Abstract
Eight hepatitis B virus (HBV) isolates of genotype G were recovered from patients and sequenced over the entire genome. Six of them had a genomic length of 3,248 bp and two had genomic lengths of 3,239 bp (USG15) and 3,113 bp (USG18) due to deletions. The 10 HBV/G isolates, including the 8 sequenced isolates as well as the original isolate (AF160501) and another isolate (B1-89), had a close sequence homology of 99.3 to 99.8% among themselves (excluding USG18 with a long deletion) but of <88.7% to any of the 68 HBV isolates of the other six genotypes with the full-length sequence known. The eight HBV/G isolates possessed an insertion of 36 bp in the core gene and two stop codons in the precore region, as did the AF160501 and B1-89 isolates. The 10 HBV/G isolates clustered on a branch separate from those bearing the other six genotypes (A through F [A-F]) in the phylogenetic tree constructed from full-length sequences of 78 HBV isolates as well as in those constructed from the core, polymerase, X, and envelope genes. Despite two stop codons in the precore region that prohibited the translation of the HBV e antigen (HBeAg), all of the eight patients with HBV/G infection possessed the HBeAg in serum. By restriction fragment length polymorphism of the surface gene, all of the eight patients were found to be coinfected with HBV of genotype A (HBV/A), which would be responsible for the expression of HBeAg in them. It is worthy of examination to determine how coinfection occurs and whether HBV/G needs HBV/A for replication.
Hepatitis B virus (HBV) persistently infects 350 million people worldwide and can induce a spectrum of acute and chronic liver diseases (12). Based on sequence divergence in the entire genome of >8%, HBV has been classified into six genotypes named with capital letters A through F (A-F) (19, 22). The six major genotypes of HBV have distinct geographic distributions (14, 15) and are associated with different clinical diseases (8, 16, 24).
Recently, a seventh HBV genotype was proposed for an HBV isolate (AF160501) recovered in France and named G, based on a sequence divergence of >11.8% from HBV isolates of the other six genotypes (26). This HBV isolate of genotype G (HBV/G) has a genomic length of 3,248 bp, a little longer than HBV isolates of the other six genotypes, which are composed of 3,182 to 3,221 bp. The longer length of the HBV/G genome is attributed to an insertion of 36 bp at codon 2 of the core (C) gene (26). Other remarkable features of HBV/G are two stop codons at positions 2 and 28 of the pre-C region that prohibit the translation of the HBV e antigen (HBeAg) (2).
For the specific detection of HBV/G, eight sera were identified, by PCR with heminested primers deduced from the 36-bp insertion, that contained HBV of this genotype. The entire nucleotide sequences were determined for the eight HBV/G isolates recovered from sera, and along with the original HBV/G isolate (AF160501) and another HBV isolate (B1-89), the genotype of which is deduced to be G (11), the HBV/G genomes were compared to one another and against 68 HBV isolates of the major six genotypes (A, 9 isolates; B, 16 isolates; C, 17 isolates; D, 18 isolates; E, 2 isolates; and F, 6 isolates). Remarkably, all of the eight patients from whom HBV/G isolates were recovered were found to be coinfected with HBV of genotype A (HBV/A).
MATERIALS AND METHODS
Patients.
Sera were obtained from patients who were persistently infected with HBV and taken care of in the Second Department of Medicine, Nagoya City University Medical School, Nagoya, Japan, and those in the Hepatology and Gastroenterology Division of the California Medical Center at San Francisco. There were eight patients from San Francisco who were found to be infected with HBV/G by PCR with specific primers that can detect 10 copies of HBV/G per test (10); HBV/G was not detected in any patients infected with HBV from Japan. The eight sera were tested for HBV markers, and HBV/G isolates in them were sequenced over the entire genome. The study design was approved by the Ethics Committee of each institution, and informed consent was obtained from each patient.
Determination of HBV/G.
Nucleic acids were extracted from serum samples (100 μl) with Smitest EX R&D (Genome Science, Fukushima, Japan) in accordance with the recommendation from the manufacturer and subjected to PCR with heminested primers designed from the 36-bp insertion in the C gene of HBV/G genomes (10). In brief, extracted nucleic acids were subjected to the first round of PCR for 40 cycles with HBHKF1 (sense, 5′-ACG GGG CGC ACC TCT CTT TAC-3′, nucleotides [nt] 1519 to 1539) and HBHKR2 that involved the 36-bp insertion characteristic of HBV/G (antisense, 5′-AGC CAA AAA GGC CAT ATG GCA-3′, nt 17 to 37 in the C gene of HBV/G) (10, 26) in the presence of AmpliTaq Gold (Applied Biosystems, Foster City, Calif.). The second round of PCR was performed for 40 cycles on the product of the first-round PCR with HBHKF2 (sense, 5′-GCA CTT CGT TTC ACC TCT GCA-3′, nt 1581 to 1601) and HBHKR2. Then the products were examined for fragments of 357 bp.
