dark gray), pCi-Neo (medium gray), or mock transfected (light gray) and then incubated for 72 h, and the number of cells was determined in a Coulter counter. The means and standard deviations (indicated by the error bars) for each triplicate (labeled Exp1 and Exp2) are shown separately. ∗, P < 0.05 compared to mock-transfected cells, analysis of variance, and Bonferonni t test.
Three possibilities could account for the observation that N protein caused a decrease in cell numbers: (i) a foreign gene is expressed from a cytomegalovirus (CMV) promoter; (ii) the N protein induces apoptosis; and (iii) the N protein delays or arrests the cell cycle. To investigate whether the expression of a second foreign gene could account for the reduction in cell numbers, we compared the number of cells after 72 h which had been either mock transfected or transfected with pTarget-CAT (a vector which expressed CAT, as well as the neomycin phosphotransferase gene). No significant difference was observed between the number of mock-transfected cells (n = 69,784) and the number of cells transfected with pCAT (n = 67,849).
N protein does not induce apoptosis.
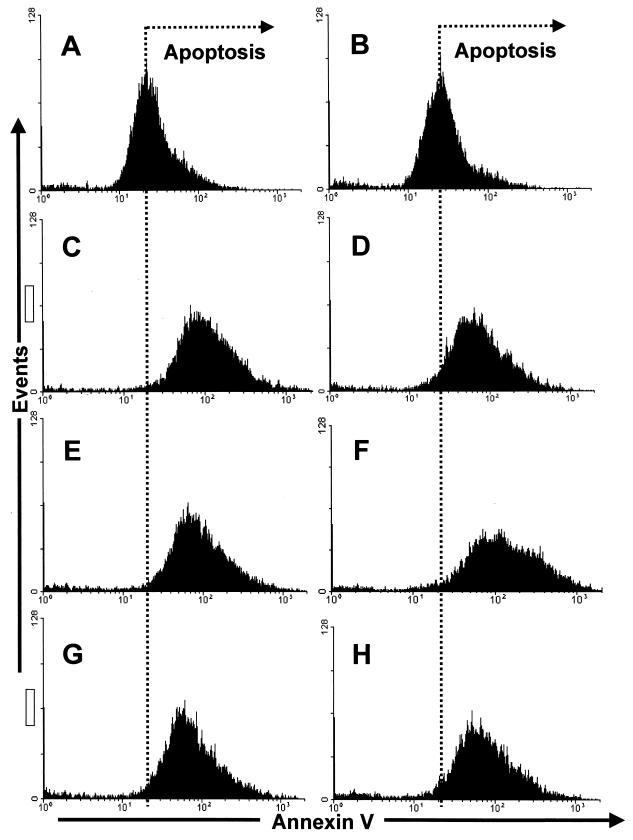
Three different coronaviruses have been shown to induce apoptosis in host cells: IBV (34), MHV (8), and TGEV (17). However, the potential role of N protein in virus-induced apoptosis was not elucidated in these studies. To investigate whether the decrease in cell number associated with the expression of N protein was a result of apoptosis, Vero cells were (i) transfected in duplicate with either pCi-IBV-N or backbone vector (pCi-Neo), (ii) mock transfected, or (iii) received no treatment. Cells were harvested 24 h after transfection, and apoptosis was measured by using Annexin V staining. Fluorescence-activated cell sorting (FACS) analysis (Fig. 10) indicated that, while there was a difference in the number of apoptotic cells observed between those cells that had been transfected and those cells that had not (compare panels A and B [no Lipofectamine treatment] with panels C to H [Lipofectamine treatment]), there was no significant difference between transfected cells that expressed N protein and those that did not. For example, compare panels E and F, for which cells were transfected with backbone vector, to panels G and H, for which cells were transfected with pCi-IBV-N. As a positive control, we induced apoptosis with etoposide (Sigma) (data not shown).
FIG. 10.
Profile of Annexin V staining in control Vero cells (A and B), mock-transfected Vero cells (C and D), Vero cells transfected with pCi-Neo (E and F), or Vero cells transfected with pCi-IBV-N (G and H). Cells were analyzed by flow cytometry for their ability to bind Annexin V 24 h after transfection in accordance with the manufacturer's guidelines (Clontech).
