Abstract
Elucidation of the host factors which influence susceptibility to human immunodeficiency virus or simian immunodeficiency virus (SIV) infection and disease progression has important theoretical and practical implications. Rhesus macaque 359, a vaccine control animal, resisted two successive intravaginal challenges with SIVmac251 and failed to seroconvert. Here, after an additional intrarectal SIVmac32H challenge, macaque 359 remained highly resistant to infection. Viral RNA (106 copies/ml) was observed in plasma only at week 2 postchallenge. Virus isolation and proviral DNA were positive only once at week eight postchallenge. The animal remained seronegative and cleared SIV in vivo. Its blood and lymph node cells obtained at 49 weeks after intrarectal challenge did not transmit SIV to a naive macaque. We found that the resistance of macaque 359 to SIV infection was not due to a high level of CD8+ suppressor activity but to an inherent resistance of its CD4+ T cells. To elucidate the basis for the unusually strong resistance of macaque 359 to SIV infection in vivo and in vitro, we investigated early events of viral infection and replication in CD4+ cells of macaque 359, including expression and mutation screening of SIV coreceptors and analysis of viral entry and reverse transcription. Mutation screening revealed no genetic alteration in SIV coreceptors. PCR analysis revealed a significant delay in production of early in vitro reverse transcription intermediates in macaque 359 cells compared to susceptible controls, but cell fusion assays showed that SIV entered the CD4+ CCR5+ cells of macaque 359 as readily as cells of macaques susceptible to SIV infection. Our results suggest that the resistance of macaque 359 to SIV infection is due to a postentry block in viral replication and implicate a cellular inhibitory mechanism in its CD4+ T cells. Identification of this host mechanism will help further elucidate the biochemistry of reverse transcription and may suggest therapeutic strategies. Determining the prevalence of this host resistance mechanism among macaques may lead to better design of SIV pathogenesis and vaccine studies.
Elucidation of host factors that modulate susceptibility to infection with human immunodeficiency virus (HIV) or simian immunodeficiency virus (SIV) and influence disease outcome will not only broaden our understanding of virus-cell interactions but will also have important practical implications. Knowledge of natural host defense mechanisms may lead to their exploitation for therapeutic or prophylactic purposes. Identification of host susceptibility factors may influence treatment decisions and further define risk factors for HIV acquisition. Clarification of host susceptibility and resistance factors in nonhuman primates should limit the variability in experimental groups and lead to improved design of preclinical studies.
A few immunologic and genetic host factors have been identified which influence HIV or SIV infection and viral replication (11). The former includes acquired immunity resulting from viral infection, as well as innate immunity involving many inducible cytokines and chemokines. Genetic factors, such as HLA haplotypes, can influence the host's immune response. Two haplotypes have been associated with rapid disease progression after HIV infection (10), whereas another has been linked to long-term nonprogression (39). A similar protective effect of the rhesus macaque Mamu A*01 genotype has been recently demonstrated (43). The most dramatic genetic influence on HIV transmission and disease progression involves a 32-bp deletion (Δ32) in the CCR5 gene, the major coreceptor for macrophage-tropic, non-syncytium-inducing HIV isolates. This deletion results in a truncated, nonfunctional gene product and is associated with protection against HIV infection in individuals homozygous for the CCR5 Δ32 allele (20, 30, 49) and with delayed disease progression and decreased CCR5 expression on T cells among heterozygous individuals (13, 45, 59). Additional CCR5 polymorphisms exist but have not been shown to modulate HIV infection (37). Polymorphisms in the CCR5 promoter have been associated with accelerated disease progression (34, 36), but the mechanisms for this are unclear. A minor coreceptor mutation, CCR2-64I, with strong linkage disequilibrium with a CCR5 promoter region mutation (25), was initially associated with delayed disease progression (53), although subsequent studies have yielded conflicting results (33, 37, 38).
Alterations in chemokine or cytokine genes or their promoters can also affect the course of HIV disease. Delayed disease progression has been attributed to a polymorphism in the RANTES promoter (29) and to an SDF-1 gene mutation, although the latter finding is controversial (19, 41, 57, 58). Polymorphisms in the interleukin-10 (IL-10) promoter have been linked to AIDS progression (52).
Identification of host factors which contribute to susceptible or resistant phenotypes is difficult in humans. Our observation, summarized below, of unusually strong resistance to SIV infection in a rhesus macaque, presented a unique opportunity for investigating novel host resistance mechanisms. In general, intravaginal infection of rhesus macaques can result in either transient or persistent viremia (40), as well as in occult systemic infection in some cases (35). In earlier preclinical vaccine studies in which rhesus macaques were challenged with infectious, pathogenic SIVmac251, we observed such variable outcomes in both controls and immunized monkeys with regard to infection and disease progression (7, 8). One naive control macaque, 359, resisted two intravaginal exposures with escalating dosages of SIVmac251. Here we report that, after an additional intrarectal challenge with SIVmac32H, macaque 359 became only transiently viremic and cleared virus from the peripheral blood. In vitro studies confirmed that the animal's peripheral blood mononuclear cells (PBMCs) were highly resistant to SIV infection, even though adequate levels of CCR5 were expressed on the surface of the cells. In order to elucidate the basis for the unusually strong resistance of macaque 359 to in vivo and in vitro SIV infection, early events in the viral infection and replication process were examined. We examined several coreceptor genes for mutational changes. Genetic polymorphisms were detected in the CCR5 gene; however, none of the changes led to amino acid substitutions. Investigation of reverse transcription events revealed significant inhibition in the accumulation of early DNA replication intermediates. However, macaque 359 cells were able to fuse with cells expressing a SIVmac251 envelope or a CCR5-tropic HIV envelope as readily as cells from macaques highly susceptible to SIV infection. Taken together, our results indicate that the resistance of macaque 359 to SIV infection is due to postentry inhibition of viral replication and implicate a host cell mechanism in this process.
MATERIALS AND METHODS
Animals.
Female rhesus macaque number 359 was nearly 7 years old at the time of the first intravaginal challenge (8) and at the time of these studies was 11 years old. She was never immunized with adenovirus type 5 host range mutant-SIV recombinants or SIV subunit proteins and was determined to be SIV and simian retrovirus negative by serologic and PCR analyses prior to any experimentation. An additional 77 rhesus macaques, which were involved in mutation screening of coreceptors, exhibited differing susceptibilities to in vivo SIV infection. Most of the animals were either immunized or control macaques from SIV vaccine studies and have been described previously. They included 11 multiparous females (7, 8) and 5 virgin females (32) challenged intravaginally; 27 macaques challenged by the intravenous route (1, 4); and 27 macaques challenged intrarectally (4, 44). An additional seven macaques challenged either intravaginally, intravenously, or intrarectally have not been previously described.
The susceptibility of the macaques to infection with SIVmac251 was categorized according to the viral load at set point and/or rate of disease progression as reported in previous publications (1, 4, 7, 8, 44). Low, medium, or high susceptibility was defined as having a viral burden at a set point of <105, >105 to <107, or ≥107 RNA copies/ml of plasma and/or a disease status of slow progressor, progressor, or rapid progressor, respectively.
Viably frozen PBMCs obtained from seven additional naive macaques (animals 724, 725, 660, 774, 789, 793, and 826) subsequently shown to be susceptible to in vivo SIV infection and three additional naive macaques (numbers E196, G79, and O58) shown to be susceptible to in vitro SIV infection were used for the in vitro infectivity studies.
Viral stocks.