Determination of the other six HBV genotypes (A-F).
The six major genotypes of HBV (A-F) were determined by restriction fragment length polymorphism of amplification products of the envelope, or surface (S), gene by a slight modification of the method reported previously (18). Since HBV/G could not be distinguished from HBV/A with the restriction patterns from the five endonucleases used in the original method (18), HinfI was introduced for their distinction. HBV/A has two recognition sites for HinfI at nt 251 to 255 and nt 530 to 534 while HBV/G possesses an additional site at nt 482 to 486. Digestion with HinfI, therefore, produced three fragments of 6, 230, and 249 bp from DNA samples of HBV/A in contrast to four fragments of 6, 48, 201, and 230 bp from those of HBV/G.
Determination of the full-length sequences of eight HBV/G isolates.
The primers used for amplifying partial sequences of HBV DNA extracted from sera of patients are shown in Table 1. The genomes of eight HBV/G isolates were sequenced on three overlapping PCR products. Two large fragments were amplified by PCR with HBGF1-HBHKR2 and HBHKF3-HBGR2. AmpliTaq Gold was activated at 96°C for 9 min, and PCR was performed for 20 cycles of 96°C for 1 min, 55°C for 1 min, and 72°C for 1.5 min followed by 25 cycles of 96°C for 1 min, 55°C for 1 min, and 72°C for 1.5 min (with an increment of 5 s in every cycle and 5 min in the last cycle). One small fragment was amplified by PCR with HBGF3-HBGR3. AmpliTaq Gold was activated by heating, and PCR was carried out for 40 cycles of 96°C for 1 min, 55°C for 1 min, and 72°C for 1 min (5 min in the last cycle). The three amplicons were sequenced directly by the dideoxy method with the BigDye Terminator cycle sequencing kit in a fluorescent 3100 DNA sequencer (Applied Biosystems).
TABLE 1.
Primers used for PCR and sequencing
| Primera | Polarity | Sequence (5′-3′) | Positions |
|---|---|---|---|
| HBGF1 | Sense | GGG TCA CCA TAT ACT TGG GAA | 2849-2870 |
| HBHKR2 | Antisense | AGC CAA AAA GGC CAT ATG GCA | 1937-1917 |
| HBHKF3 | Sense | TGC CAT ATG GCC TTT TTG GCT | 1917-1937 |
| HBGR2 | Antisense | GAA CTG GAG CCA CCA GCA GG | 75-56 |
| HBGF3 | Sense | ACG TTA CAT GGA AAC CGC CA | 1601-1620 |
| HBGR3 | Antisense | GGA AAA AAG TCA GAA GGC AA | 2009-1991 |
| HBGF4 | Sense | CCT CCT GCC TCC ACC AAT CG | 3157-3176 |
| HBGR4 | Antisense | AGA TGA GGC ATA GCA GCA GGA TG | 431-409 |
| MF2 | Sense | GTC TAG ACT CGT GGT GGA CTT CTC TC | 246-271 |
| MR2 | Antisense | AAG CCA GAC AGT GGG GGA AAG C | 730-708 |
| HBGF5 | Sense | GTT TCT CTT GGC TCA GTT TA | 659-679 |
| HBGR5 | Antisense | CAA TAC ATG AAC CTT TAC CCC GTT GCT AGG | 1158-1129 |
| HBGF6 | Sense | CTC TGC CGA TCC ATA CTG CGG AA | 1256-1278 |
| HBGR6 | Antisense | GAT TCA GCG CTG ACG GGA CGT A | 1447-1426 |
| HBGF7 | Sense | CTC GCC AAC TTA TAA GGC CTT T | 1098-1119 |
| HBGR7 | Antisense | TGC AGA TCT TCT GCG ACG CGG C | 2487-2446 |
| HBGF8 | Sense | TCA GGC AAC TAT TGT GGT TTC A | 2227-2247 |
| HBGR8 | Antisense | GGG AAA AAT CTG GCA GGC AT | 2692-2673 |
| HBGF9 | Sense | GCC GCG TCG CAG AAG ATC TGC A | 2446-2467 |
| HBGR9 | Antisense | GGA TTG AAG TCC CAA TCT GGA TT | 3020-2998 |
The first six primers listed were used for PCR and sequencing. All other primers listed were used for sequencing only.
Molecular evolutionary analyses.