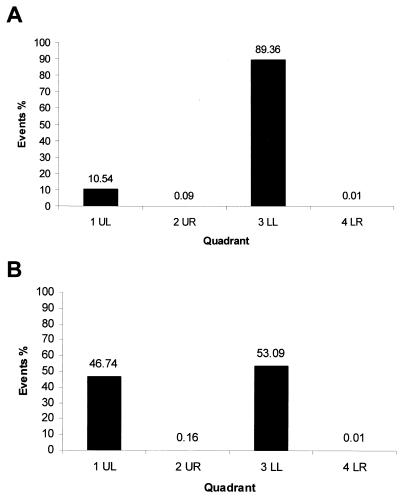
However, Annexin V staining may have reflected the possible redistribution of phosphatidylserine caused by Lipofectamine treatment rather than an induction of apoptosis. To investigate this possibility, we compared the proportion of cells that received Lipofectamine only to the proportion of cells that expressed IBV N protein (i.e., received Lipofectamine and pTriExIBVN) (Fig. 11.). N protein was detected by FACS by permeabilizing the cells and staining them with anti-N antibody and with PE as a secondary antibody. The data indicated that ca. 10% of the mock-transfected cell population was stained with PE (Fig. 11A). However, in populations of cells transfected with pTriExIBVN this value increased to ca. 46%, suggesting that ca. 36% of cells had been transfected and were expressing N protein (Fig. 11B). This result agreed with the transfection efficiency as determined by immunofluorescence (data not shown). If N protein had induced apoptosis and/or late apoptosis or necrosis, then the proportion of cells in 4LR or UR columns, respectively, would have increased. No significant increase in these numbers was observed. Taken together, these results suggested that N protein did not induce apoptosis and therefore could not account for the reduction in cell proliferation in cells that expressed IBV N protein.
FIG. 11.
Histogram of Vero cells that stain with polyclonal antibody to IBV N protein conjugated to PE in mock-transfected cells (A) and in cells which express N protein (B) (1UL). 3LL denotes the number of untransfected cells. The proportion of apoptotic cells is shown in 4LR and apoptotic-necrotic cells in 2UR. (UL, LL, UR, and LR refer to the quadrants in the dot plot generated by the CellQuest software.)
N protein disrupts cytokinesis.
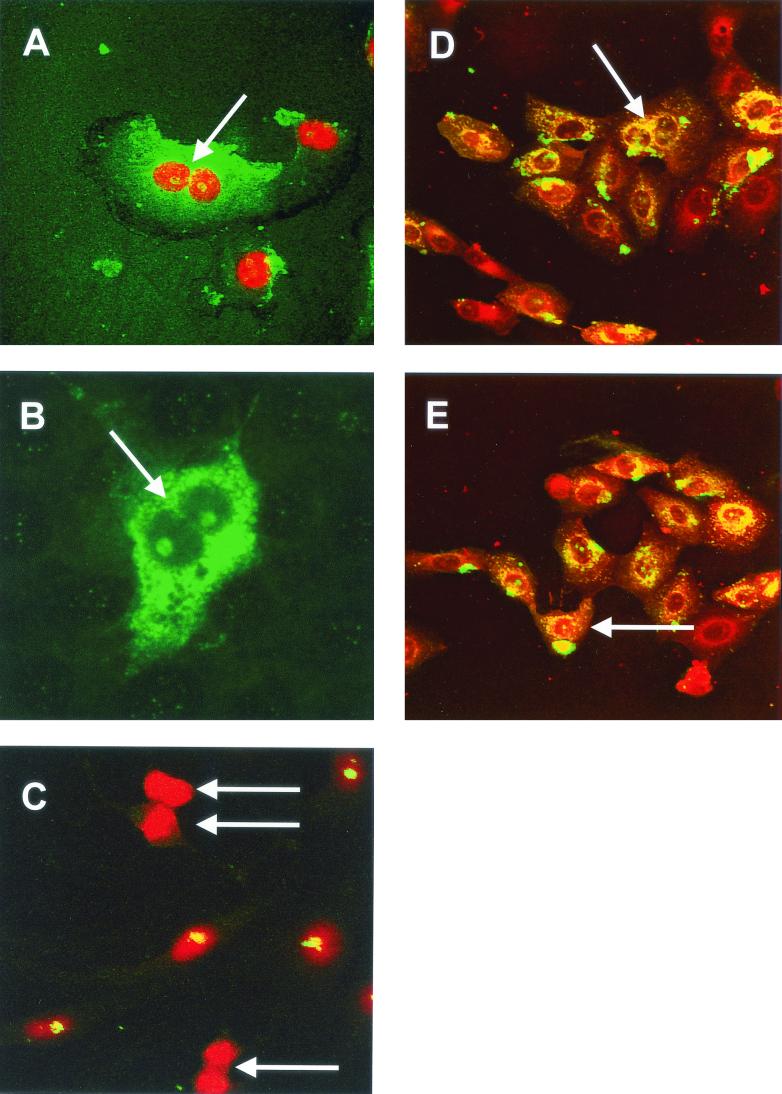
We next investigated whether the decrease in cellular proliferation caused by expression of N protein was caused by a disruption in cytokinesis. To investigate this hypothesis, we used confocal microscopy to analyze the morphology of Vero cells either transfected with pCi-IBV-N or infected with IBV. In the case of IBV-infected cells, PI was used to visualize the nucleus as described previously (26). A major sign of cytokinesis occurring is the presence of a cleavage furrow. In both infected cells (Fig. 12A and D) and in transfected cells expressing N protein (Fig. 12B), we found evidence of aberrant cell division, consistent with the fact that cytokinesis has been inhibited in these cells (ca. 19% of transfected cells exhibited this characteristic), which is also in accordance with our previously published observations of cell morphology changes in transfected cells expressing TGEV N protein (64). Both groups of cells contained a cleavage furrow (Fig. 12A and B, arrow) but also displayed a nucleolus in each nucleus, which should be absent during cell division (1). One of the main constituents of a cleavage furrow is actin, and this feature was confirmed in primary chicken kidney cells that had been infected with IBV and stained with TRITC-phalloidin to visualize actin (red); IBV proteins are shown in green, and colocalization, where it occurs, is shown in yellow (Fig. 12D, the cleavage furrow is arrowed).
FIG. 12.
