Abstract
Cellular immune responses against epitopes in conserved Gag and Pol sequences of human immunodeficiency virus type 1 have become popular targets for candidate AIDS vaccines. Recently, we used a simian-human immunodeficiency virus model (SHIV 89.6P) with macaques to demonstrate the control of a pathogenic mucosal challenge by priming with Gag-Pol-Env-expressing DNA and boosting with Gag-Pol-Env-expressing recombinant modified vaccinia virus Ankara (rMVA). Here we tested Gag-Pol DNA priming and Gag-Pol rMVA boosting to evaluate the contribution of anti-Env immune responses to viral control. The Gag-Pol vaccine raised frequencies of Gag-specific T cells similar to those raised by the Gag-Pol-Env vaccine. Following challenge, these rapidly expanded to counter the challenge infection. Despite this, the control of the SHIV 89.6P challenge was delayed and inconsistent in the Gag-Pol-vaccinated group and all of the animals underwent severe and, in most cases, sustained loss of CD4+ cells. Interestingly, most of the CD4+ cells that were lost in the Gag-Pol-vaccinated group were uninfected cells. We suggest that the rapid appearance of binding antibody for Env in Gag-Pol-Env-vaccinated animals helped protect uninfected CD4+ cells from Env-induced apoptosis. Our results highlight the importance of immune responses to Env, as well as to Gag-Pol, in the control of immunodeficiency virus challenges and the protection of CD4+ cells.
Recently, vaccines designed to raise cellular immunity have controlled virulent challenges and prevented the development of AIDS in rhesus macaques (2, 4, 5, 20, 22). These vaccines have been based on immunization with DNA adjuvanted with interleukin-2 (5), DNA immunizations boosted with recombinant modified vaccinia virus Ankara (rMVA) (DNA/rMVA vaccine) (2), vesicular stomatitis virus vectors (20), rMVA vectors (4; R. R. Amara, F. Villinger, S. I. Staprans, J. D. Altman, D. C. Montefiori, N. L. Kozyr, Y. Xu, L. Wyatt, P. L. Earl, J. G. Herndon, H. M. McClure, B. Moss, and H. L. Robinson, submitted for publication), recombinant adenovirus vectors (22), and DNA immunizations boosted with recombinant adenovirus vectors (22). All of these vaccines have raised antiviral T cells that rapidly expanded and contracted as the vaccines controlled the highly virulent simian-human immunodeficiency virus (SHIV 89.6P) challenge. Although these vaccines were designed and tested primarily for raising cellular immunity to the immunodeficiency virus Gag protein, the immunogens for all but the recombinant adenovirus trials included the viral envelope glycoprotein (Env). Env is a target for both binding and neutralizing antibodies. In the trials that included Env, the immunizations raised binding but not neutralizing antibody to Env, and the postchallenge expansion of T cells and control of viremia were simultaneous with anamnestic responses for binding antibody but preceded the appearance of neutralizing antibody. Here, we directly investigated whether immune responses to Env contribute to the protection mediated by cellular responses to Gag and Pol for the DNA/rMVA vaccine. A non-Env-containing AIDS vaccine would exhibit less sequence diversity among different human immunodeficiency virus (HIV) subtypes and have the practical advantage of allowing vaccinated populations to be monitored for infection by testing for antibodies to Env.
MATERIALS AND METHODS
DNA and rMVA immunogens.
The Gag-Pol DNA vaccine was constructed by the introduction of a stop codon and a unique EcoRI restriction site at the junction of reverse transcriptase and integrase in the Gag-Pol-Env DNA vaccine (2). The truncated Gag-Pol sequences were recombined at an EcoRI restriction endonuclease site 86 bp upstream of the Tat start site with a HIV type 1 (HIV-1) subclone that included vpu, tat, rev, and ADA env with an internal BglII deletion. The truncated env gene encoded the first 270 amino acids of Env. The Gag-Pol insert was cloned into the pGA1 expression vector (GenBank accession no. AF425297), which is identical to the pGA2 vector (GenBank accession no. AF425298) used for the Gag-Pol-Env vaccine, except that pGA1 includes intron A in the cytomegalovirus immediate-early promoter region. The levels of Gag expression for the Gag-Pol and Gag-Pol-Env vaccine DNAs were the same in transiently transfected 293T cells (data not shown). rMVA, which expressed SIV239 Gag-Pol, was the parent virus used for insertion of the HIV-1 89.6 env gene (L. S. Wyatt and B. Moss, unpublished results). Accordingly, the Gag-Pol-Env and Gag-Pol rMVA immunogens expressed equivalent levels of Gag (Wyatt and Moss, unpublished).
Immunizations and challenge.
Young adult rhesus macaques from the Yerkes breeding colony were cared for under guidelines established by the Animal Welfare Act and the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals using protocols approved by the Emory University Institutional Animal Care and Use Committee. Macaques were typed for the MAmu-A*01 allele by using PCR analyses (11). Two or more animals containing at least one MAmu-A*01 allele were assigned to each group of six animals. DNA immunizations were delivered by intradermal (i.d.) injection in phosphate-buffered saline by using a needleless jet injector (Bioject Inc., Portland, Oreg.) to deliver five 100-μl i.d. injections to each outer thigh for the 2.5-mg dose of DNA or one 100-μl i.d. injection to the right outer thigh for the 250-μg plasmid dose. rMVA boosters were administered by both i.d. and intramuscular injections with a needle for a total dose of 2 × 108 PFU. One 100-μl dose was delivered to each outer thigh for the 108-PFU i.d. dose, and one 500-μl dose was delivered to each outer thigh for the 108-PFU intramuscular dose. Control animals received vector DNA without inserts. Seven months after the rMVA booster, animals were given an intrarectal challenge with SHIV 89.6P by using a pediatric feeding tube to introduce 20 intrarectal infectious units (1.2 × 1010 copies of SHIV 89.6P RNA) 15 to 20 cm into the rectum. Animal numbers are as follows: 1, RBr-5*; 2, RIm-5*; 3, RQf-5*; 4, RZe-5; 5, ROm-5; 6, RDm-5; 13, RKw-4*; 14, RWz-5*; 15, RGo-5; 16, RLp-4; 17, RWd-6; 18, RAt-5; 25, RMb-5*; 26, RGy-5*; 27, RUs-4; 28, RPm-5; 29, RPs-4; 30, RKj-5; 31, ROv-4*; 32, RQk-5*; 33, RZo-5; 34, RFd-5; 35, RCy-4; 36, RDg-5; 37, RDv-4*; 38, RLw-4*; 39, RAv-4*; 40, RVy-4; 41, RGp-4*; 42, RCe-5. Rhesus monkeys with the A*01 allele are indicated by asterisks. Data for animals 1 to 6, 13 to 18, and 25 to 28 have, in part, been previously reported and are presented here for the sake of comparison (2).
