Abstract
The small nonstructural protein NS2 of the minute virus of mice (MVM) is required for efficient viral replication, although its mode of action is unclear. Here we demonstrate that NS2 and survival motor neuron protein (Smn) interact in vitro and in vivo. NS2 and Smn also colocalize in infected nuclei at late times following MVM infection.
Minute virus of mice (MVM) is an autonomous parvovirus consisting of a single-stranded genome of approximately 5 kb in length (7, 8). The genome contains two overlapping transcription units controlled by two viral promoters, P4 and P38 (1, 10, 15, 16). Expression of the viral nonstructural proteins, NS1 and NS2, is driven by the P4 promoter, while P38-derived transcripts encode the viral structural proteins VP1 and VP2 (1, 10, 15, 16). NS2 is a 25-kDa protein that is found both in a nonphosphorylated form in both the nucleus and the cytoplasm and a phosphorylated form found exclusively in the cytoplasm of infected cells. The function that NS2 performs during infection is still unclear. NS2-null mutants have been shown to accumulate partially assembled capsids in nonpermissive cells (5) but also show severe defects in viral replication that cannot be solely attributable to the lack of assembled capsids (5, 21). NS2 has been shown to interact with the nuclear export factor Crm1 and two of the 14-3-3 family members (3, 4). In nonpermissive cells the loss of the NS2:Crm1 interaction leads to reduced accumulated levels of genomic single-stranded DNA and the accumulation of viral capsids in the nucleus (19). The role of the NS2:14-3-3 interaction during infection remains unclear.
Recently, a novel subnuclear compartment has been identified following infection by the two highly related parvoviruses MVM and H-1 (2, 9). This structure, termed autonomous parvovirus-associated replication (APAR) bodies, was identified at early time points postinfection (up to 15 h postinfection) and was found to be distinct from most of the classically described nuclear bodies, including Cajal bodies, promyelocytic leukemia oncogenic domains (PODs), and interachromatin granules or speckles (9). At later time points during infection, however, a dramatic nuclear reorganization occurs in which formerly distinct nuclear structures, such as Cajal bodies, PODs, speckles, and APAR bodies, merge into massive structures that contain NS1. These structures are likely active sites of viral genome replication and de novo capsid assembly (27), but their role during the infection process is not yet known. In addition, the survival motor neuron (Smn) gene product is present in these large structures (hence the term SMN-associated APAR bodies or SAABs), and the Smn protein and NS1 have been shown to interact in vitro and in vivo during infection. Spinal muscular atrophy, a common neuromuscular disease, is caused by homozygous mutations or deletions of the human SMN1 gene, and a knockout of the murine Smn gene results in preembryonic lethality (14, 23). The 294-amino-acid SMN protein has recently been shown to interact with the previously identified NS1 binding partner NSAP-1/hnRNP-Q (13, 20, 22).