Vero cells were either infected with IBV (A) or transfected with pCi-IBV-N (B) and then incubated for 24 h; IBV proteins are shown in green, and nuclear DNA was visualized with PI (A). Cleavage furrows are indicated by an arrow in panels A and B. (C) Mock-transfected cells were stained for fibrillarin (green) and nuclear DNA (red) by using PI; dividing cells are indicated by an arrow. (D and E) Primary chicken kidney cells were infected with IBV for 8 h, and cells were stained for IBV (green) or actin (red). An arrow indicates a cleavage furrow in panel D and a nucleolus in E. Magnifications: A and B, ×60; C, D, and E, ×15.
The absence of nucleoli in the division of Vero cells was confirmed by examining the distribution of nucleolar proteins in mock-transfected cells undergoing division by visualizing fibrillarin (green) and nuclear DNA with PI as described above (Fig. 12C, cells undergoing division are indicated by an arrow). Fibrillarin (and by inference nucleoli) was not observed in dividing cells, whereas nondividing cells contained fibrillarin, observed as yellow structures within the nucleus of each cell. Inhibition of cytokinesis was not observed in cells that had been transfected with a plasmid expressing GFP under the control of a CMV promoter (64), indicating that overexpression of a protein from a CMV promoter did not lead to the observed effects.
DISCUSSION
The interaction of viral proteins with nucleolar antigens may account for why viral proteins have been observed in the nucleolus and may also explain the viral exploitation of nucleolar function, leading to alterations in host cell transcription, translation, and disruption of the host cell cycle to facilitate viral replication. In this study we wanted to investigate whether coronavirus N proteins interacted with two nucleolar antigens, fibrillarin and nucleolin. Interaction with either or both of these antigens might explain our previous observations that coronavirus N proteins localized to the nucleolus (26, 64). Because proteins that localize to the nucleolus have been implicated in cell growth and the cell cycle (11, 40, 41), we also wished to investigate whether coronavirus N proteins affect cell division.
Our data indicated that rather than adopting its normal (globular) Christmas tree-like appearance (5), in infected cells fibrillarin was distributed throughout the nucleolus and was possibly more concentrated in the nucleolar periphery. Transfection of either HeLa or Vero cells with plasmids that expressed either the IBV (type III coronavirus) or MHV (type II coronavirus) N proteins also resulted in a change in distribution of fibrillarin, in a similar manner to that observed in virus-infected cells. However, unlike HIV Rev protein, which localizes to the nucleolus with a pattern similar to fibrillarin (16) (determined by confocal microscopy), the coronavirus N protein localized uniformly throughout the nucleolus (see, e.g., Fig. 3F) (26, 64). The redistribution of fibrillarin, as a consequence of virus infection, is not unique to coronaviruses. Infection of cells with adenovirus also resulted in the redistribution of fibrillarin (46). To our knowledge this is the first time this has been demonstrated for an RNA virus. The reason for reorganization of fibrillarin to the perinucleolar region is unknown; however, the perinucleolar compartment has been implicated to play a role in transcription and in RNA metabolism in the host cell (28). To investigate whether the coronavirus N protein also associates with fibrillarin in the cytoplasm, experiments with a GFP-fibrillarin fusion protein indicated that both fibrillarin and IBV or MHV N protein colocalized in the perinuclear region and nucleolus (e.g., Fig. 6C and 7C, respectively).
