Abstract
We have used a reverse genetic approach to identify the viral proteins required for packaging and assembly of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV). Plasmids encoding individual LCMV proteins under the control of an RNA polymerase II promoter were cotransfected with a plasmid containing an LCMV minigenome (MG). Intracellular synthesis of the LCMV MG was driven by T7 RNA polymerase whose expression was also mediated by a Pol II promoter. The supernatant from transfected cells was passaged onto fresh cells that were subsequently infected with LCMV to provide the minimal viral trans-acting factors, NP and L, that are required for LCMV MG RNA replication and expression. Reconstitution of LCMV-specific packaging and passage was detected by expression of the chloramphenicol acetyl transferase (CAT) reporter gene present in the MG. NP and L did not direct detectable levels of MG passage. Addition of Z and GP resulted in high levels of passage of CAT activity, which could be prevented by LCMV neutralizing antibodies. Passage of LCMV MG was inhibited by omission of either GP or Z.
The prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) is an enveloped virus with a bisegmented negative-strand (NS) RNA genome. The two genomic RNA segments are designated L and S and have approximate sizes of 7.2 and 3.4 kb, respectively (21, 22, 26, 27). Each RNA segment has an ambisense coding strategy, encoding two proteins in opposite orientations, separated by an intergenic region (22, 27). The S RNA directs synthesis of the nucleoprotein (NP) (ca 63 kDa) (21, 22, 27), and the two virion glycoproteins (GP), GP-1 (40 to 46 kDa) and GP-2 (35 kDa), that are derived by posttranslational cleavage of a precursor polypeptide, GPC (75 kDa) (22, 27, 28, 32). GP-1 and GP-2 make up the spikes on the virion envelope and mediate virus interaction with host cell surface receptor (5, 7). The L RNA segment codes for the virus RNA-dependent RNA polymerase (L, ca 200 kDa) (26) and a small (11-kDa) RING finger protein (Z) (24). Both GP-1 and GP-2, as well as NP, L, and Z, are structural proteins present in virions. The NP and viral polymerase are complexed with the genomic viral RNA to form ribonucleoprotein (RNP) complexes, which are active in virus transcription and replication (11, 22, 27). As with other NS RNA viruses, this RNP is the minimum unit of LCMV infectivity.
Production of LCMV occurs by budding at the surfaces of infected cells. For most NS RNA viruses, this process is assumed to depend on the interaction between the ribonucleoprotein core and the virus-encoded transmembrane GP (12). The matrix (M) protein is thought to play an essential role in this interaction. Moreover, budding of rabies virus and vesicular stomatitis virus (VSV) does not require the presence of GP, suggesting an intrinsic budding activity of the M protein (14, 18). Nevertheless, GP can significantly enhance budding.
For a variety of NS RNA viruses, systems have been developed which permit the encapsidation, replication, and packaging of synthetic genomic RNA analogs into virus-like particles (VLPs) in cells expressing all the required viral polypeptides from plasmid. These VLPs are budded into the extracellular space and can infect new cells, where they will replicate if the required trans-acting viral proteins are also expressed. These systems have facilitated the investigation of the minimal viral protein requirements for maturation and budding of infectious VLPs. We recently described a system in which RNA synthesis, both transcription and replication, mediated by LCMV polymerase is reconstituted by intracellular coexpression of an LCMV minigenome and viral proteins from transfected plasmids (16). Using this system we showed that the 5′ and 3′ untranslated region together with the intergenic region of the S RNA are sufficient cis-acting signals to allow RNA synthesis mediated by LCMV RNA polymerase. We also demonstrated that NP and L are the minimal trans-acting viral factors required for replication and transcription of LCMV genome analogues (16). Z exhibited a strong inhibitory effect on both RNA replication and transcription in the minigenome system (8). Here we have examined the contributions of individual LCMV proteins to the formation of infectious VLPs.
The LCMV spike GP complex, composed of noncovalently linked tetramers of GP-1 and GP-2, is responsible for both viral attachment and fusion with cell membranes. Therefore, expression and correct processing of LCMV GP are expected to be required for formation of infectious LCMV-like particles (VLPs). Arenaviruses do not encode an obvious counterpart of the M protein. However, biochemical studies have suggested that Z might be the arenavirus counterpart of the matrix (M) protein found in other NS RNA viruses (22, 23). We therefore examined the role of Z and GP in the generation of LCMV-like infectious particles using the minigenome system. The prediction would be that the supernatant of cells cotransfected with the minigenome-encoding plasmid, together with plasmids encoding NP and L, as well as Z and GP, will contain infectious VLPs. We tested this hypothesis by examining the ability of the supernatant from transfected cells to drive CAT expression in fresh cells. For this, cells were infected with the VLP-containing supernatant and subsequently with helper LCMV to provide the trans-acting factors (NP and L) required for minigenome expression.