For intrarectal challenge of macaque 359, 20 monkey infectious doses of SIVmac32H (12) were administered. In vitro infections were carried out with either SIVmac251 or SIV17E-Br. The primary stock of SIVmac251 was obtained from an infected macaque and grown in rhesus PBMCs. The macrophage-tropic, neurovirulent strain, SIV17E-Br (51), was kindly provided by Janice Clements and was propagated on rhesus macaque macrophages.
Antibody assays.
SIV antibody status was assessed on serum samples diluted 1:100 by both enzyme-linked immunosorbent assay (8) and Western blotting (1) as previously described.
Infectivity assays.
Macaque PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation. The mononuclear cells were adjusted to 106 cells/ml and stimulated for 48 h with phytohemagglutinin (PHA; Murex Diagnostic, Ltd., Atlanta, Ga.) at a final concentration of 4 μg/ml in growth medium consisting of RPMI 1640 medium supplemented with 15% fetal bovine serum, 2 mM glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. The PBMCs were subsequently cultured in growth medium supplemented with human IL-2 (100 U/ml; Boehringer Mannheim Biochemicals, Indianapolis, Ind.).
Macaque PBMCs were depleted of CD8+ lymphocytes by using anti-CD8 antibody-coated immunomagnetic beads (Dynal, Lake Success, N.Y.). The remaining CD4+ cells were stimulated for 48 h with PHA as described above.
PHA-stimulated macaque PBMCs or CD8+-depleted CD4+ lymphocytes were seeded into 96-well plates at 105 cells/well. Infections were performed in quadruplicate by using seven 50% tissue culture infective doses (TCID50)/well of SIVmac251 or four TCID50/well of SIV17E-Br. After a 1-h incubation of cells and virus, 50 μl of growth medium was added and the incubation was continued at 37°C overnight. The next day, the cells were washed three times, suspended in 200 μl of growth medium containing IL-2, and cultured at 37°C. Virus replication was monitored every 3 days by analyzing SIV p27 in culture supernatants by using an antigen capture assay (Coulter, Hialeah, Fla.).
Evaluation of CD8+ T-cell antiviral activity.
PBMC samples obtained before and after intrarectal SIVmac251 challenge were assessed for CD8+-T-cell antiviral activity as described previously (27) by using endogenously infected CD4+ T cells from a seropositive rhesus macaque as targets for suppression. The method was modified by stimulating positively selected effector CD8+ cells for 3 days with goat anti-mouse immunoglobulin G immunomagnetic beads (Dynal) coated with 2 μg of anti-CD3 (a gift of Michael Rosenzweig, Harvard, Cambridge, Mass.) and 2 μg of anti-CD28 (Immunotech, Westbrook, Maine) per 107 beads. The percent suppression at effector/target cell ratios ranging from 4:1 to 0.25:1 was determined by comparing the amount of p27 produced by target cells alone with the amount produced by effector/target cell cocultures.
CCR5 expression by flow cytometry.
To assess the expression of CCR5, PBMCs were stimulated with PHA for 72 h prior to staining. A two-color flow analysis of the cells was performed. Single-cell suspensions at 106 cells/ml were incubated with fluorescein isothiocyanate-labeled anti-human CD3 (clone SP34) at 4°C for 30 min, followed by two washes. Phycoerythrin-labeled anti-human CCR5 (clone 2D7/CCR5) and anti-rhesus CD4-phycoerythrin (clone M-T477) were added to the cells which were then incubated at 4°C for 30 min, washed twice, and fixed by resuspension in 1% paraformaldehyde. All of the monoclonal antibodies used were obtained from Pharmingen, San Diego, Calif. Background controls included unstained and isotype-specific stained cells. The samples were analyzed by using a FACScan flow cytometer and CellQuest software (Becton Dickinson).
PCR-single-strand conformation polymorphism (PCR-SSCP) analysis.
Genomic DNA was isolated by using QIAamp DNA kits (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. Samples of genomic DNA (25 ng) were amplified by PCR in 50-μl mixtures containing 10 μM concentrations of sense and antisense primers, 10 mM concentrations of each deoxynucleoside triphosphate (including 2.5 μCi of [α-32P]dCTP), and 0.1 U of Taq DNA polymerase (Qiagen)/μl. Amplification was carried out for 30 cycles as follows: 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min. Approximately 5 μl of PCR product was diluted with 45 μl of 0.1% bromophenol blue and 0.05% xylene cyanole. The samples were denatured for 2 min at 95°C and separated on 6% nondenaturing polyacrylamide gels containing 10% glycerol for 8 to 10 h at 30 W of constant power. PCR was performed with the following primers complementary to the Macaca mulatta Bonzo gene (AF007858; 5′-ATGGCAGAGCATGATTACCA-3′ and 5′-CTATAACTGGAACATGCTGGTGGCG-3′), the M. mulatta CXCR4 gene (U93311; 5′-ATGGAGGGGATCAGTATATACACTT-3′ and 5′-TTAGCTGGAGTGAAAATTTGAAGA-3′), the Macaca nemestrina BOB(GPR15) gene (AF007857; 5′-ATGGACCCAGAAGAAACTTCAGTTT-3′ and 5′-TTAGAGTGACACAGACCTCTTCCTC-3′), the M. mulatta CCR3 gene (Y13776; 5′-ATGACAACCTCACTAGATACGGTTG-3′ and 5′-CTAAAACACAATAGAGAGTTCCGGC-3′), and the M. mulatta CCR5 gene (U77672): 5′-TGGTGGGCCACTAAATACTTTCTAGGGC-3′ (primer 1) and 5′-AGTCCCACTGGGCAGCAGCATAGT-3′ (primer 3), 5′-TCTCTGACCTGCTTTTCCTTCTTA-3′ (primer 4) and 5′-TGCAGGTGTAATGAAGACCTTCTC-3′ (primer 5), 5′-TGGTGGCTGTGTTTGCCTCTCTC-3′ (primer 6) and 5′-CCTGGAAGGTGTTCAGGAGAAGGAC-3′ (primer 7), and 5′-TATCTTCACCATCATGATTGTTTA-3′ (primer 8) and 5′-TCACAAGCCCACAGATATTTCCTG-3′ (primer 2). Primers 1 and 2 were used to synthesize the entire CCR5 gene. Primers 1 and 3, primers 4 and 5, primers 6 and 7, and primers 8 and 2 were used as pairs across the CCR5 gene.
DNA sequencing.
The entire CCR5 gene was synthesized by using PCR amplification as described above. DNA sequencing was performed by using the ABI Prism DNA sequencing kit (PE Applied Biosystems).
Analysis of early DNA products of SIV reverse transcription.
Frozen PBMCs of macaque 359, harvested before any SIV challenge, and of the naive control macaques 724 and 725 were thawed and stimulated for 72 h in growth medium containing PHA (10 μg/ml) and IL-2 (200 U/ml). The cells were depleted of CD8+ lymphocytes by using anti-CD8 antibody-coated Dynabeads and then cultured in growth medium containing 20% fetal bovine serum and 100 U of IL-2/ml. The CD8-depleted PBMCs (4.5 × 106) were infected with 5 × 103 TCID50 of SIVmac251 that had been pretreated with 2 μg of RNase-free DNase (Worthington)/ml for 30 min at room temperature in the presence of 0.01 M MgCl2. To eliminate virus on the surface of cells, after 2 h of incubation at 37°C, the cells were washed three times with phosphate-buffered saline (PBS), incubated with 0.25% trypsin-EDTA diluted 1:10 in PBS for 5 min at 37°C, washed with growth medium, and cultured further at 37°C. Cells (5 × 105) were harvested at various time points, including before infection, immediately after virus addition (0 h), immediately after virus washout and trypsin treatment (2 h), and at 6, 12, 24, and 72 h after infection. DNAs for PCR analysis were isolated by using QIAamp DNA blood Mini Kits according to the manufacturer's instructions.