The nucleolar functions of fibrillarin are well established and include a role in ribosome assembly (60) and, as a factor of the nucleolus, in cell cycle regulation (11). Experiments that blocked fibrillarin with antibody prevented the translocation of fibrillarin to the nucleoli and resulted in the reduction or inhibition of PolI transcription (19); by redistributing fibrillarin N protein might have a similar effect. Alternatively, by interacting with fibrillarin, N protein could potentially affect ribosomal biogenesis and, therefore, have a concomitant effect on host cell translation. During MHV infection, host cell translation is decreased, although translation of viral mRNAs is either unaffected or upregulated (24, 55). N protein may therefore interact with nucleolar components to improve translation of virus mRNAs, perhaps by sequestering ribosomes (or parts thereof) to viral mRNAs (26).
Unfortunately, from the point of view of this study, the monoclonal antibody against nucleolin, used in immunofluorescence, only recognized human nucleolin (in accordance with the manufacturer's guidelines [Leinco Technologies]), and this limited the study of possible redistribution of nucleolin by N protein to HeLa cells which expressed MHV N protein. While our data suggested that nucleolin was not reorganized in these cells, it is not possible to conclude that this does not occur in virus-infected cells or with other coronavirus N proteins expressed in species-specific cell types. For example, nucleolin is retained in the cytoplasm in poliovirus-infected cells (62), probably because cytoplasmic-nuclear trafficking is prevented (23), and adenovirus infection results in the redistribution of nucleolin to the cytoplasm (38).
Binding studies with purified immobilized phosphorylated or nonphosphorylated IBV N protein and nuclear extracts prepared from Vero cells indicated that there was a direct interaction between N protein and nucleolin (Fig. 8E, lanes 1 and 3, respectively). Two recombinant His-tagged proteins were used to investigate the specificity of nucleolin binding to N protein, HIV core (p24) protein, and DcuR, a DNA-binding protein isolated from E. coli (22). Nucleolin did not bind to either of these immobilized proteins or to the NTA beads in the absence of protein, indicating that nucleolin specifically interacted with N protein. Interestingly, the data indicated that immobilized Nphos protein bound more nucleolin than Nnonphos protein, a finding that is contrary to what would be predicted if the protein associated by electrostatic charge alone.
While mammalian nucleolin has a predicted molecular mass of ca. 77 kDa (depending on the species), the apparent molecular mass is between 100 and 110 kDa (e.g., Fig. 8D and E, arrowed) and is attributed to the amino acid composition of the N-terminal domain, which is highly phosphorylated (21). The faster-migrating products detected with anti-nucleolin antibody (Fig. 8D, lane 1, and 8E, lane 6) are most likely self-cleaved fragments of nucleolin (14). The results seen with RNase treatment suggest that nucleolin might interact with IBV N protein via a protein-protein interaction. Further, the region(s) where nucleolin interacts with N protein could be the RNA-binding domains, since these sites would be exposed in the RNase-treated sample. Alternatively, binding sites could become more accessible after RNase treatment due to a conformational change in nucleolin.
Our previous studies indicated that N protein could be actively transported to the nucleolus (26, 64). The fact that N protein colocalizes with nucleolin and fibrillarin and directly associates with nucleolin, at least in an in vitro assay, provides a possible explanation as to why the coronavirus N protein localizes to the nucleolus, as well as why some cells contain N in the nucleolus and others do not. The transport of viral antigens to the nucleolus via an interaction with nucleolin has been demonstrated for HDV antigen (33). The amount of nucleolin in a cell is cell cycle dependent (57), as is its phosphorylation state (44), which in turn affects function (21) and nucleolar localization. Therefore, N protein may only interact with nucleolin at certain stages of the cell cycle, and therefore transport to the nucleolus could be cell cycle dependent.