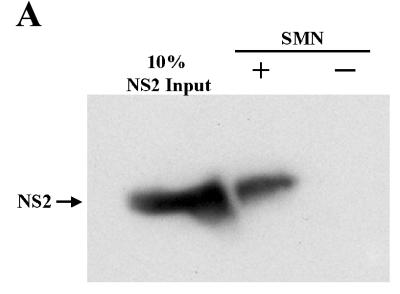
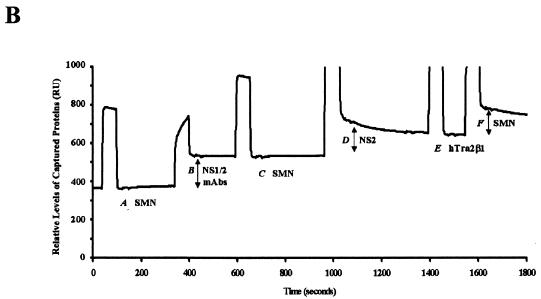
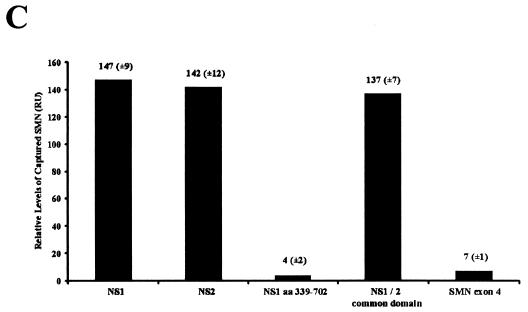
Since NS1 and NS2 share 85 amino-terminal amino acids, we sought to determine whether NS2 could interact with the murine (Smn) and human (SMN) proteins, which are nearly identical (11, 14). Recombinant glutathione-S-transferase (GST)-tagged NS2 was captured by polyhistidine (6-His)-SMN, while the resin alone failed to capture NS2 (Fig. 1A). This interaction was confirmed using biomolecular interaction analysis (BIA) (30). GST-NS2, tethered on a sensor chip using an anti-GST antibody, interacted with wild-type SMN (Fig. 1B, shift F). In the absence of NS2 protein, recombinant SMN failed to bind to the chip or the GST antibody (Fig. 1B, shifts A and C) and SMN failed to interact with two other GST-tagged peptides (Fig. 1C and see below), demonstrating the specificity of the interaction. The binding levels between SMN and NS2 were similar to those observed between SMN and full-length NS1 (27). SMN binding by NS1 and NS2 suggested that the common amino-terminal domain of the two nonstructural proteins might mediate binding to SMN. To determine whether the amino-terminal domain common to NS1 and NS2 mediated SMN binding, BIA experiments were performed with recombinant GST-NS1, NS2, the carboxy terminus of NS1, or the common amino terminus (Fig. 1C). The full-length proteins and the common domain bound efficiently to SMN, while the large carboxy terminus of NS1 did not interact appreciably with SMN (Fig. 1C). The peptide corresponding to SMN exon 4 served as a control for nonspecific binding and, as expected, did not interact with SMN (Fig. 1C). Fusion proteins were similarly soluble and stable.
FIG. 1.
SMN and NS2 interact in vitro. (A) Western blot analysis of recombinant GST-tagged NS2 captured by polyhistidine (6-His)-tagged human SMN immobilized on histidine binding resin. The blot was developed using anti-GST monoclonal antibody. (B) BIA of the SMN/NS2 complex. Bound GST monoclonal antibody (shift B), NS2 (shift D), and wild-type SMN (shift F) are indicated. Vertical shifts in the horizontal baseline represent positive interactions as response units (RU) and are indicated by arrows (shifts B, D, and F). hTra2β1 protein served as a negative control and does not bind to NS2 (E). (C) SMN binds to the N-terminal region common to NS1 and NS2. GST-tagged NS variants (NS1, NS2, amino acids 339 to 792, and the NS1 and -2 common domain) were captured using an anti-GST antibody and were then pulsed with wild-type SMN. GST-tagged SMN exon 4 was used as a negative control. Values represent binding of SMN constructs above a background control and the mean of three separate experiments.