The human β-actin gene was amplified in order to standardize input DNAs in subsequent PCRs evaluating early DNA products of reverse transcription. The β-actin primers were 5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′ (sense) and 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3′ (antisense).
PCR amplification of minus-strand strong-stop DNA, post-minus-strand transfer DNA, and the 3′ end of minus-strand DNA was done for 32 and 40 cycles in 25-μl reaction mixtures containing 5 μM each of sense and antisense primers; 10 mM dTTP, dATP, and dGTP; 8 mM dCTP; 2.5 μCi of [α-32P]dCTP; 1 U of Platinum Taq DNApol (Gibco-BRL, Grand Island, N.Y.); and 2.5 μl of buffer as specified below.
Minus-strand strong-stop DNA was amplified in a reaction mixture including Buffer #6 (Stratagene, La Jolla, Calif.) with the primers 5′-ATTGAGCCCTGGGAGGTTCTC-3′ (Forward R) and 5′-TGCTAGGGATTTTCCTGCTCCGG-3′ (Reverse U5). The cycling conditions were 94°C for 30 s, 57°C for 40 s, and 72°C for 1 min.
Post-minus-strand transfer DNA product was amplified in a reaction mixture including Taq Plus precision buffer (Stratagene) with the primers 5′-GAGGAAGATGATGACTTGGTAGGGG-3′ (Reb1) and 5′-CCAGCCAAATGTCTTTGGGTATCTA-3′ (Reb2). The cycling conditions were 94°C for 30 s, 59.3°C for 40 s, and 72°C for 1 min.
The 3′ end of the minus-strand DNA product was amplified in a reaction mixture including Buffer #10 (Stratagene) with the primers 5′-CGAACAGGACTTGAAGGAGAGTGA-3′ (Reb3) and 5′-CTGACAAGACGGAGTTTCTCGC-3′ (Reb4). The cycling conditions were 94°C for 30 s, 51.7°C for 40 s, and 72°C for 1 min.
PCR products were analyzed by electrophoresis on 5% nondenaturing polyacrylamide gels and visualized by direct autoradiography of the frozen gels for several hours.
Analysis of viral entry by cell-cell fusion assay.
293 cells were infected overnight at 31°C with the vaccinia virus V194, expressing the SIVmac251 envelope (24), or with vCB28, expressing the HIVJR-FL envelope protein (42) at a multiplicity of infection of 10, and then detached from the flask by using a cell stripper (CellQuest). The infected 293 cells (105) were mixed with an equivalent number of CD8 depleted CD4+ CCR5+ cells from macaque 359 and other macaques known to be susceptible to SIV infection. In all cases the cells were obtained prior to any SIV exposure of the macaques and stored viably frozen. Syncytia containing at least three to four fused cells were counted after 1 and/or 4 h of incubation at 37°C.
RESULTS
Resistance of macaque 359 to in vivo infection.
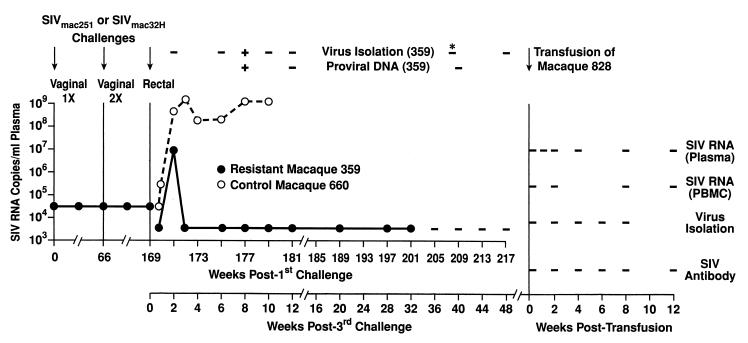
As previously published and summarized in Fig. 1, naive macaque 359 was initially challenged with >105 TCID50 of SIVmac251 by the vaginal route. No infection resulted from this challenge, as shown by the lack of proviral DNA in PBMCs and viral RNA in plasma, the inability to isolate virus from PBMCs, and the failure to seroconvert (8). Repeat assays with a sensitivity of 300 copy equivalents/ml (54) on plasma samples obtained 2, 4, and 6 weeks after virus exposure were also negative for SIV RNA. A subsequent challenge 66 weeks later was carried out by using a double dose of virus, i.e., >105 TCID50 of SIVmac251 in the morning and again in the evening of the same day (7). Again, macaque 359 did not become infected. Two years later, we attempted to infect this macaque by the intrarectal route, administering 20 monkey infectious doses of SIVmac32H. As illustrated in Fig. 1, we detected a modest level of SIV RNA in the plasma of macaque 359 at a single time point 2 weeks postchallenge. Thereafter, all RNA analyses were negative, although on one occasion we were able to isolate virus and detect proviral DNA in PBMCs obtained 8 weeks after the challenge. The animal failed to become SIV seropositive, as determined by both enzyme-linked immunosorbent assay (not shown) and Western blotting (Fig. 2). Naive macaque 660, challenged at the same time, became readily infected, exhibiting peak viral RNA levels of >109 copies/ml of plasma. Macaque 660 subsequently rapidly developed AIDS and was euthanized at 11 weeks postchallenge.
FIG. 1.
In vivo resistance of rhesus macaque 359 to mucosal SIV infection and clearance of virus. The first and second vaginal challenges and the failure of macaque 359 to become infected have been previously described in detail (7, 8). The third intrarectal exposure is detailed in Materials and Methods. SIV RNA in plasma was quantitated by nucleic acid sequence-based amplification (47). The sensitivity of SIV RNA detection was 50,000 copies/ml during the monitoring after the first and second intravaginal challenges. Repeat assays failed to detect RNA at a level of <300 copy equivalents (54). After the third challenge, the sensitivity of detection was 5,000 copies/ml in a quantitative assay and <500 copies/ml of plasma in a qualitative assay. A negative result is indicated by a dash. Attempts to isolate virus and detect proviral DNA were carried out on PBMCs except for one sample of bone marrow, marked by an asterisk. Macaque 660 was euthanized as a result of AIDS-related complications at 11 weeks postchallenge. Blood and lymph node cells obtained from macaque 359 49 weeks after intrarectal challenge were transfused into naive macaque 828, which was then monitored for 12 weeks for evidence of viral infection.
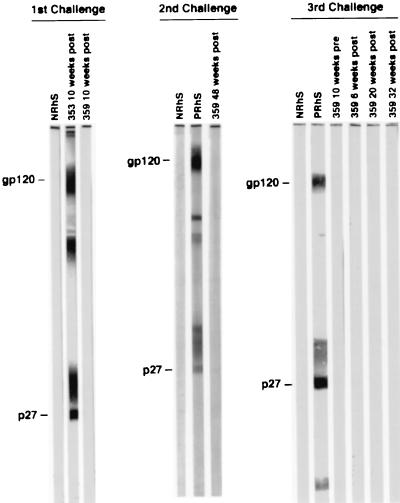
FIG. 2.
Western blot analysis of representative macaque 359 sera obtained after successive mucosal SIV challenges; the first and second challenges were intravaginal, and the third was intrarectal. NRhS and PRhS, SIV antibody-negative and -positive rhesus macaque sera, respectively. SIV antibody-positive macaque 353 received the first vaginal challenge at the same time as macaque 359.