The nucleolus and associated proteins are also implicated in the regulation of the cell cycle (11). For example, at a gross level, mammalian nucleoli are only present during interphase (1); more specifically, the redistribution of fibrillarin can be dependent on the cell cycle (7), and CDK2-cyclin E localizes to Cajal bodies (which are associated with nucleoli) in a cell cycle-dependent manner (35). We have shown previously that cytokinesis is abrogated in cells transfected with plasmids that express TGEV N protein (64), leading to our hypothesis that cell growth would be inhibited or delayed. (However, nucleolar localization of N protein and a role in the regulation of cell growth may be independent of each other.) In our present study, we further investigated the effect of N protein on cell growth and/or division. Experiments comparing the growth of cells transfected with plasmids that expressed the IBV N protein indicated that these cells grew more slowly than cells transfected with control plasmids (Fig. 9). Three possibilities were investigated: (i) that expression of a foreign gene from a CMV promoter caused a decrease in cell numbers, (ii) that N protein induced apoptosis, and (iii) that N protein delayed cell division. The data indicated that expression of a foreign gene (CAT) from a CMV promoter did not cause a significant reduction in transfected cell numbers.
Previous studies have shown that the coronavirus envelope protein induces apoptosis in MHV (3) but not in IBV (34). For TGEV, MHV, and IBV, apoptosis was shown to be caspase dependent (3, 17, 34). The possible role of N protein in virally induced apoptosis was not investigated. To assay for the possible induction of apoptosis by N protein, we measured the binding of Annexin V-FITC, which binds to phosphatidylserine and provides an early marker for apoptosis (18, 61) (Fig. 10 and 11). The data indicated that, while Lipofectamine treatment caused a general increase in apoptosis, N protein did not induce apoptosis with any greater frequency than apoptosis induced in cells transfected with backbone vector alone.
Morphological analysis of primary cells infected with IBV (Fig. 12A) or Vero cells transfected with pCi-IBV-N (Fig. 12B) indicated that cell division had been delayed, probably due to the inhibition of cytokinesis, as observed by the presence of a cleavage furrow and a nucleolus in each nucleus, as previously described for TGEV N protein (64). As expected, nucleoli were absent in dividing mammalian cells (Fig. 12C). The presence of a cleavage furrow, which is composed of actin, was confirmed by using TRITC-phalloidin that binds to actin (Fig. 12D, arrowed). Fibrillarin would also appear to be concentrated to the nuclear region, at least in cells transfected with plasmids that express IBV N, and this cell was identified as undergoing aberrant cell division due to the presence of only one nucleolus (arrowed) and a cleavage furrow (Fig. 5B). The actin stain also detects nucleoli (e.g., Fig. 12E, arrowed). This is possible because both actin and spectrin (an actin-binding protein) have been proposed to function to keep nucleoli attached to the inner nucleolar membrane (12). Thus, IBV N protein appears to abrogate cellular division in infected or in cells transfected with a plasmid capable of expressing N protein. Similar to TGEV N protein (64), IBV N protein also appeared to promote endoreduplication (cells with two or more nuclei), which leads to multinucleated cells.
During MHV infection, actin mRNA levels are reduced (30), possibly as a result of a decrease in host cell translation (24, 55, 59). It seems reasonable, therefore, to expect that the cellular concentration of actin will also be reduced during infection. This may provide an explanation for the inhibition of cytokinesis in infected cells since, in order for a cell to undergo cytokinesis, a contractile ring of actin and myosin II must form beneath the plasma membrane in late anaphase. However, when actin is in limiting amounts, a contractile ring will not completely form, preventing cytokinesis (1). Interestingly, HIV-1 viral protein R (Vpr) expression arrests the cell cycle in G2 (27, 48) and induces the formation of aberrant multipolar spindles as a result of abnormal centrosome duplication with a subsequent inhibition of cytokinesis (63). These observations are morphologically similar to those in cells expressing IBV N protein or in IBV-infected primary cells (Fig. 12). Coronaviruses might therefore maintain cells in interphase in order to promote conditions for translation of viral proteins and virus assembly.
The coronavirus N protein has been implicated in a variety of functions, including formation of the viral core, virus translation, transcription, and replication (32). Here we provide evidence that N protein may interact with the host cell in hitherto-unexpected ways. By interacting with fibrillarin and nucleolin N protein may be disrupting the normal functions of these proteins, namely, in rRNA processing, ribosome biogenesis, and control of cell growth. The degree of this interaction may be determined by the phosphorylation state of N protein. Nucleolin has been shown to interact with the 3′ untranslated region of the poliovirus genome (62) and the IRES to promote translation (29), and N protein may be recruiting such nucleolar antigens as accessories for replication, transcription, and/or translation of viral RNAs.
Acknowledgments
H.C. and T.W. contributed equally to this study.
This work was supported by a BBSRC project grant (45/S12883) to J.A.H. T.W. was supported by the Research Endowment Trust Fund awarded to J.A.H.
