Abstract
Covalently closed circular DNA (cccDNA) is a crucial intermediate in the replication of hepadnaviruses. We inhibited the replication of duck hepatitis B virus in congenitally infected ducks with a combination of lamivudine and a dideoxyguanosine prodrug. Inhibition of viral replication should prevent renewal of the cccDNA pool, and its decay was measured in liver biopsy samples collected over a 5-month period. In three ducks, the cccDNA pools declined exponentially, with half-lives ranging from 35 to 57 days. In two others, the pools declined exponentially for about 70 days but then stabilized at about 6 copies/diploid genome. The selection of drug-resistant virus mutants is an unlikely explanation for this unexpected stabilization of cccDNA levels. Liver sections stained for the cell division marker PCNA showed that animals in which cccDNA loss was continuous had significantly greater numbers of PCNA-positive nuclei than did those animals in which cccDNA levels had plateaued.
The hepadnaviruses include the human pathogen Hepatitis B virus (HBV) and animal viruses such as Duck hepatitis B virus (DHBV) and Woodchuck hepatitis B virus (WHV), which are important models for studying hepadnavirus biology. These viruses are characterized by small, circular, partially double-stranded DNA genomes. The minus strand of the viral DNA is covalently linked to protein at its 5′ end and contains a nick at its 3′ end. The plus strand is only partially complete (for reviews, see references 10 and 21). A crucial event in the replication of these viruses is the conversion of this form of the viral genome into a covalently closed circular DNA (cccDNA) form within the nucleus of a newly infected hepatocyte (15, 17, 26). cccDNA is the template for viral transcripts encoding structural proteins and the viral polymerase. It is also the template for the pregenomic RNA which is the precursor of the viral genome. Early in infection, the pool of nuclear cccDNA is amplified to about 20 molecules/cell (13, 14, 24). This is accomplished when an intracellular circuit of pregenomic RNA is encapsidated and converted to viral DNA in the cytoplasm and subsequently enters the nucleus and converts to cccDNA (29). Once a pool of cccDNA is established, capsids containing newly replicated viral DNA are enveloped and exported from the cell rather than entering the nucleus. This change in the fate of nucleocapsids is thought to be caused by rising levels of the pre-S envelope protein resulting from an increasing pool of cccDNA template (22, 23).
Attempts to cure hepadnavirus infections by treatment with antiviral drugs suggest that the cccDNA pool is very stable. If cccDNA turnover were rapid, then inhibitors of the viral reverse transcriptase would prevent replenishment of the cccDNA pool, leading to its elimination after a short course of treatment. However, human HBV carriers treated with lamivudine for 3 months usually relapse quickly when treatment is discontinued, indicating that cccDNA persists (5). If treatment is continued for 1 to 2 years, drug-resistant mutants often arise, indicating that cccDNA must have persisted for at least as long as the treatment period (2, 11). Similar results are seen in animal model systems. In cultured woodchuck hepatocytes, WHV cccDNA is maintained over several weeks of antiviral therapy (4, 18). DHBV-infected ducks treated with lamivudine for many months nevertheless show a rebound in viremia as soon as drug treatment ceases (unpublished observations). Measurement of the half-life of the cccDNA pool would aid in the design of more effective antiviral treatment regimens.
The first attempt to measure the stability of the cccDNA pool of a hepadnavirus was described by Civitico and Locarnini (3). They measured the turnover rate of the endogenous cccDNA pool when primary duck hepatocytes were transferred to a medium containing bromodeoxyuridine. This nucleoside was incorporated into newly synthesized cccDNA and increased its buoyant density. In this pulse-chase experiment, unlabeled cccDNA of normal buoyant density turned over very rapidly, with a half-life of only 3 to 5 days. This result is difficult to reconcile with the evident stability of the cccDNA pool in vivo. The cccDNA pool is known to rise rapidly as cultured hepatocytes dedifferentiate in vitro to levels that are 50 to 100 times higher than that found in an intact liver. In addition, the bromodeoxyuridine density label used in this experiment inhibits DHBV transcription and replication (3, 26). A combination of these effects may have led to an underestimate of the cccDNA half-life in an intact liver.
Zhu et al. used a different strategy to estimate the half-lives of the cccDNA pools in the livers of woodchucks infected with WHV (31). Synthesis of new cccDNA was suppressed withl-FMAU (l-2′-deoxy-2′fluoro-5-methyl-1-β-d-arabinosylura-cil), a chain-terminating nucleoside analog which specifically inhibits the viral polymerase. The sizes of the cccDNA pools were measured in liver tissues retrieved by biopsy at various times after drug treatment was initiated. The cccDNA pools decayed, with half-lives of 33 days in two animals and 50 days in a third animal. These results are consistent with the reported difficulty in eliminating cccDNA by long-term treatments of HBV, DHBV, and WHV infections with viral polymerase inhibitors. Using a similar strategy, we sought to measure the half-lives of the cccDNA pools in the livers of ducks congenitally infected with DHBV.