Measurement of T-cell responses.
For tetramer analyses, approximately 106 peripheral blood mononuclear cells (PBMC) were surface stained with antibodies to CD3 (FN-18; Biosource International, Camarillo, Calif.), CD8 (SK1; Becton Dickinson, San Jose, Calif.), and Gag-CM9 (CTPYDINQM)-MAmu-A*01 tetramer conjugated to different fluorochromes (2). For gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assays, anti-human IFN-γ antibody (clone B27; Pharmingen, San Diego, Calif.) was used for capture and biotinylated anti-human IFN-γ antibody (clone 7-86-1; Diapharma Group Inc., West Chester, Ohio), followed by avidin-horseradish peroxidase (Vector Laboratories Inc., Burlingame, Calif.), was used for detection (for details, see reference 2). For intracellular cytokine assays, approximately 106 PBMC were stimulated in 5-ml polypropylene tubes in RPMI medium containing 0.1% bovine serum albumin, anti-human CD28 antibody, anti-human CD49d antibody (each at 1 μg/ml; Pharmingen, Inc., San Diego, Calif.), and Gag peptide pools (each peptide at 100 μg/ml) in a volume of 100 μl. After 2 h, 900 μl of RPMI containing 10% fetal bovine serum and monensin (10 μg/ml) was added and the cells were cultured for an additional 4 h at 37°C at an angle of 5°. Cells were stained and analyzed as previously described (2).
Quantitation of SHIV copy number.
SHIV copy number was determined by using a quantitative real-time PCR as previously described. (2, 8). All specimens were extracted and amplified in duplicate, and the mean results are reported.
Intracellular p27 staining.
Approximately 106 PBMC were fixed and permeabilized with Cytofix/Cytoperm solution (Pharmingen, Inc.) and stained sequentially with anti-SIV Gag antibody (clone FA-2; obtained from the NIH AIDS Reagent Program) and phycoerythrin-conjugated anti-mouse immunoglobulin (Pharmingen, Inc.) in Perm/Wash buffer for 30 min at 4°C. Cells were washed twice with perm wash and incubated in perm wash solution with antibodies to human CD3 (clone FN-18; Biosource International, Camarillo, Calif.) and CD8 (clone SK1; Becton Dickinson) conjugated to fluorescein isothiocyanate and PerCP, respectively. Approximately 150,000 lymphocytes were acquired on a FACScalibur and analyzed with FloJo software.
Assays for antibody.
Enzyme-linked immunosorbent assays (ELISAs) for total anti-Gag antibody and anti-Env antibody were carried out as previously described (2). Standard curves for Gag and Env ELISAs were produced by using serum from a SHIV 89.6-infected macaque with known amounts of anti-Gag or anti-Env immunoglobulin G. Sera were assayed at threefold dilutions in duplicate wells. Standard curves were fitted, and sample concentrations were interpolated as micrograms of antibody per milliliter of serum with SOFTmax 2.3 software (Molecular Devices, Sunnyvale, Calif.). For more details, see reference 2. The titers of neutralizing antibodies for SHIV 89.6 and SHIV 89.6P were determined by using MT-2 cell killing and neutral red staining as previously described (15).
Statistical analyses.
To examine the effects of the dose and immunogen upon viral loads, CD4 levels, and antibody and T-cell responses, analyses of variance for repeated measures were performed on log-transformed values (S-Plus 6 Statistical Package; Insightful Corp., Seattle, Wash.). We used the interaction term (group × week) to compare the rate of viral control as defined by the slopes of the log viral concentration over the period during which a decline occurred (weeks 2 to 5 postchallenge). Rates of CD4 decline were examined similarly by comparing the prechallenge and week 2 postchallenge slopes. Consistency of the steady-state level of viral RNA in plasma was compared between groups by evaluating the analysis of variance main effect for group differences in log viral load from weeks 8 to 20 postchallenge. Similarly, steady-state CD4 levels were compared between groups at weeks 8 to 20 postchallenge.
RESULTS
Immunization.
To investigate the role of anti-Env immune responses in the control of an immunodeficiency virus challenge, Gag-Pol vaccines were constructed and tested for the ability to control the SHIV 89.6P challenge. As in our previous Gag-Pol-Env trial, sequences from SHIV 89.6 were used to construct DNA and rMVA vaccines and the highly pathogenic virus SHIV 89.6P was administered as a mucosal challenge (2, 9). The 89.6 Env protein primes binding but not neutralizing antibody for the 89.6P Env protein (14). Immunizations again tested high (2.5 mg)- and low (250 μg)-dose DNA priming at 0 and 8 weeks, followed by 2 × 108 PFU rMVA boosting at 24 weeks and an intrarectal challenge administered 7 months after the rMVA booster.
Similar patterns of postvaccination Gag-specific T cells.