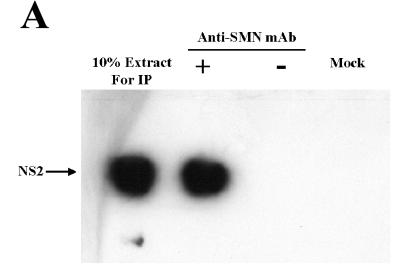
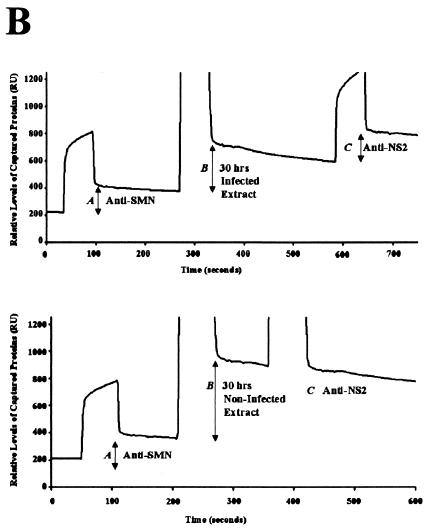
To determine whether the NS2/Smn complex formed in vivo, coimmunoprecipitations were performed in cellular extracts generated from A92L cells transiently expressing HA-tagged NS2. NS2:Smn complexes were immunoprecipitated with an anti-SMN monoclonal antibody (MANSMA3) that recognizes human and murine SMN proteins (30). Western blot analysis with an anti-NS2 rabbit polyclonal antibody identified NS2 as an SMN binding partner in vivo (Fig. 2A). BIA was used to determine whether Smn and NS2 form a complex during MVM infection by analyzing extracts from A92L cells harvested 30 h postrelease from a highly synchronized MVM infection (12). Consistent with the results obtained in vitro, NS2 was detected in a complex with endogenous Smn (Fig. 2B; top panel). As expected, NS2 was not coprecipitated with Smn from synchronized, noninfected A92L cells (Fig. 2B, bottom panel; compare the horizontal plateaus for peak C in top and bottom panels) nor from infected cells at 0 h postrelease (data not shown).
FIG. 2.
SMN and NS2 interact in vivo. (A) Anti-SMN (MANSMA3) coprecipitates endogenous SMN and hemagglutinin-tagged NS2 from A92L cells transiently expressing NS2. Anti-NS2 rabbit polyclonal sera were used to detect NS2. NS2 was not precipitated in the absence of MANSMA3 (−) or from nontransfected A92L cells (mock). mAb, monoclonal antibody; IP, immunoprecipitation. (B) BIA of NS2 coimmunoprecipitated with endogenous Smn from synchronized MVM-infected A92L cells (top panel). Total protein extracts from A92L cells 30 h postinfection were used. Anti-SMN (MANSMA3) coprecipitated SMN, and anti-NS2 rabbit polyclonal sera detected NS2 complexed with SMN. Captured levels of anti-SMN antibody (shift A), SMN/NS2 complex (shift B), and anti-NS2 rabbit polyclonal antibody (shift C) are indicated. NS2 is not coprecipitated with SMN from double-blocked, infected A92L cells 0 h postrelease (bottom panel). Total protein extracts were used from A92L cells 30 h after being released from the blocking process. Anti-SMN (MANSMA3)-precipitated SMN and anti-NS2 rabbit polyclonal sera detected no NS2 complexed with SMN. Captured levels of anti-SMN antibody (shift A), SMN (shift B), and anti-NS2 rabbit polyclonal antibody (shift C) are indicated as response units (RU).
