Abstract
We have previously shown that the 301-amino-acid herpes simplex virus tegument protein VP22 exhibits a range of subcellular localization patterns when expressed in isolation from other virus proteins. By using live-cell analysis of cells expressing green fluorescent protein (GFP)-tagged VP22 we have shown that when VP22 is first expressed in the cell it localizes to the cytoplasm, where, when present at high enough concentrations, it can assemble onto microtubules, causing them to bundle and become highly stabilized. In addition we have shown that when a cell expressing VP22 enters mitosis, the cytoplasmic population of VP22 translocates to the nucleus, where it efficiently binds mitotic chromatin. Here we have investigated the specific regions of the VP22 open reading frame required for these properties. Using GFP-VP22 as our starting molecule, we have constructed a range of N- and C-terminal truncations and analyzed their localization patterns in live cells. We show that the C-terminal 242 residues of VP22 are sufficient to induce microtubule bundling. Within this subregion, the C-terminal 89 residues contain a signal for cytoplasmic localization of the protein, while a larger region comprising the C-terminal 128 residues of the VP22 protein is required for mitotic chromatin binding. Furthermore, a central 100-residue domain of VP22 maintains the ability to bind microtubules without inducing bundling, suggesting that additional regions flanking this microtubule binding domain may be required to alter the microtubule network. Hence, the signals involved in dictating the complex localization patterns of VP22 are present in overlapping regions of the protein.
The herpes simplex virus type 1 (HSV-1) structural protein VP22 is a major component of the virus tegument,which is the virion compartment located between the capsid and the envelope (4), and in spite of a number of studies carried out on VP22, its role in virus infection is unclear. In previous studies of VP22, we and others examined in detail the subcellular targeting of the protein by immunofluorescence of transiently expressing cells (6, 8, 9). More recently, we have used VP22 tagged at its N terminus with green fluorescent protein (GFP) to investigate the localization and trafficking of VP22 in live cells. By using a combination of studies on fixed and live cells we have shown that, when expressed in isolation from other virus proteins, VP22 localizes to the cytoplasm of expressing cells (7-9). Furthermore, by using a virus that expresses GFP-tagged VP22 in place of native VP22, we have shown that GFP-VP22 also localizes predominantly to the cytoplasm of infected cells, confirming that VP22 cytoplasmic targeting is a feature of virus infection (10).
When VP22 is present at high levels in the cytoplasm of transfected cells, it has the capacity to bind, reorganize, and stabilize cellular microtubules (MTs) in a manner similar to that demonstrated for cellular microtubule associated proteins (MAPs) (7, 8). However, in cells infected with HSV-1 the pronounced accumulation of VP22 onto MTs is not observed (10), leading us to speculate that the heavily bundled MTs detected in transiently transfected cells are a result of overexpression of VP22 (7). Indeed, such MT bundling has been shown to occur when a number of cellular MAP proteins are overexpressed in cells by transient transfection (3, 15, 17). Nevertheless, as with cellular MAP proteins, where MT bundling due to overexpression is a consequence of a relevant MT interaction, we believe that the VP22-induced MT bundles observed in transfected cells is indicative of a VP22-MT interaction that may occur in HSV-1-infected cells.
In addition to these cytoplasmic properties of VP22, we have further shown that it has the ability to translocate from the cytoplasm to the nucleus during cell division via an association with mitotic chromatin (7). These events occur in cells expressing VP22 either by transient transfection or by virus infection, confirming that such a chromatin interaction is relevant to virus replication (7). Thus, VP22 exhibits at least three distinct properties in relation to its subcellular localization: cytoplasmic accumulation, MT interaction and reorganization, and binding of mitotic chromatin that leads to nuclear retention. The combination of these three localizations makes the subcellular targeting of VP22 a complex issue.
HSV-1-encoded VP22 is a 38-kDa protein that is highly basic, rich in proline residues, and highly phosphorylated in infected cells (11, 12). The UL49 gene that encodes HSV-1 VP22 is conserved throughout the alphaherpesvirus subfamily, and several recent studies on the bovine herpes virus type 1 and Marek's disease virus homologues of VP22 have suggested that at least some of the above-mentioned properties of HSV-1 VP22 are conserved in these other viral proteins (5, 14, 20). Hence, it is possible that there are conserved motifs present within the structure of these proteins that could be responsible for the various interactions of VP22 with the cellular architecture. In this report we have constructed a range of N- and C-terminal truncations of VP22 expressed as GFP fusion proteins and have investigated the localization of these GFP-tagged proteins in relation to their ability to preferentially localize to the cytoplasm, bind MTs, and bind mitotic chromatin. We have identified the regions of VP22 required for each of these functions, and we show that they exist as a set of overlapping motifs contained within a larger domain that is sufficient for the reorganization of MTs. The delineation of these various motifs within the structure of VP22 has enabled us to design a model for the mechanism of VP22 interaction with MTs and should now help us to investigate the requirements for cytoplasmic localization, MT binding, and chromatin binding in HSV-1 infection.
MATERIALS AND METHODS
Plasmids.