The failure of macaque 359 to exhibit a robust infection after three separate exposures to SIV and its apparent clearance of the virus was highly unusual. To confirm its in vivo resistance and clearance of SIV, we conducted a transfusion experiment. A 9-ml portion of blood and a suspension of inguinal lymph node cells were obtained from macaque 359 at 49 week after intrarectal challenge and transfused into naive macaque 828. As shown in Fig. 1, macaque 828 was monitored for 12 weeks posttransfusion but failed to give any evidence of infection. It remained negative for SIV RNA, virus isolation, and SIV antibodies. Taken together, macaque 359 resisted SIV infection in vivo and was able to clear the virus.
Resistance of macaque 359 T cells to in vitro infection.
Mucosal transmission of SIV is more complex than intravenous transmission. Lack of or low-level infection by the mucosal route could be due to physical barriers imposed by mucus, failure of the appropriate cell type to be exposed to the infectious virus, or the lack of appropriate primary or secondary receptors. To address these questions, we first assessed the ability of macaque 359 PBMCs obtained prior to the intrarectal SIV challenge to be infected by SIV in vitro. PBMCs of macaque 660, which had been shown to be highly susceptible to SIVmac251 infection in vivo (Fig. 1), and of macaques 724 and 725, which had previously exhibited susceptibility to SIVmac251 infection in vitro, served as controls.
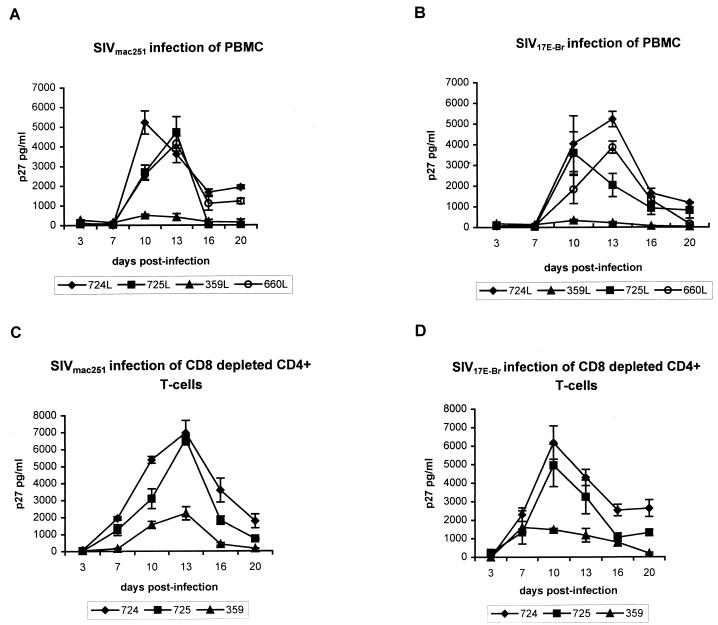
PBMCs of the control macaques were readily infected by SIVmac251, exhibiting peak p27 levels by 10 to 13 days postinfection (Fig. 3A). In contrast, PBMCs of macaque 359 showed very little virus infection. Similarly, macaque 359 PBMCs were able to resist in vitro infection by the macrophage-tropic, neurovirulent strain, SIV17E-Br, while all control PBMCs exhibited high levels of virus replication within the same time period (Fig. 3B).
FIG. 3.
In vitro susceptibility of rhesus macaque PBMCs to SIV infection. The kinetics of replication of SIVmac251 (A) and SIV17E-Br (B) in unfractionated PBMCs and of SIVmac251 (C) and SIV17E-Br (D) in the CD8+-depleted CD4+ T cells of the macaques are plotted as a function of p27 production in culture supernatants.
We also tested the susceptibility to SIV infection of CD8+-depleted CD4+ T cells of macaque 359 obtained 4 months after intrarectal exposure. The results shown in Fig. 3C and D were very similar to those observed with unfractionated macaque PBMCs (Fig. 3A and B). Although there was some SIV production in the CD4+ cells of macaque 359, the level of SIV replication in the CD4+ T cells of control macaques 724 and 725 was >3-fold higher than that of macaque 359. These results suggested that the resistance of 359 to SIV infection could not be explained solely by the high levels of innate suppressor factor activity.
To explore this possibility further, macaque 359 positively selected CD8+ T cells obtained 17 weeks before and 6 weeks after the intrarectal challenge were assessed for functional antiviral activity by using chronically SIV-infected CD4 cells as targets. Prior to challenge 0% suppression was observed, and after challenge only 9% suppression was observed. In contrast, although CD8+ T cells obtained at the same time points of the susceptible macaque 660 exhibited no antiviral activity before challenge (3.8% suppression), after challenge their antiviral activity rose, exhibiting 58% suppression. Thus, this innate immune response did not appear to be responsible for the resistance of macaque 359 to SIV infection.
Taken together, the in vitro data suggested that the resistance of macaque 359 in vivo might be due to an inherent resistance of 359 cells to SIV infection rather than to physical barriers or inadequate virus exposure. The ability of SIV to infect target cells is dependent on both the viral strain and the availability and expression levels of the appropriate primary receptor and coreceptors (60). Macaque 359 has consistently exhibited normal CD4 T-cell numbers throughout the 4 years during which she has been monitored. Since macaque 359 PBMCs are also resistant to infection by SIV17E-Br, a CD4 independent isolate, we considered that coreceptor expression or variability, especially of CCR5, the major coreceptor utilized by SIV for infection, might contribute to the resistance of macaque 359 to SIV infection. The cell surface expression of CCR5 was therefore examined.
Analysis of CCR5 on the surface of macaque 359 and 660 T cells.
CCR5 expression on PBMCs of macaques 359 and 660 obtained prior to the intrarectal SIVmac251 exposure was assessed by flow cytometry as described in Materials and Methods. As indicated in Table 1, both resistant macaque 359 and susceptible macaque 660 had similar numbers of CD4+ cells and CD4+ cells expressing CCR5. Notably, the density of CCR5 on the surface of CD4+ CCR5+ cells, as estimated by the mean fluorescent intensity, was somewhat higher on the cells of macaque 359 than on those of macaque 660. Although increased CCR5 density has been associated with greater infection (46), the slightly higher level shown here did not lead to better in vitro susceptibility of macaque 359 T cells to SIV. Because CCR5 expression was similar on the CD4 cells of both macaques, we considered the possibility that the CCR5 gene of the resistant macaque 359 was altered.
TABLE 1.
CCR5 expression on CD4 cells of resistant macaque 359 and susceptible macaque 660a
| Macaque | % Positive
|
Mean fluorescent intensity (CD4+ CCR5+ cells) | |
|---|---|---|---|
| CD4+ cells | CD4+ CCR5+ cells | ||
| 359 | 46 | 18 | 72 |
| 660 | 43 | 16 | 43 |
Macaque PBMCs obtained 6 and 4 weeks prior to the intrarectal challenge for macaques 359 and 660, respectively, were stimulated with PHA and stained as described in Materials and Methods. Background staining was subtracted from the percent positive cells.
Analysis of SIV coreceptors by PCR-SSCP.
