Abstract
The 293 cell line that was generated by transforming human embryonic kidney cells with human adenovirus type 5 (HAV5) early region 1 (E1) sequences is an excellent host for generating and growing HAV5 recombinants with E1 deleted, but it does not support the replication of bovine adenovirus type 3 (BAV3). Madin-Darby bovine kidney (MDBK), an established bovine cell line, is an excellent host for growing and plaquing BAV3. For the purpose of combining the unique characteristics of these two cell lines (293 and MDBK), we generated a number of bovine × human hybrid (BHH) cell lines. Comparison of three BHH hybrid clones—BHH3, BHH8, and BHH2C—with 293-Puro (puromycin-resistant 293 cells) and MDBK-Neo (G418-resistant MDBK cells) cell lines for total cellular DNA content, species-specific surface markers, isoenzyme analysis, and karyotyping indicate that they are hybrid in nature. BHH clones constitutively expressed the E1 proteins (E1A, E1B-21kDa, and E1B-55kDa) of HAV5 and efficiently supported the replication of both wild-type and replication-incompetent bovine or human adenoviruses. Transient gene expression experiments with a plasmid encoding the bacterial β-galactosidase gene demonstrated that BHH cell hybrids seem to have better transfection efficiencies than either of the parental cell lines. These cell lines will be useful for isolating and growing replication-competent human or bovine adenovirus recombinants with E1 deleted and for the study of cellular or viral factors important for viral replication. The development of somatic cell hybrids appears to be a simple way of combining some of the desirable characteristics present separately in two parental cell lines.
The most widespread use of somatic cell hybrids is the formation of hybridomas for monoclonal antibody production. They are generated by the fusion of antibody-producing lymphocytes from a mouse or rat immunized with an antigen of interest with a myeloma cell line (1, 29). Production of inter- and intraspecific cell hybrids by means of polyethylene glycol (PEG)-induced fusion for extending the life of mammalian cells has been demonstrated (42). Somatic cell hybrids have also been used as tools for studying the chromosomal assignment of genes (12, 26, 27), mapping normal as well as tumor-related genes (47), and localizing the site-specific integration of adeno-associated proviral DNA into human chromosome 19 (31, 49). Hybridization between bovine × rodent, mouse, or hamster were exploited for the mapping of bovine genes (51), and bovine × mouse hybrid cells were used for studying the replication of mengo virus (7, 30, 55).
Adenoviral early region 1 (E1) genes are the first set of the genes that are expressed and are necessary for virus replication (22, 50). Adenovirus recombinants with E1 deleted are routinely produced and grown in a permissive cell line that constitutively expresses E1 proteins (17). The 293 cell line constitutively expresses E1 proteins and was derived from human embryonic kidney cells after transformation with human adenovirus type 5 (HAV5) DNA (17). It is widely used for the propagation of HAV5 recombinants with E1 deleted (5, 16). In addition to HAVs, a number of nonhuman adenoviruses, such as bovine adenovirus type 3 (BAV3) (37), canine adenovirus type 2 (28), porcine adenovirus type 3 (45), and ovine adenovirus (23), have been developed as vectors for producing recombinant vaccines and also for their potential use in gene therapy.
Functional trans-complementation between BAV3 E1A and HAV5 E1A was initially demonstrated by showing the transactivation of HAV5 E2 and E3 promoters (56) and subsequently by replicating a BAV3 with deletions of E1A and E3 (BAV3ΔE1AE3) (44) in a cell line derived from fetal bovine retinal cells (FBRT), VIDO R2 (44), or FBRT-HE1 (52) that constitutively express HAV5 E1 proteins. It was also observed that the level of complementation between the homologous (BAV3 and BAV3 E1) or the heterologous (BAV3 and HAV5 E1) system was similar (52).
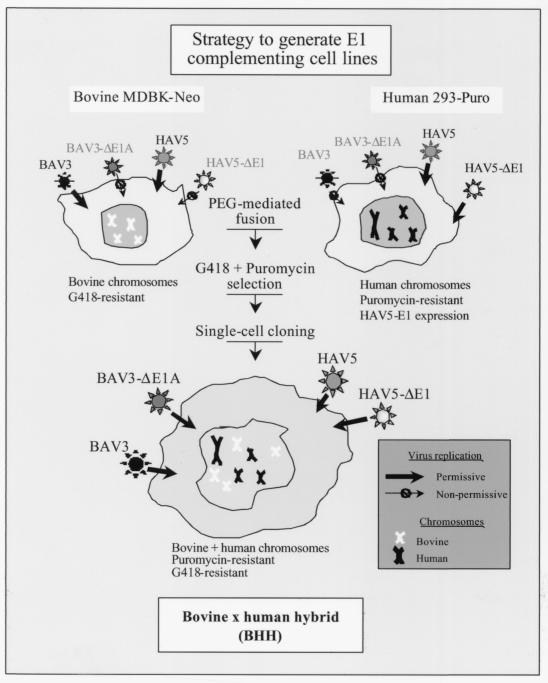
On the basis of the observations that (i) 293 cells express HAV5 E1 proteins but do not support the replication of wild-type (wt) BAV3 (43), (ii) Madin-Darby bovine kidney (MDBK) cell line is an excellent host for growing and titrating wt BAV3, and (iii) there is efficient functional trans-complementation between BAV3 E1A and HAV5 E1A, we hypothesized that the concept of somatic cell hybrids could be easily tested by generating hybrids that support replication of BAV3ΔE1AE3. The rationale for generating bovine × human hybrid (BHH) cell lines is presented in Fig. 1. We describe here the generation and characterization of three BHH cell lines and discuss the usefulness of the somatic cell hybridization technique for generating cell lines with desired characteristics and for studying some aspects of viral replication.
FIG. 1.
Strategy for the generation of somatic cell hybrids. MDBK cells support the replication of wt BAV3 and wt HAV5 but do not complement viruses with E1 deleted, whereas 293 cells that constitutively express HAV5 E1 proteins support the growth of HAV5 with E1 deleted and the wt. Some of the unique characteristics of both of the parental cell lines (MDBK and 293) could be combined by generating BHH cell lines. Some of the BHH clones should assist the replication of both HAV5 and/or BAV3 with E1 deleted and the wt.
MATERIALS AND METHODS
Cells and viruses.
MDBK, 293, and A549 cells were maintained in complete minimal essential medium (MEM) supplemented with 10% fetal calf serum (FetalClone III; HyClone) and 50 μg of gentamicin/ml. The cell lines 293-Puro and MDBK-Neo were maintained in complete MEM containing 2 μg of puromycin (Sigma Chemical)/ml and 400 μg of G418 (Geneticin; Life Technologies)/ml, respectively. Bovine × human somatic cell hybrids were maintained in complete MEM containing 2 μg of puromycin and 400 μg of G418/ml. The WBR-1 strain of BAV3 was obtained from the American Type Culture Collection (ATCC) and was propagated and titrated in MDBK cells. BAV3 with E1A deleted, BAV3ΔE1AE3 (44), was grown and titrated in FBRT-HE1 (a cell line derived from fetal bovine retinal cells transfected with HAV5 E1) (52). HAV5 and AdCA36lacZ (HAV5 recombinant with E1 deleted containing the bacterial β-galactosidase [lacZ] gene) (2) were grown and titrated in 293 or BHH2C cells (generated in this study). BAV3ΔE1AE3 and AdCA36lacZ are referred to as BAV3-ΔE1A and HAV5-ΔE1, respectively, in this study. Virus stocks were maintained in phosphate-buffered saline (PBS; pH 7.2) containing 0.01% MgCl2, 0.01% CaCl2, and 10% glycerol and stored at −70°C. The virus purification and DNA extraction were carried out as previously described (18, 38).