The plasmids expressing GFP (pEGFPC1 and pEGFPC2) and GFP-tubulin (pEGFP-tub) were obtained from Clontech. The construction of the expression vector pGE155, containing the human cytomegalovirus immediate-early promoter driving the VP22 open reading frame fused to the C terminus of enhanced GFP, has been described previously (7). To aid the construction of the VP22 truncation mutants, we utilized a range of insertion mutants based on the plasmid pUL49ep (16). These insertion mutants contained 9-bp insertions at positions 60, 119, 159, 172, and 192 in the VP22 open reading frame, resulting in the introduction of a BglII site at each of the insertion sites (see Fig. 2A). For the C-terminal truncations, plasmids pAM6, pAM7, and pAM8, containing the VP22 residues 1 to 191 (residue 1-191), 1-172, and 1-159, respectively, were constructed by inserting the relevant BglII-pPuMI fragments from the pUL49ep insertion series (see Fig. 2A) into BamHI-pPuMI-digested pGE155. Plasmid pAM17 containing residue 1-212 of VP22 fused to GFP was constructed by digesting pGE155 with NsiI and PstI, followed by religation. For the N-terminal truncations, plasmids pGE170, pGFP160-301, and pGE171, containing the VP22 residues 60-301, 160-301, and 174-301, respectively, were constructed from the same range of insertion mutants based on pUL49ep as that mentioned above. Plasmids pGE170 and pGE171, consisting of GFP fused to VP22 residues 60-301 and 174 to 301, respectively, were constructed by transferring the relevant BglII/BamHI fragments into BglII-cut pEGFPC1, while pGFP160-301 was constructed by transferring the relevant BglII/BamHI fragment into BglII/BamHI-cut pEGFPC2. Plasmids pGE191 and pGE192, consisting of VP22 residues 213-301 and 192-301 fused to GFP, were constructed by PCR amplification of these regions, which were subsequently cloned as HindIII-BamHI fragments in pEGFPC1. Plasmid pAM18, encoding GFP fused to residue 108-301 of VP22, was constructed by inserting the BspEI -BamHI fragment from pUL49ep (see Fig. 2A) into BspEI -BamHI-digested pEGFPC1. For the construction of the GFP-tagged N- and C-terminal VP22 deletion mutant encoding GFP fused to residue 108-212 of VP22, BspEI-digested plasmid pAM17 was religated to form plasmid pAM19.
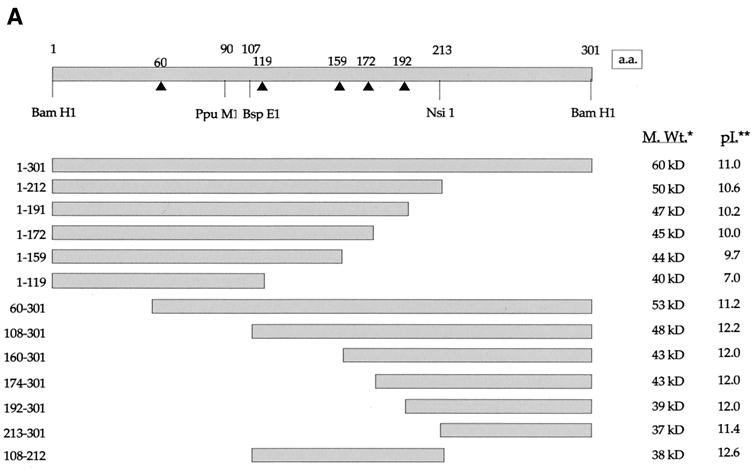
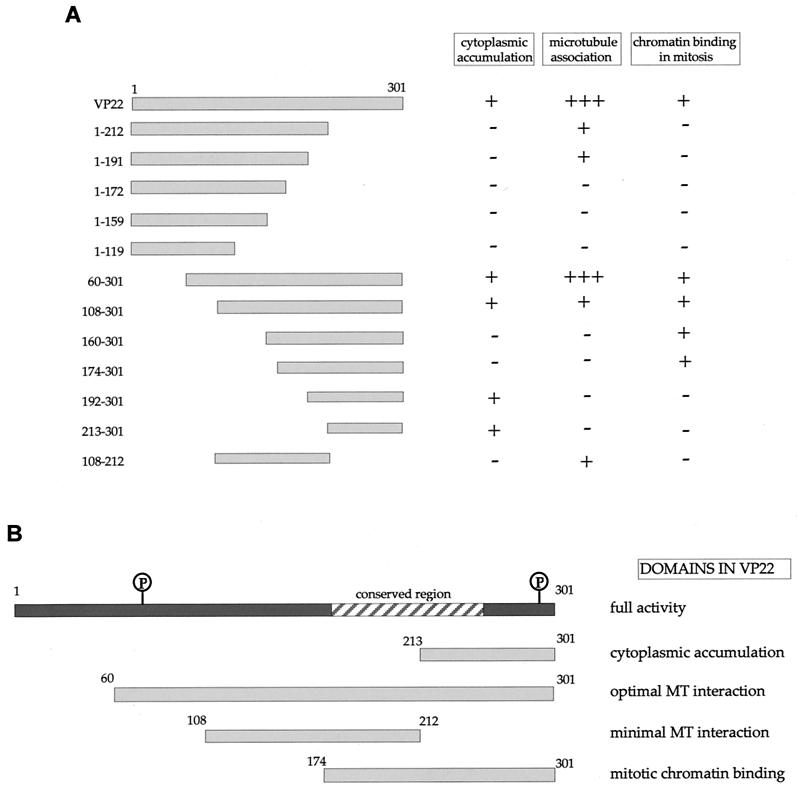
FIG. 2.
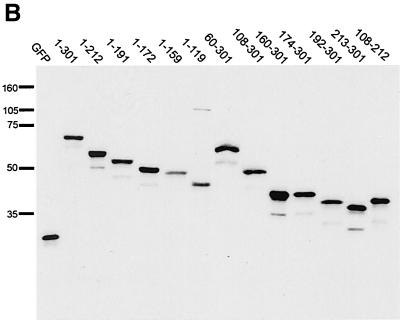
Construction and expression of VP22 truncations. (A) A range of N- and C-terminal truncations of VP22 were constructed either with restriction sites present within the VP22-encoding gene sequence (UL49) or with BglII sites introduced by insertion mutagen-esis (represented by triangles). Each truncated VP22 was fused at its N terminus to GFP to produce the panel of proteins shown. In all cases the numbers refer to amino acid (a.a.) position within the VP22 open reading frame. *, Predicted molecular size (M. Wt.) of the GFP fusion proteins. **, Predicted pI of the VP22 moiety of the fusion protein. (B) Expression vectors for the full-length and truncated VP22s, to-gether with the GFP expression vector, were all transfected into COS-1 cells, and cell extracts were analyzed by Western blotting with an anti-GFP monoclonal antibody. Note that the proteins 160-301 and 174-301 have similar molecular sizes because of the presence of an extra polylinker sequence in the 174-301 construct.