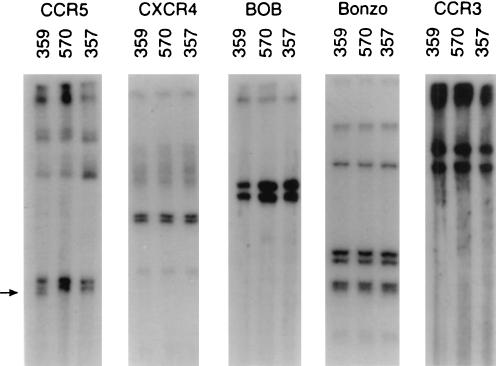
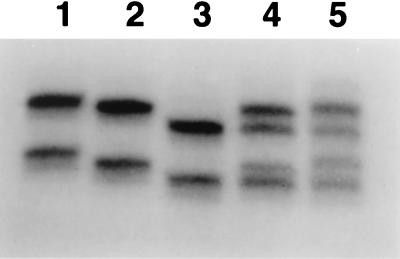
The majority of SIV isolates prefer to use CCR5 for viral entry. However, SIV and the closely related HIV type 2 isolates can also use alternate coreceptors, including Bonzo, BOB(GPR15), CCR3, and CXCR4, for viral entry (3, 14, 16, 28). To clarify the roles of all potential SIV coreceptors in the resistance of macaque 359 to SIV infection, mutations in these five coreceptors were investigated by using PCR-SSCP methods. Genomic DNA of macaque 359 was extracted, and the coding sequences of the five coreceptors were individually amplified by PCR with specific primers for the entire open reading frames. After digestion with a single restriction enzyme, the resultant DNA fragments were denatured. The mobility (conformation) of the single-stranded DNA PCR products was then analyzed by electrophoresis on a nondenaturing polyacrylamide gel. As controls we used genomic DNAs from macaques 357 and 570. Macaque 357 was a vaccinated macaque which exhibited a high level of persistent viremia after an intravaginal challenge with SIVmac251 (8). Macaque 570, initially treated with a microbicide, was readily infected with SIVmac251 after a single intravaginal exposure and exhibited continuous plasma viral RNA. The results of the SSCP analysis showed no shifts in the CXCR4, CCR3, Bonzo, or BOB(GPR15) banding patterns of macaque 359 compared to those of macaques 357 and 570 (Fig. 4). However, a distinct band shift was observed in the CCR5 gene of macaque 359 (Fig. 4), indicating a possible mutation(s) in this gene and suggesting that further investigation was warranted.
FIG. 4.
Analysis of CCR5, CXCR4, BOB, Bonzo, and CCR3 of resistant macaque 359 and susceptible macaques 570 and 357. The coreceptor coding sequences were amplified by PCR with the primers specified in Materials and Methods. The resultant PCR products were digested with the restriction enzymes PstI (CCR5), BamHI (CXCR4), NdeI (BOB), EcoRV (Bonzo), or KpnI (CCR3); denatured; and analyzed on polyacrylamide gels. The arrow marks the altered banding pattern observed in the CCR5 gene of macaque 359.
Mutation screening of CCR5 by PCR-SSCP analysis.
Although the observed resistance of macaque 359 was highly unusual, SIVmac251 infection in general results in variable viral burdens and rates of disease progression. Thus, among both naive and immunized macaques, low viral loads and slow disease progression are seen at one end of the spectrum and high viral burdens and rapid disease progression are observed at the other end. Therefore, in addition to an in-depth analysis of the CCR5 gene of macaque 359, we also investigated the CCR5 gene of 77 additional macaques to determine whether disease outcome after SIV infection is associated with mutations within the CCR5 gene. The additional macaques are described in Materials and Methods.
Each of the 78 genomic DNA samples was amplified with four pairs of sense and antisense primers in order to generate DNA products of 290 to 360 bp which, in total, spanned the entire open reading frame of CCR5. Subsequently, the single-stranded DNA mobility of the resulting PCR products was analyzed by electrophoresis on a nondenaturing polyacrylamide gel.
Among the 78 macaque DNA samples, five different mobility patterns were observed, all amplified by primers 8 and 2 at the 3′ end of the CCR5 gene. Macaque 359 exhibited a four-band pattern, as did 34.6% of the 78 macaques. Four additional patterns were observed as illustrated in Fig. 5: a condensed four-band pattern (10.3% of macaques) and three two-band patterns with different mobilities, which we have called high two-band (a) and high two-band (b) (38.5% of macaques for a and b together) and a low two-band (16.7% of macaques) (Fig. 5). The distribution of patterns among the 78 macaques within vaccinated or unvaccinated groups showed no correlation with viral load after infection or rate of disease progression (not shown).
FIG. 5.
SSCP analysis of rhesus macaque CCR5. Representative banding patterns observed after analysis of 78 macaque CCR5 genes are illustrated in lanes 1 through 5: high two-band (a), high two-band (b), low two-band, condensed four-band, and four-band. Macaque 359 is represented in lane 5.
Subsequently, the entire CCR5 genes of macaques representative of each of the five SSCP patterns were sequenced. The results are summarized in Table 2. Different DNA polymorphisms involving nucleotide substitutions at positions 786 and 897 were identified for each of the five banding patterns. The four-band patterns represented heterozygous alleles, and the two-band patterns represented homozygous alleles. None of the DNA polymorphisms, however, led to deduced changes in amino acids.
TABLE 2.
DNA polymorphisms in the rhesus macaque CCR5 genea
| Band type | Nucleotide sequence change |
|---|---|
| Four band | A-786, G-786, C-897 |
| Condensed four band | A-786, G-786, T-897 |
| High two band (a) | A-786, C-897 |
| High two band (b) | A-786, T-897 |
| Low two band | G-786, C-897 |
The CCR5 genes of macaques representing each of the banding patterns indicated were sequenced as described in Materials and Methods. Nucleotides present at the two positions where changes were identified are summarized.
Analysis of postentry viral replication events.
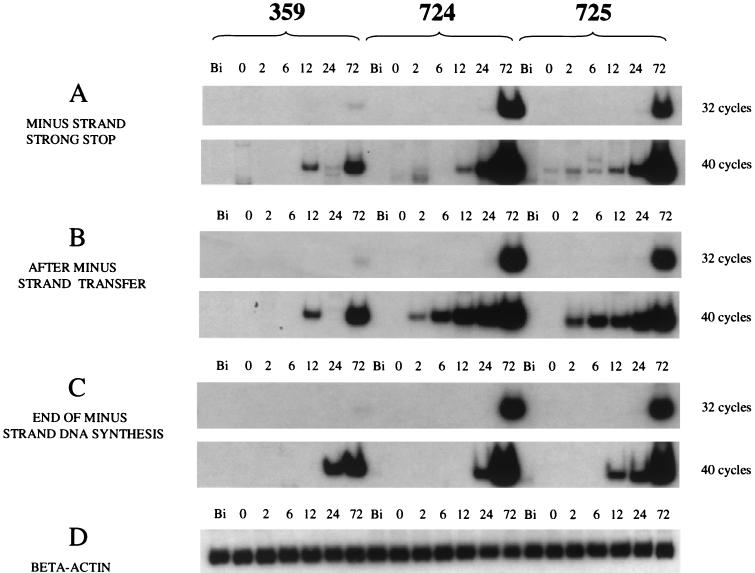
Because an alteration in the CCR5 coreceptor could not explain the resistance of macaque 359 to SIV infection, we examined postentry events of viral reverse transcription by PCR analysis. Viably frozen PBMCs of macaque 359 obtained before any mucosal exposures to SIV were depleted of CD8 cells and infected in vitro with SIVmac251. Genomic DNA was extracted before infection and at 6 time points from 0 to 72 h postinfection and amplified by PCR for 32 cycles with primer pairs specific for minus-strand strong-stop DNA, the DNA product after minus-strand transfer, and the product at the end of minus-strand DNA synthesis. As shown in Fig. 6, reverse transcription in 359 cells was not completely blocked, but the accumulation of all three reverse transcription DNA intermediates was significantly inhibited compared to the DNA intermediates synthesized in cells of susceptible control macaques 724 and 725. In order to visualize intermediate products produced in a single round of reverse transcription, PCR amplification was carried out for 40 cycles (Fig. 6). Cells of all three macaques were negative for SIV DNA before in vitro infection. Production of minus-strand strong-stop DNA and DNA after minus-strand transfer was evident in the control 724 and 725 cells by 2 h postinfection but not until 12 h in the cells of macaque 359. These results suggested that the viral replication cycle was inhibited at a very early stage in the cells of macaque 359.