Six-week-old Pekin ducks were used in our study and were maintained according to the regulations of the Canadian Council on Animal Care. At 6 weeks of age, a duck's liver mass and cccDNA pool size are stable (30). All of the ducks were infected with the Alberta strain of DHBV type 16. Some were congenitally infected by their mothers; others were infected in ovo (25) with high-titer infectious duck serum. Initial virus levels in serum, ascertained by dot blot analysis, ranged from 2 × 108 to 5 × 109 viral genome equivalents (vge)/ml (Tables 1 and 2). DHBV replication was strongly inhibited by a combination of 40 mg of lamivudine/kg of body weight (a gift of Glaxo Wellcome, Research Triangle Park, N.C.) and 20 mg of 2-amino-6-methoxypurine-2′,3′-dideoxyriboside/kg. Both drugs were administered twice per day by intramuscular injection. The latter is a dideoxyguanosine (ddG) prodrug and was synthesized according to the method of Robbins et al. (20). The ducks were treated with two drugs, because ddG can inhibit both the protein priming of DHBV replication (12) and chain elongation, the target of lamivudine action. We assumed that the combined drug treatment would more profoundly inhibit DHBV replication than would treatment with either drug alone. In addition, preliminary studies have shown that polymerase mutations that are resistant to lamivudine remain sensitive to ddG (D. L. J. Tyrrell, M. Robbins, L. Condrey, and K. P. Fischer, Abstr. 48th Meet. Am. Soc. Liver Dis., abstr. 1207, 1997). Combined drug therapy should therefore reduce the selection of drug-resistant viral mutants over the course of the study.
TABLE 1.
Characteristics of control ducks
| Duck | Sex | Mode of infection | Initial serum virus level (109 vge/ml) | Mean cccDNA copy no./diploid genome (± SD) | ASTa (IU/liter) |
|---|---|---|---|---|---|
| 17 | Female | In ovo | 1.0 | 27.0 (± 8.5) | 24 |
| 48 | Male | In ovo | 1.0 | 28.0 (± 7.3) | 22 |
| 77 | Female | Congenital | 4.5 | 23.2 (± 6.8) | 22 |
Serum AST activity level on day 152.
TABLE 2.
Characteristics of ducks treated with lamivudine and 2-amino-6-methoxypurine-2′,3′-dideoxyriboside for 155 days
| Duck | Sex | Mode of infection | Initial serum virus level (109 vge/ml) | cccDNA pool half-life (days) | r2b | ASTa (IU/liter) |
|---|---|---|---|---|---|---|
| 50 | Male | In ovo | 0.2 | 38c | 0.85 | 10 |
| 65 | Male | In ovo | 4.8 | 47c | 0.96 | 55 |
| 68 | Male | In ovo | 0.6 | 57 | 0.94 | 5 |
| 992 | Male | Congenital | 2.7 | 35 | 0.97 | 14 |
| 998 | Female | Congenital | 1.1 | 48 | 0.98 | 7 |
Serum AST activity level on day 152.
r2, coefficient of determination of exponential regression analysis.
Over the first 80 days of treatment before cccDNA levels stabilized in these animals.
Our strategy for measuring the half-life of the cccDNA pool was predicated on a rapid and thorough inhibition of new cccDNA synthesis. In preliminary experiments, the effectiveness of the two drugs at inhibiting DHBV replication was determined by measuring the kinetics of the loss of DHBV DNA from the sera of treated animals. Two congenitally infected ducks (ducks 33 and 19) were treated with the drug combination and were bled at frequent intervals over a 4-day period. Virus levels in serum were measured by dot blot hybridization and phosphorimager analysis. The decline in the virus levels followed a complex course, as shown in Fig. 1A and B. The curves in Fig. 1A and B suggest that a loss in viral DNA, effected by the drugs, was superimposed on pulses of virus released into the serum. To test this possibility, four untreated, congenitally infected ducks (ducks 6, 26, 42, and 979) were bled on the same schedule as were the ducks described above. As shown in Fig. 1C, the virus levels in their sera oscillated, declining to a minimum over the first 12 h and subsequently fluctuating, with peaks at 18 and 42 h after the experiment began. The peaks occurred at midnight, in the middle of the animals' “dark period,” and were remarkably coherent for all four animals. Subsequently, the fluctuations in the animals' virus levels began to lose their periodicity and coherence. To our knowledge, this is the first report of such oscillations in DHBV levels in serum. We suggest that this response was induced by the stress experienced by the animals when being handled and bled and that its period was tied to the diurnal rhythm of the animals. The periodicity in serum virus levels was lost as the animals became habituated to handling. This conjecture is reasonable, since young ducks show a marked diurnal rhythm in the release of the stress hormone corticosterone (28). In pigeons, frequent handling increases the corticosterone level but not the rhythm of its release (27). Finally, corticosteroids are known to stimulate DHBV replication (9). We think that in our experiment, inhibition of viral replication by the drug treatment was partially overcome by a stimulation of replication induced by the stress experienced by the animals during handling; the combination of these antagonistic effects resulted in the curves seen in Fig. 1A and B. Despite these complications, it was evident that the antiviral drug combination resulted in the immediate and rapid inhibition of DHBV replication that is required for a cccDNA half-life study.