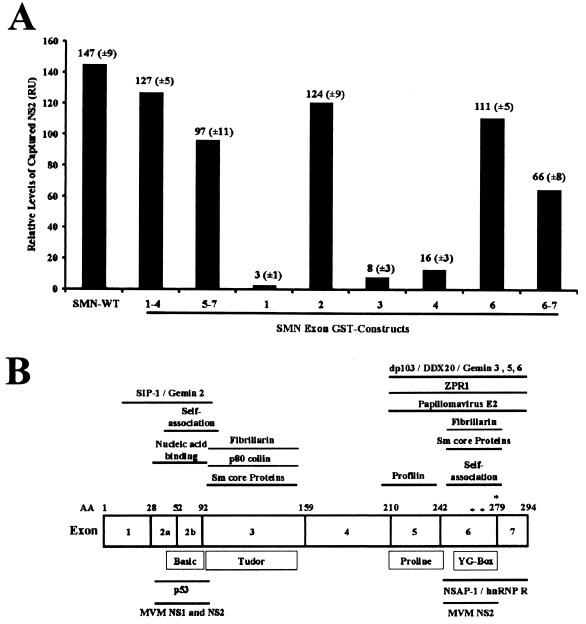
To identify the region(s) of the SMN protein that mediated the NS2 interaction, binding experiments were performed with a panel of GST-SMN subdomains. All the SMN peptides were abundantly expressed, stable, and soluble, and an equivalent molar amount of each protein was used in each assay (data not shown). Previous studies have shown that these constructs refold to form native epitopes (17, 26, 30). Recombinant NS2 immobilized a sensor chip with an anti-SMN antibody-bound wild-type SMN, SMN exons 1 to 4, and SMN exon 2 but not SMN exon 1, 3, or 4; NS2 also bound fusion-containing SMN exons 5 to 7, 6 and 7, and 6 alone (Fig. 3A). These results suggest that two separate NS2 binding sites are present within SMN, since NS2 efficiently captured GST fusion constructs that contained the SMN peptides encoded by exon 2 or 6. The proteins of wild-type SMN, SMN exons 1 to 4, exons 5 to 7, and exons 2, 6, and 6 to 7 displayed similar NS2 binding affinities under increasing salt concentrations (data not shown). In parallel experiments, none of the SMN constructs directly interacted with the anti-NS2 antibody or the sensor chip alone, demonstrating the specificity of the interactions (data not shown). The SMN domains that mediated NS2 binding have previously been characterized as regions essential to SMN biochemical activities and in mediating protein interactions (Fig. 3B).
FIG. 3.
(A) SMN exons 2 and 6 independently mediate NS2 binding. GST-NS2 immobilized on a rabbit anti-mouse sensor chip using anti-NS2 antibody was pulsed with GST-SMN fusion proteins (full-length SMN [SMN-FL], SMN exons 1 to 4, 5 to 7, 1, 2, 3, 4, and 6) and His exons 6 to 7 alone. Values represent binding of SMN constructs above a background control as response units (RU) and the mean of three separate experiments. (B) A schematic representation of the SMN protein. Previously reported physical and functional domains, amino acid positions, exon borders, and the relative positions of the three missense mutations (∗) analyzed in Fig. 4 are indicated.
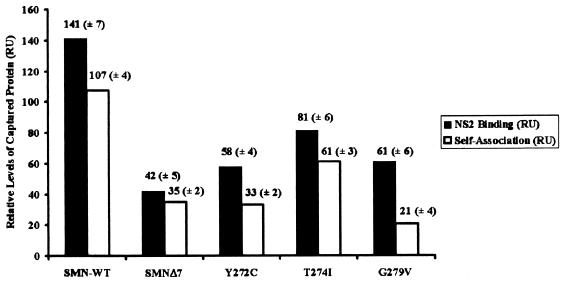
Functional SMN exists in at least a dimeric state, and the ability of the human SMN protein to self associate correlates with spinal muscular atrophy disease severity (18, 29). Several groups have previously shown that defects in SMN self-association significantly reduce the ability of SMN to interact with numerous SMN-interacting proteins (18, 22, and 30 and references within). To determine whether disease-causing SMN mutations that disrupt SMN self-association also disrupt the NS2:SMN complex, three patient-derived SMN single-amino-acid substitution mutations (Y272C, T274I, and G279V) were assayed for their ability to interact with NS2. Additionally, we assessed the ability of SMNΔ7 to interact with NS2. SMNΔ7 is a naturally occurring, alternatively spliced isoform produced at high levels in spinal muscular atrophy patients. All recombinant mutant SMN proteins were soluble and stable at levels similar to those in the wild type, and an equivalent amount of purified recombinant proteins was used in each assay (data not shown). Compared to wild-type SMN, the SMN mutations and SMNΔ7 displayed a dramatically reduced ability to bind NS2 (Fig. 4, black bars). The defect in SMN self-association observed with the SMN mutations directly correlates with the extent of the NS2 binding defect (Fig. 4, white bars), suggesting that the dimerized form of SMN interacts with NS2. While the mutations that we analyzed are in the human form of the protein, murine and human SMN proteins are greater than 90% identical and share even greater degrees of identity in the self-association domain and the exon 2 region (11, 14).