FIG. 6.
PCR analysis of intermediate products of reverse transcription. Times of extraction of genomic DNA are indicated as Bi (before infection) and 0 to 72 h postinfection. (A to C) Intermediate products of SIV replication amplified for 32 or 40 cycles with specific primer pairs. (D) Standardization of input DNA amounts by amplification with β-actin-specific primers.
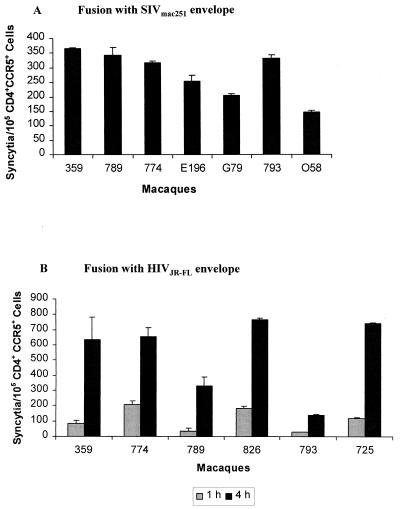
To further define the nature of the block in the replication cycle, we examined the ability of macaque 359 cells to support viral entry by using a cell-cell fusion assay. As illustrated in Fig. 7, CD4+ T cells of macaque 359 and control macaques were assessed for their ability to form syncytia with 293 cells expressing SIVmac251 or HIVJR-FL envelope. After 4 h of incubation, cells of macaque 359 formed syncytia with 293 cells expressing SIVmac251 envelope as readily as did the cells of other macaques highly susceptible to SIV infection (Fig. 7A). The ability of macaque 359 cells to support the entry of another CCR5-tropic virus, HIVJR-FL, was also investigated (Fig. 7B). A range of syncytia was observed after 1 to 4 h of incubation among cells of several different macaques. However, the cells of macaque 359 were again as susceptible to syncytia formation as were the cells of the other macaques susceptible to SIV infection. Based on the results of these fusion experiments, we concluded that the block in early SIV replication events in macaque 359 cells occurs postentry.
FIG. 7.
Analysis of ability of macaque 359 cells to support virus entry using a cell-cell fusion assay. Syncytium formation assays of CD4+ CCR5+ macaque T cells with 293 cells expressing SIVmac251 envelope (A) or HIVJR-FL envelope (B) were carried out in duplicate. The error bars indicate the standard deviation. In panel B, macaque 359 cells were evaluated on two separate occasions, and the results reflect the mean of both assays.
DISCUSSION
To elucidate the basis for the unusually strong resistance of macaque 359 to in vivo and in vitro SIV infection, we investigated early events in the viral infection and replication process. Initially, we examined the coding sequences of coreceptors used by SIV for mutations. We found no alterations in CCR3, Bonzo, BOB(GPR15), or CXCR4 genes in either resistant or susceptible macaques. On the other hand, genetic polymorphisms, confirmed by sequencing, were detected in the CCR5 gene; however, none of the changes led to amino acid substitutions. Thus, coreceptor alterations that might prevent viral entry could not account for the resistance of macaque 359 to SIV infection. Further, CD4 cells of macaque 359 in comparison to those of the highly susceptible macaque 660 expressed similar levels of CCR5 on the cell surface, both in terms of the percent positive cells and the fluorescence intensity as a measure of coreceptor density (Table 1). Thus, a block at the level of cell entry seemed unlikely.
We therefore investigated postentry events in the viral replication cycle, focusing on early DNA intermediates of reverse transcription. The reverse transcription process is highly complex (55), involving two strand transfer reactions. We focused on transcription of the minus strand, including the initial minus-strand strong-stop DNA, the DNA product made just after the first-strand transfer, and the DNA product at the end of minus-strand synthesis. All three of these steps were significantly inhibited after in vitro infection of macaque 359 CD4 T cells with SIVmac251, indicating an early block in viral replication. The inhibition of early reverse transcription events would occur if viral entry were inhibited, as well as from a postentry mechanism. We therefore revisited the question of viral entry. A cell-cell fusion assay was used since it specifically evaluates entry. Other systems, such as pseudotyped viruses carrying the luciferase gene, involve several stages of the viral life cycle in addition to entry for expression of the marker gene. Here, the cell-cell fusion assays effectively demonstrated that SIV can enter cells of the macaque 359 as efficiently as cells of macaques highly susceptible to SIV infection. Therefore, the inhibition in viral replication is due to a postentry block in the replication cycle.
Since the replication of our SIVmac251 stock proceeded efficiently in the cells of control macaques, our results implicate a host cell mechanism in inhibition of viral replication in macaque 359 cells. Others have previously shown that the susceptibility of the macaque CD4+ T cells to in vitro SIV infection reliably predicts viral replication in vivo (18, 50). Importantly, these earlier studies suggested that the susceptibility phenotype was an intrinsic property of the CD4 T cells themselves rather than a consequence of immune responses acquired after viral infection. The cellular basis for the various susceptibilities was not elucidated, although Goldstein et al. suggested that postentry mechanisms might be involved (18). Here the investigation of a highly resistant macaque which was able to clear its viral infection allowed us to demonstrate that at least one mechanism of innate host resistance is a postentry block in early viral reverse transcription events. It will be critically important to determine the prevalence of this phenomenon in the rhesus macaque population and also to identify the cellular inhibitory mechanism.
A number of cellular host factors that participate in the retroviral replication process have been identified. Several of these interact at the level of integration of proviral DNA. The product of the well-known murine Fv-1 gene, a derivative of an endogenous retroviral gag gene (5), limits the replication of certain murine leukemia virus strains to mice that carry particular Fv-1 alleles (21). Its inhibitory effect is associated with inefficient viral genome integration. Other cellular proteins positively influence the integration of proviral DNA, including the barrier to autointegration factor protein (26); INI1, a homolog to a yeast transcription factor (22); and proteins associated with integration-competent nucleoprotein complexes such as HMG I (2) and HMG I(Y) (15).
Other host proteins that function earlier in the viral replication cycle include components of the cytoskeleton and cyclophilin A. The latter, although not required for SIV replication (6), binds the HIV type 1 capsid protein (31) and is necessary for viral entry or uncoating (17, 56). A comparable cellular protein could fulfill the role of this host protein with chaperone-like activity for the SIV capsid. The early interaction of HIV with the cellular cytoskeleton is also intriguing. Actin microfilaments have been implicated in the formation of reverse transcription complexes necessary for initiating and completing the process of HIV reverse transcription (9). Other unidentified cellular factors in the cytoskeleton compartment may also be important for reverse transcription (9). Recently, a double-stranded RNA-binding protein, NF90, was shown to inhibit early events in HIV reverse transcription in GHOST cells transiently transfected with NF90 or stably expressing the protein (E. Agbottah, A. Spruill, and A. Kumar, unpublished data). Although NF90 is a nuclear protein, this observation suggests another category of cellular proteins that could potentially influence viral replication events.
The postentry block in SIV infection observed in vitro provides an explanation for the significant resistance of macaque 359 seen in vivo as well. The initial intravaginal SIV exposures which did not result in infection nevertheless elicited a low-level cellular immune response (B. Peng et al., unpublished data). Similar observations have been described for highly exposed but persistently seronegative women (23, 48). Although macaque 359 became infected after intrarectal SIV exposure, its low-level immunity, presumably helped by the host resistance mechanism inhibiting SIV replication, was sufficient to control infection and eventually clear the virus, as demonstrated by the transfusion experiment.