FIG. 1.
(A to C) Serum DHBV DNA levels decline rapidly in ducks treated with lamivudine and a ddG prodrug. The drug treatment and test bleeds of all ducks began at 6 a.m., and antiviral drugs were administered by intramuscular injection every 12 h thereafter. The ducks were initially bled every 6 h, then every 8 h, and finally every 12 h as indicated by the symbols on the graphs. Serum virus levels were measured by dot blot hybridization to a 32P-labeled DHBV probe. Radioactivity was measured with a Fuji phosphorimager, and viremia is reported in arbitrary units (PI units). The birds were housed in a room with a cycle of 10 h of light (commencing at 8 a.m.) followed by 14 h of darkness. (A) Duck 33 was treated with lamivudine and ddG prodrug, and its serum virus level was monitored over 3.5 days. (B) Duck 19 was treated identically to duck 33. (C) Four control ducks (⧫, duck 6; ▪, duck 26; ▴, duck 42; ✖, duck 979) were bled on the same schedule as ducks 33 and 19 but received no drug treatment. (D) Dot blot assay of viremia in ducks. Ten microliters of serum from each duck in the cccDNA half-life study was obtained at each of the indicated time points, applied to a Hybond-N membrane (Amersham), hybridized to a 32P-labeled DHBV probe, and autoradiographed. Ducks 17, 48, and 77 were left untreated, and ducks 50, 65, 68, 992, and 998 were treated with lamivudine and a ddG prodrug. (E) DNA was isolated from serum samples from treated duck 992 at the indicated time points, and DHBV sequences were amplified by PCR. The amount of amplicon for each time point was estimated by measuring the intensity of its band on a stained agarose gel. The amounts of amplicon produced by the indicated serial dilutions of DNA isolated on day 0 are marked by horizontal lines. (F) Quantitative Southern blot of cccDNA present in liver DNA at the start of the half-life study. One microgram of DNA from duck liver tissue (Duck) or uninfected duck erythrocyte doped with known amounts of a plasmid carrying the DHBV genome (Copies DHBV × 106) was digested with EcoRI endonuclease, separated on an agarose gel, transferred to a Hybond-N+ membrane, and probed with 32P-labeled DHBV DNA. A Fuji phosphorimager was used to quantify the hybridized probe. The cccDNA copy numbers inferred from the Southern blot are shown and are compared to the copy numbers derived from the competitive PCR assay.
The ducks were bled weekly over the course of the study. The viremia of the treated ducks became undetectable by dot blot assay within the first week (Fig. 1D). The detection limit of this assay was only about 0.5% of the initial serum virus levels. A more sensitive PCR assay was used to estimate lower virus levels. Amplification of the region between positions 1039 and 1945 of the DHBV sequence was performed on DNA isolated from 20-μl serum samples (primers CTCAAGAGATTCCTCAGCC and GTCATACCATTCTCCTACT; 40 cycles consisting of 95°C for 30s, 50°C for 30s, and 72°C for 60s). Samples collected during the entire course of drug treatment were analyzed for each animal. The benchmarks for estimating virus levels were established by amplifying 10-fold serial dilutions of serum DNA from day 0 (before treatment began). The PCR products were separated by electrophoresis and stained with ethidium bromide, and the amount of 906-bp amplicon was measured with a video image capture system and NIH Image Gauge software. Figure 1E shows the results obtained for duck 992; similar results were obtained for the other animals. Over the first week of treatment, the virus level was reduced to 10−2 to 10−3 of its initial level; the virus level continued to fall over the next several weeks and plateaued at or below 10−5 of the initial level after 5 to 8 weeks of treatment. The inhibition of replication we observed was fourfold more rapid than that reported by Zhu et al. in their study of the half-life of WHV cccDNA (31). The rapid and potent inhibition of viral replication produced by our drug treatment regimen was thought to be sufficient to prevent renewal of the cccDNA pool and permit analysis of its decay over time.