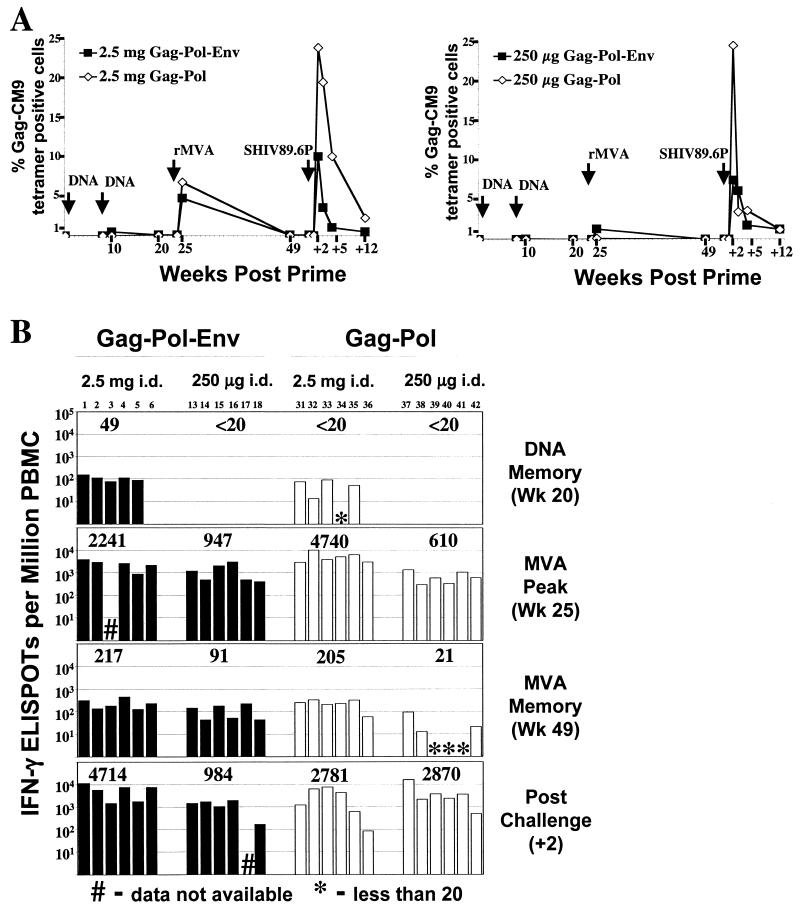
The Gag-Pol vaccine raised patterns of anti-Gag T cells that were similar to those raised by the Gag-Pol-Env vaccine (Fig. 1). The frequencies of Gag-CM9 epitope-specific CD8+ T cells were assessed by using Mamu-A*01 tetramers (1), and the frequencies of T cells against epitopes throughout Gag were assessed by using pools of overlapping peptides and an ELISPOT assay (10, 18). Gag-CM9 tetramer analyses were restricted to macaques that expressed the Mamu-A*01 histocompatibility type, whereas ELISPOT responses did not depend on a specific histocompatibility type. Following the second DNA inoculation, A*01 macaques had low frequencies of CD8+ cells for the A*01-restricted Gag-CM9 epitope. By 1 week after the rMVA booster, these cells had expanded to frequencies as high as 22% of the total number of CD8+ T cells and had geometric mean frequencies of 7% in the high-dose DNA-primed group and 0.1% in the low-dose DNA-primed group (Fig. 1A). These values were comparable to the geometric means in the previous high- and low-dose Gag-Pol-Env groups of 5 and 1%, respectively. A similar temporal pattern occurred for peptide-stimulated IFN-γ-producing cells detected by ELISPOT analyses. In the memory phase of the DNA-primed response, only the animals primed with high-dose Gag-Pol-Env DNA had detectable ELISPOT levels. Following the rMVA booster, these memory responses expanded to geometric frequencies between 610 and 4,740 IFN-γ-producing cells per 106 PBMC (Fig. 1B). The IFN-γ ELISPOT frequency of the high-dose Gag-Pol group was higher than that of the high-dose Gag-Pol-Env group (P = 0.05); the two low-dose groups had similar IFN-γ ELISPOT frequencies (P = 0.47). At the time of the challenge at 7 months after the booster, the IFN-γ ELISPOTs had contracted into memory and were present at frequencies 20 times lower than at the peak response (Fig 1B).
FIG. 1.
Temporal frequencies of Gag-specific T cells. (A) Gag-specific CD8+ T-cell responses raised by DNA priming and rMVA booster immunizations. The schematic presents the geometric mean of Gag-CM9 tetramer binding for the A*01 animals in each group. (B) Gag-specific IFN-γ ELISPOTs in Gag-Pol-Env-vaccinated (solid bars) and Gag-Pol-vaccinated (open bars) macaques at various times prechallenge and at 2 weeks postchallenge. Three pools of 10 to 13 Gag peptides (22-mers overlapping by 12 amino acids) were used for the analyses. The values above the bars are the geometric means of the ELISPOTs of the groups. The numbers above the graphs designate individual animals. #, data not available; ∗, <20 ELISPOTs per 106 PBMC. Data for the Gag-Pol-Env-vaccinated groups are reprinted from reference 2 with permission of the publisher. Wk, week.