We thank Ian Jones for help with the development of the baculovirus expression system and FACS analysis. We thank Steve Poutney for assistance with the confocal microscopy, Rosa Casais and Tereza Cristina Cardoso for help with infection of the chicken kidney cells, and Jane Harper for help with the Coulter Counter analysis.
REFERENCES
- 1.Alberts, B., D. Bray, J. Lewis, M. Raff, K. Roberts, and J. D. Watson. 1994. Molecular biology of the cell, 3rd ed., p. 381-382. Garland Publishing, New York, N.Y.
- 2.Allain, F. H.-T., P. Bouvet, T. Dieckmann, and J. Feigon. 2000. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 19:6870-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, S., C.-J. Chen, X. Yu, J. L. Leibowitz, and S. Makino. 1999. Induction of apoptosis in murine coronavirus-infected cultured cells and demonstration of E protein as an apoptosis inducer. J. Virol. 73:7853-7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen, J. S., C. E. Lyon, A. H. Fox, A. K. L. Leung, Y. W. Lam, H. Steen, M. Mann, and A. I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1-11. [DOI] [PubMed] [Google Scholar]
- 5.Aris, J. P., and G. Blobel. 1991. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc. Natl. Acad. Sci. USA 88:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 7.Azum-Gelade, M.-C., J. Noaillac-Depeyre, M. Caizergues-Ferrer, and N. Gas. 1994. Cell cycle redistribution of U3 snRNA and fibrillarin. J. Cell Sci. 107:463-475. [DOI] [PubMed] [Google Scholar]
- 8.Belyavskyi, M., E. Belyavskaya, G. A. Levy, and J. L. Leibowitz. 1998. Coronavirus MHV-3-induced apoptosis in macrophages. Virology 250:41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boursnell, M. E. G., T. D. K. Brown, I. J. Foulds, P. F. Green, F. M. Tomley, and M. M. Binns. 1987. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 68:57-77. [DOI] [PubMed] [Google Scholar]
- 10.Brierley, I., M. E. G. Boursnell, M. M. Binns, B. Bilimoria, V. C. Blok, T. D. K. Brown, and S. C. Inglis. 1987. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 6:3779-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carmo-Fonseca, M., L. Mendes-Soares, and I. Campos. 2000. To be or not to be in the nucleolus. Nat. Cell Biol. 2:E107-E112. [DOI] [PubMed] [Google Scholar]
- 12.Carotenuto, R., G. Maturi, V. Infante, T. Capriglione, T. C. Petrucci, and C. Campanella. 1997. A novel protein cross-reacting with antibodies against spectrin is localised in the nucleoli of amphibian oocytes. J. Cell Sci. 110:2683-2690. [DOI] [PubMed] [Google Scholar]
- 13.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 14.Chen, C.-M., S.-Y. Chiang, and N.-H. Yeh. 1991. Increased stability of nucleolin in proliferating cells by inhibition of its self-cleaving activity. J. Biol. Chem. 266:7754-7758. [PubMed] [Google Scholar]
- 15.Chen, D., and S. Huang. 2001. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153:169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dundr, M., G. H. Lena, M. L. Hammarskjold, D. Rekosh, C. Helga-Maria, and M. O. Olson. 1995. The roles of nucleolar structure and function in the subcellular localisation of the HIV-1 rev protein. J. Cell Sci. 108:2811-2823. [DOI] [PubMed] [Google Scholar]
- 17.Eleouet, J.-F., S. Chilmonczyk, L. Besnardeau, and H. Laude. 1998. Transmissible gastroenteritis coronavirus induces programmed cell death in infected cells through a caspase-dependent pathway. J. Virol. 72:4918-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fadok, V. A., D. Voelker, P. A. Campbell, J. J. Cohen, D. L. Bratton, and P. M. Henson. 1992. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 148:2207-2215. [PubMed] [Google Scholar]
- 19.Fomproix, N., J. Gebrane-Younes, and D. Hernandez-Verdun. 1998. Effects of anti-fibrillarin antibodies on building of functional nucleoli at the end of mitosis. J. Cell Sci. 111:359-372. [DOI] [PubMed] [Google Scholar]
- 20.Ginisty, H., F. Amalric, and P. Bouvet. 1998. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 17:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ginisty, H., H. Sicard, B. Roger, and P. Bouvet. 1999. Structure and functions of nucleolin. J. Cell Sci. 112:761-772. [DOI] [PubMed] [Google Scholar]