FIG. 4.
Patient-derived SMN mutations decrease NS2 binding. Truncated and point-mutated SMN displays a reduced affinity for NS2. SMN wild type (WT), SMN point mutations (type I mutations, 274 and 279; type II mutation, 272), or SMNΔ7 was immobilized on a sensor chip using an anti-SMN monoclonal antibody and probed with recombinant NS2. Truncated and point-mutated SMN displays a reduced ability to self associate (open bars). Wild-type monomeric SMN covalently bound to a sensor chip was exposed to truncated and point-mutated SMN to determine the various proteins' ability to dimerize. Binding values are represented as response units (RU).
NS2 and Smn colocalize in SAABs.
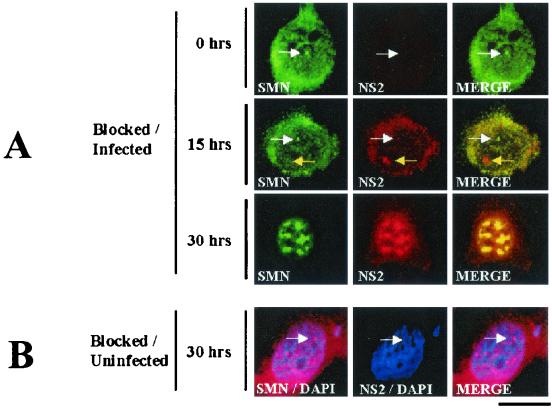
Typically, nuclear SMN accumulates within Cajal bodies and the nucleolus (28, 29). To determine whether NS2 and Smn colocalized during MVM infection, double-label immunofluorescence assaying was performed on highly synchronized A92L cells infected with MVM. Primary antibodies used were 3G2 (an anti-NS2 monoclonal antibody that does not recognize NS1) and a purified rabbit polyclonal antibody against SMN (6, 28). Cells were harvested at multiple time points postrelease into S phase (0, 15, and 30 h) (27). As expected at 0 h postrelease, Smn is present in Cajal bodies and NS2 is not detectable (Fig. 5). At 15 h postrelease, a significant amount of NS2 is found in the cytoplasm as previously described (3, 5, 6). At this time point, nuclear NS2 mainly localizes in bodies lacking Smn, although a low level is present in Cajal bodies with Smn (Fig. 5A). It is presently unclear whether the NS2 nuclear structures observed at 15 h postrelease are APAR bodies or novel nuclear structures. At 30 h postrelease, however, Smn and the majority of NS2 colocalized in large nuclear SAABs (Fig. 5A). SAAB structures were confirmed by the presence of previously described SAAB components, including NS1 and p80 coilin (data not shown; P. J. Young et al., unpublished data). Preabsorption of the anti-NS2 sera with purified recombinant NS2 abolished the NS2 staining pattern, and prior to release, other typical nuclear structures displayed a normal distribution pattern (data not shown). A normal distribution of Smn was observed in blocked, uninfected cells at 30 h postrelease, demonstrating that the relocalization observed in infected cells was not a consequence of the blocking procedure (Fig. 5B).
FIG. 5.