Eight ducks were used in the cccDNA half-life study. Three control ducks were injected with 0.5 ml of phosphate-buffered saline, and five ducks were treated with lamivudine and the ddG prodrug as described above. At eight points over a 155-day period, liver tissue from each animal was obtained by needle biopsy, total DNA was isolated without proteinase treatment, and duplicate competitive PCR assays selective for DHBV cccDNA were performed as previously described (1). The accuracy of the PCR assay at the start of the study, before drug treatment had eliminated other viral replicative intermediates, was confirmed by quantitative Southern blotting. An autoradiogram of the Southern blot along with a comparison of the cccDNA copy numbers inferred from the two methods is shown in Fig. 1F. The two assays yielded very similar estimates of the initial cccDNA copy numbers. At the end of the study, the cccDNA levels of each animal, expressed as the copies of cccDNA per diploid genome, were plotted against time. Where appropriate, the data were fitted to an exponential decay curve by regression analysis (Microsoft Excel). These curves are shown in Fig. 2. The cccDNA copy numbers in samples taken on day 43 of the experiment were much lower than expected for both the control and treated animals. We think that these results are anomalous and possibly due to the selective loss of cccDNA from the DNA samples isolated on that day. While the results of the day 43 assays are shown on the graphs, they were not included in the regression analyses used to calculate the half-lives of cccDNA pools.
FIG. 2.
DHBV cccDNA has a half-life of 35 to 57 days in ducks treated with lamivudine and ddG prodrug. Drug treatment was performed as described in the legend to Fig. 1. Control ducks were injected with a placebo of phosphate-buffered saline twice per day. At the indicated time points, liver tissue was sampled by needle biopsy, total cellular DNA was isolated, and the cccDNA copy number was measured by competitive PCR. Data from day 43 were anomalous and are shown but were not used in the construction of the curves (see the text). (A) cccDNA copy numbers in untreated ducks (◊, duck 17; □, duck 48; ▵, duck 77); (B to F) decline in cccDNA copy numbers in ducks treated with antiviral drugs.
The sizes of the cccDNA pools in untreated control ducks are shown in Fig. 2A. The cccDNA levels at the beginning and the end of the experiment were similar, but the most notable features of these data are the fluctuations in the cccDNA levels over time, which are reflected in the large standard deviations shown in Table 1. We think that these fluctuations are real and not due to experimental error, since the data for the treated ducks show much less random variation than the data for the controls. The patterns of fluctuation were similar for all three animals, suggesting that the variation is driven by some environmental factor. Virus levels in serum also fluctuated over the course of the study, but there was no correlation, either positive or negative, between cccDNA levels and serum virus levels (data not shown).
The cccDNA pool sizes in treated ducks 998, 992, and 68 decayed in a simple pattern, as shown in Fig. 2B to D. These data fit exponential decay curves very well, as reflected by the coefficient of determination values that are close to 1 (Table 2). The half-lives of the cccDNA pools in the three animals were quite different: 48 days for duck 998, 35 days for duck 992, and 57 days for duck 68. Both the magnitude of and the animal-to-animal variation in the cccDNA half-lives in these ducks were similar to those reported for WHV cccDNA in woodchuck livers (half-lives of 33 and 50 days) by Zhu et al. (31).
The cccDNA pools in the two remaining treated animals, ducks 65 and 50, showed an unexpected pattern of decay. As shown in Fig. 2E and F, the cccDNA pools decayed exponentially, with half-lives of 47 and 38 days, respectively, for about the first 70 days of treatment. Then the cccDNA levels abruptly plateaued at about 8 and 6 copies of cccDNA/diploid genome, respectively. The reason for the stabilization of the cccDNA pools in these animals is not clear. All the animals in the study had normal serum aspartic acid transaminase (AST) levels on day 152 (Tables 1 and 2), suggesting that impaired liver function could not provide an explanation for the unexpected pattern of cccDNA loss in ducks 50 and 65. The emergence of a drug-resistant mutant virus would explain these results. The selection of such mutants occurs frequently upon treatment of infected woodchucks and humans (7, 16, 31). However, we think this explanation is unlikely. Viral DNA could not be detected by dot blotting in the sera of these ducks at any point during drug treatment (Fig. 1D). The more sensitive PCR assay did not detect any increase in viral DNA in serum from the very low levels typical of all treated animals late in the study. Had a drug-resistant mutant arisen, a resurgent viremia should have resulted. PCR amplification of the low levels of virus (under the conditions described above) in serum collected on day 155 permitted the sequencing of viral DNA encoding the YMDD and upstream FLL motifs that can mutate to confer lamivudine resistance (8). The amplicons from ducks 50 and 65 were sequenced, and the sequences of the polymerase reading frame encoding amino acids 484 to 594, a region containing both the FLL and YMDD motifs, were found to be wild type. Thus, the viral polymerase likely remained sensitive to inhibition by lamivudine. It is unlikely that the emergence of a drug-resistant virus prevented ducks 50 and 65 from continuing to lose cccDNA.