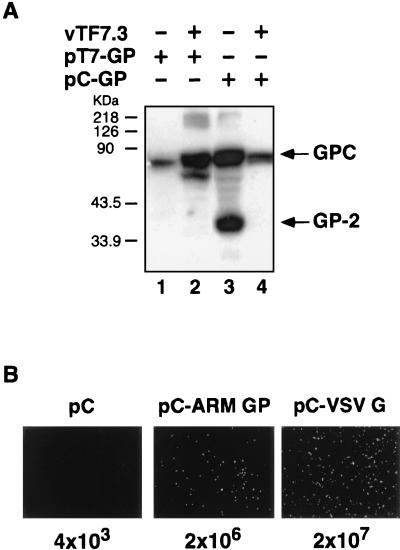
Our previously described LCMV minigenome system was based on the use of infection with the recombinant vaccinia virus vTF7-3 (10) to supply the T7 RNA polymerase required for plasmid-mediated expression of both minigenome RNA and viral trans-acting factors. We found, unexpectedly, that although the GPC precursor was efficiently expressed using the vTF7-3 system, its processing was severely impaired (Fig. 1A, lane 2). In contrast, we observed correct processing of GPC into GP-1 and GP-2 when GP was expressed using pCAGGS (referred to as pC), an expression vector based on the chicken â-actin promoter (19) (Fig. 1A, lane 3). However, processing of GPC was inhibited in cells transfected with pC-GP and also infected with vTF7-3 (Fig. 1A, lane 4), indicating that vaccinia virus infection actively interferes with the processing of LCMV GP. To overcome this problem, we expressed the viral trans-acting factors, as well as Z and GP, under the control of an RNA polymerase II promoter using the pC expression vector. We also included an additional plasmid (pC-T7) expressing T7 RNA polymerase to allow for T7-mediated intracellular synthesis of LCMV ARM S-segment minigenome (ARM/S-MG) RNA.
FIG. 1.
(A) Effect of vTF7-3 infection on the processing of LCMV GPC. BHK-21 cells, left uninfected (lanes 1 and 3) or infected with vTF7-3 (multiplicity of infection of 3) (lanes 2 and 4), were transfected with 0.5 μg of each pT7-GP (lanes 1 and 2) or pC-GP (lanes 3 and 4) by using Lipofectamine as described previously (8, 20). Cell lysates prepared at 36 h posttransfection were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and assayed for expression of GP by a Western blot. Detection of LCMV GPC and GP-2 was done using a mouse monoclonal antibody to GP-2 (32). The positions of the GPC and GP-2 proteins are indicated on the right. The GP-2 monoclonal antibody detected a nonspecific protein in lysates of uninfected control cells (white asterisk). (B) Generation of infectious VSVΔG* pseudotype particles by complementation with LCMV GP. 293T cell monolayers in six-well plates (80% confluent) were transfected with 2 μg of the plasmid DNA indicated at the top of each panel by using Lipofectamine. Thirty-two hours after transfection, cells were infected with VSVΔG* at a multiplicity of infection of 3 PFU/cell. After a 60-min adsorption period, the inoculum was removed, cells were extensively washed with DMEM, and fresh culture medium was added. Twenty hours postinfection, supernatants were collected and clarified by low-speed centrifugation, and their infectivity was analyzed on Vero cells. Representative fields of Vero cells infected with each type of pseudotype virus are shown. Viral titers are indicated at the bottom of each panel.
Since single amino acid substitutions can affect trafficking and binding properties of LCMV GP (2), we tested whether the GP protein encoded by pC-GP was functional in receptor recognition and cell entry. For this, we used a pseudotype approach based in a recently developed recombinant VSV in which the gene for green fluorescent protein (GFP) was substituted for the VSV G protein gene (VSVΔG*) (29). Complementation of VSVΔG* with pC-GP, but not with empty pC, resulted in infectious VSVΔG* pseudotypes (Fig. 1B).