Our observation that a host cell mechanism inhibits early SIV replication events, thereby contributing to resistance to SIV infection, has several important implications. At a fundamental level, identification of the mechanism will expand our knowledge and understanding of the biochemical steps involved in reverse transcription. For clinical applications, identification of a natural cellular inhibitor of the reverse transcription process may be able to be exploited in novel therapeutic regimens. Finally, in a practical sense, an enhanced ability to screen rhesus macaques for cellular resistance to infection will greatly improve the macaque model for study of SIV pathogenesis and for vaccine development. It will be important to determine the prevalence of macaques that possess this type of cellular resistance. Subsequent identification of the mechanism in an outbred population will be difficult. However, macaques could be sorted based on the outcomes of mucosal challenges and subsequently assessed for levels of early reverse transcription products in vitro. Ultimately, for purposes of vaccine studies, macaques could be selected for susceptibility to infection, reducing the viral dose necessary to ensure the infection of all control animals and bringing challenge experiments to more realistic levels that are comparable to the natural exposures that occur in the human population.
Acknowledgments
We thank Janice Clements for providing the SIV17E-Br isolate, Judith Levin for helpful suggestions concerning analysis of the reverse transcriptase intermediate DNA products, Tongmao Zhao for advice on the SSCP methodology, L. Jean Patterson for preparation of the SIV17E-Br grown on rhesus macaque macrophages, Jeffrey Lifson for some real-time PCR analyses for SIV RNA, and Julio Melgar and Michael Justice for expert technical assistance.
REFERENCES
- 1.Abimiku, A. G., M. Robert-Guroff, J. Benson, J. Tartaglia, E. Paoletti, P. D. Markham, and G. Franchini. 1997. Long-term survival of SIVMac251-infected macaques previously immunized with NYVAC-SIV vaccines. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 15(Suppl. 1):S78-S85. [Google Scholar]
- 2.Aiyar, A., P. Hindmarsh, A. M. Skalka, and J. Leis. 1996. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J. Virol. 70:3571-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkhatib, G., F. Liao, E. A. Berger, J. M. Farber, and K. W. C. Pedan. 1997. A new SIV co-receptor, STRL33. Nature 388:238.. [DOI] [PubMed] [Google Scholar]
- 4.Benson, J., C. Chougnet, M. Robert-Guroff, D. Montefiori, P. Markham, G. Shearer, R. C. Gallo, M. Cranage, E. Paoletti, K. Limbach, D. Venzon, J. Tartaglia, and G. Franchini. 1998. Recombinant vaccine-induced protection against the highly pathogenic simian immunodeficiency virus SIVmac251: dependence on route of challenge exposure. J. Virol. 72:4170-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 6.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIVCPZGAB but not group O HIV-1 or other primate immunodeficiency viruses. J. Virol. 70:4220-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buge, S. L., L. Murty, K. Arora, V. S. Kalyanaraman, P. D. Markham, E. S. Richardson, K. Aldrich, L. J. Patterson, C. J. Miller, S.-M. Cheng, and M. Robert-Guroff. 1999. Factors associated with slow disease progression in macaques immunized with an adenovirus-simian immunodeficiency virus (SIV) envelope priming-gp120 boosting regimen and challenged vaginally with SIVmac251. J. Virol. 73:7430-7440. (Erratum, J. Virol. 73:9692.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buge, S. L., E. Richardson, S. Alipanah, P. Markham, S. Cheng, N. Kalyan, C. J. Miller, M. Lubeck, S. Udem, J. Eldridge, and M. Robert-Guroff. 1997. An adenovirus-simian immunodeficiency virus env vaccine elicits humoral, cellular, and mucosal immune responses in rhesus macaques and decreases viral burden following vaginal challenge. J. Virol. 71:8531-8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukrinskaya, A., B. Brichacek, A. Mann, and M. Stevenson. 1998. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J. Exp. Med. 188:2113-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hooks, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-CW*04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 11.Cohen, O. J., A. Kinter, and A. S. Fauci. 1997. Host factors in the pathogenesis of HIV disease. Immunol. Rev. 159:31-48. [DOI] [PubMed] [Google Scholar]
- 12.Cranage, M. P., N. Cook, P. Johnstone, P. J. Greenaway, P. A. Kitchin, E. J. Stott, N. Almond, and A. Baskerville. 1990. SIV infection of rhesus macaques: in vivo titration of infectivity and development of an experimental vaccine, p. 103-113. In H. Schellekens and M. C. Horzinek (ed.), Animal models in AIDS. Elsevier, Amsterdam, The Netherlands.
- 13.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Confield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, S. J. O'Brien, et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 14.Deng, H., D. Unutmaz, V. N. KewalRamani, and D. R. Littman. 1997. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296-300. [DOI] [PubMed] [Google Scholar]
- 15.Farnet, C. M., and F. D. Bushman. 1997. HIV-1 cDNA integration: requirement of HMG 1(Y) protein for function of preintegration complexes in vitro. Cell 88:483-492. [DOI] [PubMed] [Google Scholar]
- 16.Farzan, M., H. Choe, K. Martin, L. Marcon, W. Hofman, G. Karlson, Y. Sun, P. Barrett, N. Marchand, N. Sullivan, N. Gerard, C. Gerard, and J. Sodroski. 1997. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immune deficiency virus infection. J. Exp. Med. 186:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franke, E. K., H. E. H. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein, S., C. R. Brown, H. Dehghani, J. D. Lifson, and V. M. Hirsch. 2000. Intrinsic susceptibility of rhesus macaque peripheral CD4+ T cells to simian immunodeficiency virus in vitro is predictive of in vivo viral replication. J. Virol. 74:9388-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendel, H., N. Henon, H. Lebuanec, A. Lachgar, H. Poncelet, S. Caillat-Zucman, C. A. Winkler, M. W. Smith, L. Kenefic, S. O'Brien, W. Lu, J. M. Andrieu, D. Zagury, F. Schachter, J. Rappaport, and J. F. Zagury. 1998. Distinctive effects of CCR5, CCR2, and SDF-1 genetic polymorphisms in AIDS progression. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:381-386. [DOI] [PubMed] [Google Scholar]
- 20.Huang, Y., W. A. Paxton, S. M. Wolinsky, A. U. Neumann, L. Zhang, S. Kang, D. Ceradini, Z. Jin, K. Yazdanbakhsh, K. Kunstman, D. Erikson, I. Dragon, N. R. Landau, J. Phair, D. D. Ho, and R. A. Koup. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 11:1240-1243. [DOI] [PubMed] [Google Scholar]
- 21.Jolicoeur, P. 1979. The Fv-1 gene of the mouse and its control of murine leukemia virus replication. Curr. Top. Microbiol. Immunol. 86:68-122. [DOI] [PubMed] [Google Scholar]
- 22.Kalpana, G. V., S. Marmon, W. Wang, G. R. Crabtree, and S. P. Goff. 1994. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002-2006. [DOI] [PubMed] [Google Scholar]