The difference in the kinetics of cccDNA loss between the two groups of treated ducks prompted us to examine sections of liver tissue recovered at the end of the study for histopathological differences. The liver tissue was fixed in 4% buffered paraformaldehyde, embedded in paraffin, sectioned, and stained with eosin and hematoxylin. The three untreated animals showed mild to moderate portal inflammation and mild piecemeal and focal necrosis. Ducks 17 and 77 showed fatty changes in their livers, which were severe in the case of duck 17 and mild in the case of duck 77. The ducks treated with antiviral drugs had milder hepatitis than did the control animals. Duck 65 showed no evidence of hepatitis. The four other treated ducks showed mild portal inflammation. Necrosis was absent in all but duck 68, which showed mild focal necrosis. No histological difference was seen between animals in which cccDNA levels stabilized at a low level and those in which the cccDNA pools continued to decline over the course of the study. These histological findings were corroborated by tests for liver enzymes in serum. The serum AST levels, shown in Tables 1 and 2, were in the normal range at the end of the study, as were those of the other liver enzymes tested (alanine aminotransferase, bilirubin, and γ-glutamyl aminotransferase; data not shown). Immunohistochemical staining of liver sections for DHBV core and pre-S proteins was similarly uninformative. Sections from treated animals showed weaker staining than did sections from untreated animals, which was expected since treatment had reduced the levels of cccDNA, the template for viral transcription.
We speculated that the killing or natural turnover of hepatocytes might be an important mechanism by which cccDNA is eliminated from the livers of the treated animals. Loss of hepatocytes would be accompanied by the cell division needed for liver regeneration. The accumulation of PCNA (proliferating cell nuclear antigen) is a marker for dividing cells. Levels of this highly conserved protein rise at the end of the G1 and the beginning of the S phases of the cell cycle. In addition to its crucial role in DNA replication, PCNA is also involved in several DNA repair pathways, DNA methylation, and chromatin assembly. Thus, we sought to measure the loss of hepatocytes indirectly by examining liver tissue sections for nuclei in S phase that were marked by PCNA expression. Sections of liver tissue recovered at the end of the study were stained for PCNA by indirect immunohistochemistry with mouse monoclonal anti-PCNA antibodies (Novocastra NC-PCNA and Zymed 13-3900) bridged with an anti-mouse immunoglobulin G-biotin conjugate (Sigma B-7274) to an avidin-peroxidase (Sigma E-8386) detection reagent. PCNA-positive nuclei stained brown with the peroxidase substrate 3,3′-diaminobenzidine; all other nuclei stained blue with the hematoxylin counterstain. There were significant differences among the ducks in the staining results for PCNA. The results for control animals differed from those for the treated animals both qualitatively and quantitatively. As illustrated in Fig. 3, PCNA staining was weaker in tissues from control animals than in tissues from animals treated with the combination of antiviral drugs. Positive nuclei were pale brown, with none of the almost black nuclei seen in sections from the treated ducks. The basis for this qualitative difference in staining results between the control and treated animals is not known. The liver samples from all the animals were processed identically; therefore, the states and accessibilities of the PCNA antigen were unlikely to differ between the treated and untreated groups. It is possible that the nucleoside analogs used in the study increased PCNA accumulation in hepatocyte nuclei. Incorporation of nucleoside analogs into cellular DNA during replication may lengthen the S phase of the cell cycle and lead to increased PCNA accumulation or induce DNA repair pathways, of which PCNA is a component.
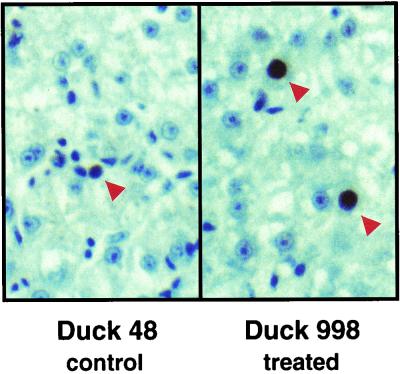
FIG. 3.
Comparison of PCNA staining in control and treated ducks. Liver tissue sections from control duck 48 and treated duck 998 recovered on day 155 of the study were immunostained for the presence of PCNA. PCNA-positive nuclei are indicated by arrowheads. Magnification, ×400.