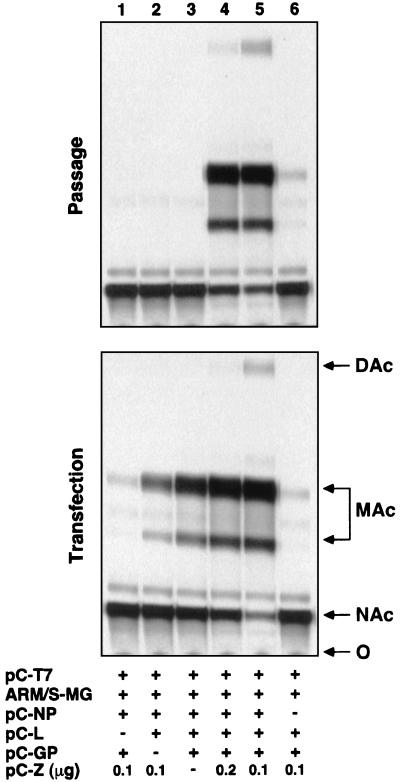
We then examined the requirements of viral proteins for production of LCMV infectious VLPs. The absence of Z or GP in the transfection mix did not affect levels of CAT activity in cell lysates of transfected cells but prevented CAT expression in fresh cells coinfected with the supernatant of transfected cells and helper LCMV (Fig. 2). These findings suggest that efficient assembly, budding, and passaging of VLPs requires both Z and GP. However, the individual contributions of GP and Z to these processes remain to be established.
FIG. 2.
Passage of CAT activity to a fresh monolayer of BHK-21 cells. 293T cell monolayers in six-well plates (80% confluent) were transfected with pC-T7 (1 μg), ARM/S-MG (0.5 μg), pC-NP (0.8 μg), pC-L (0.2 μg), pC-GP (0.5 μg), and pC-Z (0 to 0.2 μg) using the combinations indicated in the chart. Forty-eight hours posttransfection, supernatants (1.5 ml) were saved and cell lysates were prepared for CAT assay (8). Aliquots (0.5 ml) of supernatants from transfected cells were used to infect fresh monolayers of BHK-21 cells in six-well plates. After adsorption of the supernatants for 2 h, LCMV helper virus (ARM strain) was added to a multiplicity of infection of 2 PFU/cell, and adsorption was continued for another 90 min. After this, the inoculum was removed and fresh culture medium was added. Seventy-two hours postinfection, cell lysates were prepared for CAT assay. Aliquots from lysates of 293T (transfection) and BHK-21 (passage) cells were assayed for CAT activity (8). O, origin; Cm, chloramphenicol; MAc, monoacetylated chloramphenicol; DAc, diacetylated chloramphenicol.
The requirement of GP for passaging of LCMV MG was expected based on the proposed role of GP-1 in virus receptor recognition (5, 7). Consistent with this, LCMV GP was sufficient to mediate cell attachment and entry of VSVΔG* pseudotypes (Fig. 1B). Besides its role in receptor interaction and cell entry, GP may also participate in assembly and budding. Z is not required for replication and expression of ARM/S-MG or formation of functionally active RNPs (8). Hence, the requirement of Z for production of infectious VLPs likely reflects a role of Z in assembly and budding. Nevertheless, we cannot presently rule out a possible participation of Z in early steps of virus entry.
Direct biochemical characterization of produced VLPs would be very informative. Unfortunately, we were unable to biochemically characterize VLPs by detecting LCMV proteins in the supernatant of transfected cells. Nevertheless, using reverse transcription (RT)-PCR, we detected ARM/S-MG RNA associated with infectious VLPs in the supernatant of cells transfected with Z and GP in addition to L and NP plasmids. The low level of VLP production may be related to the relatively low level of virion production, 5 to 10 infectious particles per cell, characteristically found in LCMV-infected cells.
We conducted control experiments to confirm that the CAT activity detected in the passage was mediated by GP-containing VLPs. Incubation of the supernatant of transfected cells with anti-LCMV GP-1 neutralizing antibodies, but neither with an anti-VSV serum nor with RNase, inhibited CAT activity in the passage (Fig. 3A). The LCMV neutralizing monoclonal antibodies that were used did not have a significant effect on the infectivity associated with LCMV RNP (Fig. 3B). These results indicate that CAT expression detected in the passage was mediated by GP-containing VLPs and not by RNP complexes or free CAT RNA molecules. Moreover, in the absence of infection with helper virus, we did not detect CAT activity in the passage (Fig. 3A, lane 8), indicating that LCMV VLPs enclosed a CAT RNA rather than CAT enzyme.