Cells and transfections.
COS-1 cells were maintained in Dulbecco's modified minimal essential medium containing 10% newborn calf serum. For transfection, COS-1 cells were plated at a density of 105 cells per well of a 2-well coverglass chamber (LabTek) and were transfected 24 h later with 200 ng of plasmid DNA. Transfections were carried out by the calcium phosphate precipitation technique, modified with BES [N,N-bis(2-hydroxyl)-2-aminoethanesulfonic acid]-buffered saline in place of HEPES-buffered saline. Transfected cells were analyzed by live-cell microscopy at various times up to 48 h after transfection. To stabilize MTs in transfected COS-1 cells, the cells were incubated for 2 h prior to analysis in medium containing taxol (Sigma) at a concentration of 50 μg/ml. To depolymerize MTs in transfected cells, the cells were incubated for 2 h prior to analysis in medium containing nocodazole (Sigma) at a concentration of 10 μg/ml. To arrest transfected cells in mitosis, the cells were incubated for 16 h prior to analysis in medium containing nocodazole at a concentration of 50 ng/ml.
Live-cell microscopy.
All live-cell microscopy of cells expressing GFP-tagged proteins was carried out with a Zeiss LSM 410 inverted confocal microscope, with resulting images processed with Adobe Photoshop software. Cells for short-term live analysis were examined directly in the 2-well coverglass chambers in which they were grown.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis.
Solubilized proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the gels were transferred to nitrocellulose filters. The filters were incubated with a monoclonal anti-GFP antibody (Clontech) at an appropriate concentration followed by incubation with a horseradish peroxidase-linked secondary conjugate. Reactive bands were visualized with the enhanced chemiluminescence (ECL) detection reagents (Amersham).
RESULTS
The MT stabilizing drug taxol enhances the bundling of MTs by GFP-VP22.
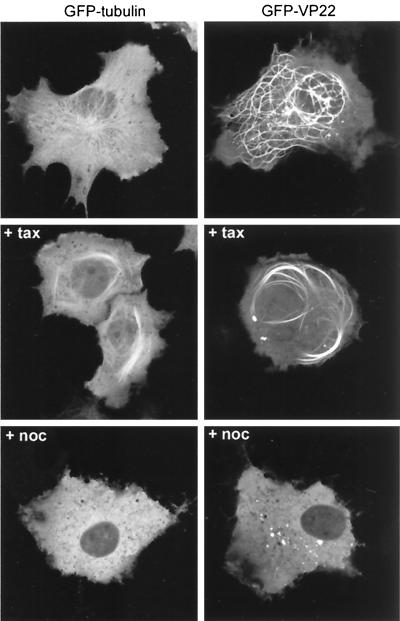
We have previously shown that VP22-induced MT bundles exhibit an enhanced bundling in the presence of the MT stabilizing drug taxol (8). We therefore wished to assess the behavior of our GFP-VP22 protein when expressed by transient transfection with regard to its localization in the absence and the presence of taxol. As a comparison, we also investigated the behavior of cellular MTs in live cells by expressing GFP-tagged α-tubulin in the same manner. Thus, COS-1 cells were transiently transfected with either the GFP-VP22-expressing plasmid pGE155 or a plasmid expressing GFP-tubulin, and 20 h later cells were either left untreated or taxol was added. Two hours later the cells were examined live, and representative images of GFP-tubulin and GFP-VP22 in the absence or presence of taxol were acquired. In the absence of taxol, GFP-tubulin could be seen outlining the already-existing MT filaments in the cytoplasm of COS-1 cells (Fig. 1, GFP-tubulin, untreated). A high proportion of GFP-tubulin was also present as a diffuse cytoplasmic background. By contrast, the vast majority of GFP-VP22 in expressing cells was assembled into thick MT bundles, the organization of which was quite different from that of unbundled MTs (Fig. 1, GFP-VP22, untreated). In addition, the MT organizing center was absent from cells containing VP22-bundled MTs and, as we have previously observed, there was little VP22 present in the nuclei of these expressing cells (7, 8). The addition of taxol to GFP-tubulin-expressing cells resulted in the reorganization of GFP-tubulin containing MTs into shorter thick bundles (Fig. 1, GFP-tubulin, + tax). However, the effect of taxol on GFP-VP22 containing MTs was much more pronounced, with thick whorls of GFP-VP22 fluorescence visible in the cytoplasm (Fig. 1, GFP-VP22, + tax).
FIG. 1.
Comparison of GFP-tubulin and GFP-VP22 localization in live cells. COS-1 cells were transfected with expression vectors for either GFP-tubulin or GFP-VP22. Twenty hours later the cells were left untreated (top panels), incubated in medium containing taxol (+ tax), or incubated in medium containing nocodazole (+ noc). Live expressing cells were then imaged 2 h later by confocal microscopy.