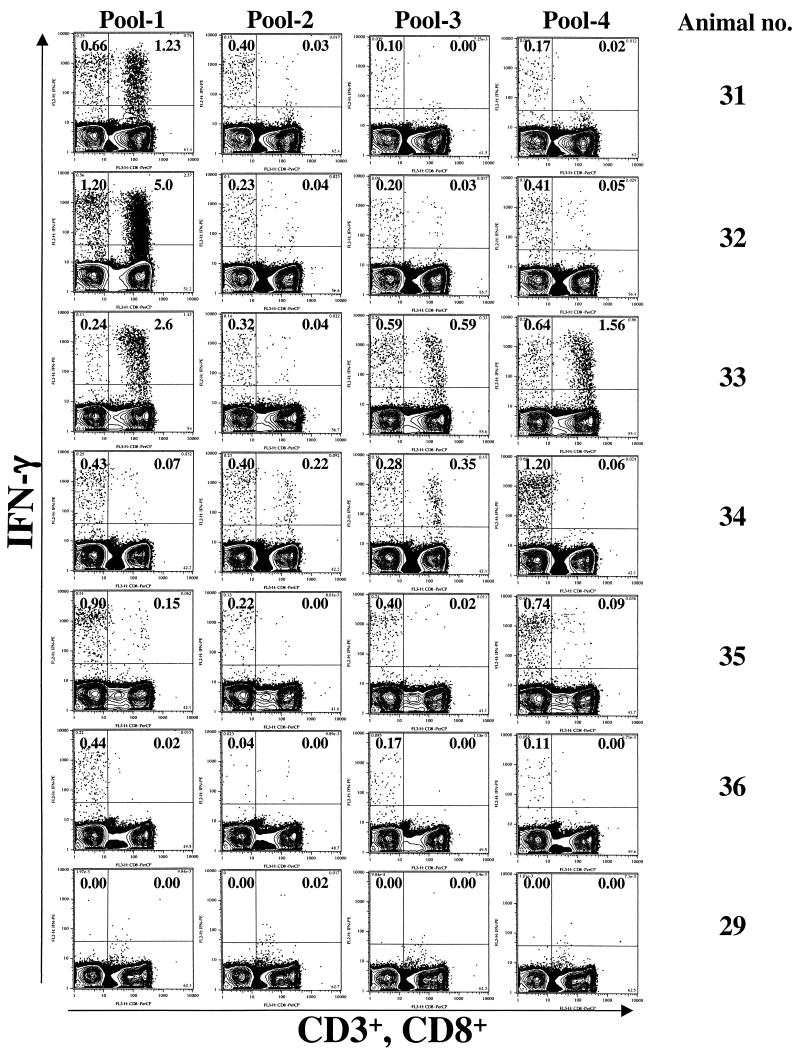
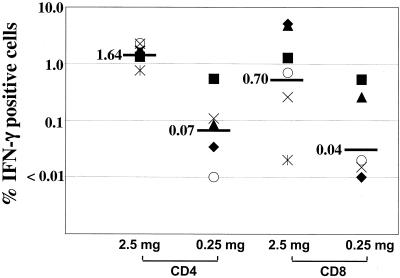
Intracellular cytokine analyses revealed that the Gag-Pol immunizations had raised high frequencies of both CD4+ and CD8+ T cells (Fig. 2 and 3) (12). The CD4 responses were both broader and more uniform in magnitude than the CD8 responses. Each of the Gag-Pol-immunized animals had responded to CD4 epitopes in each of the four peptide pools tested. In contrast, macaque 36 had no detectable CD8 response to any of the Gag pools whereas macaques 33 and 34 had CD8 responses to three pools. CD8 responses ranged from a high of 5% of the total number of CD8+ cells (see macaque 32, which was stimulated with peptide pool 1) to lows of <0.1% of the total number of CD8+ cells (see macaque 34, which was stimulated with peptide pool 1), whereas CD4 responses ranged from a high of 1.2% of the total number of CD4+ cells (see macaque 32, which was stimulated with peptide pool 1) to a low of 0.1% of the total number of CD4+ cells (see macaque 31, which was stimulated with peptide pool 3). A dose response was observed for both CD4 and CD8 (Fig. 3). The geometric mean frequencies of IFN-γ-producing CD4+ cells were 0.07% for the low dose and 1.64% for the high dose. Similarly, the geometric mean frequencies of IFN-γ-producing CD8+ cells were 0.04% for the low dose and 0.76% for the high dose.
FIG. 2.
Intracellular cytokine analyses with Gag peptide pools in high-dose Gag-Pol-vaccinated animals at 1 week after administration of the rMVA booster. PBMC were stimulated with four Gag pools (22-mers overlapping by 12 amino acids) for 6 h, fixed, and stained for CD3, CD8, and IFN-γ. Cells were gated on lymphocytes, followed by CD3, and analyzed for CD8 and IFN-γ. Cells in the left quadrants represent CD4+ (CD3+ CD8−). The frequencies in the upper quadrants are IFN-γ-producing cells as percentages of the total number of CD4+ (left quadrants) or CD8+ (right quadrants) cells.
FIG. 3.
Gag-specific T-cell responses at 1 week after administration of the rMVA booster. Gag-specific CD4 and CD8 responses were determined for both high-dose and low-dose DNA-primed animals at 1 week after administration of the rMVA booster by intracellular cytokine analyses. The sum of responses for the four Gag pools that represent the total Gag response was used for the comparison. Each symbol represents an individual animal. Symbols for animals are the same as in Fig. 4. Horizontal bars represent the geometric means for each group.
Inconsistent control of viremia and loss of CD4+ cells.
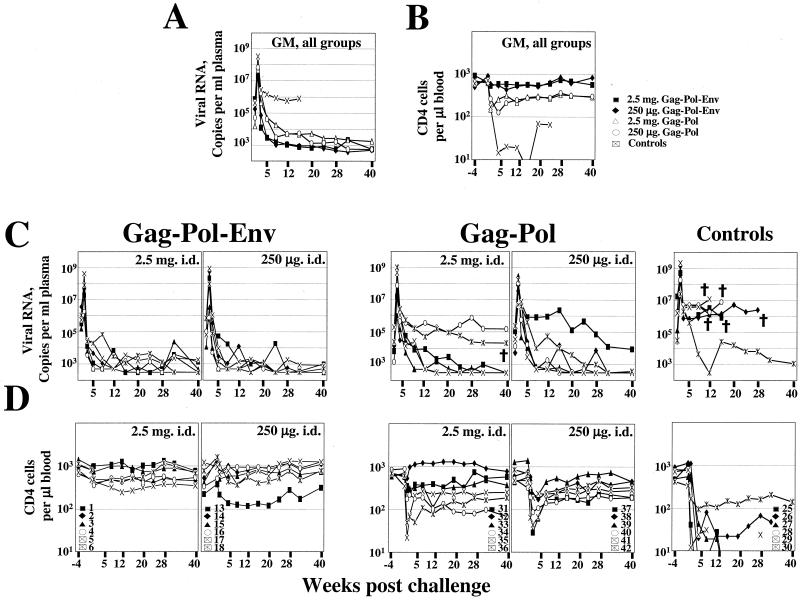
In contrast to the Gag-Pol-Env groups, in which all of the animals rapidly controlled the challenge infection, the Gag-Pol-immunized animals were both slow and inconsistent in their control of the challenge (Fig. 4A and C). By 12 weeks postchallenge, 12 of 12 Gag-Pol-Env-immunized animals, but only 7 of 12 Gag-Pol-immunized animals, had contained their infections to 1,000 copies of viral RNA per ml of plasma (Fig. 4C). None of the six control animals managed to control its infection. Discouragingly, the Gag-Pol groups also lost CD4+ cells, with only 1 of the 12 vaccinated animals regaining its prechallenge level of CD4+ cells (Fig. 4B and D). This loss was not as severe as in the control group but much more severe than that in the Gag-Pol-Env groups, which suffered relatively minor and transient losses of CD4+ cells. Differences between the Gag-Pol-Env- and Gag-Pol-vaccinated animals for the rate of virus control (P < 0.001), the consistency of the steady-state level of viral RNA in plasma (P = 0.002), the rate of CD4+ cell loss (P = 0.007), and the level of CD4+ cells once the infection had reached a steady state (P = 0.02) were all significant. By 46 weeks postchallenge, one of the Gag-Pol-immunized animals, macaque 36, had progressed to disease and was euthanized. In contrast, all of the Gag-Pol-Env-immunized animals have controlled their infections at the background of detection for more than 1.5 years and have not shown signs of CD4 loss or AIDS.