FIG. 3.
(A) Passage of CAT activity is inhibited by treatment with anti-LCMV neutralizing antibodies but not by RNase treatment. Lanes 1 and 2 correspond to 293T cells transfected with the combination of plasmids indicated in lanes 5 and 1, respectively, of Fig. 2. Forty-eight hours posttransfection, supernatants were saved and cell lysates were prepared for CAT assay. Supernatant from cells transfected with the complete set of plasmids (lane 1) was subjected to the treatments indicated in the chart prior to being used for infection of BHK-21 cells. In all cases, except lane 8, BHK-21 cells were subsequently infected with helper LCMV at a multiplicity of infection of 2 PFU/cell. Seventy-two hours postinfection with helper LCMV, cell lysates were prepared for CAT assay. Aliquots of lysates from transfection (lanes 1 and 2) and passage (lanes 3 to 9) samples were assayed for CAT activity. (B) Monoclonal antibodies 36.1 and 197.1 to LCMV GP-1 (31) neutralized virion-associated, but not RNP-associated, LCMV infectivity. Aliquots of purified LCMV virions (200 PFU) and RNP (equivalent to 100 PFU) prepared from cytosolic extracts of LCMV-infected BHK-21 cells as described previously (8), were treated as indicated in the chart, and their infectivities were tested by standard plaque assay (virions) or transfection (RNP) under agarose overlay. Infectivity of untreated samples was considered to be 100% in each case; these values were used to normalize the infectivity of samples treated as indicated in the chart.
When we compared levels of CAT activity, normalized on a per-cell basis, they were much higher in the transfected cell population than in the passage cell population. Based on this finding, one would predict that the percentage of cells positive for reporter gene expression would be significantly lower in the passage than the transfection. Measurement of CAT activity provides information about the efficiency of transcription and replication of an LCMV RNA analog containing the CAT reporter gene in the population of transfected or passage cells, but it does not allow determination of the frequency of cells capable of carrying out the synthetic events within the cell population. To address this issue, we used for our assay an ARM/S-MG encoding the GFP in place of CAT. This allowed us to visualize directly the fraction of cells transcribing the minigenome in both the transfected and passage cell populations. In several independent experiments, we observed that only 1 to 3% of the cells expressed levels of GFP that were readily detected by direct epifluorescence upon transfection with NP and L and in the presence or absence of Z and/or GP. Consistent with the CAT results (Fig. 2), after passage and superinfection with LCMV, GFP foci were observed only in cells incubated with the supernatant of cells transfected with all four L, NP, GP, and Z LCMV proteins (Fig. 4). Intriguingly, we found that infection with LCMV up to 90 min prior to infection with VLPs prevented successful passage of the reporter gene activity. We obtained the same results with both the GFP- and CAT-based ARM/S-MG. We do not have presently an explanation for this phenomenon, but it could be a reflection of the apparent replicative disadvantage of the ARM/S-MG RNP with respect to the bona fide viral RNP as indicated by the rapid decrease seen in CAT activity over serial passages. Thus, we did not detect CAT activity in cell extracts from the third and later serial passages of LCMV VLPs (data not shown). In the case of the GFP ARM/S-MG, we could not detect gene reporter activity in passage P2. Moreover, in passage P1 we observed a decrease in the number of GFP-positive cells over time. This finding most likely reflects a high level of sensitivity of the CAT assay compared to detection of GFP expression by epifluorescence. However, it is also possible that cell toxicity due to constitutive high expression levels of GFP contributed to these results (17).
FIG. 4.
Expression of GFP ARM/S-MG in transfected and passage cell populations. 293T cells in six-well plates (80% confluent) were transfected with pMG-GFP (1 μg), pC-T7 (1 μg), pC-NP (0.8 μg), pC-L (0.2 μg), pC-GP (0.3 μg), and pC-Z (0.1 μg). Cells were analyzed for GFP expression 48 h after transfection (lower panels). Supernatants were collected 48 h after transfection, and aliquots (0.3 ml) were used to infect fresh monolayers of BHK-21 cells seeded on M24 plates. Cells were incubated with supernatants for 4 h before adding helper LCMV (MOI of 2 PFU/cell). After 90 min, the inoculum was removed and fresh medium was added. Sixty hours postinfection the passage culture was examined for GFP expression (top panels). GFP-positive foci (one to five per M24 well) were only detected in the passage of cells transfected with GP and Z in addition to L and NP.