SMN and NS2 colocalize in SAABs at 30 h post-MVM infection. (A) SMN (primary, anti-SMN rabbit polyclonal sera; secondary, fluorescein isothiocyanate/green) and NS2 (primary: mouse monoclonal antibody; secondary: tetramethyl rhodamine isothiocyanate/red) are indicated. Synchronized infections were obtained by performing an isoleucine-aphidicolin double block on A92L cells prior to infection. Immunofluorescence experiments were performed on MVM-infected cells 0, 15, and 30 h postrelease (27). Nuclear Cajal bodies (white arrows) and NS2-positive/SMN-negative nuclear bodies (yellow arrows) are indicated. (B) SAAB formation is a consequence of viral infection. Smn localization was examined in synchronized, noninfected A92L cells 30 h postrelease. SMN (primary: anti-SMN rabbit polyclonal sera; secondary: tetramethyl rhodamine isothiocyanate/red) and NS2 (primary: mouse monoclonal antibody; secondary: fluorescein isothiocyanate/green) are indicated. Cell nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (blue). The bar represents ≈30 μm.
Recently, several viral nonstructural proteins have been identified as direct or indirect SMN-interacting factors, including bovine papillomavirus E2, Epstein-Barr virus LMP1, and MVM NS1 (24, 25, 27). We recently demonstrated an interaction between the MVM large nonstructural protein NS1 and Smn (27). This activity also mapped to the region encoded by Smn exon 2 (27). The Smn protein is the first reported example of a protein that interacts with NS1 and NS2. Consistent with this, the SMN-interaction domains are localized within the common amino termini of NS1 and NS2 (Fig. 1C).
The human and murine SMN proteins are highly conserved and are greater than 90% identical and share even greater degrees of identity in regions such as their self-association domains (exons 6 and 2b), the nucleic acid binding region (exon 2), and the NS2-interacting region (exon 2) (11, 14). The mutations within Smn exon 6 that inhibit binding to NS2 may do so through two mechanisms: (i) direct disruption of the exon 6 NS2 binding domain or (ii) inhibition of SMN self-association, thereby preventing the proper quaternary structure required for the formation of the NS2 binding domain. The active form of the SMN protein is a higher-order complex, consisting of at least a dimer (18, 30). SMN self-association is a prerequisite for SMN biochemical properties, including SMN protein stability and efficient interaction with nearly all SMN-interacting proteins (references 18, 22, and 30 and references within). Consistent with this, single-amino-acid substitution mutations that disrupt SMN self-association disrupted the efficiency of NS2:SMN complex formation. The degree to which self-association was decreased was reflected in the levels of NS2 binding, suggesting that the NS2:Smn interaction is not simply a nonspecific interaction.
The NS2 polypeptides are 25-kDa phosphoproteins. Nonphosphorylated forms are present in the nucleus and cytoplasm of MVM-infected cells, while phosphorylated NS2 isoforms are found exclusively in the cytoplasm (3, 5, 6). Phosphorylation of NS2 has previously been shown to influence its ability to interact with cellular factors: nonphosphorylated NS2 binds Crm1, while 14-3-3 isoforms exclusively bind phosphorylated NS2 (3, 4). Our results suggest that Smn can at least bind the nonphosphorylated NS2. Smn efficiently interacted with the nonphosphorylated recombinant NS2 that was expressed and purified from bacteria, and at 30 h postrelease, when Smn and NS2 are nearly exclusively nuclear, an NS2/Smn complex was detected. Further work will be required to determine whether Smn preferentially binds to nonphosphorylated NS2 and to characterize the temporal accumulation of the NS2:Smn complex. Future work is aimed at identifying the role of the NS2:Smn interaction during the viral life cycle. Although we have identified SAABs as sites of genome replication and capsid assembly (27), it is not known whether these structures are required for efficient viral replication or, alternatively, whether they are a manifestation of a cellular response to the late stages of viral infection. A better understanding of the nature of the parvovirus:cell interaction at later times of infection may help clarify this issue.
Acknowledgments
We thank Glenn Morris and Angus Lamond for antibodies, the W. M. Keck BioImaging Laboratory for confocal imaging, and Constance Chamberlain for technical assistance.
P.J.Y. was supported by a fellowship from Families of SMA, and K.T.J. was supported by a postdoctoral fellowship from the University of Missouri Life Science Mission Enhancement Program. Funding for these studies was provided by Families of SMA (C.L.L.), Andrew's Buddies (C.L.L.), the Muscular Dystrophy Association (C.L.L), and the National Institutes of Health (C.L.L., RO1 NS41584-01; and D.J.P., RO1 AI21302 and RO1 AI46458).