FIG. 4.
Temporal viral loads and CD4+ cell counts postchallenge of vaccinated and control animals. Panels: A, geometric mean viral loads; B, geometric mean CD4+ cell counts; C, viral loads; D, CD4+ cell counts of individual animals in the vaccine and control groups. The key to animal numbers is presented in panel D. Assays for the first 12 weeks for the Gag-Pol-Env-immunized groups had a background of 1,000 copies of RNA per ml of plasma. Animals with loads of less than 1,000 copies of RNA per ml of plasma were scored with a load of 500. For all other assays, the background for detection was 300 copies of RNA per ml of plasma and animals with levels of virus below 300 copies of RNA per ml of plasma up to 12 weeks were scored at 500 copies of RNA per ml of plasma and the rest were scored at 300 copies of RNA per ml of plasma. The symbol † represents the death of an animal. Data points that are not connected reflect missing data. Data for the Gag-Pol-Env-immunized groups are reproduced in part from reference 2 with permission of the publisher.
Postchallenge T-cell responses.
Postchallenge, Gag-Pol-vaccinated animals, like the Gag-Pol-Env-immunized animals, underwent a rapid expansion of Gag-specific CD8+ T cells (Fig. 1). In keeping with their slower control of viral loads, there was a higher frequency of Gag-CM9-specific T-cell mobilization in the Gag-Pol-immunized animals than in the Gag-Pol-Env-immunized animals (∼24% as opposed to ∼10% of the total number of CD8+ cells in the high-dose groups and ∼25% as opposed to ∼7% of the total number of CD8+ cells in the low-dose groups, respectively) (Fig. 1A). The contraction of the Gag-CM9 CD8+ T-cell response also was less rapid in the Gag-Pol-immunized animals than in the Gag-Pol-Env-immunized animals, requiring up to 12 weeks to return to a steady state. The ELISPOTs also underwent strong anamnestic responses in the Gag-Pol groups that were comparable to those previously observed in the Gag-Pol-Env groups (Fig. 1B) (P > 0.1). Thus, the poor control of the infection in the Gag-Pol groups was not due to the absence of a vigorous T-cell response against Gag.
Antibody response.
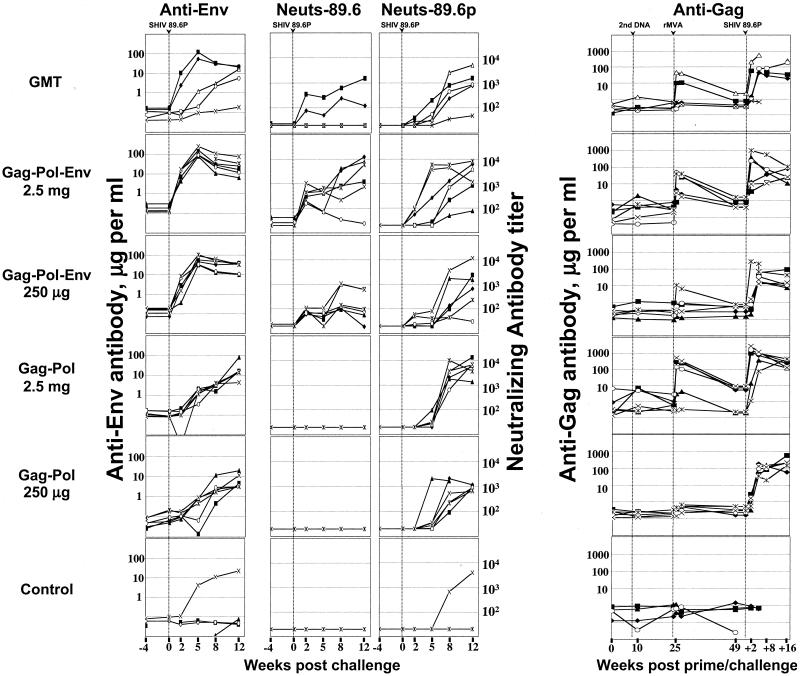
The Gag-Pol-immunized animals underwent a primary antibody response for Env; binding antibodies for 89.6 appeared by 5 weeks postchallenge, and neutralizing antibodies for 89.6P appeared between 5 and 8 weeks postchallenge. This was in contrast to the Gag-Pol-Env-vaccinated animals, which had a secondary antibody response to Env, producing both binding and neutralizing antibody for the 89.6 immunogen by 2 weeks postchallenge and neutralizing antibody for the 89.6P challenge virus by 2 to 5 weeks postchallenge (Fig. 5). At 2 weeks postchallenge, the difference in the appearance of the binding antibody was highly significant (P < 0.001) whereas the difference in the appearance of neutralizing antibody did not achieve significance (P = 0.15). Neutralizing antibodies for 89.6 were not raised in the Gag-Pol group, consistent with studies demonstrating that 89.6 and 89.6P do not raise cross-neutralizing antibodies early after infection (14). Only one of the six control animals developed neutralizing antibody. This animal (macaque 30) had moderate levels of ELISPOT responses at 2 weeks postchallenge (378 spots per 106 PBMC), developed good titers of neutralizing antibody for 89.6P by 12 weeks postchallenge (titer of 3,977), and exhibited transient control of the viral infection (Fig. 4C).