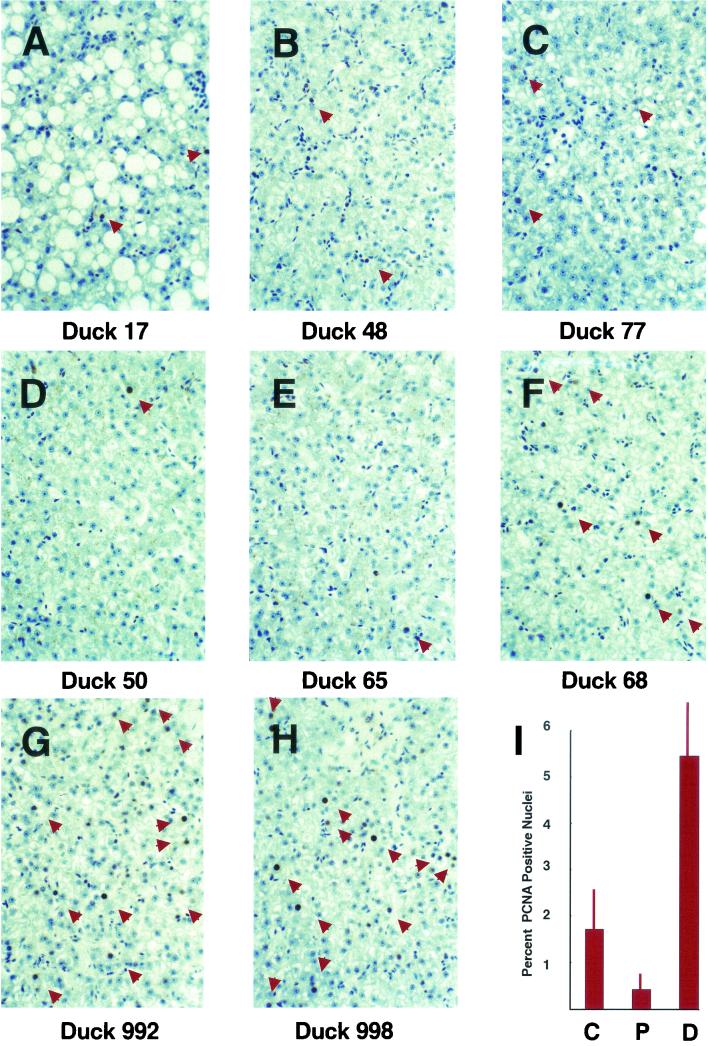
The frequency of PCNA-positive liver nuclei varied among the animals in the cccDNA half-life study. Figure 4 shows the PCNA staining of liver sections from each of these animals. Infrequent, weakly PCNA-positive nuclei were seen in sections from the control animals (Fig. 4A to C). Among the treated animals, two classes could be distinguished. Ducks 50 and 65 showed rare but intensely PCNA-positive nuclei (Fig. 4D and E). These were ducks in which cccDNA levels initially fell but then stabilized at a low level. Intensely PCNA-positive hepatocyte nuclei were abundant in ducks 68, 992, and 998 (Fig. 4F to H). These were the animals in which cccDNA levels showed a continuous decline.
FIG. 4.
Immunohistochemical staining of duck liver sections with anti-PCNA monoclonal antibodies. Liver samples recovered on day 155 of the study were fixed, sectioned, and stained for the presence of PCNA. PCNA-positive nuclei are marked by arrowheads. Sections from control ducks (A to C), ducks in which cccDNA levels plateaued after an initial decline (D and E), and ducks in which cccDNA levels declined through the course of the study (F to H) are shown. The average frequency of PCNA-positive hepatocyte nuclei in three random fields of sections from each duck in each of the three groups (C, control; P, cccDNA plateaud; D, cccDNA declined) is also shown (I). Magnifications, ×160 (all panels)
To quantify these differences, PCNA-positive nuclei in three random fields from liver sections from each duck were counted and divided by the total number of hepatocytes in each field to yield a PCNA expression index. The average indexes for the control animals, for those in which cccDNA levels plateaued, and for those in which cccDNA levels declined continuously are shown in Fig. 4I. The differences among these averages are statistically significant (P < 0.001, Student's t test). A continuous decline in cccDNA levels correlates with a high PCNA expression index (5.5% ± 1.2% [mean ± standard deviation] of hepatocytes), while an initial loss of cccDNA followed by maintenance or a much slower loss of cccDNA correlates with a much lower frequency of PCNA-positive hepatocytes (0.28% ± 0.3%). The PCNA expression index of 1.6% ± 1.1% for the control animals lies between these values and is similar to the 1 to 2% reported by Nicholl et al. (19).
Our data suggest that the turnover of hepatocytes, which is indicated by a high frequency of PCNA-positive nuclei, is associated with the continued loss of cccDNA from the liver, as seen in ducks 68, 992, and 998. The cause of this enhanced turnover rate is not known. If the immune system is destroying infected cells, then it seems to be doing so in a drug-dependent manner since the hepatocyte turnover rate is significantly lower in the control animals than in the treated animals. The low hepatocyte turnover rate seen in ducks 50 and 65 is associated with a stable or very slowly decaying cccDNA pool. A direct relationship between the hepatocyte turnover rate and the rate of cccDNA loss is suggested but cannot be established on the basis of our data. First, the number of animals in the study was small and the correlation might be coincidental. Second, it would have been informative to have liver tissue samples from early in the treatment period, when the cccDNA pools of ducks 50 and 65 were declining at rates similar to those of the other treated ducks. A high PCNA staining index at that time would have strengthened the argument that hepatocyte loss is required for cccDNA loss. Our study was designed simply to measure the half-lives of cccDNA pools in treated animals. We did not anticipate that some of the animals would cease to lose cccDNA at an appreciable rate during the course of drug treatment.