Generation of parental cell lines resistant to antibiotic selection.
Semiconfluent cell monolayers of 293 were grown in complete MEM in 60-mm culture dishes and transformed with a plasmid (pJ6Ω-puro) containing the puromycin resistance (puromycin-N-acetyltransferase) gene (11) by using Lipofectin (Life Technologies). A number of puromycin-resistant clones were isolated; one of these clones was named 293-Puro and was used as one of the fusion partners for the generation of BHH cells.
Similarly, MDBK cells were transfected with plasmid pcDNA3 (Invitrogen) containing the neomycin resistance (aminoglycoside acetyltransferase) gene (48). Several G418-resistant clones were isolated; one of these clones was named MDBK-Neo and was used for fusion experiments. Except for the antibiotic-resistant phenotype, there were no morphological or functional differences between 293 and 293-Puro or between the MDBK and MDBK-Neo cell lines.
Formation of BHH somatic cell hybrids by using PEG-induced fusion.
The fusion protocol described by Davidson and Gerald (10) was used with the following modifications. Briefly, 293-Puro and MDBK-Neo cells were grown in complete MEM supplemented with 2 μg of puromycin and 400 μg of G418/ml, respectively. Cells were trypsinized, counted, and plated in 60-mm dishes in the amounts indicated in Table 1 and incubated at 37°C for 24 h. The culture media were removed, and the cells were overlaid with a 50% solution of PEG 1450 (molecular weight, 1,450; Sigma Chemical) in 1× MEM and incubated at the room temperature. After 1 min, the PEG solution was aspirated off, followed by three quick rinses with MEM. The cells were cultured in a 5% CO2 incubator for 24 h in complete MEM. The cells were trypsinized and plated at lower densities (ca. 5 × 105 per 60-mm dish) in complete MEM. Dual antibiotic selection (complete MEM containing 600 μg of G418 and 2 μg of puromycin/ml) started 72 h after fusion. Individual clones that appeared after 3 weeks of selection were collected by using cloning cylinders, transferred to 12-well tissue culture plates, subsequently propagated in 60-mm dishes, and aliquoted as frozen stocks for future use. The cells remaining in the 60-mm dishes after the collection of individual clones were also trypsinized and collected as a pool. These pooled cells were named BHH-P/N and were used for single cell cloning by using limiting dilutions from which clones BHH3 and BHH8 were selected for further analysis. The clone BHH2C was isolated as an independent cell colony. Hybrid cell lines between passage levels 20 and 30 were used for various assays described in this study.
TABLE 1.
Ratios of 293-Puro and MDBK-Neo cells used for PEG-induced fusiona
| No. of 293- Puro cells | No. of MDBK- Neo cells | 293-Puro/MDBK- Neo ratio |
|---|---|---|
| 5 × 105 | 5 × 105 | 1:1 |
| 1 × 106 | 1 × 105 | 10:1 |
| 1 × 106 | 1 × 104 | 100:1 |
| 1 × 106 | 1 × 103 | 1,000:1 |
| 1 × 106 | 0 | Neob |
| 1 × 105 | 1 × 106 | 1:10 |
| 1 × 104 | 1 × 106 | 1:100 |
| 1 × 103 | 1 × 106 | 1:1,000 |
| 0 | 1 × 106 | Puroc |
Various numbers of cells of each parental cell line were mixed, seeded in 60-mm dishes, and incubated for 24 h before PEG-induced fusion.
Neo, neomycin selection control.
Puro, puromycin selection control.
Cytometric analysis of nuclear DNA content.
The protocol for the measurement of DNA content of propidium iodide-stained cells has been described elsewhere (46). Briefly, exponentially growing cultures of 293-Puro, MDBK-Neo, BHH2C, BHH3, and BHH8 cells were trypsinized, and the cell pellets were washed with ice-cold PBS. The cells (106) were mixed with an equal volume of Vindelov's propidium iodide stain (for 100 ml, we used 100 mM Tris, 10 mM NaCl, 1 mg of RNase, 5 mg of propidium iodide, and 0.1 ml of Nonidet P-40, adjusted pH to 8.0 and filtered through 0.45-μm [pore-size] membrane filter) and incubated for 30 min. Cell suspensions were passed through a 30-μm mesh to eliminate non-single cells. The samples were analyzed at the Purdue Cytometry Laboratory by using a Coulter XL-MCL cytometer at 488 nm.
Species-specific immunofluorescence and isoenzyme analysis.
Parental (293-Puro and MDBK-Neo) and hybrid (BHH2C, BHH3, and BHH8) cell lines were tested for species-specific markers by immunofluorescence with anti-human, anti-bovine, and anti-mouse sera (41).
BHH3, BHH8, BHH2C, MDBK-Neo, and 293-Puro cells extracts were analyzed for glucose-6-phosphate dehydrogenase (G6PD), malate dehydrogenase (MDH), purine nucleoside phosphorylase (NP), and lactate dehydrogenase (LDH) isoenzymes. Each enzyme was analyzed by using a separate electrophoresis universal-agarose film. Samples were applied to the slots of the agarose film. The films were placed in an electrophoresis chamber and run for 25 min and stained by using colorimetric reactions according to a protocol presented elsewhere (19). The cell extract from a cell line of mouse origin was used as a negative control.
Immunofluorescence assays and isoenzyme analyses were done commercially at the Cell Culture Identification Service, The Children's Hospital of Michigan, Detroit.
Cytogenetic analysis.
Cytogenetic analysis of clones BHH3, BHH8, and BHH2C was performed by Applied Genetics Laboratory, Melbourne, Fla. After colcemid treatment the cells were stained with Wright's solution, and Giemsa-banded methaphase chromosomes were photographed and karyotyped (24).
Radiolabeling of proteins and immunoprecipitation.
Immunoprecipitations of radiolabeled cell extracts were performed essentially as described previously (36). Mock- or virus-infected cells were labeled with Trans[35S]-label containing methionine and cysteine (ICN Pharmaceuticals), and the labeled cell lysates were immunoprecipitated with E1-specific antibodies. HAV5 E1-specific antibodies were purchased as follows: HAV2 E1A (13S-5; Santa Cruz), HAV5 E1B-21kDa (1G11; Calbiochem), and HAV5 E1B-55kDa (9C10; Calbiochem). Immunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. At the end of the run, the gels were dried under vacuum, and the immunoprecipitated bands were captured by using a phosphor-storage imaging system (Cyclone; Packard BioScience).
Transient-transfection assay.
Plasmid pMvOBAV3ΔE1lacZ (ca. 37 kb) was generated from pMvOBAV3 (53) that contains the entire genome of BAV3 by replacing the E1 region with the lacZ gene under the control of the mouse cytomegalovirus immediate-early gene promoter. Hybrid (BHH3, BHH8, and BHH2C) and parental (293-Puro and MDBK-Neo) cell monolayers in 60-mm culture dishes were separately transfected with PacI-digested pMvOBAV3ΔE1lacZ (5 μg/dish) to release the BAV3 genome from the plasmid by using liposome-mediated transfection (Lipofectin; Life Technologies) according to the manufacturer's recommendations. At 72 h posttransfection, the cells were analyzed for LacZ expression by using histochemical staining and LacZ assay.
LacZ histochemical staining.
Cells expressing LacZ were stained in situ with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) in a histochemical staining reaction (8) as described previously (4). Cells expressing LacZ contained blue precipitates and were photographed and counted. The total number of blue cells present in 3 to 10 photographic images per cell line was counted and the results are presented as the mean value ± the standard deviation.