Generation of VLPs, as well as recombinant viruses, containing alternative, or additional, heterologous GP that can be used as attachment proteins has been documented for several NS RNA viruses (15, 25, 29). Investigations to delineate the requirements for receptor recognition and cell entry, as well as antibody-mediated neutralization, may facilitate the development of effective antiviral therapies for highly pathogenic arenaviruses, such as Lassa fever virus. The ability to generate LCMV VLPs expressing heterologous GPs can overcome some of the biosafety issues associated with these studies. Our finding that Lassa fever virus GP could efficiently substitute for LCMV GP in the generation of VLPs (data not shown) supports the feasibility of this experimental approach.
Ample evidence indicates that LCMV GP-1 is responsible for receptor recognition and cell entry. Thus, monoclonal antibodies to GP-1, but not to other viral polypeptides, have been shown to have strong neutralizing activity and to protect mice against lethal intracraneal infection with LCMV (1). Therefore, as predicted, expression and correct processing of GPC was required for the formation of LCMV infectious VLPs. In contrast, the role of Z during the natural course of LCMV infection is poorly understood. We previously showed that Z is not required for replication and transcription of ARM/S-MG mediated by the LCMV polymerase. Z has been shown to interact with several host cell proteins. The association of Z with the eukaryotic initiation factor 4E has been implicated in repression of protein synthesis in a eukaryotic initiation factor 4E-dependent manner (6). In addition, Z interacts also with the promyelocytic leukemia protein, leading to the relocation of promyelocytic leukemia protein nuclear bodies to the cytoplasm, which has been proposed to be responsible for the noncytolytic nature of LCMV (3, 4). Together, these observations raise intriguing questions about a spectrum of potential functions played by the Z protein in the biology of arenaviruses. The requirement of Z for formation of LCMV infectious VLPs raises the possibility that Z could also play a role in budding similar to that assigned to the M proteins of other NS viruses. Interestingly, the C termini of arenavirus Z proteins contain a PPxY motif similar to that found in the N terminus of the M protein of VSV and other rhabdoviruses as well as filoviruses. This PPxY sequence motif closely resembles the PY motif in the late domain in the Gag protein of Rous sarcoma virus and other retroviruses (30, 33). Biochemical and genetic evidence indicates that the PPxY motif present in the M protein of several mononegaviruses is involved in promoting virus budding (9, 13, 14). We are currently investigating the possible role of the PPxY motif present in the arenavirus Z proteins in virus assembly and budding.
Acknowledgments
We are indebted to M. Buchmeier for the monoclonal antibodies to LCMV GP-1 and to M. Whitt for the monoclonal antibody to VSV G.
This work was supported by NIH grant AI47140 (J.C.T.).
Footnotes
This is publication 14640-NP from The Scripps Research Institute.
REFERENCES
- 1.Baldridge, J. R., and M. J. Buchmeier. 1992. Mechanisms of antibody-mediated protection against lymphocytic choriomeningitis virus infection: mother-to-baby transfer of humoral protection. J. Virol. 66:4252-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyer, W. R., H. Miletic, W. Ostertag, and D. von Laer. 2001. Recombinant expression of lymphocytic choriomeningitis virus strain WE glycoproteins: a single amino acid makes the difference. J. Virol. 75:1061-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borden, K. L., E. J. Campbell Dwyer, and M. S. Salvato. 1998. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J. Virol. 72:758-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borden, K. L., E. J. Campbell Dwyer, and M. S. Salvato. 1997. The promyelocytic leukemia protein PML has a pro-apoptotic activity mediated through its RING domain. FEBS Lett. 418:30-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrow, P., and M. B. Oldstone. 1994. Mechanism of lymphocytic choriomeningitis virus entry into cells. Virology 198:1-9. [DOI] [PubMed] [Google Scholar]