REFERENCES
- 1.Ahn, J. K., Z. W. Pitluk, and D. C. Ward. 1992. The GC box and TATA transcription control elements in the P38 promoter of the minute virus of mice are necessary and sufficient for transactivation by the nonstructural protein NS1. J. Virol. 66:3776-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bashir, T., J. Rommelaere, and C. Cziepluch. 2001. In vivo accumulation of cyclin A and cellular replication factors in autonomous parvovirus minute virus of mice-associated replication bodies. J. Virol. 75:4394-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodendorf, U., C. Cziepluch, J. C. Jauniaux, J. Rommelaere, and N. Salome. 1999. Nuclear export factor CRM1 interacts with nonstructural proteins NS2 from parvovirus minute virus of mice. J. Virol. 73:7769-7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brockhaus, K., S. Plaza, D. J. Pintel, J. Rommelaere, and N. Salome. 1996. Nonstructural proteins NS2 of minute virus of mice associate in vivo with 14-3-3 protein family members. J. Virol. 70:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotmore, S. F., A. M. D'Abramo, Jr., L. F. Carbonell, J. Bratton, and P. Tattersall. 1997. The NS2 polypeptide of parvovirus MVM is required for capsid assembly in murine cells. Virology 231:267-280. [DOI] [PubMed] [Google Scholar]
- 6.Cotmore, S. F., and P. Tattersall. 1990. Alternate splicing in a parvoviral nonstructural gene links a common amino-terminal sequence to downstream domains which confer radically different localization and turnover characteristics. Virology 177:477-487. [DOI] [PubMed] [Google Scholar]
- 7.Cotmore, S. F., and P. Tattersall. 1987. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 33:91-174. [DOI] [PubMed] [Google Scholar]
- 8.Cotmore, S. F., and P. Tattersall. 1986. Organization of nonstructural genes of the autonomous parvovirus minute virus of mice. J. Virol. 58:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cziepluch, C., S. Lampel, A. Grewenig, C. Grund, P. Lichter, and J. Rommelaere. 2000. H-1 parvovirus-associated replication bodies: a distinct virus-induced nuclear structure. J. Virol. 74:4807-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deleu, L., A. Pujol, S. Faisst, and J. Rommelaere. 1999. Activation of promoter P4 of the autonomous parvovirus minute virus of mice at early S phase is required for productive infection. J. Virol. 73:3877-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiDonato, C. J., X. N. Chen, D. Noya, J. R. Korenberg, J. H. Nadeau, and L. R. Simard. 1997. Cloning, characterization, and copy number of the murine survival motor neuron gene: homolog of the spinal muscular atrophy-determining gene. Genome Res. 7:339-352. [DOI] [PubMed] [Google Scholar]
- 12.Hardt, N., C. Dinsart, S. Spadari, G. Pedrali-Noy, and J. Rommelaere. 1983. Interrelation between viral and cellular DNA synthesis in mouse cells infected with the parvovirus minute virus of mice. J. Gen. Virol. 64:1991-1998. [DOI] [PubMed] [Google Scholar]
- 13.Harris, C. E., R. A. Boden, and C. R. Astell. 1999. A novel heterogeneous nuclear ribonucleoprotein-like protein interacts with NS1 of the minute virus of mice. J. Virol. 73:72-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefebvre, S., L. Burglen, S. Reboullet, O. Clermont, P. Burlet, L. Viollet, B. Benichou, C. Cruaud, P. Millasseau, M. Zeviani, et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155-165. [DOI] [PubMed] [Google Scholar]
- 15.Lorson, C., J. Pearson, L. Burger, and D. J. Pintel. 1998. An Sp1-binding site and TATA element are sufficient to support full transactivation by proximally bound NS1 protein of minute virus of mice. Virology 240:326-337. [DOI] [PubMed] [Google Scholar]