FIG. 5.
Temporal antibody responses. Amounts (micrograms) of total SIV239 Gag or 89.6 Env antibody were determined by using ELISAs. Neutralization titers are the reciprocal of the serum dilution giving 50% neutralization of the indicated viruses grown in human PBMC. Symbols for animals are the same as in Fig. 4. Data points that are not connected reflect missing data. Data for the Gag-Pol-Env-immunized groups are, in part, reproduced from reference 2. Neuts, neutralizing antibody; GMT, geometric mean titer.
The rMVA booster raised antibodies to Gag in both of the groups primed with a high DNA dose (Fig. 5). The titers of these responses were fairly similar in the Gag-Pol-vaccinated animals (geometric mean of 45 μg/ml) and the Gag-Pol-Env-vaccinated animals (geometric mean of 10 μg/ml) (P = 0.3). Postchallenge, anamnestic antibody responses to Gag reached titers of up to 1 mg/ml in both groups. The Gag-Pol groups sustained both higher and longer-lasting antibody responses to Gag than did the Gag-Pol-Env group (P < 0.001) due to the greater viral loads and longer persistence of virus in the Gag-Pol group. None of the control animals developed detectable levels of anti-Gag antibodies by 12 weeks postchallenge.
CD4+ T-cell loss.
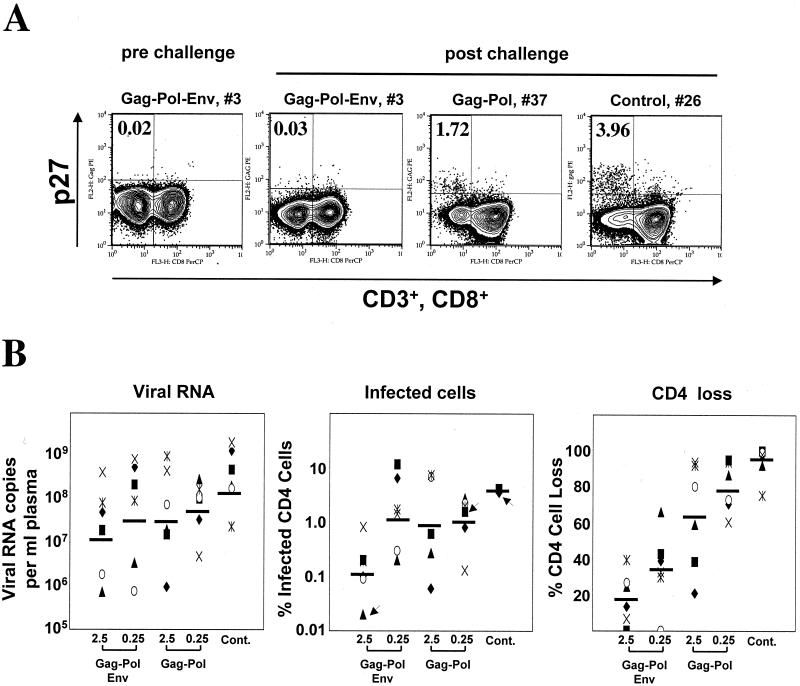
The greater CD4+ T-cell loss in the Gag-Pol-vaccinated animals than in the Gag-Pol-Env-vaccinated animals did not correlate with higher levels of infected cells in the blood (Fig. 6). Acute-phase levels of virus-infected CD4+ cells were measured by means of intracellular p27 staining at 2 weeks postchallenge (Fig. 6A). The Gag-Pol-vaccinated animals in the high- and low-dose DNA-primed groups had geometric mean frequencies of infected cells of 0.87 and 1.11%, respectively. These levels were comparable to those in animals vaccinated with a low Gag-Pol-Env DNA dose (geometric mean frequency of 1.48%) and about 10-fold-higher than those in the animals vaccinated with a high Gag-Pol-Env DNA dose (geometric mean frequency of 0.13%). Similar results were obtained when cells were scored in cocultivation assays with uninfected macaque PBMC (data not shown). Frequencies of infected cells in lymph nodes reflected those in the peripheral blood (data not shown). These differences in the frequency of infected cells did not correlate with the much greater loss of CD4+ cells in the Gag-Pol-vaccinated animals than in the Gag-Pol-Env-vaccinated animals (Fig. 6B). Loss of uninfected CD4+ cells has previously been reported for HIV-1 infections in humans. (6, 13).
FIG. 6.
Viral loads, infected cells, and CD4+ cell loss in the peripheral blood at 2 weeks postchallenge. (A) Intracellular p27 staining. PBMC were fixed and stained for intracellular Gag, CD3, and CD8. Cells were gated on lymphocytes, followed by CD3, and analyzed for CD8 and Gag. Cells in the left quadrants represent CD4+ cells (CD3 positive, CD8 negative). The frequencies in the upper left quadrant are Gag-positive cells as the percentages of the total number of CD4+ cells (left quadrants). Data are shown for animal 3 prechallenge and animals 3, 37, and 26 postchallenge. Animals were chosen to represent the range of the observed values. Slight differences in the gates in the different panels represent different photomultiplier tube settings for assays run at different times. (B) Comparison of viral loads, numbers of infected cells, and CD4+ cell losses at 2 weeks postchallenge. Geometric means of viral RNA copies and percent infected CD4+ cells and arithmetic means of percent CD4+ cell loss are represented as horizontal bars on the respective graphs. Arrows indicate symbols for the animals whose results are shown in panel A. Cont., control.