We have previously demonstrated that prior to GFP-VP22 assembling onto MTs the protein localizes in a diffuse cytoplasmic pattern, with little or no protein in the nucleus (7). We wished to investigate the possibility that the depolymerization of MTs would somehow allow the import of GFP-VP22 into the nucleus. Thus, we treated both GFP-tubulin and GFP-VP22 expressing cells with the MT depolymerizing drug nocodazole, using a concentration that we know is high enough to disassemble the GFP-VP22 stabilized MTs (8). As expected, GFP-tubulin localized in a diffuse cytoplasmic pattern following the depolymerization of GFP-tubulin containing MTs (Fig. 1, GFP-tubulin, + noc). However, disassembly of GFP-VP22 containing MTs also resulted in diffuse cytoplasmic fluorescence with no significant nuclear import, indicating that the depolymerization of MTs does not allow the nuclear localization of GFP-VP22 (Fig. 1, GFP-VP22, +noc). This analysis of the GFP-VP22 interaction with cellular MTs confirmed that GFP-VP22 behaves in an identical manner to that of untagged VP22, which we have previously analyzed by immunofluorescence (8), and hence validates GFP-VP22 as a suitable system for investigating the subcellular targeting of VP22.
The cytoplasmic localization of VP22 requires a motif present between residues 213 and 301 of the protein.
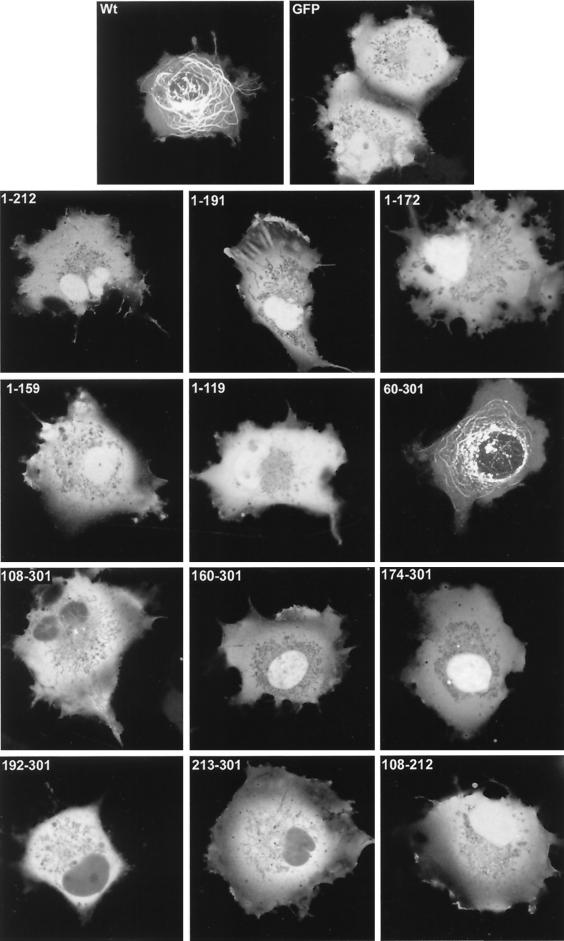
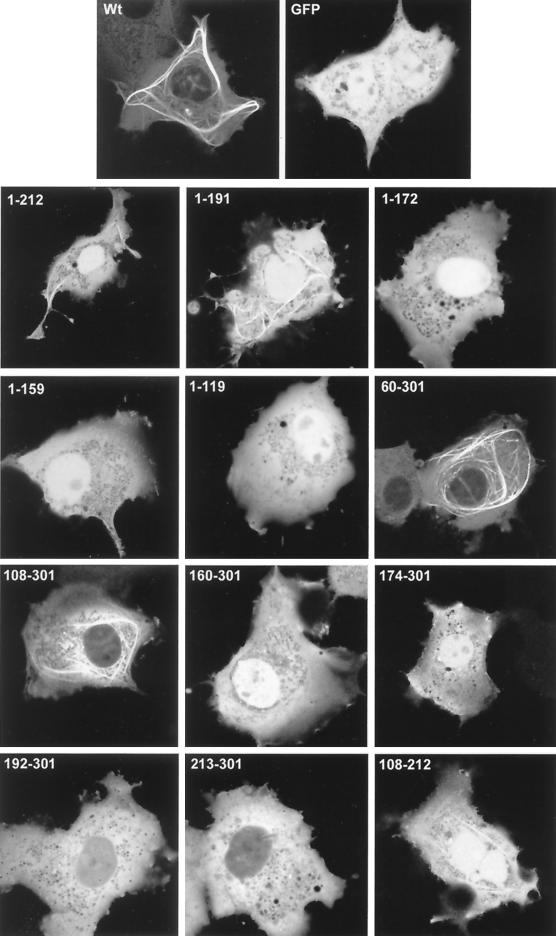
We wished to determine if discrete domains of VP22 were responsible for its characteristic properties of cytoplasmic accumulation, MT binding, and mitotic chromatin binding. Hence, we constructed a range of N- and C-terminal truncations of VP22, all of which were fused at their N termini to GFP (Fig. 2A). Western blotting of cells transfected with each of these constructs confirmed that the fusion proteins were all expressed in similar amounts and were of approximately the correct molecular size (Fig. 2B). It is noteworthy that GFP-VP22 together with all the C-terminal truncations had a slightly slower mobility than that predicted, a result that has been noted previously for full-length and for C-terminal truncations of native VP22 (12). We next investigated the subcellular compartmentalization of the truncated versions of GFP-VP22 by transfecting equal amounts of each plasmid into COS-1 cells and examining the live cells 24 h later for the presence of GFP in the cytoplasm and nuclei of expressing cells (Fig. 3, results summarized in Fig. 6A). The localization of the C-terminal truncations indicated that the removal of 89 residues from the C terminus of VP22 resulted in GFP-VP22 localizing in a pattern similar to that of unfused GFP, with fluorescence present in both the cytoplasm and nucleus of expressing cells (Fig. 3, compare GFP and 1-212) and no obvious MT bundling (see below). Thus, removal of the C terminus of VP22 altered the localization of VP22 from a cytoplasmic location to both a nuclear and cytoplasmic location (Fig. 3, compare Wt with 1-212). Further truncations from residue 212 resulted in GFP fusion proteins with localization patterns similar to those of residue 1-212 (Fig. 3, 1-191, 1-172, 1-159, and 1-119). By contrast, removal of either 59 or 107 residues from the N terminus of VP22 had little effect on the cytoplasmic accumulation of GFP-VP22 (Fig. 3, compare Wt with 60-301 and 108-301). Moreover, fusion of the C-terminal region in the form of residue 192-301 or 213-301 to GFP also resulted in a protein that preferentially localized to the cytoplasm (Fig. 3, 192-301 and 213-301). Taken in isolation, we interpreted these results to mean that the C terminus of VP22 contains a signal sufficient for cytoplasmic localization. However, this interpretation was further complicated by the observation that GFP fused to either residue 160-301 or 174-301, which incorporates an additional N-terminal region onto the cytoplasmic localizing C terminus, and both localize efficiently to the nucleus (Fig. 3, 160-301 and 174-301). A possible explanation for the nuclear accumulation of these two proteins is provided later (see Discussion).