LacZ assay.
The colorimetric assay was performed essentially as previously described (4, 40). Purified Escherichia coli LacZ was used as a standard. The results were described as micrograms of LacZ per 106 cells.
RESULTS
Generation of BHH cell lines.
For the purpose of developing BHH cell lines from two established cell lines (293 and MDBK), a suitable selection system was needed to force the selective amplification of hybrid cells. Therefore, we first isolated MDBK-Neo (MDBK cells expressing the neomycin-resistant gene) and 293-Puro (293 cells expressing the puromycin-resistant gene) so that BHH clones could be easily identified in the presence of G418 and puromycin. Different cell ratios were used to increase the frequency of fusion events between the two cell lines, thereby leading to the formation of heterokaryons. This is particularly important where one of the cell lines has a considerably faster rate of fusion and hence causes the formation of massive multinucleated cells, which precludes or drastically reduces the interspecific cell fusion events (10).
MDBK-Neo and 293-Puro in various ratios were mixed and subjected to PEG-mediated fusion. After a 3-week-period of dual antibiotic selection, more than 80 clones were observed, trypsinized, and transferred to 12-well plates for propagation. All clones except for the plates containing 293-Puro:MDBK-Neo in a ratio of 1,000:1 or 1:1,000 were collected. Controls containing only one of the parental cell lines (293-Puro or MDBK-Neo) did not survive dual antibiotic selection. Fused cells were characterized by a marked increase in size with respect to the parental cells; many had multiple nuclei and appeared to grow slower, resulting in a high number of dead cells and rapid acidification of the culture media. This crisis period lasted for approximately 2 weeks, and 31 independent clones survived. The clone BHH2C was one of the 31 clones. The cells remaining in the 60-mm dishes after the collection of individual clones were trypsinized and collected as a pool and were given the name BHH-P/N. Sixty clones, including BHH3 and BHH8, were obtained after subcloning of the BHH-P/N pool by limiting dilutions. Different clones were tested for their ability to support the replication of either or both BAV3 and HAV5ΔE1, and thus BHH2C, BHH3, and BHH8 were selected for further characterization.
Confirmation of hybrid nature of BHH cell lines.
In order to ascertain the hybrid character of BHH cells, the cells were tested for the total DNA content, species-specific immunofluorescence, isoenzyme analysis, and karyotyping.
Amounts of DNA content in BHH clones.
Flow cytometric analysis of propidium iodide-stained cells allows a rapid assessment of the cellular DNA content and provides a preliminary measurement of the relative number of chromosomes present in a hybrid cell line with respect to the parental cells. In exponentially growing cells, the frequency distribution of the DNA content per cell plotted in a histogram could be used to identify the percentage of the cell population that is going through a particular phase of the cell cycle (e.g., G1, S, and G2/M) (32).
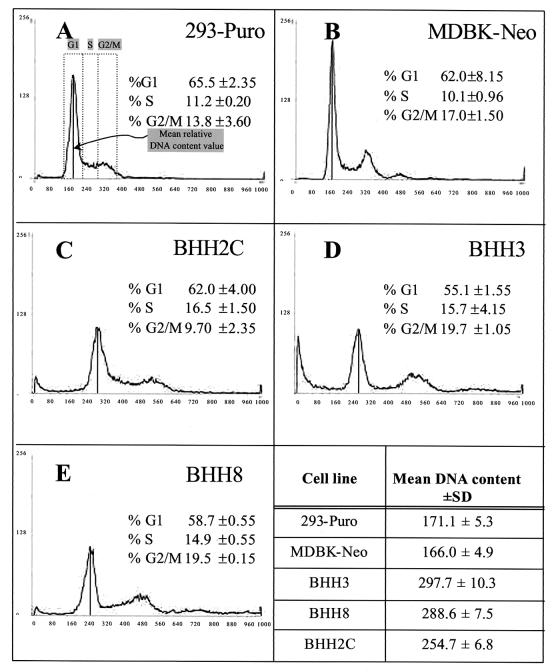
For flow cytometric analysis, exponentially growing cultures of BHH3, BHH8, and BHH2C were stained with propidium iodide and compared with the parental cell lines (293-Puro and MDBK-Neo). The values obtained for BHH3, BHH8, BHH2C, 293-Puro, and MDBK-Neo were 297.7, 288.6, 254.7, 171.1, and 166.0, respectively. The percent increases in the DNA contents of BHH3, BHH8, and BHH2C relative to the average value calculated for the parental cell lines were 76.6, 71.2, and 51.1%, respectively, suggesting that all hybrid clones had increased DNA content relative to the parental cell lines. The similar percentages of cell distribution frequencies in the different phases of the cell cycle (G1, S, and G2/M) of 293-Puro, MDBK-Neo, BHH3, BHH8, and BHH2C (Fig. 2) suggest that these are nonsenescent, actively dividing cell lines with a similar degree of transformation.
FIG. 2.
Flow cytometric analysis of propidium iodide-stained BHH cells. Panels A to E present the histograms showing the distribution frequencies of cells in different phases of the cell cycle. The area under the curve represents the number of cells in each phase of the cell cycle (G1, S, and G2/M) at a given time. The vertical solid line connecting the peak of the G1 curve with the x axis indicates the mean relative DNA content value. (A) 293-Puro; (B) MDBK-Neo; (C) BHH2C; (D) BHH3; (E) BHH8.
Species-specific immunofluorescence of BHH cells.
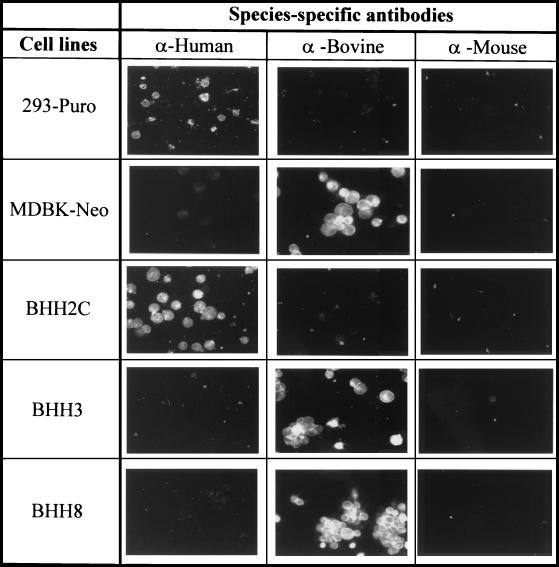
The species of origin of a cell line can easily be determined by detection of species-specific surface antigens (41). To determine whether hybrid clones predominantly possess bovine- or human-specific surface antigens, the BHH2C, BHH3, and BHH8 cell lines were analyzed by species-specific immunofluorescence assays. BHH3 and BHH8 cells showed extensive fluorescence with an anti-bovine fluorescein-conjugated serum, whereas BHH2C cells demonstrated positive staining with an anti-human fluorescein-conjugated serum (Fig. 3). No significant fluorescence was observed in BHH3 and BHH8 cells with either human- or mouse-specific antibodies. Similarly in BHH2C cells, no significant fluorescence was observed with either bovine- or mouse-specific antibodies. As expected, the parental cell lines, MDBK-Neo and 293-Puro, were stained with either bovine- or human-specific antibodies, respectively. The mouse-specific antibody was used as a negative control. These results suggested that BHH3 and BHH8 cells retained bovine-specific surface antigens, whereas BHH2C cells maintained human-specific antigens.
FIG. 3.