DISCUSSION
We have compared the protection of macaques against a mucosal challenge following immunization with Gag-Pol-Env- or Gag-Pol-expressing DNA/rMVA vaccines. All of the animals immunized with Gag-Pol-Env-expressing vaccine rapidly controlled the challenge infection, whereas some of the Gag-Pol-immunized animals failed to control the challenge (Fig. 4). The slow and erratic control of the challenge in the Gag-Pol-immunized group was not due to the absence of cellular responses to Gag; indeed, Gag-specific T cells underwent an even higher level of mobilization against the challenge infection in Gag-Pol-vaccinated animals than in Gag-Pol-Env-vaccinated animals (Fig. 1A). The importance of both Env and Gag for protective immunity has been suggested in previous studies by using recombinant poxviruses for priming and recombinant poxviruses or proteins for boosting. (16, 17). Our immunizations extended these studies to a DNA/rMVA vaccine regimen that is highly effective at raising virus-specific T cells (19, 21) and thus might render the need for antibody responses to Env less important.
In addition to providing better control of the challenge virus, the Gag-Pol-Env immunizations also provided better protection of CD4+ cells (Fig. 4B and D). Interestingly, differences in the protection of CD4+ cells appeared to be due primarily to differences in the protection of uninfected CD4+ cells (Fig. 6). At 2 weeks postchallenge, the two low-dose groups had very similar numbers of infected cells (∼1% of the total number of CD4+ cells) but substantial differences in CD4+ cell loss (approximately 35% for the Gag-Pol-Env-immunized group, as opposed to approximately 80% for the Gag-Pol-immunized group). In AIDS infections, much of the early loss of CD4+ cells reflects the apoptotic destruction of noninfected cells (13). We suggest that anti-Env binding antibody that rapidly appeared postchallenge in the Gag-Pol-Env-immunized groups contributed to the early protection of CD4+ cells by binding shed envelope glycoproteins and thus limiting the apoptotic effects of Env on uninfected cells (Fig. 5) (3, 23). This phenomenon could be particularly marked for the 89.6P challenge virus, which has an envelope glycoprotein that is unusually cytopathic for CD4+ cells (7). Effects of Env also could have been due to increased killing of infected cells through antibody-dependent cytotoxicity or through CD8+ targets in Env. We were not able to assess the contributions of these phenomena because neither was measured early after infection.
In contrast to our Gag-Pol immunogens, adenovirus-vectored Gag immunogens have effectively controlled a SHIV 89.6P challenge (22). At the time of challenge, animals in the adenovirus-vectored trials had higher levels of Gag-CM9-specific CD8+ cells than did our DNA/rMVA groups. These higher levels of CD8+ cells, in part, reflected the adenovirus vector raising prolonged peaks of specific CD8+ cells that frequently had two peaks rather than the single sharp peak of specific CD8+ cells raised by DNA/rMVA immunizations. The presence of two peaks after booster administration in the adenovirus-vectored trial could be consistent with some spread of the replication-defective vector in the immunized macaques. Animals in the adenovirus-vectored trial were also challenged relatively soon after the last immunization, before T-cell responses had completely fallen into memory (6 to 12 weeks after booster administration, as opposed to 55 weeks after booster administration).
Despite the superiority of our Gag-Pol-Env immunogens, our Gag-Pol immunogens did provide considerable protection to vaccinated animals. This was presumably mediated by virus-specific T-cell responses (Fig. 1 and 2). Whereas none of 6 control animals controlled their infections to background levels, 7 of 12 Gag-Pol-immunized animals contained their challenges to the level of detection (Fig. 4). By 46 weeks postchallenge, 5 of 6 control animals but only 1 of 12 Gag-Pol-vaccinated animals had developed AIDS. The virus-specific T-cell responses in the Gag-Pol-immunized animals also preserved the ability of these animals to mount a de novo antibody response to Env, which had appeared by 8 weeks postchallenge (Fig. 5). Thus, our results demonstrate that immune responses to Gag and Pol can control an immunodeficiency virus challenge but that immune responses to Env synergize with those to Gag and Pol in achieving efficient and consistent control of the challenge. Our results are also provocative in that they suggest that binding antibody to Env, in the presence of a strong T-cell response, may help protect against the early apoptotic loss of uninfected CD4+ T cells.
Acknowledgments
This study was supported by Integrated Preclinical/Clinical AIDS Vaccine Development program project P01 AI 43045, the Emory/Atlanta Center for AIDS Research (P30 DA 12121), Yerkes Regional Primate Research Center base grant P51 RR00165, a Centers for Disease Control and Prevention postdoctoral fellowship, and National Institute of Allergy and Infectious Diseases contract AI 85343 to D. Montefiori.
We thank J. Sodroski for molecularly cloned SHIV 89.6; L. Ratner for molecularly cloned HIV-1 ADA sequences; K. Reimann for the SHIV 89.6P seed stock; D. Pauza for plasmid pGEX27; L. Frampton for help in preparation of the rMVA; the NIH AIDS Research and Reference Reagent Program for anti-SIV p27 monoclonal antibody clone FA-2; J. Pohl and the Emory Microchemical Facility for synthesis of peptides; R. Polavarapu and the Emory DNA Sequence Facility for DNA sequencing; B. Grimm, S. Sharma, M. Patel, S. Patel, and D. Campbell for excellent technical help; and H. Drake-Perrow for outstanding administrative support. We are grateful to The Yerkes Division of Research Resources for the consistent excellence of veterinary care and pathology support.