FIG. 3.
Localization patterns of the truncated VP22 proteins. Expression vectors for GFP, full-length VP22, and the truncated VP22s were transfected into COS-1 cells grown in coverslip chambers. Twenty hours later live expressing cells were analyzed by confocal microscopy and representative images were acquired. Wt, wild type.
FIG. 6.
Summary of localization patterns for the VP22 truncations. (A) Each of the truncation mutants analyzed has been scored for cytoplasmic accumulation (+ or −), microtubule association (+++ for bundling in absence of taxol, + for bundling only in the presence of taxol, or − for no detectable bundling), and chromatin binding (+ or −). (B)The results shown in panel A were used to delineate specific domains within the VP22 open reading frame. Phosphorylation sites (P) within the VP22 protein are circled.
Interaction of the individual VP22 truncations with MTs.
Our original assessment of the VP22 truncations indicated that only one of the panel of GFP-VP22 fusion proteins assembled onto MTs in a manner similar to that of wild-type GFP-VP22, namely residue 60-301 (Fig. 3, 60-301). These results suggested that the optimal MT binding domain of VP22 encompassed residue 60-301 of the protein. However, we reasoned that some of the other VP22 truncations may still be able to interact with MTs but may not have the capability of ordered assembly onto and bundling of the MT network. Hence, a potential VP22-MT interaction could be masked by the large concentration of soluble GFP-VP22 in the cytoplasm of the expressing cells. Thus, to see if we could detect an interaction between the other VP22 truncations and MTs, we stabilized and bundled the MTs of transfected cells with taxol, as described for Fig. 1. The presence of taxol had a dramatic effect on the detection of GFP-VP22 containing MT bundles, with several of the apparently non-MT-interacting truncations now easily detected in cytoplasmic bundles (Fig. 4, results summarized in Fig. 6A). The C-terminal truncated proteins 1-212 and 1-191 both formed fluorescent bundles in the presence of taxol, while 1-172, 1-159, and 1-119 remained in a diffuse pattern (Fig. 4). Likewise, the N-terminal truncated proteins 60-301 and 108-301 also clearly bundled in the presence of taxol, whereas 160-301, 174-301, 192-301, and 213-301 did not (Fig. 4). Taken together, these results suggest that while the optimal MT bundling region encompasses residue 60-301, the minimal region sufficient for MT interaction may be refined to within residue 108-192.
FIG. 4.
Localization patterns of the truncated VP22 proteins in the presence of the microtubule stabilizing drug taxol. Expression vectors for GFP, full-length VP22, and the truncated VP22s were transfected into COS-1 cells grown in coverslip chambers. Eighteen hours later taxol was added to the media and cells were incubated for a further 2 h. Live expressing cells were then analyzed by confocal microscopy, and representative images were acquired. Wt, wild type.
In an attempt to delineate a minimal MT binding domain, we constructed a further GFP-VP22 fusion protein that incorporated only residue 108-212 of VP22, thereby removing both the N- and C-terminal ends of the protein (Fig. 2B). This protein localized to both the cytoplasm and nuclei of expressing cells, as predicted from the localization of its parental constructs, and did not appear to interact with MTs in the absence of taxol (Fig. 3, 108-212). However, in the presence of taxol, this GFP fusion protein assembled into bundles within the cytoplasm, albeit slightly less efficiently than either of the parental proteins 108-301 or 1-212 (Fig. 4, 108-212). Immunofluorescence of these cells with an antibody against α-tubulin confirmed that these bundles represented VP22-induced MT bundles (data not shown). Thus, in contrast to the N-terminal region (residue 1-119) and the C-terminal region (residue 213-301), the central region of the VP22 protein (residue 108-212) maintains the potential for interaction with MTs.
The VP22 motif for mitotic chromatin binding overlaps the MT binding domain.