Identification of species-specific markers on hybrid cell lines. 293-Puro, MDBK-Neo, BHH2C, BHH3, or BHH8 cell monolayers were harvested and fixed on glass slides. Cells were reacted with fluorescein-conjugated anti-human, anti-bovine, or anti-mouse sera in a direct fluorescent assay and examined under a fluorescent microscope.
Isoenzyme analysis of BHH clones.
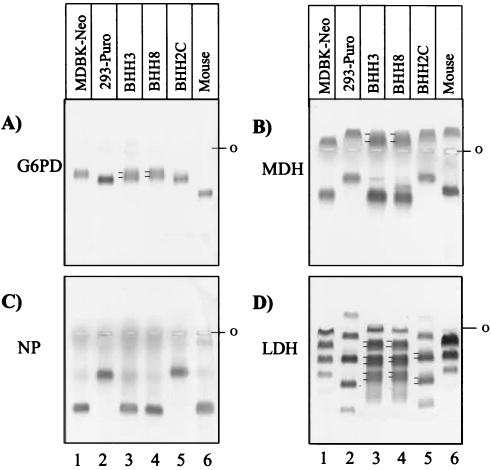
Isoenzymes are multimeric proteins that usually aggregate in pairs or, rarely, in triplets. An isoenzyme exhibits interspecies and intraspecies polymorphisms that can be detected by differences in the electrophoretic mobility patterns of the isoenzyme subunits. By using enzyme-specific colorimetric reactions, it is possible to determine the host species for a test sample by comparing it with known controls. In addition to the banding pattern similar to that of a particular isoenzyme observed in one of the parental cell lines, a hybrid cell line would contain one or more extra bands due to formation of heteromeric proteins, thus confirming that both of the subunits were produced in the same cell. To determine the hybrid nature of BHH3, BHH8, and BHH2C cell lines, cells were analyzed for electrophoretic mobility patterns for four isoenzymes (G6PD, MDH, NP, and LDH). MDBK-Neo and 293-Puro cells lysates were used as parental cell controls, and a mouse cell lysate served as a negative control. The G6PD (Fig. 4A, lane 5), MDH (Fig. 4B, lane 5), and NP (Fig. 4C, lane 5) isoenzyme patterns for BHH2C cells were comparable to those obtained with 293-Puro (Fig. 4A to C, lane 2). Interestingly, the banding pattern of LDH in the BHH2C (Fig. 4D, lane 2) demonstrated three extra bands that were not observed with 293-Puro (Fig. 4D, lane 2), indicating that BHH2C cells were a hybrid clone. The NP mobility patterns for BHH3 (Fig. 4C, lane 3) and BHH8 (Fig. 4C, lane 4) cells were similar to that obtained with MDBK-Neo cells (Fig. 4C, lane 1). The presence of one or more additional bands in the G6PD (Fig. 4A, lane 3), MDH (Fig. 4B, lane 3), and LDH (Fig. 4D, lane 3) isoenzyme patterns for BHH3 cells indicate the hybrid nature of this clone. The G6PD (Fig. 4A, lane 4), MDH (Fig. 4B, lane 4), and LDH (Fig. 4D, lane 4) isoenzyme patterns for BHH8 cells were similar to those patterns produced with BHH3 cells.
FIG. 4.
Isoenzyme analyses of BHH cell lines. The electrophoretic mobilities of G6PD (A), MDH (B), purine NP (C), and LDH (D) were determined for MDBK-Neo, 293-Puro, BHH3, BHH8, BHH2C, and a mouse cell line (control) extracts. The position of the double band, including the band generated due to aggregation of heteromeric subunits, is depicted by two lines on the left side of the band. ο, position of the wells where the samples were loaded.
Cytogenetic analysis of BHH clones.
For the purpose of determining the genetic make-up of BHH clones and to obtain a preliminary determination of the number of chromosomes originating from 293-Puro or MDBK-Neo cells, cytogenetic examination of the BHH3, BHH8, and BHH2C cell lines was conducted. The karyotypes (2N) reported by the ATCC for 293 (ATCC number CRL-1573) and MDBK (ATCC number CCL-22) are 62 and 60, respectively.
The number of normal human chromosomes, bovine chromosomes, and unidentified chromosomes observed in the karyotypes of the three hybrid cell lines are presented in Table 2. Unidentified chromosomes comprise all unidentified chromosomes and chromosomal rearrangements. BHH3, BHH8, and BHH2C cell lines retained 48, 47, and 4 bovine chromosomes and 27, 19, and 71 normal human chromosomes, respectively. In addition to normal human and bovine chromosomes, BHH3, BHH8, and BHH2C cells also contained 38, 41, and 22 unidentified chromosomes, respectively.
TABLE 2.
Karyotype of BHH cell hybridsa
| Human chromosome no. | No. of copies of normal human chromosomes in cell line:
|
||
|---|---|---|---|
| BHH3 | BHH8 | BHH2C | |
| 1 | 0 | 0 | 4 |
| 2 | 0 | 0 | 5 |
| 3 | 0 | 1 | 4 |
| 4 | 1 | 1 | 2 |
| 5 | 2 | 1 | 1 |
| 6 | 1 | 0 | 6 |
| 7 | 1 | 2 | 3 |
| 8 | 1 | 0 | 2 |
| 9 | 1 | 0 | 2 |
| 10 | 0 | 0 | 3 |
| 11 | 1 | 1 | 3 |
| 12 | 1 | 0 | 4 |
| 13 | 2 | 0 | 2 |
| 14 | 0 | 1 | 2 |
| 15 | 0 | 2 | 3 |
| 16 | 1 | 3 | 3 |
| 17 | 2 | 3 | 3 |
| 18 | 1 | 1 | 3 |
| 19 | 3 | 2 | 4 |
| 20 | 2 | 1 | 5 |
| 21 | 3 | 0 | 3 |
| 22 | 3 | 0 | 3 |
| X | 1 | 0 | 1 |
| Y | 0 | 0 | 0 |
| Total | 27 | 19 | 71 |
The total numbers of unidentified chromosomes for the cells lines BHH3, BHH8, and BHH2C were 38, 41, and 22, respectively. The total numbers of bovine chromosomes for the cell lines BHH3, BHH8, and BHH2C were 48, 47, and 4, respectively. The total numbers of human plus bovine plus unidentified chromosomes for the cell lines BHH3, BHH8, and BHH2C were 113, 103, and 97, respectively.
E1 protein expression in BHH clones.
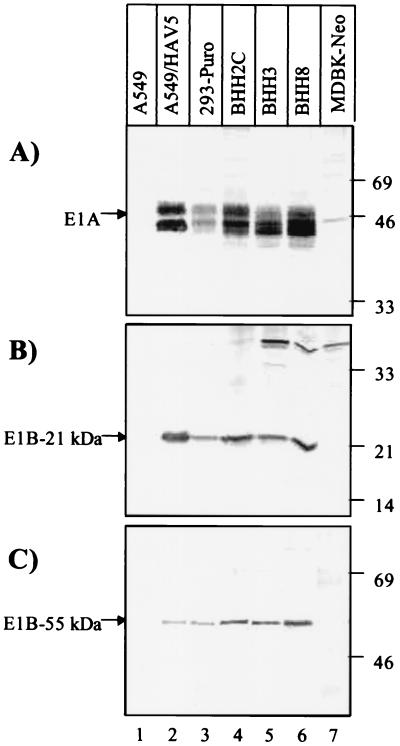
The main objective for generating BHH clones was to isolate E1-expressing cell lines that support the replication of BAV3 with E1A deleted. It was important to know whether these cell lines constitutively express E1 proteins similar to that of 293 cells. For this purpose, 35S-labeled extracts of BHH3, BHH8, and BHH2C cells were analyzed by using radioimmunoprecipitation of HAV5 E1A, E1B-21kDa, and E1B-55kDa proteins with HAV2 E1A-, HAV5 E1B-21kDa-, and HAV5 E1B-55kDa-specific antibodies, respectively. The results were compared with the expression E1 proteins in HAV5-infected A549 cells or mock-infected 293-Puro cells. The lysates from BHH3, BHH8, and BHH2C cells produced patterns of bands for E1A (Fig. 5A), E1B-21kDa (Fig. 5B) and E1B-55kDa (Fig. 5C) proteins that were similar to those observed for 293-Puro or HAV5-infected A549 cells (Fig. 5), suggesting that these hybrid clones effectively expressed E1 proteins. Mock-infected A549 and MDBK-Neo cells were used as negative controls (Fig. 5, lanes 1 and 7, respectively).