- 6.Campbell Dwyer, E. J., H. Lai, R. C. MacDonald, M. S. Salvato, and K. L. Borden. 2000. The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J. Virol. 74:3293-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, W., M. D. Henry, P. Borrow, H. Yamada, J. H. Elder, E. V. Ravkov, S. T. Nichol, R. W. Compans, K. P. Campbell, and M. B. Oldstone. 1998. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282:2079-2081. [DOI] [PubMed] [Google Scholar]
- 8.Cornu, T. I., and J. C. de la Torre. 2001. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J. Virol. 75:9415-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuller-Pace, F. V., and P. J. Southern. 1989. Detection of virus-specific RNA-dependent RNA polymerase activity in extracts from cells infected with lymphocytic choriomeningitis virus: in vitro synthesis of full-length viral RNA species. J. Virol. 63:1938-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garoff, H., R. Hewson, and D. J. E. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74:9818-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahn, J. S., M. J. Schnell, L. Buonocore, and J. K. Rose. 1999. Recombinant vesicular stomatitis virus expressing respiratory syncytial virus (RSV) glycoproteins: RSV fusion protein can mediate infection and cell fusion. Virology 254:81-91. [DOI] [PubMed] [Google Scholar]
- 16.Lee, K. J., I. S. Novella, M. N. Teng, M. B. Oldstone, and J. C. de La Torre. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 74:3470-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, H. S., M. S. Jan, C. K. Chou, P. H. Chen, and N. J. Ke. 1999. Is green fluorescent protein toxic to the living cells? Biochem. Biophys. Res. Commun. 260:712-717. [DOI] [PubMed] [Google Scholar]
- 18.Mebatsion, T., M. Konig, and K. K. Conzelmann. 1996. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell 84:941-951. [DOI] [PubMed] [Google Scholar]
- 19.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 20.Perez, M., M. Watanabe, M. A. Whitt, and J. C. de la Torre. 2001. N-terminal domain of Borna disease virus G (p56) protein is sufficient for virus receptor recognition and cell entry. J. Virol. 75:7078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riviere, Y., R. Ahmed, P. J. Southern, M. J. Buchmeier, F. J. Dutko, and M. B. Oldstone. 1985. The S RNA segment of lymphocytic choriomeningitis virus codes for the nucleoprotein and glycoproteins 1 and 2. J. Virol. 53:966-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salvato, M. S. 1993. The Arenaviridae, 1st ed., vol. 1. Plenum Press, New York, N.Y.
- 23.Salvato, M. S., K. J. Schweighofer, J. Burns, and E. M. Shimomaye. 1992. Biochemical and immunological evidence that the 11 kDa zinc-binding protein of lymphocytic choriomeningitis virus is a structural component of the virus. Virus Res. 22:185-198. [DOI] [PubMed] [Google Scholar]
- 24.Salvato, M. S., and E. M. Shimomaye. 1989. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology 173:1-10. [DOI] [PubMed] [Google Scholar]
- 25.Schnell, M. J., L. Buonocore, E. Kretzschmar, E. Johnson, and J. K. Rose. 1996. Foreign glycoproteins expressed from recombinant vesicular stomatitis viruses are incorporated efficiently into virus particles. Proc. Natl. Acad. Sci. USA 93:11359-11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh, M. K., F. V. Fuller-Pace, M. J. Buchmeier, and P. J. Southern. 1987. Analysis of the genomic L RNA segment from lymphocytic choriomeningitis virus. Virology 161:448-456. [DOI] [PubMed] [Google Scholar]
- 27.Southern, P. J. 1996. Arenaviridae: the viruses and their replication, p. 1505-1551. In D. M. K. Bernard, N. Fields, and Peter M. Howley (ed.), Fields virology, 3rd edition ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 28.Southern, P. J., M. K. Singh, Y. Riviere, D. R. Jacoby, M. J. Buchmeier, and M. B. Oldstone. 1987. Molecular characterization of the genomic S RNA segment from lymphocytic choriomeningitis virus. Virology 157:145-155. [DOI] [PubMed] [Google Scholar]
- 29.Takada, A., C. Robison, H. Goto, A. Sanchez, K. G. Murti, M. A. Whitt, and Y. Kawaoka. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA 94:14764-14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright, K. E., M. S. Salvato, and M. J. Buchmeier. 1989. Neutralizing epitopes of lymphocytic choriomeningitis virus are conformational and require both glycosylation and disulfide bonds for expression. Virology 171:417-426. [DOI] [PubMed] [Google Scholar]
- 32.Wright, K. E., R. C. Spiro, J. W. Burns, and M. J. Buchmeier. 1990. Post-translational processing of the glycoproteins of lymphocytic choriomeningitis virus. Virology 177:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]