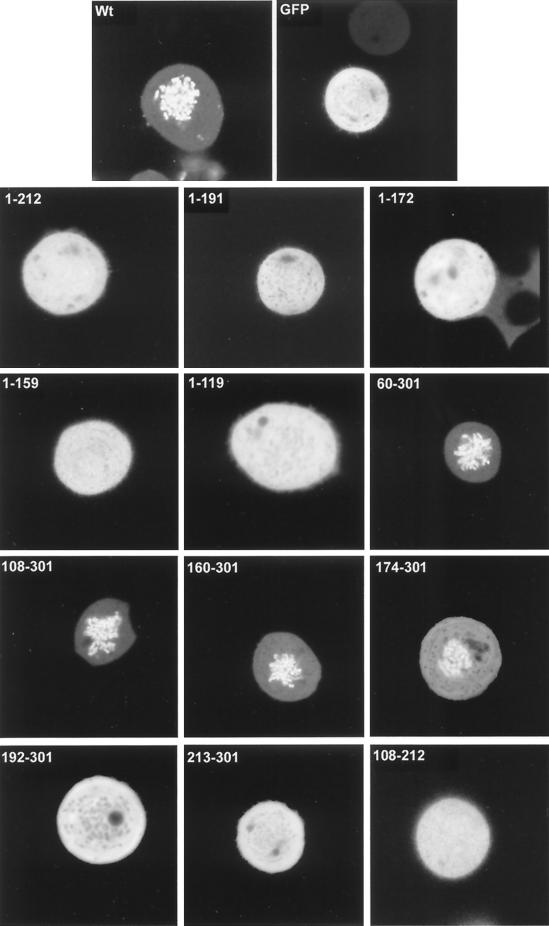
We next wished to investigate if there was a relationship between the two properties of MT binding and mitotic chromatin binding; hence, we examined the localization of all the VP22 truncations during mitosis (Fig. 5, results summarized in Fig. 6A). To facilitate this analysis we blocked the transfected cells in mitosis by incubation in a low concentration of nocodazole. With the wild-type GFP-VP22, such treatment resulted in highly fluorescent chromosomes in the rounded up cells, with only low levels of GFP-VP22 present in the surrounding cytosol (Fig. 5, Wt). By contrast, unfused GFP showed no association with condensed chromatin (Fig. 5, GFP), confirming that this property was conferred by the VP22 region of the fusion protein. Removal of only the C-terminal 89 residues of VP22 abolished the association of the protein with mitotic chromatin (Fig. 5, 1-212), such that these mitotic cells appeared identical to those expressing GFP alone. Likewise, the mitotic cells containing residue 1-192, 1-172, 1-159, or 1-119 showed a homogeneous localization of fluorescence throughout the cells with no obvious concentration around the chromosomes (Fig. 5, 1-192, 1-172, 1-159, and 1-119). However, at the N terminus of VP22 it was possible to remove up to 159 residues from the protein without in any way affecting the ability of the protein to assemble onto mitotic chromatin (Fig. 5, compare Wt with 60-301, 108-301, and 160-301). The removal of a further 14 residues from the N terminus slightly reduced the chromatin binding of VP22 (Fig. 5, compare 160-301 with 174-301), while the removal of another 18 or 39 residues resulted in a pattern of fluorescence that was indistinguishable from that of GFP (Fig. 5, 192-301 and 213-301). Hence, chromatin binding by VP22 requires the C-terminal residue 174-301 of the protein, and this domain overlaps the domains for both cytoplasmic accumulation and MT interaction (Fig. 6B).
FIG. 5.
Localization patterns of the truncated VP22 proteins in COS-1 cells arrested in mitosis. Expression vectors for GFP, full-length VP22, and the truncated VP22s were transfected into COS-1 cells grown in coverslip chambers. Twenty-four hours later nocodazole was added to the media, and cells were incubated for a further 16 h. Live expressing cells were then analyzed by confocal microscopy, and representative images were acquired. Wt, wild type.
DISCUSSION
During recent years it has become clear that the subcellular localization properties of the HSV tegument protein VP22 are complex. By rigorously examining the cellular compartmentalization of VP22 under different conditions we have devised a model explaining this complexity of VP22 localization in expressing cells, which we have presented previously (7). Briefly, this model involves VP22 localizing to the cytoplasm when it is first expressed, where the protein then has one of two fates: it either assembles onto and bundles MTs, which become highly stabilized, or it remains diffuse within the cytoplasm. In the case of cells containing VP22-stabilized MTs, these cells can no longer undergo cell division (7). However, when cells containing diffuse cytoplasmic VP22 enter mitosis, VP22 binds condensed chromatin very efficiently with virtually complete recruitment of the VP22 content onto chromosomes (7). The result of this association is that chromatin-bound VP22 remains in the nuclei of the two daughter cells once the nuclear envelope has reformed, and hence VP22 undergoes a mitosis-induced cytoplasm-to-nucleus translocation (7). Such a mitotic-dependent cytoplasm-to-nucleus movement has several precedents, including the cellular protein cyclin B1 (13, 18). An inescapable consequence of this cell-cycle-dependent compartmentalization is that with increasing time after the start of expression, the number of cells containing VP22 in their nuclei will necessarily increase as expressing cells undergo mitosis. Nonetheless, by using the powerful tool of GFP-VP22 we have shown that it is possible to follow this series of events from early after the start of expression through to mitosis and into the next cell cycle (7).
In this report we have used a range of N- and C- terminal truncations of VP22 in the background of GFP-VP22 to show that up to 59 residues can be removed from the N terminus of the protein without affecting any of the above-mentioned characteristics of VP22. Within this shorter VP22 molecule we have identified three domains of the protein (Fig. 6B) which are sufficient and necessary for cytoplasmic localization (213-301), mitotic chromatin binding (174-301), and MT interaction without inducing bundling (108-212). Interestingly, all three domains overlap with a region of VP22 that is conserved in the alphaherpesvirus homologues (Fig. 6B), suggesting that some of these activities may be conserved in the other VP22 molecules. Indeed, the bovine herpesvirus type 1 homologue has been shown to both bundle MTs and bind chromatin in mitosis (14, 20). Furthermore, our identification of truncation 174-301 as the chromatin-binding domain further refines a similar result that was published recently (1).