- 22.Golby, P., S. Davies, D. J. Kelly, J. R. Guest, and S. C. Andrews. 1999. Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli. J. Bacteriol. 181:1238-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gustin, K. E., and P. Sarnow. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex formation. EMBO J. 20:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hilton, A., L. Mizzen, G. Macintyre, S. Cheley, and R. Anderson. 1986. Translational control in murine hepatitis virus infection. J. Gen. Virol. 67:923-932. [DOI] [PubMed] [Google Scholar]
- 25.Hiscox, J. A. Brief review. The nucleolus: a gateway to viral infection? Arch. Virol., in press. [DOI] [PMC free article] [PubMed]
- 26.Hiscox, J. A., T. Wurm, L. Wilson, D. Cavanagh, P. Britton, and G. Brooks. 2001. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 75:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hrimech, M., X.-J. Yao, P. E. Branton, and E. A. Cohen. 2000. Human immunodeficinecy virus type 1 Vpr-mediated G2 cell cycle arrest: Vpr interferes with cell cycle signaling cascades by interacting with the B subunit of serine/threonine protein phosphatase 2A. EMBO J. 19:3956-3967. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Huang, S., T. J. Deerinck, M. H. Ellisman, and D. L. Spector. 1998. The perinucleolar compartment and transcription. J. Cell Biol. 143:35-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izumi, R. E., B. Valdez, R. Banerjee, M. Srivastava, and A. Dasgupta. 2001. Nucleolin stimulates viral internal ribosome entry site-mediated translation. Virus Res. 76:17-29. [DOI] [PubMed] [Google Scholar]
- 30.Kyuwa, S., M. Cohen, G. Nelson, S. Tahara, and S. Stohlman. 1994. Modulation of cellular macromolecular synthesis by coronavirus: implication for pathogenesis. J. Virol. 68:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamond, A. I., and W. C. Earnshaw. 1998. Structure and function in the nucleus. Science 280:547-553. [DOI] [PubMed] [Google Scholar]
- 32.Laude, H., and P. S. Masters. 1995. The coronavirus nucleocapsid protein, p. 141-163. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, Inc., New York, N.Y.
- 33.Lee, C. H., S. C. Chang, C. J. Chen, and M. F. Chang. 1998. The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J. Biol. Chem. 273:7650-7656. [DOI] [PubMed] [Google Scholar]
- 34.Liu, C., Y. Xu, and D. X. Liu. 2001. Induction of caspase-dependent apoptosis in cultured cells by the avian coronavirus infectious bronchitis virus. J. Virol. 75:6402-6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, J.-L., M. B. Hebert, Y. Ye, D. J. Templeton, H.-J. King, and A. G. Matera. 2000. Cell cycle-dependent localization of the CDK2-cyclin E complex in Cajal (coiled) bodies. J. Cell Sci. 113:1543-1552. [DOI] [PubMed] [Google Scholar]
- 36.Liu, J.-L., L. F. Lee, Y. Ye, Z. Qian, and H.-J. King. 1997. Nucleolar and nuclear localization properties of a herpesvirus bZIP oncoprotein, MEQ. J. Virol. 71:3188-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lutz, P., F. Puvion-Dutilleul, Y. Lutz, and C. Kedinger. 1996. Nucleoplasmic and nucleolar distribution of the adenovirus IVa2 gene product. J. Virol. 70:3449-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matthews, D. A. 2001. Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J. Virol. 75:1031-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matthews, D. A., and W. C. Russell. 1998. Adenovirus core protein V is delivered by the invading virus to the nucleolus of the infected cell and later in infection is associated with nucleoli. J. Gen. Virol. 79:1671-1675. [DOI] [PubMed] [Google Scholar]
- 40.Olson, M. O., M. Dundr, and A. Szebeni. 2000. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10:189-196. [DOI] [PubMed] [Google Scholar]
- 41.Pederson, T. 1998. The plurifunctional nucleolus. Nucleic Acids Res. 26:3871-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peeples, M. E., C. Wang, and N. Coleman. 1992. Nuclear entry and nucleolar localization of the Newcastle disease virus (NDV) matrix protein occur early in infection and do not require other NDV proteins. J. Virol. 66:3263-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Penzes, Z., K. Tibbles, K. Shaw, P. Britton, T. D. K. Brown, and D. Cavanagh. 1994. Characterisation of a replicating and packaged defective RNA of avian coronavirus infectious bronchitis virus. Virology 203:8660-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peter, M., J. Nakagawa, M. Doree, J. C. Labbe, and E. A. Nigg. 1990. Identification of major nucleolar proteins as candidate mitotic substrates of cdc2 kinase. Cell 60:791-801. [DOI] [PubMed] [Google Scholar]