The number of nucleotide substitutions per site was estimated by using the six-parameter method (5), and phylogenetic trees were constructed by using the neighbor-joining method (25) and the numbers of substitutions. To confirm the reliability of the phylogenetic tree, bootstrap resampling tests were carried out 100 times (4). These analyses were conducted with the ODEN program of the National Institute of Genetics (Mishima, Japan) (6).
Nucleotide sequence accession numbers.
The sequences of the eight HBV/G isolates have been deposited in the DDBJ/GenBank/EMBL database as follows: isolate USG15, accession no. AB056514; isolate USG16, accession no. AB056515; isolate USG17, accession no. AB056513; isolate USG18, accession no. AB056516; isolate USG769, accession no. AB064310; isolate USG825, accession no. AB064311; isolate USG916, accession no. AB064312; and isolate USG2638, accession no. AB064313.
RESULTS
Eight HBV/G isolates.
The entire nucleotide sequences of the eight HBV/G isolates have been deposited in the DDBJ/GenBank/EMBL database. They were compared to one another and against the sequences of two other HBV/G strains, i.e., AB160501 (26) and B1-89 (11). Six of the eight HBV/G isolates examined had a genomic length of 3,248 bp, which is identical to that of AB160501 and B1-89. USG15 had a shorter length of 3,239 bp because of a deletion of 9 bp in the pre-S2 region, and USG18 had a length of 3,113 bp due to a deletion of 87 bp in the pre-S1 and pre-S2 regions as well as a deletion of 48 bp in the C gene, together accounting for 135 bp. As was the case for AB160501 and B1-89, the eight HBV/G isolates had 29 amino acids (aa) encoded by the pre-C region, 195 aa encoded by the C gene, 842 aa encoded by the polymerase (P) gene (839 aa for USG15 and 805 aa for USG18), 154 aa encoded by the X gene, and 118 aa encoded by the pre-S1 region (99 aa for USG18). The subtype of the HBsAg was deduced to be adw in all of the eight HBV/G isolates by the presence of lysine residues at positions 122 and 160 (21); it was also adw in isolates AB160501 and B1-89 (11, 26). The YMDD motif that is prone to mutation during lamivudine therapy (3, 13) was identified at aa 548 to 551 (aa 545 to 548 in USG15 and aa 511 to 514 in USG18), with methionine at position 549 (position 546 in USG15 and position 512 in USG18).
Including the two reported isolates (AB160501 and B1-89), a total of 10 HBV/G isolates were compared to one another. They were 99.3% (85.3% when USG18, with a long deletion, was included) to 99.8% homologous within the full-length sequence. The 10 HBV/G isolates were examined for homology to the 68 HBV isolates of six distinct genotypes (A-F), the full-length sequences of which are deposited in the database (Table 2). They showed sequence homologies of 83.4 to 88.7% in the entire genome, 80.6 to 90.0% in the C gene, 73.1 to 90.8% in the P gene, 80.9 to 87.3% in the X gene, 78.1 to 92.0% in the large S gene (pre-S1 and pre-S2 regions included), and 88.9 to 97.5% in the S gene.
TABLE 2.
Sequence homology of 10 HBV/G isolates to previously reported HBV isolates of the other six genotypes
| Part of HBV | Range of % homology to HBV isolates of genotype (n):
|
|||||
|---|---|---|---|---|---|---|
| A (9) | B (16) | C (17) | D (18) | E (2) | F (6) | |
| Full genome | 87.3-88.1 | 84.9-86.8 | 86.1-87.3 | 85.2-87.5 | 88.2-88.7 | 83.4-85.0 |
| C gene | 81.5-89.6 | 82.9-89.7 | 81.9-89.7 | 80.6-89.3 | 82.3-90.0 | 81.0-88.9 |
| P gene | 76.2-89.8 | 77.8-88.4 | 75.5-88.9 | 75.3-89.3 | 75.5-90.8 | 73.1-86.5 |
| X gene | 84.9-87.3 | 84.3-86.5 | 80.9-85.8 | 84.3-86.2 | 85.2-86.0 | 83.7-84.9 |
| Large S gene | 81.7-91.7 | 79.6-89.7 | 78.1-90.9 | 80.5-92.0 | 80.0-92.2 | 78.6-87.8 |
| S gene | 94.6-97.5 | 93.4-94.7 | 88.9-94.3 | 94.6-95.7 | 94.6-95.2 | 92.2-93.4 |
Phylogenetic differences between 10 HBV/G isolates and 68 HBV isolates of the other six genotypes.