FIG. 5.
Immunoprecipitation of HAV5 E1 proteins from extracts of BHH cell lines with HAV5 E1-specific antibodies. A549, A549-infected with HAV5, 293-Puro, BHH2C, BHH3, and BHH8 cells were labeled with Trans[35S]-label, and cell extracts were immunoprecipitated with anti-HAV2 E1A (13S-5; Santa Cruz) (A), anti-HAV2 E1B-21kDa (3D11; Calbiochem) (B), and anti-HAV5 E1B-55kDa (9C10; Calbiochem) (C) antibodies. Each HAV5 E1 protein is indicated by an arrow. The position of the molecular weight marker is shown on the right side of the panel.
Replication of BAV3 with E1A deleted or HAV5 with E1 deleted in BHH cell lines.
The size and shape of the individual cells in monolayers formed by BHH clones demonstrated noticeable differences compared to either of the parental cell lines (Fig. 6A -1 to E-1). These differences were maintained even after growth for 10 passages without antibiotic selection, suggesting that these clones were fairly stable (data not shown).
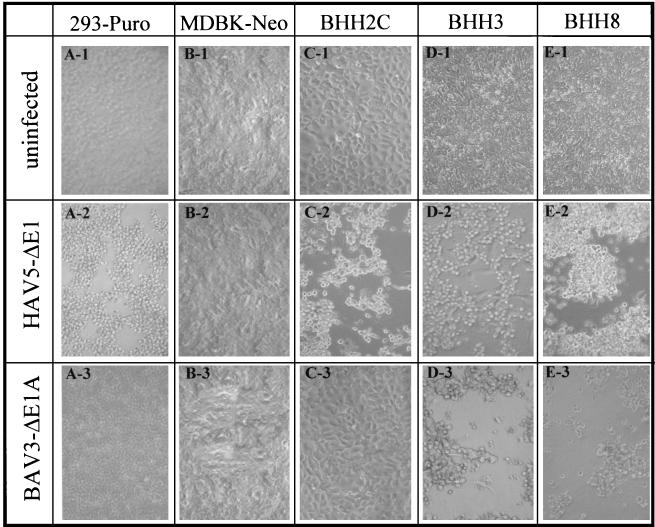
FIG. 6.
Susceptibility of BHH cell lines to HAV5 or BAV3 with E1 deleted. Cell monolayers of 293-Puro (A), MDBK-Neo (B), BHH2C (C), BHH3 (D), or BHH8 (E) in 60-mm dishes were either left uninfected (top row) or infected with HAV5-ΔE1 (AdCA36lacZ) (middle row) or BAV3-ΔE1A (BAV3ΔE1AE3) (bottom row) at a multiplicity of infection of 5 PFU/cell. At 48 h postinfection, monolayers were observed under an inverted microscope at a magnification of ×40, and the images were captured by using a Kodak DC290 digital camera.
To determine the permissiveness of BHH clones to HAV5 with E1 deleted or BAV3 with E1A deleted, BHH2C, BHH3, and BHH8 cells were infected with either HAV5-ΔE1 (AdCA36lacZ) or BAV3-ΔE1A (BAV3ΔE1AE3) and examined daily for cytopathic effect (CPE) (Fig. 6). All of these hybrid cell lines produced a CPE with HAV5-ΔE1 similar to that seen in 293 cells. BHH3 and BHH8 cells infected with BAV3-ΔE1A resulted in CPE, but no CPE was observed in BHH2C even 4 days postinfection.
Development of wt HAV5 or HAV5-ΔE1 plaques in BHH2C was usually 1 to 2 days earlier than in 293 cells (data not shown), suggesting that BHH2C could be an excellent host for growing and titrating both wt HAV5 and HAV5-ΔE1.
Replication kinetics of wt or human or bovine adenoviruses with E1 deleted in E1-expressing BHH cell lines.
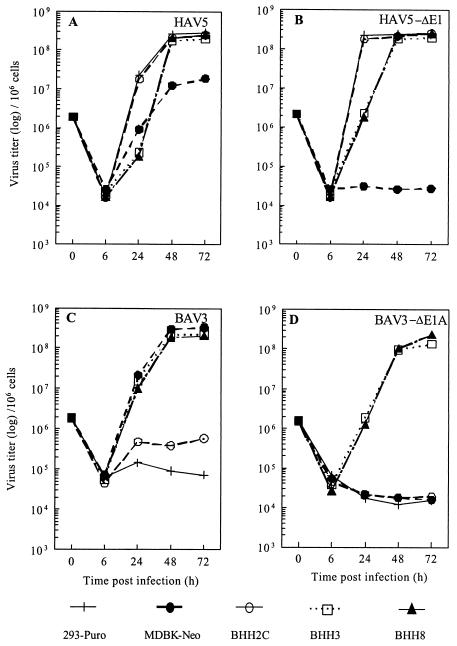
To determine the efficiency of replication of wt or human or bovine adenoviruses with E1 deleted in E1-expressing BHH clones, BHH2C, BHH3, or BHH8 cells were infected with wt HAV5 (Fig. 7A), HAV5-ΔE1 (Fig. 7B), wt BAV3 (Fig. 7C), or BAV3-ΔE1 (Fig. 7D) for the single-step growth curves. The kinetics of wt HAV5 or HAV5-ΔE1 in BHH2C were similar to those obtained with 293-Puro cells, and virus replication in BHH3 and BHH8 cells was delayed but reached titers similar to those obtained in 293-Puro or BHH2C cells at 48 h postinfection and onward. MDBK-Neo cells supported the replication of HAV5, but the titers were approximately 1 log lower than those of 293-Puro cells. As expected, MDBK-Neo cells did not support replication of HAV5-ΔE1. The wt BAV3 or BAV3-ΔE1 replicated to similar titers in BHH3 and BHH8 cells. BAV3 titers in MDBK-Neo were similar to those obtained with BHH3 and BHH8, but there was no indication of BAV3-ΔE1A replication in MDBK-Neo cells. A slight increase in the BAV3 titers in BHH2C (Fig. 7C) was observed, whereas BAV3-ΔE1 did not replicate in 293-Puro, MDBK-Neo, or BHH2C cells. These results suggested that all three BHH cell lines efficiently supported the replication of wt HAV5, HAV5-ΔE1, and wt BAV3, whereas BHH3 and BHH8 cells supported the replication of BAV3-ΔE1A.
FIG. 7.
Replication kinetics of HAV5 (A), HAV5-ΔE1 (B), BAV3 (C), or BAV3-ΔE1A (D) in hybrid cell lines. 293-Puro, MDBK-Neo, BHH2C, BHH3, or BHH8 cell monolayers in 60-mm dishes were infected with HAV5, HAV5-ΔE1 (AdCA36lacZ), BAV3, or BAV3-ΔE1A (BAV3ΔE1AE3) at a multiplicity of infection of 2 PFU per cell. After virus adsorption, virus-infected cell monolayers were washed with PBS and incubated at 37°C in MEM containing 2% FetalClone III. At 6, 24, 48, and 72 h postinfection, cells were harvested along with the medium and freeze-thawed three times, and virus titrations were done by plaque assays.