- 45.Platani, M., I. Goldberg, J. R. Swedlow, and A. I. Lamond. 2000. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J. Cell Biol. 151:1561-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puvion-Dutilleul, F., and M. E. Christensen. 1993. Alterations of fibrillarin distribution and nucleolar ultrastructure induced by adenovirus infection. Eur. J. Cell Biol. 61:168-176. [PubMed] [Google Scholar]
- 47.Pyper, J. M., J. E. Clements, and M. C. Zink. 1998. The nucleolus is the site of Borna disease virus RNA transcription and replication. J. Virol. 72:7697-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Re, F., D. Braaten, E. K. Franke, and J. Luban. 1995. Human immunodefiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J. Virol. 69:6859-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowland, R. R., R. Kerwin, C. Kuckleburg, A. Sperlich, and D. A. Benfield. 1999. The localisation of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 64:1-12. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 51.Scheer, U., and R. Hock. 1999. Structure and function of the nucleolus. Curr. Opin. Cell Biol. 11:385-390. [DOI] [PubMed] [Google Scholar]
- 52.Schmid, I., C. H. Uittenbogaart, and J. V. Giorgi. 1991. A gentle fixation and permeabilization method for combined cell surface and intracellular staining with improved precision in DNA quantification. Cytometry 12:279-285. [DOI] [PubMed] [Google Scholar]
- 53.Schwab, M. S., and C. Dreyer. 1997. Protein phosphorylation sites regulate the function of the bipartite NLS of nucleolin. Eur. J. Cell Biol. 73:287-297. [PubMed] [Google Scholar]
- 54.Shih, K.-N., and S. J. Lo. 2001. The HDV large-delta antigen fused with GFP remains functional and provides for studying its dynamic distribution. Virology 285:138-152. [DOI] [PubMed] [Google Scholar]
- 55.Siddell, S., H. Wege, A. Barthel, and V. ter Meulen. 1981. Intracellular protein synthesis and the in vitro translation of coronavirus JHM mRNA. Adv. Exp. Med. Biol. 142:193-207. [DOI] [PubMed] [Google Scholar]
- 56.Siomi, H., H. Shida, H. M. Maki, and M. Hatanaka. 1990. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J. Virol. 64:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sirri, V., P. Roussel, M. C. Gendron, and D. Hernandez-Verdun. 1997. Amount of the two major Ag-NOR proteins, nucleolin and protein B23 is cell-cycle dependent. Cytometry 28:147-156. [PubMed] [Google Scholar]
- 58.Snaar, S., K. Wiesmeijer, A. G. Jochemsen, H. J. Tanke, and R. W. Dirks. 2000. Mutational analysis of fibrillarin and its mobility in living human cells. J. Cell Biol. 151:653-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tahara, S. M., T. A. Dietlin, C. C. Bergmann, G. W. Nelson, S. Kyuwa, R. P. Anthony, and S. A. Stohlman. 1994. Coronavirus translational regulation: leader affects mRNA efficiency. Virology 202:621-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tollervey, D., H. Lehtonen, R. Jansen, H. Kern, and E. C. Hurt. 1993. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72:443-457. [DOI] [PubMed] [Google Scholar]
- 61.Vermes, I., C. Haanen, H. Steffens-Nakken, and C. Reutelingsperger. 1995. A novel assay for apoptosis: flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein-labeled Annexin V. J. Immunol. Methods 184:39-43. [DOI] [PubMed] [Google Scholar]
- 62.Waggoner, S., and P. Sarnow. 1998. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 72:6699-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watanabe, N., T. Yamaguchi, Y. Akimoto, J. Rattner, H. Hirano, and H. Nakauchi. 2000. Induction of M-phase arrest and apoptosis after HIV-1 Vpr expression through uncoupling of nuclear and centrosomal cycle in HeLa cells. Exp. Cell Res. 258:261-269. [DOI] [PubMed] [Google Scholar]
- 64.Wurm, T., H. Chen, P. Britton, G. Brooks, and J. A. Hiscox. 2001. Localization to the nucleolus is a common feature of coronavirus nucleoproteins and the protein may disrupt host cell division. J. Virol. 75:9345-9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zatsepina, O. V., A. Rousselet, P. K. Chan, M. O. J. Olson, E. G. Jordan, and M. Bornens. 1999. The nucleolar phosphoprotein B23 redistributes in part to the spindle poles during mitosis. J. Cell Sci. 112:455-466. [DOI] [PubMed] [Google Scholar]