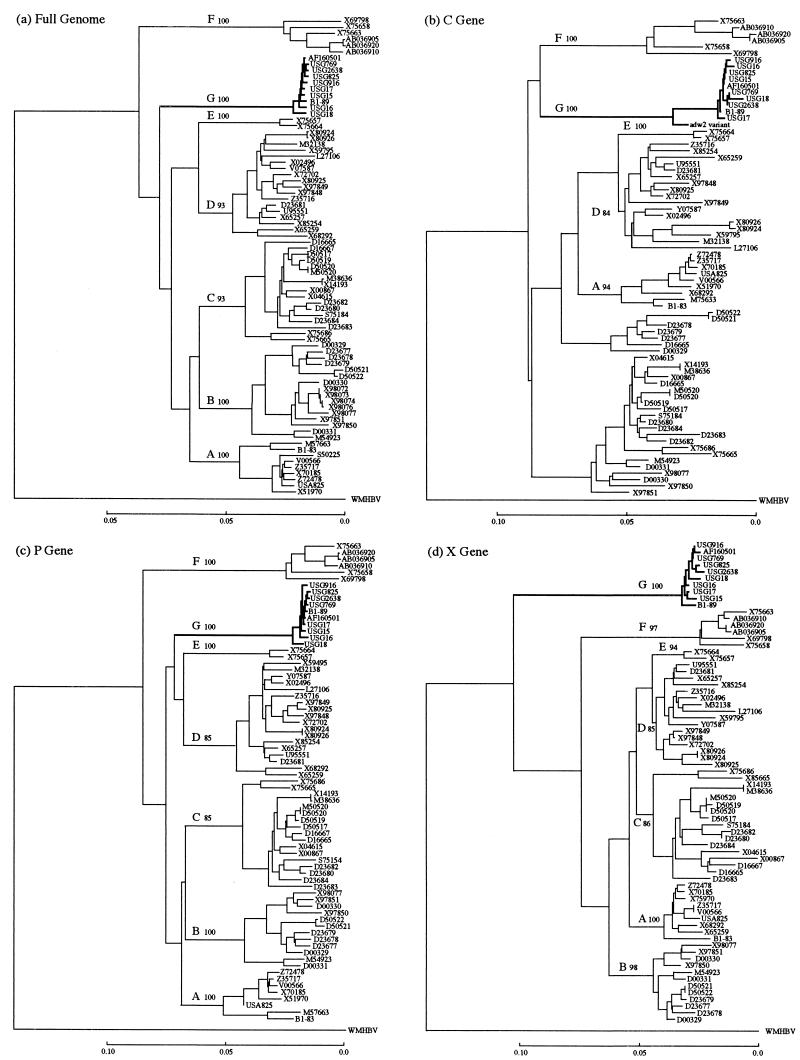
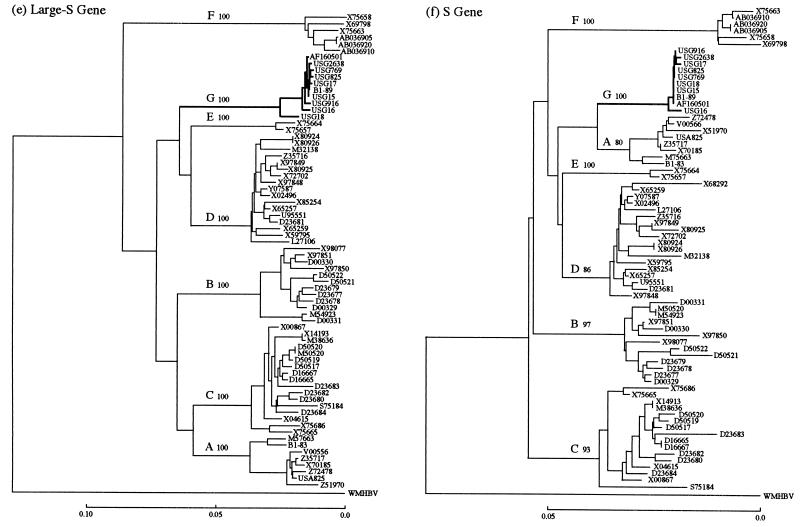
Phylogenetic trees were constructed based on the entire genome of 78 HBV isolates (including the 10 HBV/G isolates) as well as individual genes (C, X, P, large S, and S) (Fig. 1a through f). The 10 HBV/G isolates clustered on a branch separate from those bearing the other six genotypes in all six of the phylogenetic trees. Of the 10 HBV/G isolates, all except USG18 were particularly close to each other, with a homology of 99.3 (excluding USG18 with a long deletion) to 99.8% in the entire genomic sequence (Fig. 1a). A close relatedness of the nine HBV/G isolates (USG18 excluded) was maintained within the C, P, X, large S, and S genes as well (Fig. 1b through f). The sequence of the C gene is available for an additional HBV/G isolate (adw2 variant) (27), and it was most divergent among the 11 C gene sequences of HBV/G isolates (Fig. 1b). HBV isolates of genotypes B and C did not cluster on two separate branches in the phylogenetic analysis of the C gene, unlike in phylogenetic analyses of the entire genome or of the other genes (Fig. 1b). Notably, a clear branching of the seven genotypes (A-F and G) in the entire genomic sequence (Fig. 1a) was reproduced only in the P gene (Fig. 1c). Primary and secondary nodes distinct from those in the phylogenetic tree of the entire genome were obvious in the trees for the C, X, and S genes (Fig. 1b, d, and f).