The two responses of the DHBV cccDNA pools to inhibitors of DNA replication—a slow uninterrupted decline in copy numbers and a decline followed by stabilization—may also occur when HBV infections are treated with antiviral drugs. Long-term treatment of chronic HBV infections with lamivudine seems to follow two courses (6, 7). Some patients eventually stop producing HBeAg, which is perhaps a reflection of the loss of the cccDNA pool in the liver. These patients have the best chance of escaping relapse when drug therapy is discontinued. Other patients continue to produce HBeAg even after prolonged therapy and inevitably relapse if therapy is stopped. In these patients, the cccDNA pool may have stabilized. The invasiveness of liver biopsy will make the testing of this hypothesis with humans difficult. Further work with the duck model system is required to identify the mechanism by which cccDNA is lost during prolonged antiviral therapy and to understand how that mechanism can fail to eliminate the cccDNA pool in some cases.
Nucleotide sequence accession number
The sequence of the Alberta strain of DHBV type 16 has been deposited in GenBank under accession no. AF047045.
Acknowledgments
We acknowledge Gerald Lachance for expert care of the animals and thank Karl Fischer for useful comments on the manuscript.
This research was supported by Glaxo Wellcome Canada.
REFERENCES
- 1.Addison, W. R., W. W. S. Wong, K. P. Fischer, and D. L. J. Tyrrell. 2000. A quantitative competitive PCR assay for the covalently closed circular form of the duck hepatitis B virus. Antivir. Res. 48:27-37. [DOI] [PubMed] [Google Scholar]
- 2.Chyama, K., Y. Suzuki, M. Kobayashi, M. Kobayashi, A. Tsubota, M. Hashimoto, Y. Miyano, H. Koike, M. Kobayashi, I. Koida, Y. Arase, S. Saitoh, N. Murashima, K. Ikeda, and H. Kumada. 1998. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology 27:1711-1716. [DOI] [PubMed] [Google Scholar]
- 3.Civitico, G. M., and S. A. Locarnini. 1994. The half-life of duck hepatitis B virus supercoiled DNA in congenitally infected primary hepatocyte cultures. Virology 203:81-89. [DOI] [PubMed] [Google Scholar]
- 4.Dandri, M., M. R. Burda, H. Will, and J. Petersen. 2000. Increased hepatocyte turnover and inhibition of woodchuck hepatitis B virus replication by adefovir in vitro do not lead to reduction of the closed circular DNA. Hepatology 32:139-146. [DOI] [PubMed] [Google Scholar]
- 5.Dienstag, J. L., R. P. Perrillo, E. R. Schiff, M. Bartholomew, C. Vicary, and M. Rubin. 1995. A preliminary trial of lamivudine for chronic hepatitis B infection. N. Engl. J. Med. 333:1657-1661. [DOI] [PubMed] [Google Scholar]
- 6.Dienstag, J. L., E. R. Schiff, T. L. Wright, R. P. Perillo, H.-W. L. Hann, Z. Goodman, L. Crowther, L. Condreay, M. Woessner, M. Rubin, and N. A. Brown. 1999. Lamivudine as initial treatment for chronic hepatitis B in the United States. N. Engl. J. Med. 341:1266-1273. [DOI] [PubMed] [Google Scholar]
- 7.Fischer, K. P., K. S. Gutfreund, and D. L. J. Tyrrell. 2001. Lamivudine: resistance mechanisms and their clinical implications. Drug Resist. Updates 4:118-128. [DOI] [PubMed] [Google Scholar]
- 8.Fischer, K. P., and D. L. J. Tyrrell. 1996. Generation of duck hepatitis B virus polymerase mutants through site-directed mutagenesis which demonstrate resistance to lamivudine [(−)-β-l-2′,3′-dideoxy-3′-thiacytidine] in vitro. Antimicrob. Agents Chemother. 40:1957-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuda, R., S. Okinaga, S. Akagi, M. Hidaka, N. Ono, S. Fukumoto, and Y. Shimada. 1988. Alteration of infection pattern of duck hepatitis B virus by immunomodulatory drugs. J. Med. Virol. 27:387-396. [DOI] [PubMed] [Google Scholar]
- 10.Ganem, D., and H. E. Varmus. 1987. The molecular biology of hepatitis B viruses. Annu. Rev. Biochem. 56:651-693. [DOI] [PubMed] [Google Scholar]
- 11.Gutfreund, K. S., M. Williams, R. George, V. Bain, M. M. Ma, E. M. Yoshida, J.-P. Villeneuve, K. P. Fischer, and D. L. J. Tyrrell. 2000. Genotypic succession of mutations of the hepatitis B virus polymerase associated with lamivudine resistance. J. Hepatol. 33:469-475. [DOI] [PubMed] [Google Scholar]
- 12.Howe, A. Y. M., M. J. Robbins, J. S. Wilson, and D. L. J. Tyrrell. 1996. Selective inhibition of the reverse transcription of duck hepatitis B virus by binding of 2′,3′-dideoxyguanosine-5′-triphosphate to the viral polymerase. Hepatology 23:87-96. [DOI] [PubMed] [Google Scholar]