Efficiency of transfection of BHH clones.
One of the important applications of BHH cell lines will be the isolation of adenovirus recombinants with E1 deleted. Adenovirus recombinants can be produced either by intracellular recombination between two plasmids (for a review, see reference 9) or by recombination in bacteria, followed by transfection of a suitable cell line with the genome length linear DNA (6, 21). Either of these approaches will involve transfection of cells with one or two fairly large DNA. To determine the efficiency of transfection of BHH cell lines with a DNA fragment of approximately the size of adenoviral genome, transient-transfection experiments with a 37-kb linearized plasmid encoding the lacZ gene (pMvOBAV3ΔE1lacZ) were performed and were compared with the results obtained with MDBK-Neo or 293-Puro cells. Cells were cultured for 72 h after transfection and were analyzed either in situ for LacZ expression histochemistry or for LacZ assay.
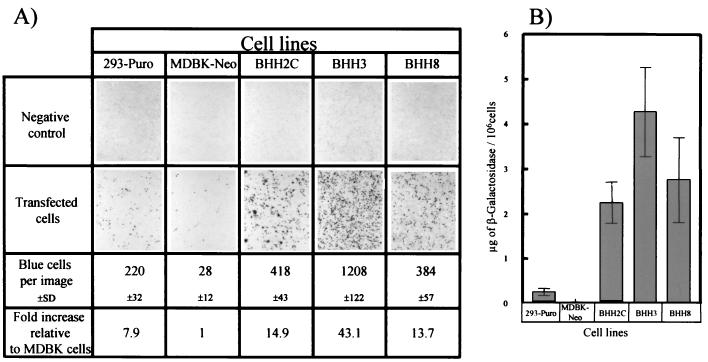
The numbers of LacZ-expressing cells per photographic image in 293-Puro, MDBK-Neo, BHH2C, BHH3, and BHH8 cells were 220 ± 32, 28 ± 12, 418 ± 43, 1,208 ± 122, and 384 ± 57, respectively, as determined by LacZ histochemistry (Fig. 8A). The transfection efficiencies of 293-Puro, BHH2C, BHH3, and BHH8 cells were ca. 8-, 15-, 43-, and 14-fold higher, respectively, than that of MDBK-Neo cells.
FIG. 8.
Transfection efficiency of BHH cell lines. 293-Puro, MDBK-Neo, BHH2C, BHH3, or BHH8 cell monolayers in 60-mm dishes were transfected with 5 μg of PacI-digested pMvOBAV3ΔE1lacZ by using Lipofectin-mediated transfection protocol (Life Technologies). At 48 h posttransfection, the cells were prepared for LacZ histochemistry (A) or LacZ assay (B). The assays were done in triplicate, and the results are presented as mean ± the standard deviation.
LacZ expression in 293-Puro, MDBK-Neo, BHH2C, BHH3, and BHH8 cells was 0.26 ± 0.04, 0.03 ± 0.01, 2.25 ± 0.07, 4.27 ± 0.17, and 2.76 ± 0.12 μg/106 cells, respectively, by LacZ assay (Fig. 8B). The LacZ expression in BHH2C, BHH3, and BHH8 cells was ca. 8-, 16-, and 10-fold higher than that obtained in 293-Puro cells. These results indicate that all three BHH clones exhibit higher transfection efficiencies than either of the parental cell lines.
DISCUSSION
Somatic cell hybrids between bovine and mouse or hamster have been used for gene mapping, genetic linkage, and gene expression studies (3, 13, 35, 51, 54). We planned to develop somatic cell hybrids for the purpose of combining some of the desirable characteristics of two parental cell lines, human 293 and MDBK. Human 293 cells express the E1 proteins (22), support the replication of wt and HAV5 with E1 deleted (14), are not permissive to BAV3 replication (39), require extra care during handling (particularly for plaque assay), and have a DNA transfection efficiency better than MDBK (44). On the other hand, bovine MDBK cells support the replication of BAV3 and are resistant and resilient to adverse culture conditions; also, as mentioned above, their DNA transfection efficiency is poor compared to 293 cells.
When the three hybrid clones—BHH2C, BHH3, and BHH8—were tested for the total genomic DNA content, species-specific immunofluorescence, isoenzyme analysis, and karyotype, it became evident that BHH2C retained more characteristics from the 293-Puro cell line, whereas BHH3 and BHH8 were closer to the MDBK-Neo cell line. Species-specific immunofluorescence (15, 41), isoenzyme analysis (7, 20), and karyotyping (33) are usually used for species identification, hybrid characterization, and determining the origin and number of chromosomes per cell. In our fusion experiment it appears that the segregation of chromosomes did not follow a hierarchical species-based pattern of chromosomal elimination which was observed during the formation of interspecies somatic cell hybrids between bovine × mouse (7) and human × hamster or mouse (1), where invariably the bovine and human chromosomes are preferentially eliminated. Clone BHH2C retained only four normal bovine chromosomes but clones BHH3 and BHH8 retained 48 and 47 normal bovine chromosomes, respectively.
293 cells contain the left end of the HAV5 genome from nucleotides (nt) 1 to 4344 (including the E1 region), and this insertion is located in the pregnancy-specific β-1-glycoprotein 4 (PSG4) on chromosome 19 (19q13.2) (34). Since the BHH2C, BHH3, and BHH8 cell lines have the chromosome 19 originating from 293 cells, it was assumed that HAV5 E1 sequences were intact in these hybrid clones. Therefore, it was not surprising to determine that these clones efficiently expressed E1A, E1B-21kDa, and E1B-55kDa as detected by immunoprecipitation.
The real test to define the success of somatic cell hybridization in acquiring desirable characteristics from two parental cell lines was to determine whether BAV3-ΔE1A could replicate in hybrid cell lines. Clones BHH3 and BHH8 efficiently supported the replication of BAV3-ΔE1A to titers close to those of wt BAV3 in MDBK-Neo cells. We earlier demonstrated that BAV3-ΔE1A replicated efficiently in FBK-34 (a cell line expressing BAV3 E1 proteins that was derived from fetal bovine kidney cells) or FBRT-HE1 (a cell line that expresses HAV5 E1 proteins and was derived from fetal bovine retinal cells) (52). BAV3-ΔE1A titers in BHH3 and BHH8 cell lines were similar to those obtained in FBK-34 or FBRT-HE1 cells. These hybrid cell lines also replicated both wt and HAV5 with E1 deleted efficiently but with an initial delay in virus replication. This may be because the BHH3 and BHH8 cell lines have a closer resemblance to the bovine progenitor. The replication of BAV3-ΔE1A in BHH3 and BHH8 cell lines supports our hypothesis that the traits present in parental cell lines could be brought together by somatic cell hybridization.
However, BHH2C was an excellent cell line for growing and titrating wt or HAV5 with E1 deleted, but it was not permissive to the replication of wt or BAV3 with E1 deleted, presumably because it lacked some of the essential cellular factors required for BAV3 replication. BHH2C appears to have certain advantages over 293 cells, such as growth characteristics like monolayer cultures and growth and titration of both wt and HAV5 with E1 deleted.
The phenotypic differences among the three BHH clones are most likely the result of differences in their genetic makeup. BAV3 does not replicate in 293 cells, and this block is not at the level of virus attachment and penetration (25). This strongly suggests that there are one or more bovine cellular factors that are required for BAV3 replication. Development of somatic cell hybrids could be an excellent tool for determining the cellular and viral components that are important in virus replication.