FIG. 1.
Phylogenetic trees constructed for HBV isolates of seven genotypes on the full genome (a) and on the C (b), P (c), X (d), large S (e), and S (f) genes. Confidence values calculated by bootstrap analysis are indicated after each genotype.
Insertion and deletion of nucleotides in the C gene and pre-S1 region as well as two stop codons in the pre-C region.
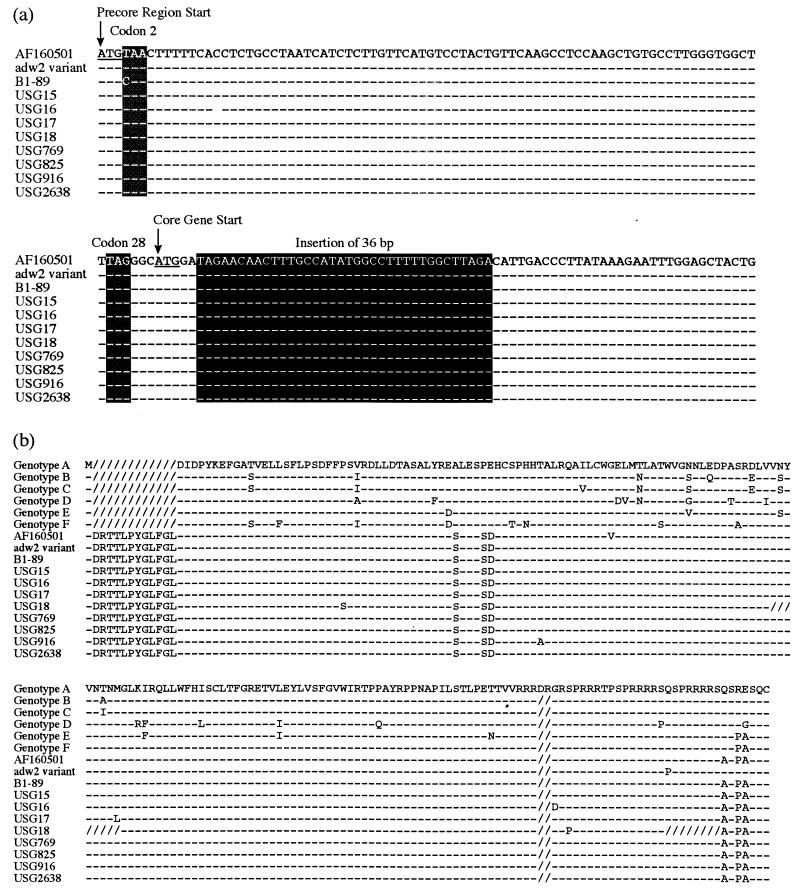
The eight HBV/G isolates possessed an insertion of 36 nt at the fifth nucleotide in the C gene (Fig. 2a) as in the original HBV/G isolate (AF160501) (26) and B1-89 (11). Hence the 36-bp insertion would be the hallmark of genotype G, distinguishing it from the other six genotypes. Likewise, a 2-aa deletion at the carboxy terminus of the product of the C gene was preserved in all 10 of the HBV/G isolates (Fig. 2) as in any HBV isolates except for HBV/A isolates.
FIG. 2.
(a) Nucleotide sequences of the eight HBV/G isolates within the pre-C region and a 5′-terminal part of the C gene. Sequences of the original HBV/G strain (AF160501 [25]) and an isolate of retrospective genotype G (adw2 variant [27]) are shown for comparison. The 36-bp insertion in the C gene and two stop codons in the pre-C region are shaded. (b) Amino acid sequence of the C gene product in the eight HBV/G isolates. Sequences of the other six genotypes (A-F) in representative strains and the original HBV/G strain (AF160501 [26]) as well as an isolate with retrospective genotype G (adw2 variant [27]) are shown for comparison.