At the early stages of VP22 expression and prior to assembly of VP22 onto MTs, the protein is predominantly cytoplasmic. Moreover, depolymerization of VP22-induced MT bundled with a high concentration of nocodazole results in VP22 becoming diffuse in the cytoplasm with no migration of the protein to the nucleus, a result that is true for both unfused VP22 (8) and for GFP-VP22. The differing abilities of the various truncated VP22 molecules to enter the nucleus cannot be explained by simple size or charge comparisons (Fig. 2A) but appears to be dictated by the presence or absence of a signal located in the C terminus of VP22 that confers cytoplasmic localization. This is suggested by the result that removal of residue 213-301 from either full-length VP22 or VP22 truncated at its N terminus (residues 108-301) alters the localization of either of these proteins from being predominantly cytoplasmic to being both nuclear and cytoplasmic. Furthermore, the fusion of this region of VP22 to the C terminus of GFP alters the subcellular localization of GFP from a homogeneous nuclear/cytoplasmic pattern to a predominantly cytoplasmic pattern. Although we do not know how this region of VP22 functions to direct either itself or GFP to the cytoplasm, it is likely to either interact with a cytoplasmic component to effect retention or act as a nuclear export signal to drive the protein out of the nucleus.
Our description of a VP22 cytoplasmic signal was complicated by the observation that two of the proteins containing this region, namely 160-301 and 174-301, did not accumulate in the cytoplasm but were efficiently targeted to the nucleus. However, among the panel of mutants that we analyzed, these were the only two proteins that bound chromatin but did not interact with MTs. Furthermore, our results showed that the region between residues 160 and 191 was absolutely required for chromatin binding, and once this region was removed from the 160-301 protein (i.e., to make 192-301), the resulting protein accumulated in the cytoplasm once more. Therefore, it is possible that the removal of the N-terminal half of the protein unmasked a potential signal between residues 160 and 191 that is capable of targeting the truncated protein to the nucleus but is not functional in the full-length protein.
We have previously shown that VP22 colocalizes with cellular MTs and has multiple effects on the structure of the MT network, including the loss of the cellular MT organizing center; the reorganization of MTs into bundles; the stabilization of MTs (such that they are resistant to depolymerizing agents, such as cold and nocodazole); and the hyperacetylation of these stabilized MTs (8). In spite of this comprehensive range of effects, all of which are routinely utilized as characteristics of MT-interacting proteins, a recent report by Blouin and Blaho has suggested that our VP22-MT interaction is simply an artifact of the fixation technique employed (2). However, as we have already shown in a previous report (7) and go on to study in more detail here, the necessity for evoking a fixation-induced MT interaction of VP22 is rendered needless by the demonstration that GFP-VP22 is easily observed to bundle MTs in live cells. Indeed, by the technology of time-lapse microscopy it is possible to simply view and record GFP-VP22 assembling onto MTs and inducing their bundling. Hence, the evidence for a VP22-MT interaction in the absence of other virus proteins is irrefutable.
We have shown here that removal of either the C terminus (residue 213-301) or the N terminus (residue 1-107) of VP22 abolishes the ability of VP22 to form bundles. In the case of the C terminus, the interaction with MTs may be reduced simply by virtue of this protein's increased ability to accumulate in the nucleus. However, removal of the N terminus from VP22 does not affect the cytoplasmic location of VP22, and thus the inability of this particular protein to bundle MTs cannot be explained by an alteration in its location. When taxol was added to cells expressing the VP22 mutants, it became apparent that a number of the molecules that were unable to bundle MTs alone were now able to assemble MT bundles that were qualitatively different from both full-length VP22-containing bundles and GFP-tubulin-containing bundles. We interpret these results to mean that this subclass of VP22 mutants (1-212, 1-191, 108-301, and 108-212) is actually able to associate with MTs in the absence of taxol. However, this interaction does not affect the organization of MTs, and the proteins are not able to nucleate assembly along the length of the MTs. In the presence of taxol, the additional increased stability and bundling of MTs provided by taxol would be sufficient to enhance a weak interaction by this range of mutants.
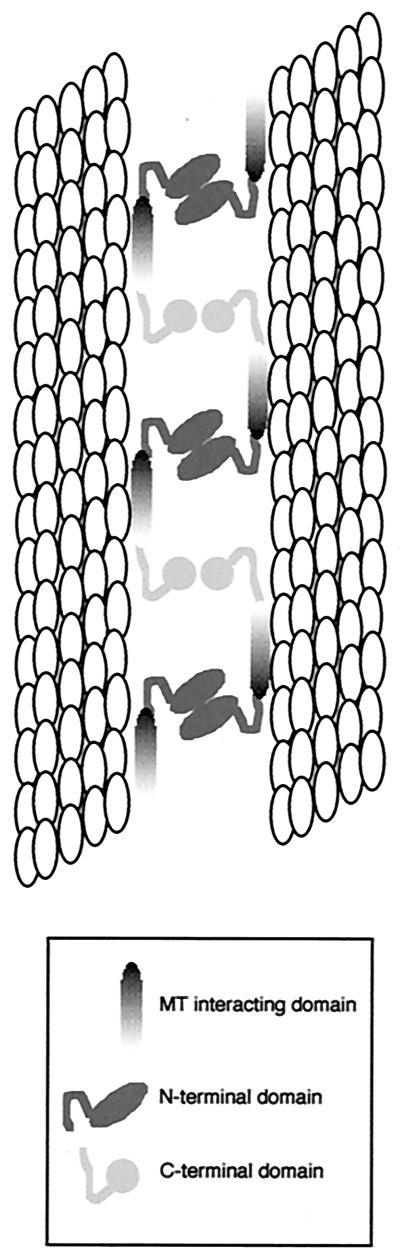
It has previously been suggested that cellular MAP proteins, such as tau, MAP4, and MAP2, have two MT binding domains, neither of which can bundle MTs when expressed in isolation (19). However, these domains are joined by a bridge of repeat sequences that in itself exhibits no MT binding but, in conjunction with the MT binding domains, induces the assembly of MTs and the formation of bundles. In the case of VP22, neither the N-terminal third (residue 1-119) nor the C-terminal third (residue 213-301) of VP22 are able to associate with MTs when expressed in isolation even in the presence of taxol, whereas the central third (residue 108-212) retains MT interaction. Thus, by analogy with other MAPs, we suggest that the middle region of VP22 is directly involved in MT interaction, while the C and N termini are more likely to be involved in cross-linking MTs to each other to induce bundles by, for instance, dimerization (Fig. 7). Furthermore, the efficient assembly of VP22 onto MTs displayed by full-length and 60-301 proteins suggests that in these cases there may be an additional step of polymerization along the length of the MTs such that a high proportion of soluble protein assembles into these bundles (Fig. 7). It is noteworthy that of the two major sites of phosphorylation which we have previously identified in VP22 (11), both fall into the N- and C-terminal domains (Fig. 6B), and hence phosphorylation may play a role in regulating VP22 assembly onto MTs.