With regard to the enhanced transfection efficiency of the hybrid clones compared to either of the parental cell lines, we speculate that the gene products that act as positive and/or negative regulators of this phenomenon were segregated or eliminated favorably during the hybrids' formation. The characterization of an expanded panel of these somatic cell hybrids should help to further investigate this hypothesis. This characteristic, regardless of the mechanism, has certain advantages. For example, the generation of recombinant adenoviruses requires transfection or cotransfection with relatively large, linear or circular DNA molecules into a suitable cell line. Although there are many factors that govern the efficiency of recombinant virus production, it is reasonable to argue that augmentation of the DNA transfection ability would certainly have a positive impact.
Acknowledgments
We thank F. L. Graham, Departments of Biology and Pathology, McMaster University, Hamilton, Ontario, Canada, for providing AdCA36lacZ and S. K. Tikoo, Veterinary Infectious Disease Organization, University of Saskatchewan, Saskatoon, Saskatchewan, Canada, for providing plasmid pFBAV500 for the generation of BAV3ΔE1AE3.
This work was supported by Public Health Service grant GM5516 from NIH/NIGMS to S.K.M.
REFERENCES
- 1.Abbott, C., and S. Povey. 1995. Somatic cell hybrids: the basics. IRL Press at Oxford University Press, Oxford, England.
- 2.Addison, C. L., M. Hitt, D. Kunsken, and F. L. Graham. 1997. Comparison of the human versus murine cytomegalovirus immediate early gene promoters for transgene expression by adenoviral vectors. J. Gen. Virol. 78:1653-1661. [DOI] [PubMed] [Google Scholar]
- 3.Adkison, L. R., L. C. Skow, T. L. Thomas, M. Petrash, and J. E. Womack. 1988. Somatic cell mapping and restriction fragment analysis of bovine genes for fibronectin and gamma crystallin. Cytogenet. Cell Genet. 47:155-159. [DOI] [PubMed] [Google Scholar]
- 4.Aggarwal, N., H. HogenEsch, P. Guo, A. North, M. Suckow, and S. K. Mittal. 1999. Biodegradable alginate microspheres as a delivery system for naked DNA. Can. J. Vet. Res. 63:148-152. [PMC free article] [PubMed] [Google Scholar]
- 5.Berkner, K. L., and P. A. Sharp. 1984. Expression of dihydrofolate reductase and of the adjacent EIb region in an Ad5-dihydrofolate reductase recombinant virus. Nucleic Acids Res. 12:1925-1941. [DOI] [PMC free article] [PubMed]
- 6.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinchar, V. G., A. D. Floyd, G. D. Chinchar, and M. W. Taylor. 1979. Characterization of hybrids between bovine (MDBK) and mouse (L-cell) cell lines. Biochem. Genet. 17:133-148. [DOI] [PubMed] [Google Scholar]
- 8.Dannenberg, A., and M. Suga. 1981. Histochemical stains for macrophages, cell smears, and tissue sections: beta-galactosidase, acid phosphatase, nonspecific esterase, succinic dehydrogenase, and cytochrome oxidase, p. 375-396. In D. O. Adams, P. J. Edelson, and H. S. Koren (ed.), Methods for studying mononuclear phagocytes. Academic Press, New York, N.Y.
- 9.Danthinne, X., and M. J. Imperiale. 2000. Production of first generation adenovirus vectors: a review. Gene Ther. 7:1707-1714. [DOI] [PubMed] [Google Scholar]
- 10.Davidson, R. L., and P. S. Gerald. 1976. Improved techniques for the induction of mammalian cell hybridization by polyethylene glycol. Somat. Cell Genet. 2:165-176. [DOI] [PubMed] [Google Scholar]
- 11.de la Luna, S., I. Soria, D. Pulido, J. Ortin, and A. Jimenez. 1988. Efficient transformation of mammalian cells with constructs containing a puromycin-resistance marker. Gene 62:121-126. [DOI] [PubMed] [Google Scholar]
- 12.Deng, A. Y., L. Gu, J. P. Rapp, C. Szpirer, and J. Szpirer. 1994. Chromosomal assignment of 11 loci in the rat by mouse-rat somatic hybrids and linkage. Mamm. Genome 5:712-716. [DOI] [PubMed] [Google Scholar]
- 13.Dietz, A. B., and J. E. Womack. 1989. Somatic cell mapping of the bovine somatostatin gene. J. Hered. 80:410-412. [DOI] [PubMed] [Google Scholar]
- 14.Fallaux, F. J., O. Kranenburg, S. J. Cramer, A. Houweling, O. H. Van, R. C. Hoeben, and A. J. van der Eb. 1996. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum. Gene Ther. 7:215-222. [DOI] [PubMed] [Google Scholar]
- 15.Gallagher, T., Y. Chardonnet, J. Viac, J. Patet, and A. Lefevre. 1989. Antigenic immunostaining patterns in somatic hybrids of human HeLa cells and mouse fibroblasts 3T3.4E propagated in conventional medium and delipidized serum. Virchows Arch. B Cell Pathol. Mol. Pathol. 57:339-347. [DOI] [PubMed] [Google Scholar]
- 16.Graham, F. L., and L. Prevec. 1992. Adenovirus expression vectors and recombinant vaccines, p. 363-390. In R. W. Ellis (ed.), Vaccines: new approaches to immunological problems. Butterworth-Heinemann, Boston, Mass. [DOI] [PubMed]
- 17.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 18.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 19.Halton, D. M., W. D. Peterson, and B. Hukku. 1983. Cell culture quality control by rapid isoenzymatic characterization. In Vitro 19:16-24. [DOI] [PubMed] [Google Scholar]
- 20.Hamers, M. N., A. Westerveld, M. Khan, and J. M. Tager. 1977. Characterization of alpha-galactosidase isoenzymes in normal and Fabry human-Chinese hamster somatic cell hybrids. Hum. Genet. 36:289-297. [DOI] [PubMed] [Google Scholar]
- 21.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hitt, M. M., and F. L. Graham. 1990. Adenovirus E1A under the control of heterologous promoters: wide variation in E1A expression levels has little effect on virus replication. Virology 179:667-678. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann, C., P. Loser, G. Cichon, W. Arnold, G. W. Both, and M. Strauss. 1999. Ovine adenovirus vectors overcome preexisting humoral immunity against human adenoviruses in vivo. J. Virol. 73:6930-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hozier, J., J. Sawyer, M. Moore, B. Howard, and D. Clive. 1981. Cytogenetic analysis of the L5178Y/TK+/− leads to TK−/− mouse lymphoma mutagenesis assay system. Mutat. Res. 84:169-181. [DOI] [PubMed] [Google Scholar]
- 25.Iwasaki, A., C. C. Dela, A. R. Young, and B. H. Barber. 1999. Epitope-specific cytotoxic T lymphocyte induction by minigene DNA immunization. Vaccine 17:2081-2088. [DOI] [PubMed] [Google Scholar]
- 26.Kelsell, D. P., L. Rooke, D. Warne, M. Bouzyk, L. Cullin, S. Cox, L. West, S. Povey, and N. K. Spurr. 1995. Development of a panel of monochromosomal somatic cell hybrids for rapid gene mapping. Ann. Hum. Genet. 59:233-241. [DOI] [PubMed] [Google Scholar]