The two stop codons at positions 2 and 28 in the pre-C region of the original HBV/G isolate (26) were invariably preserved in the eight HBV/G isolates (Fig. 2). The stop codon at position 2 (TAA) is converted to the codon for glutamine (CAA) in B1-89 (11) while the stop codon at position 28 (TAG) is retained in it.
The 1-aa deletion at position 3 in the pre-S1 region in the original HBV/G isolate (AF160501) (26) was possessed in common by the eight HBV/G isolates examined and B1-89 (11); it is shared by HBV isolates of genotype E (19).
HBeAg detected in sera from eight patients who were coinfected with HBV/G and HBV/A.
The eight patients in San Francisco from whom HBV/G isolates were recovered were predominantly Caucasian and male (six patients were male and six patients were Caucasian). They were 27 to 49 years of age. Except for one patient, from whom USG2638 was recovered, they all had elevated levels of alanine aminotransferase in the serum (>50 U/liter). Remarkably, although the HBV/G isolates had two stop codons in the pre-C region, HBeAg was detected in sera from all eight of the patients (Fig. 2).
For evaluating the possibility of coinfection with HBV of the other genotypes with an HBeAg-positive phenotype, HBV DNA samples from sera of the eight patients were genotyped by restriction fragment length polymorphism of the S gene (18). The pattern of HBV/A was detected in all of them; it was distinguished from that of HBV/G (see Materials and Methods). Hence, HBV/A coinfecting the patients with HBV/G would have been coding for HBeAg and responsible for serum HBeAg in all of them.
DISCUSSION
A seventh genotype of HBV (G) has been proposed for a French HBV isolate named AF160501 (26) based on a sequence divergence of >8.0% from the entire genome of HBV isolates of the six major genotypes (A-F) (19, 22). A search for sequence homology in the DNA database identified another HBV/G isolate (B1-89) previously reported from France (11), albeit a particular genotype was not assigned for it. In order to confirm the genotype G as a separate entity from the six major genotypes (19, 22) and characterize it virologically, the entire genomes of eight additional HBV/G isolates were sequenced and features specific to this genotype were sorted out.
In addition to the two documented isolates (AF160501 and B1-89), eight HBV/G isolates were compared with 68 HBV isolates of the major six genotypes (A-F), the sequences of which are retrievable from DNA databases. The 10 HBV/G isolates were >99.3% (>85.7% when USG18 with a long deletion was included) homologous along the entire genomic sequence but differed from 68 HBV isolates of the other genotypes by >11.3%, which cleared the threshold of >8% separating HBV genotypes (19, 22). Taken along with the two previously reported results (11, 26), therefore, the eight HBV/G isolates examined in the present study would establish G as a seventh genotype in addition to the major six genotypes (A-F).
Two remarkable traits have been pointed out in the original HBV/G isolate (AF160501) that are not shared by any HBV isolates of the other six genotypes (A-F). Most remarkably, HBV/G has an insertion of 36 bp at the fifth nucleotide in the C gene (26), which is exhibited by another isolate (11) and shared by all eight of the HBV/G isolates studied herein (Fig. 2). The peculiar 36-bp insertion has been taken advantage of, in detecting HBV/G isolates, by use of its sequence as a type-specific primer in PCR (10).
The second conspicuous brand of HBV/G isolates are two stop codons in the pre-C region at positions 2 and 28, either of which prohibits the translation of the HBeAg precursor (2, 23). The two stop codons were preserved in the original HBV/G isolate and all eight isolates examined in the present study; the one at position 2 is missing in B1-89 while the one at position 28 is retained (11). Thus, all 10 of the HBV/G isolates would have an HBeAg-negative phenotype, which influences the replication and disease-inducing capacity of HBV (17). HBV/G infection would need to be looked into with special reference to the acute and chronic liver diseases it induces, in view of HBV genotypes associated with the severity of hepatitis B (8, 16, 24).
HBV/G may have a distinct geographic distribution just as the major six genotypes do (14, 15). Originally, HBV/G was identified in 11 of 82 (13%) HBV carriers in Georgia and 2 of 39 (5%) carriers from France (26). It does not appear to be a coincidence that the B1-89 isolate (11) is reported to be from France, the genotype of which is deduced to be G because of the 36-bp insertion in the C gene, and that an adw variant of probable genotype G, judged by the peculiar 36-bp insertion, was isolated from a homosexual carrier from San Francisco also infected with human immunodeficiency virus type 1 (1). Furthermore, all eight of the HBV/G isolates in the present study were recovered from patients living in San Francisco. By contrast, HBV/G was not detected in any HBV carriers from Nagoya, Japan. It seems as if HBV/G prevails in restricted areas in the world, possibly via particular routes of transmission.