- 13.Jilbert, A. R., T.-T. Wu, J. M. England, P. M. Hall, N. Z. Carp, A. P. O'Connell, and W. S. Mason. 1992. Rapid resolution of duck hepatitis B virus infections occurs after massive hepatocellular involvement. J. Virol. 66:1377-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kajino, K., A. R. Jilbert, J. Saputelli, C. E. Aldrich, J. Cullen, and W. S. Mason. 1994. Woodchuck hepatitis virus infections: very rapid recovery after a prolonged viremia and infection of virtually every hepatocyte. J. Virol. 68:5792-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köck, J., and H.-J. Schlicht. 1993. Analysis of the earliest steps of hepadnavirus replication: genome repair after infectious entry into hepatocytes does not depend on viral polymerase activity. J. Virol. 67:4867-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mason, W. S., J. Cullen, G. Moraleda, J. Saputelli, C. E. Aldrich, D. S. Miller, B. Tennant, L. Frick, D. Averett, L. D. Condreay, and A. R. Jilbert. 1998. Lamivudine therapy of WHV-infected woodchucks. Virology 245:18-32. [DOI] [PubMed] [Google Scholar]
- 17.Miller, R. H., and W. S. Robinson. 1984. Hepatitis B virus DNA forms in nuclear and cytoplasmic fractions of infected human liver. Virology 137:390-399. [DOI] [PubMed] [Google Scholar]
- 18.Moraleda, G., J. Saputelli, C. E. Aldrich, D. Averett, L. Condreay, and W. S. Mason. 1997. Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J. Virol. 71:9392-9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholl, A. J., P. W. Angus, S. T. Chou, C. A. Luscombe, R. A. Smallwood, and S. A. Locarnini. 1997. Demonstration of duck hepatitis B virus in bile duct epithelial cells: implications for pathogenesis and persistent infection. Hepatology 26:463-469. [DOI] [PubMed] [Google Scholar]
- 20.Robins, M. J., J. S. Wilson, D. Madej, D. L. J. Tyrrell, W. P. Gati, R. J. Lindmark, and S. F. Wnuk. 2001. Nucleic acid related compounds. 114. Synthesis of 2,6-(disubstituted)purine-2′,3′-dideoxynucleosides and selected cytotoxic, anti-hepatitis B, and adenosine deaminase substrate activities. J. Heterocycl. Chem. 38:1297-1306. [Google Scholar]
- 21.Seeger, C., J. Summers, and W. Mason. 1991. Viral DNA synthesis. Curr. Top. Microbiol. Immunol. 168:41-59. [DOI] [PubMed] [Google Scholar]
- 22.Summers, J., P. M. Smith, and A. L. Horwich. 1990. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J. Virol. 64:2819-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Summers, J., P. M. Smith, M. Huang, and M. Yu. 1991. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J. Virol. 65:1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tencza, M. G., and J. E. Newbold. 1997. Heterogeneous response for a mammalian hepadnavirus infection to acyclovir: drug-arrested intermediates of minus-strand viral DNA synthesis are enveloped and secreted from infected cells as virion-like particles. J. Med. Virol. 51:6-16. [PubMed] [Google Scholar]
- 25.Tsiquaye, K. N., M. Rapicetta, T. F. McCaul, and A. J. Zuckerman. 1985. Experimental in ovo transmission of duck hepatitis B virus. J. Virol. Methods 11:49-57. [DOI] [PubMed] [Google Scholar]
- 26.Tuttleman, J. S., C. Pourcel, and J. Summers. 1986. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47:451-460. [DOI] [PubMed] [Google Scholar]
- 27.Westerhof, I., J. A. Mol, W. E. Van den Brom, J. T. Lumeij, and A. Rijnberk. 1994. Diurnal rhythms of plasma corticosterone concentrations in racing pigeons (Columba livia domestica) exposed to different light regimens, and the influence of frequent blood sampling. Avian Dis. 38:428-434. [PubMed] [Google Scholar]
- 28.Wilson, S. C., F. J. Cunningham, and T. R. Morris. 1982. Diurnal changes in the plasma concentrations of corticosterone, luteinizing hormone and progesterone during sexual development and the ovulatory cycle of Khaki Campbell ducks. J. Endocrinol. 93:267-277. [DOI] [PubMed] [Google Scholar]
- 29.Wu, T.-T., L. Coates, C. E. Aldrich, J. Summers, and W. S. Mason. 1990. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology 175:255-261. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, Y.-Y., and J. Summers. 2000. Low dynamic state of viral competition in a chronic avian hepadnavirus infection. J. Virol. 74:5257-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu, Y., T. Yamamoto, J. Cullen, J. Saputelli, C. E. Aldrich, D. S. Miller, S. Litwin, P. A. Furman, A. R. Jilbert, and W. S. Mason. 2001. Kinetics of hepadnavirus loss from the liver during inhibition of viral DNA synthesis. J. Virol. 75:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]