- 27.Kelsell, D. P., and N. K. Spurr. 1997. Gene mapping using somatic cell hybrids. Methods Mol. Biol. 68:45-52. [DOI] [PubMed] [Google Scholar]
- 28.Klonjkowski, B., P. Gilardi-Hebenstreit, J. Hadchouel, V. Randrianarison, S. Boutin, P. Yeh, M. Perricaudet, and E. J. Kremer. 1997. A recombinant E1-deleted canine adenoviral vector capable of transduction and expression of a transgene in human-derived cells and in vivo. Hum. Gene Ther. 8:2103-2115. [DOI] [PubMed] [Google Scholar]
- 29.Kohler, G., and C. Milstein. 1976. Derivation of specific antibody-producing tissue culture and tumor lines by cell fusion. Eur. J. Immunol. 6:511-519. [DOI] [PubMed] [Google Scholar]
- 30.Kostia, S., J. Vilkki, M. Pirinen, J. E. Womack, W. Barendse, and S. L. Varvio. 1997. SINE targeting of bovine microsatellites from bovine/rodent hybrid cell lines. Mamm. Genome 8:365-367. [DOI] [PubMed] [Google Scholar]
- 31.Kotin, R. M., M. Siniscalco, R. J. Samulski, X. D. Zhu, L. Hunter, C. A. Laughlin, S. McLaughlin, N. Muzyczka, M. Rocchi, and K. I. Berns. 1990. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 87:2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishan, A. 1975. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J. Cell Biol. 66:188-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lillie, J. W., and M. R. Green. 1989. Transcription activation by the adenovirus E1a protein. Nature 338:39-44. [DOI] [PubMed] [Google Scholar]
- 34.Louis, N., C. Evelegh, and F. L. Graham. 1997. Cloning and sequencing of the cellular-viral junctions from the human adenovirus type 5 transformed 293 cell line. Virology 233:423-429. [DOI] [PubMed] [Google Scholar]
- 35.Matiz, K., K. Ursu, B. Harrach, Z. Zadori, and M. Benko. 1998. Sequencing and phylogenetic analysis of the protease gene, and genetic mapping of bovine adenovirus type 10 define its relatedness to other bovine adenoviruses. Virus Res. 55:29-35. [DOI] [PubMed] [Google Scholar]
- 36.Mittal, S. K., M. R. McDermott, D. C. Johnson, L. Prevec, and F. L. Graham. 1993. Monitoring foreign gene expression by a human adenovirus-based vector using the firefly luciferase gene as a reporter. Virus Res. 28:67-90. [DOI] [PubMed] [Google Scholar]
- 37.Mittal, S. K., D. M. Middleton, S. K. Tikoo, and L. A. Babiuk. 1995. Pathogenesis and immunogenicity of bovine adenovirus type 3 in cotton rats (Sigmodon hispidus). Virology 213:131-139. [DOI] [PubMed] [Google Scholar]
- 38.Mittal, S. K., L. Prevec, L. A. Babiuk, and F. L. Graham. 1992. Sequence analysis of bovine adenovirus type 3 early region 3 and fibre protein genes. J. Gen. Virol. 73:3295-3300. (Erratum, 74:2825, 1993.) [DOI] [PubMed] [Google Scholar]
- 39.Mittal, S. K., L. Prevec, F. L. Graham, and L. A. Babiuk. 1995. Development of a bovine adenovirus type 3-based expression vector. J. Gen. Virol. 76:93-102. [DOI] [PubMed] [Google Scholar]
- 40.Norton, P. A., and J. M. Coffin. 1985. Bacterial β-galactosidase as a marker of Rous sarcoma virus gene expression and replication. Mol. Cell. Biol. 5:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson, W. D., Jr., W. F. Simpson, and B. Hukku. 1979. Cell culture characterization: monitoring for cell identification. Methods Enzymol. 58:164-178. [DOI] [PubMed] [Google Scholar]
- 42.Pontecorvo, G. 1976. Polyethylene glycol (PEG) in the production of mammalian somatic cell hybrids. Cytogenet. Cell Genet. 16:399-400. [DOI] [PubMed] [Google Scholar]
- 43.Rasmussen, U. B., M. Benchaibi, V. Meyer, Y. Schlesinger, and K. Schughart. 1999. Novel human gene transfer vectors: evaluation of wild-type and recombinant animal adenoviruses in human-derived cells. Hum. Gene Ther. 10:2587-2599. [DOI] [PubMed] [Google Scholar]
- 44.Reddy, P. S., N. Idamakanti, Y. Chen, T. Whale, L. A. Babiuk, M. Mehtali, and S. K. Tikoo. 1999. Replication-defective bovine adenovirus type 3 as an expression vector. J. Virol. 73:9137-9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reddy, P. S., N. Idamakanti, B. H. Hyun, S. K. Tikoo, and L. A. Babiuk. 1999. Development of porcine adenovirus-3 as an expression vector. J. Gen. Virol. 80:563-570. [DOI] [PubMed] [Google Scholar]
- 46.Robinson, J. P. 1993. Handbook of flow cytometry methods. Wiley-Liss, New York, N.Y.
- 47.Ryan, J., P. E. Barker, K. Shimizu, M. Wigler, and F. H. Ruddle. 1983. Chromosomal assignment of a family of human oncogenes. Proc. Natl. Acad. Sci. USA 80:4460-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Southern, P. J., and P. Berg. 1982. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J. Mol. Appl. Genet. 1:327-341. [PubMed] [Google Scholar]
- 49.Szpirer, C., J. Szpirer, M. Riviere, S. Ingvarsson, B. Vennstrom, M. Q. Islam, and G. Levan. 1991. Chromosomal assignment of five cancer-associated rat genes: two thyroid hormone receptor (ERBA) genes, two ERBB genes and the retinoblastoma gene. Oncogene 6:1319-1324. [PubMed] [Google Scholar]
- 50.Takayesu, D., J. G. Teodoro, S. G. Whalen, and P. E. Branton. 1994. Characterization of the 55K adenovirus type 5 E1B product and related proteins. J. Gen. Virol. 75:789-798. [DOI] [PubMed] [Google Scholar]
- 51.Vaiman, D., D. Mercier, K. Moazami-Goudarzi, A. Eggen, R. Ciampolini, A. Lepingle, R. Velmala, J. Kaukinen, S. L. Varvio, and P. Martin. 1994. A set of 99 cattle microsatellites: characterization, synteny mapping, and polymorphism. Mamm. Genome 5:288-297. [DOI] [PubMed] [Google Scholar]
- 52.van Olphen, A. L., S. K. Tikoo, and S. K. Mittal. 2002. Characterization of bovine adenovirus type 3 E1 proteins and isolation of E1-expressing cell lines. Virology 295:108-118. [DOI] [PubMed] [Google Scholar]
- 53.van Olphen, A. L., and S. K. Mittal. 1999. Generation of infectious genome of bovine adenovirus type 3 by homologous recombination in bacteria. J. Virol. Methods 77:125-129. [DOI] [PubMed] [Google Scholar]
- 54.Womack, J. E., J. S. Johnson, E. K. Owens, C. E. Rexroad III, J. Schlapfer, and Y. P. Yang. 1997. A whole-genome radiation hybrid panel for bovine gene mapping. Mamm. Genome 8:854-856. [DOI] [PubMed] [Google Scholar]
- 55.Womack, J. E., and Y. D. Moll. 1986. Gene map of the cow: conservation of linkage with mouse and man. J. Hered. 77:2-7. [DOI] [PubMed] [Google Scholar]
- 56.Zheng, B., S. K. Mittal, F. L. Graham, and L. Prevec. 1994. The E1 sequence of bovine adenovirus type 3 and complementation of human adenovirus type 5 E1A function in bovine cells. Virus Res. 31:163-186. [DOI] [PubMed] [Google Scholar]