In the original report, HBeAg was detected in the serum from a carrier infected with HBV/G, in spite of an HBeAg-negative phenotype deduced from the two pre-C region stop codons (26). It may strike one as a big surprise that all eight of the HBV/G carriers examined in the present study possessed HBeAg in the serum as well. Moreover, HBeAg was detected in sera from two individuals from whom HBV/G isolates (B1-89 and an adw variant) were recovered (1, 11).
HBV/A with an HBeAg-positive phenotype was invariably detected, by restriction fragment length polymorphism, in the S gene (18) of the eight patients from San Francisco from whom HBV/G isolates were recovered. The coinfection with HBV/G and HBV/A is also deduced in an HBeAg-positive individual in France from whom the B1-89 isolate was obtained (11). Taken altogether, coinfection of HBV/G with HBV/A would be frequent. HBeAg in the sera of individuals infected with HBV/G examined in previous studies (11, 26) or in the present study, therefore, would be attributed to the HBV/A with which they were coinfected.
In a previous report (9), the sera of four of the eight patients (corresponding to isolates USG15, USG16, USG17, and USG18) from whom HBV/G isolates, whose entire genomes were sequenced in the present study, were recovered were examined for partial HBV DNA clones with special reference to HBeAg and antibody to HBeAg in serum. The clones of HBV/G and those of HBV/A were recovered from them in various proportions. In addition, HBV DNA clones representing the recombination between HBV/G and HBV/A, within a partial sequence from the X gene to the C gene, were recovered from three of them (USG15, USG17, and USG18). In one of them (USG17), a shift from the clones of HBV/A to those of HBV/G occurred along with seroconversion from HBeAg to antibody to HBeAg in serum. These results would be taken as evidence to ascribe the presence of HBeAg to the HBV/A coinfecting them with HBV/G, although transfection experiments are required to ascertain that HBV/G has an HBeAg-negative phenotype.
Given strong evidence for frequent coinfection of HBV/G with HBV/A, one wonders whether HBV/G is replication competent by itself or if it represents a defective virion in need of HBV/A to persistently infect hosts. A close association between HBV/G and HBV/A genomes manifested itself with high homology within the S gene sequence up to 94.6 to 97.5% that is closer than the divergence of >4% in the S gene separating the six major genotypes (A-F) (20). The similarity may well be due to coinfection with HBV/G and HBV/A and the recombination between them.
Replication competence of the HBV/G genome can be evaluated in transfection of HepG2 or Huh7 cell lines by it alone or with HBV/A and, better still, in experimental transmission to chimpanzees. The 36-bp insertion changes the conformation of the ɛ-encapsidation signal (7) and gives rise to an additional Watson-Crick pair between A1914 (the last nucleotide inserted) and U1817 (the fourth nucleotide in the pre-C region) that can consolidate it theoretically. Whether or not it might give HBV/G an advantage to overgrow HBV/A and thrive in some infected individuals (9) would be a another matter of virological interest.
REFERENCES
- 1.Bhat, R. A., P. P. Ulrich, and G. N. Vyas. 1990. Molecular characterization of a new variant of hepatitis B virus in a persistently infected homosexual man. Hepatology 11:271-276. [DOI] [PubMed] [Google Scholar]
- 2.Carman, W. F., M. R. Jacyna, S. Hadziyannis, P. Karayiannis, M. J. McGarvey, A. Makris, and H. C. Thomas. 1989. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet ii:588-591. [DOI] [PubMed]
- 3.Chayama, K., Y. Suzuki, M. Kobayashi, A. Tsubota, M. Hashimoto, Y. Miyano, H. Koike, I. Koida, Y. Arase, S. Saitoh, N. Murashima, K. Ikeda, and H. Kumada. 1998. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology 27:1711-1716. [DOI] [PubMed] [Google Scholar]
- 4.Felsenstein, J. 1985. Confidence limits on phylogenesis: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 5.Gojobori, T., K. Ishii, and M. Nei. 1982. Estimation of average number of nucleotide substitutions when the rate of substitution varies. J. Mol. Evol. 28:414-423. [DOI] [PubMed] [Google Scholar]
- 6.Ina, Y. 1994. ODEN: a program package for molecular evolutionary analysis and database search of DNA and amino acid sequences. Comput. Appl. Biosci. 10:11-12. [DOI] [PubMed] [Google Scholar]