REFERENCES
- 1.Allen, T. M., T. U. Vogel, D. H. Fuller, B. R. Mothe, S. Steffen, J. E. Boyson, T. Shipley, J. Fuller, T. Hanke, A. Sette, J. D. Altman, B. Moss, A. J. McMichael, and D. I. Watkins. 2000. Induction of AIDS virus-specific CTL activity in fresh, unstimulated peripheral blood lymphocytes from rhesus macaques vaccinated with a DNA prime/modified vaccinia virus Ankara boost regimen. J. Immunol. 164:4968-4978. [DOI] [PubMed] [Google Scholar]
- 2.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H.-L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 3.Banda, N. K., J. Bernier, D. K. Kurahara, R. Kurrle, N. Haigwood, R. P. Sekaly, and T. H. Finkel. 1992. Crosslinking CD4 by human immunodeficiency virus gp120 primes T cells for activation-induced apoptosis. J. Exp. Med. 176:1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch, D. H., S. Santra, M. J. Kuroda, J. E. Schmitz, R. Plishka, A. Buckler-White, A. E. Gaitan, R. Zin, J. H. Nam, L. S. Wyatt, M. A. Lifton, C. E. Nickerson, B. Moss, D. C. Montefiori, V. M. Hirsch, and N. L. Letvin. 2001. Reduction of simian-human immunodeficiency virus 89.6P viremia in rhesus monkeys by recombinant modified vaccinia virus Ankara vaccination. J. Virol. 75:5151-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, and M. G. Lewis. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 6.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 7.Etemad-Moghadam, B., D. Rhone, T. Steenbeke, Y. Sun, J. Manola, R. Gelman, J. W. Fanton, P. Racz, K. Tenner-Racz, M. K. Axthelm, N. L. Letvin, and J. Sodroski. 2001. Membrane-fusing capacity of the human immunodeficiency virus envelope proteins determines the efficiency of CD+ T-cell depletion in macaques infected by a simian-human immunodeficiency virus. J. Virol. 75:5646-5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann-Lehmann, R., R. K. Swenerton, V. Liska, C. M. Leutenegger, H. Lutz, H. M. McClure, and R. M. Ruprecht. 2000. Sensitive and robust one-tube real-time reverse transcriptase-polymerase chain reaction to quantify SIV RNA load: comparison of one- versus two-enzyme systems. AIDS Res. Hum. Retrovir. 16:1247-1257. [DOI] [PubMed] [Google Scholar]
- 9.Karlsson, G. B., M. Halloran, J. Li, I.-W. Park, R. Gomila, K. A. Reimann, M. K. Axthelm, S. A. Iliff, N. L. Letvin, and J. Sodroski. 1997. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J. Virol. 71:4218-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kern, F., I. P. Surel, N. Faulhaber, C. Frömmel, J. Schneider-Mergener, C. Schönemann, P. Reinke, and H.-D. Volk. 1999. Target structures of the CD8+-T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J. Virol. 73:8179-8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knapp, L. A., E. Lehmann, M. S. Piekarczyk, J. A. Urvater, and D. I. Watkins. 1997. A high frequency of Mamu-A*01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens 50:657-661. [DOI] [PubMed] [Google Scholar]
- 12.Maecker, H. T., H. S. Dunn, M. A. Suni, E. Khatamzas, C. J. Pitcher, T. Bunde, N. Persaud, W. Trigona, T. M. Fu, E. Sinclair, B. M. Bredt, J. M. McCune, V. C. Maino, F. Kern, and L. J. Picker. 2001. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J. Immunol. Methods 255:27-40. [DOI] [PubMed] [Google Scholar]
- 13.McCune, J. M. 2001. The dynamics of CD4+ T-cell depletion in HIV disease. Nature 410:974-979. [DOI] [PubMed] [Google Scholar]
- 14.Montefiori, D. C., K. A. Reimann, M. S. Wyand, K. Manson, M. G. Lewis, R. G. Collman, J. G. Sodroski, D. P. Bolognesi, and N. L. Letvin. 1998. Neutralizing antibodies in sera from macaques infected with chimeric simian-human immunodeficiency virus containing the envelope glycoproteins of either a laboratory-adapted variant or a primary isolate of human immunodeficiency virus type 1. J. Virol. 72:3427-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montefiori, D. C., W. E. Robinson, Jr., S. S. Schuffman, and W. M. Mitchell. 1988. Evaluation of antiviral drugs and neutralizing antibodies to human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J. Clin. Microbiol. 26:231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ourmanov, I., M. Bilska, V. M. Hirsch, and D. C. Montefiori. 2000. Recombinant modified vaccinia virus Ankara expressing the surface gp120 of simian immunodeficiency virus (SIV) primes for a rapid neutralizing antibody response to SIV infection in macaques. J. Virol. 74:2960-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polacino, P. S., V. Stallard, J. E. Klaniecki, S. Pennathur, D. C. Montefiori, A. J. Langlois, B. A. Richardson, W. R. Morton, R. E. Benveniste, and S.-L. Hu. 1999. Role of immune responses against the envelope and the core antigens of simian immunodeficiency virus SIVmne in protection against homologous cloned and uncloned virus challenge in macaques. J. Virol. 73:8201-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Power, C. A., C. L. Grand, N. Ismail, N. C. Peters, D. P. Yurkowski, and P. A. Bretscher. 1999. A valid ELISPOT assay for enumeration of ex vivo, antigen-specific, IFNγ-producing T cells. J. Immunol. Methods 227:99-107. [DOI] [PubMed] [Google Scholar]
- 19.Ramshaw, I. A., and A. J. Ramsay. 2000. The prime-boost strategy: exciting prospects for improved vaccination. Immunol. Today 21:163-165. [DOI] [PubMed] [Google Scholar]
- 20.Rose, N. F., P. A. Marx, A. Luckay, D. F. Nixon, W. J. Moretto, S. M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J. K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539-549. [DOI] [PubMed] [Google Scholar]
- 21.Schneider, J., S. C. Gilbert, C. M. Hannan, P. Degano, E. G. Sheu, M. Plebanski, and A. V. S. Hill. 1999. Induction of CD8+ T cells using heterologous prime-boost immunisation strategies. Immunol. Rev. 170:29-38. [DOI] [PubMed] [Google Scholar]
- 22.Shiver, J. W., T.-M. Fu, L. Chen, D. Casimiro, M. E. Davies, R. K. Evans, Z.-Q. Zhang, A. J. Adam, W. Trigona, S. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. Persaud, L. Guan, K. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Rohannon, D. B. Volkin, D. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 23.Westendorp, M. O., R. Frank, C. Ochsenbauer, K. Stricker, J. Dhein, H. Walczak, K. M. Debatin, and P. H. Krammer. 1995. Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature 375:497-500. [DOI] [PubMed] [Google Scholar]