- 16.Lorson, C., and D. J. Pintel. 1997. Characterization of the minute virus of mice P38 core promoter elements. J. Virol. 71:6568-6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorson, C. L., and E. J. Androphy. 1998. The domain encoded by exon 2 of the survival motor neuron protein mediates nucleic acid binding. Hum. Mol. Genet. 7:1269-1275. [DOI] [PubMed] [Google Scholar]
- 18.Lorson, C. L., J. Strasswimmer, J. M. Yao, J. D. Baleja, E. Hahnen, B. Wirth, T. Le, A. H. Burghes, and E. J. Androphy. 1998. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet. 19:63-66. [DOI] [PubMed] [Google Scholar]
- 19.Miller, C. L., and D. J. Pintel. 2002. Interaction between parvovirus NS2 protein and nuclear export factor Crm1 is important for viral egress from the nucleus of murine cells. J. Virol. 76:3257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mourelatos, Z., L. Abel, J. Yong, N. Kataoka, and G. Dreyfuss. 2001. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 20:5443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naeger, L. K., J. Cater, and D. J. Pintel. 1990. The small nonstructural protein (NS2) of the parvovirus minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J. Virol. 64:6166-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossol, W., A. K. Kroning, U. M. Ohndorf, C. Steegborn, S. Jablonka, and M. Sendtner. 2002. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum. Mol. Genet. 11:93-105. [DOI] [PubMed] [Google Scholar]
- 23.Schrank, B., R. Gotz, J. M. Gunnersen, J. M. Ure, K. V. Toyka, A. G. Smith, and M. Sendtner. 1997. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA 94:9920-9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strasswimmer, J., C. L. Lorson, D. E. Breiding, J. J. Chen, T. Le, A. H. Burghes, and E. J. Androphy. 1999. Identification of survival motor neuron as a transcriptional activator-binding protein. Hum. Mol. Genet. 8:1219-1226. [DOI] [PubMed] [Google Scholar]
- 25.Voss, M. D., A. Hille, S. Barth, A. Spurk, F. Hennrich, D. Holzer, N. Mueller-Lantzsch, E. Kremmer, and F. A. Grasser. 2001. Functional cooperation of Epstein-Barr virus nuclear antigen 2 and the survival motor neuron protein in transactivation of the viral LM:1 promoter. J. Virol. 75:11781-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young, P. J., P. M. Day, J. Zhou, E. J. Androphy, G. E. Morris, and C. L. Lorson. 2001. A direct interaction between the survival motor neuron protein and p53 and its relationship to spinal muscular atrophy. J. Biol. Chem. 9:2852-2859. [DOI] [PubMed] [Google Scholar]
- 27.Young, P. J., K. T. Jensen, L. R. Burger, D. J. Pintel, and C. L. Lorson. 2002. Minute virus of mice NS1 interacts with the SMN protein and they colocalize in novel nuclear bodies induced by parvovirus infection. J. Virol. 76:3892-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young, P. J., T. T. Le, M. Dunckley, T. M. Nguyen, A. H. Burghes, and G. E. Morris. 2001. Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp. Cell Res. 265:252-261. [DOI] [PubMed] [Google Scholar]
- 29.Young, P. J., T. T. Le, N. thi Man, A. H. Burghes, and G. E. Morris. 2000. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell Res. 256:365-374. [DOI] [PubMed] [Google Scholar]
- 30.Young, P. J., N. T. Man, C. L. Lorson, T. T. Le, E. J. Androphy, A. H. M. Burghes, and G. E. Morris. 2000. The exon 2b region of the spinal muscular atrophy protein, SMN, is involved in self-association and SIP1 binding. Hum. Mol. Genet. 9:2869-2877. [DOI] [PubMed] [Google Scholar]