- 7.Junker-Niepmann, M., R. Bartenschlager, and H. Schaller. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 9:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 2000. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 118:554-559. [DOI] [PubMed] [Google Scholar]
- 9.Kato, H., E. Orito, R. G. Gish, N. Bzowej, M. Newsom, F. Sugauchi, S. Suzuki, R. Ueda, Y. Miyakawa, and M. Mizokami. 2002. Hepatitis B e antigen in sera from individuals infected with hepatitis B virus of genotype G. Hepatology 35:922-929. [DOI] [PubMed] [Google Scholar]
- 10.Kato, H., E. Orito, F. Sugauchi, R. Ueda, R. G. Gish, S. Usuda, Y. Miyakawa, and M. Mizokami. 2001. Determination of hepatitis B virus genotype G by polymerase chain reaction with hemi-nested primers. J. Virol. Methods 98:153-159. [DOI] [PubMed] [Google Scholar]
- 11.Kremsdorf, D., F. Garreau, F. Capel, M. A. Petit, and C. Brechot. 1996. In vivo selection of a hepatitis B virus mutant with abnormal viral protein expression. J. Gen. Virol. 77:929-939. [DOI] [PubMed] [Google Scholar]
- 12.Lee, W. M. 1997. Hepatitis B virus infection. N. Engl. J. Med. 337:1733-1745. [DOI] [PubMed] [Google Scholar]
- 13.Leung, N. 2000. Liver disease-significant improvement with lamivudine. J. Med. Virol. 61:380-385. [PubMed] [Google Scholar]
- 14.Lindh, M., A. S. Andersson, and A. Gusdal. 1997. Genotypes, nt 1858 variants, and geographic origin of hepatitis B virus—large-scale analysis using a new genotyping method. J. Infect. Dis. 175:1285-1293. [DOI] [PubMed] [Google Scholar]
- 15.Magnius, L. O., and H. Norder. 1995. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology 38:24-34. [DOI] [PubMed] [Google Scholar]
- 16.Mayerat, C., A. Mantegani, and C. Frei. 1999. Does hepatitis B virus (HBV) genotype influence the clinical outcome of HBV infection? J. Viral Hepat. 6:299-304. [DOI] [PubMed] [Google Scholar]
- 17.Miyakawa, Y., H. Okamoto, and M. Mayumi. 1997. The molecular basis of hepatitis B e antigen (HBeAg)-negative infections. J. Viral Hepat. 4:1-8. [DOI] [PubMed] [Google Scholar]
- 18.Mizokami, M., T. Nakano, E. Orito, Y. Tanaka, H. Sakugawa, M. Mukaide, and B. H. Robertson. 1999. Hepatitis B virus genotype assignment using restriction fragment length polymorphism patterns. FEBS Lett. 450:66-71. [DOI] [PubMed] [Google Scholar]
- 19.Norder, H., A. M. Courouce, and L. O. Magnius. 1994. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 198:489-503. [DOI] [PubMed] [Google Scholar]
- 20.Norder, H., B. Hammas, S. D. Lee, K. Bile, A. M. Courouce, I. K. Mushahwar, and L. O. Magnius. 1993. Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen. J. Gen. Virol. 74:1341-1348. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto, H., M. Imai, F. Tsuda, T. Tanaka, Y. Miyakawa, and M. Mayumi. 1987. Point mutation in the S gene of hepatitis B virus for a d/y or w/r subtypic change in two blood donors carrying a surface antigen of compound subtype adyr or adwr. J. Virol. 61:3030-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamoto, H., F. Tsuda, H. Sakugawa, R. I. Sastrosoewignjo, M. Imai, Y. Miyakawa, and M. Mayumi. 1988. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J. Gen. Virol. 69:2575-2583. [DOI] [PubMed] [Google Scholar]
- 23.Okamoto, H., S. Yotsumoto, Y. Akahane, T. Yamanaka, Y. Miyazaki, Y. Sugai, F. Tsuda, T. Tanaka, Y. Miyakawa, and M. Mayumi. 1990. Hepatitis B viruses with precore region defects prevail in persistently infected hosts along with seroconversion to the antibody against e antigen. J. Virol. 64:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orito, E., M. Mizokami, H. Sakugawa, K. Michitaka, K. Ishikawa, T. Ichida, T. Okanoue, H. Yotsuyanagi, S. Iino, and Japan HBV Genotype Research Group. 2001. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Hepatology 33:218-223. [DOI] [PubMed] [Google Scholar]
- 25.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 26.Stuyver, L., S. De Gendt, C. Van Geyt, F. Zoulim, M. Fried, R. F. Schinazi, and R. Rossau. 2000. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J. Gen. Virol. 81:67-74. [DOI] [PubMed] [Google Scholar]
- 27.Tran, A., D. Kremsdorf, F. Capel, C. Housset, C. Dauguet, M. A. Petit, and C. Brechot. 1991. Emergence of and takeover by hepatitis B virus (HBV) with rearrangements in the pre-S/S and pre-C/C genes during chronic HBV infection. J. Virol. 65:3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]