FIG. 7.
A model for VP22 bundling of microtubules. Based on the results depicted in Fig. 6, we propose that the central region of VP22 binds the MT filaments within the cell while the N and C termini cross-link VP22 molecules across the space between the two filaments.
In summary, we have identified regions within VP22 that are responsible for its complicated pattern of localization when expressed in isolation from other virus proteins. We can now use the VP22 mutants described in this report to investigate the roles of cytoplasmic localization, MT interaction, and chromatin binding in virus infection, and to determine the part played by VP22 in the replication of HSV.
Acknowledgments
We thank Hans van Leeuwen for the GFP160-301 construct and Neil Brewis for critical reading of the manuscript.
This work was funded by Marie Curie Cancer Care.
REFERENCES
- 1.Aints, A., H. Guven, G. Gahrton, C. I. Smith, and M. S. Dilber. 2001. Mapping of herpes simplex virus-1 VP22 functional domains for inter- and subcellular protein targeting. Gene Ther. 8:1051-1056. [DOI] [PubMed] [Google Scholar]
- 2.Blouin, A., and J. A. Blaho. 2001. Assessment of the subcellular localization of the herpes simplex virus structural protein VP22 in the absence of other viral gene products. Virus Res. 81:57-68. [DOI] [PubMed] [Google Scholar]
- 3.Burgin, K. E., B. Ludin, J. Feralli, and A. Matus. 1994. Bundling of microtubules in transfected cells does not involve an autonomous dimerization site on the MAP2 molecule. Mol. Cell. Biol. 5:511-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dargin, D. 1986. The structure and assembly of herpes viruses, p. 359-437. In J. R. Harris and R. W. Horne (ed.), Virus structure, vol. 5. Academic Press, London, United Kingdom. [Google Scholar]
- 5.Dorange, F., S. El Mehdaoui, C. Pichon, P. Coursaget, and J. F. Vautherot. 2000. Marek's disease virus (MDV) homologues of herpes simplex virus type 1 UL49 (VP22) and UL48 (VP16) genes: high-level expression and characterization of MDV-1 VP22 and VP16. J. Gen. Virol. 81:2219-2230. [DOI] [PubMed] [Google Scholar]
- 6.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott, G., and P. O'Hare. 2000. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J. Virol. 74:2131-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott, G., and P. O'Hare. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J. Virol. 72:6448-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 10.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, G., D. O'Reilly, and P. O'Hare. 1999. Identification of phosphorylation sites within the herpes simplex virus tegument protein VP22. J. Virol. 73:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott, G., D. O'Reilly, and P. O'Hare. 1996. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology 226:140-145. [DOI] [PubMed] [Google Scholar]
- 13.Hagting, A., M. Jackman, K. Simpson, and J. Pines. 1999. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr. Biol. 9:680-689. [DOI] [PubMed] [Google Scholar]
- 14.Harms, J. S., X. Ren, S. C. Oliveira, and G. A. Splitter. 2000. Distinctions between bovine herpesvirus 1 and herpes simplex virus type 1 VP22 tegument protein subcellular associations. J. Virol. 74:3301-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanai, Y., R. Takemura, T. Oshima, H. Mori, Y. Ihara, M. Yanangisawa, T. Masiki, and N. Hirokawa. 1989. Expression of multiple isoforms and microtubule bundle formation in fibroblasts transfected with a single tau cDNA. J. Cell Biol. 109:1173-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leslie, J., F. J. Rixon, and J. McLauchlan. 1996. Overexpression of the herpes simplex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology 220:60-68. [DOI] [PubMed] [Google Scholar]
- 17.Olson, K., J. McIntosh, and J. Olmsted. 1995. Analysis of MAP4 function in living cells using green fluorescent protein (GFP) chimeras. J. Cell Biol. 130:639-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pines, J., and T. Hunter. 1991. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol. 115:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preuss, U., J. Biernat, E.-M. Mandelkow, and E. Mandelkow. 1997. The ‘jaws’ model of tau-microtubule interaction examined in CHO cells. J. Cell Sci. 110:789-800. [DOI] [PubMed] [Google Scholar]
- 20.Ren, X., J. S. Harms, and G. A. Splitter. 2001. Bovine herpesvirus 1 tegument protein VP22 interacts with histones, and the carboxyl terminus of VP22 is required for nuclear localization. J. Virol. 75:8251-8258. [DOI] [PMC free article] [PubMed] [Google Scholar]