- 23.Kaul, R., F. A. Plummer, J. Kimani, T. Dong, P. Kiama, T. Rostron, E. Njagi, K. S. MacDonald, J. J. Bwayo, A. J. McMichael, and S. L. Rowland-Jones. 2000. HIV-1-specific mucosal CD8+ lymphocyte responses in the cervix of HIV-1-resistant prostitutes in Nairobi. J. Immunol. 164:1602-1611. [DOI] [PubMed] [Google Scholar]
- 24.Koenig, S., V. M. Hirsch, R. A. Olmsted, D. Powell, W. Maury, A. Rabson, A. S. Fauci, R. H. Purcell, and P. R. Johnson. 1989. Selective infection of human CD4+ cells by simian immunodeficiency virus: productive infection associated with envelope glycoprotein-induced fusion. Proc. Natl. Acad. Sci. USA 86:2443-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kostrikis, L. G., Y. Huang, J. P. Moore, S. M. Wolinsky, L. Zhang, Y. Guo, L. Deutsch, J. Phair, A. U. Neumann, and D. D. Ho. 1998. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat. Med. 4:350-353. [DOI] [PubMed] [Google Scholar]
- 26.Lee, M. S., and R. Craigie. 1998. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. USA 95:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leno, M., L. Carter, D. J. Venzon, J. Romano, P. D. Markham, K. Limbach, J. Tartaglia, E. Paoletti, J. Benson, G. Franchini, and M. Robert-Guroff. 1999. CD8+ lymphocyte antiviral activity in monkeys immunized with SIV recombinant poxvirus vaccines: potential role in vaccine efficacy. AIDS Res. Hum. Retrovir. 15:461-470. [DOI] [PubMed] [Google Scholar]
- 28.Liao, F., G. Alkhatib, K. W. C. Peden, G. Sharma, E. A. Berger, and J. M. Farber. 1997. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J. Exp. Med. 185:2015-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, H., D. Chao, E. E. Nakayama, H. Taguchi, M. Goto, X. Xin, J.-K. Takamatsu, H. Saito, Y. Ishikawa, T. Akaza, T. Juji, Y. Takebe, T. Ohishi, K. Fukutake, Y. Maruyama, S. Yashiki, S. Sonoda, T. Nakamura, Y. Nagai, A. Iwamoto, and T. Shioda. 1999. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc. Natl. Acad. Sci. USA 96:4581-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. Macdonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 co-receptor accounts for resistance of some multiply exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 31.Luban, J., K. A. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 32.Margolis, L., S. Glushakova, C. Chougnet, G. Shearer, P. Markham, S. L. Buge, M. Robert-Guroff, R. Benveniste, C. J. Miller, M. Cranage, V. Hirsch, and G. Franchini. 2000. Replication of simian immunodeficiency virus (SIV) in ex vivo lymph nodes as a means to assess susceptibility of macaques in vivo. Virology 275:391-397. [DOI] [PubMed] [Google Scholar]
- 33.Mariani, R. S. Wong, L. C. F. Mulder, D. A. Wilkinson, A. L. Reinhart, G. LaRosa, R. Nibbs, T. R. O'Brien, N. L. Michael, R. I. Connor, M. Macdonald, M. Busch, R. A. Koup, and N. R. Landau. 1999. CCR2-64I polymorphism is not associated with altered CCR5 expression or coreceptor function. J. Virol. 73:2450-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin, M. P., M. Dean, M. W. Smith, C. Winkler, B. Gerrard, N. L. Michael, B. Lee, R. W. Doms, J. Margolick, S. Buchbinder, J. J. Goedert, T. R. O'Brien, M. W. Hilgartner, D. Vlahov, S. J. O'Brien, and M. Carrington. 1998. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science 282:1907-1911. [DOI] [PubMed] [Google Scholar]
- 35.McChesney, M. B., J. R. Collins, D. Lu, X. Lu, J. Torten, R. L. Ashley, M. W. Cloyd, and C. J. Miller. 1998. Occult systemic infection and persistent simian immunodeficiency virus (SIV)-specific CD4+-T-cell proliferative responses in rhesus macaques that were transiently viremic after intravaginal inoculation of SIV. J. Virol. 72:10029-10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDermott, D. H., P. A. Zimmerman, F. Guignard, C. A. Kleeberger, S. F. Leitman, and P. M. Murphy for the Multicenter AIDS Cohort Study (MACS). 1998. CCR5 promoter polymorphism and HIV-1 disease progression. Lancet 352:866-870. [DOI] [PubMed] [Google Scholar]
- 37.Michael, N. L. 1999. Host genetic influences on HIV pathogenesis. Curr. Opin. Immunol. 11:466-474. [DOI] [PubMed] [Google Scholar]
- 38.Michael, N. L., L. G. Louie, A. L. Rohrbaugh, K. A. Schultz, D. E. Dayhoff, C. E. Wang, and H. W. Sheppard. 1997. The role of CCR5 and CCR2 polymorphisms in HIV-1 transmission and disease progression. Nat. Med. 3:1160-1162. [DOI] [PubMed] [Google Scholar]
- 39.Migueles, S. A., M. C. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, C. J., M. Marthas, J. Torten, N. J. Alexander, J. P. Moore, G. F. Doncel, and A. G. Hendrickx. 1994. Intravaginal inoculation of rhesus macaques with cell-free simian immunodeficiency virus results in persistent or transient viremia. J. Virol. 68:6391-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mummidi, S., S. S. Ahuja, E. Gonzalez, S. A. Anderson, E. N. Santiago, K. T. Stephan, F. E. Craig, P. O'Connell, V. Tryon, R. A. Clark, M. J. Dolan, and S. K. Ahuja. 1998. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat. Med. 4:786-793. [DOI] [PubMed] [Google Scholar]
- 42.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pal, R., D. Venzon, N. L. Letvin, S. Santra, D. C. Montefiori, N. R. Miller, E. Tryniszewska, M. G. Lewis, T. C. VanCott, V. Hirsch, R. Woodward, A. Gibson, M. Grace, E. Dobratz, P. D. Markham, Z. Hel, J. Nacsa, M. Klein, J. Tartaglia, and G. Franchini. 2002. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J. Virol. 76:292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patterson, L. J., F. Robey, A. Muck, K. Van Remoortere, K. Aldrich, E. Richardson, W. G. Alvord, P. D. Markham, M. Cranage, and M. Robert-Guroff. 2001. A conformational C4 peptide polymer vaccine coupled with live recombinant vector priming is immunogenic but does not protect against rectal SIV challenge. AIDS Res. Hum. Retrovir. 17:837-849. [DOI] [PubMed] [Google Scholar]
- 45.Paxton, W. A., R. Liu, S. Kang, L. Wu, T. R. Gingeras, N. R. Landau, C. R. Mackay, and R. A. Koup. 1998. Reduced HIV-1 infectability of CD4+ lymphocytes from exposed-uninfected individuals: association with low expression of CCR5 and high production of β-chemokines. Virology 244:66-73. [DOI] [PubMed] [Google Scholar]
- 46.Reynes, J., P. Portales, M. Segondy, V. Baillat, P. Andre, B. Reant, O. Avinens, G. Coudere, M. Benkirane, J. Clot, J.-F. Eliaou, and P. Corbeau. 2000. CD4+ T-cell surface CCR5 density as a determining factor of virus load in persons infected with human immunodeficiency virus type 1. J. Infect. Dis. 181:927-932. [DOI] [PubMed] [Google Scholar]
- 47.Romano, J. W., B. van Gemen, and T. Kievits. 1996. A novel, isothermal detection technology for qualitative and quantitative HIV-1 RNA measurements. Clin. Lab. Med. 16:89-103. [PubMed] [Google Scholar]
- 48.Rowland-Jones, S. L., T. Dong, K. R. Fowke, J. Kimani, P. Krausa, H. Newell, T. Blanchard, K. Ariyoshi, J. Oyugi, E. Ngugi, J. Bwayo, K. S. MacDonald, A. J. McMichael, and F. A. Plummer. 1998. Cytotoxic T-cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J. Clin. Investig. 102:1758-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sampson, M., F. Libert, B. L. Dorang, J. Rucker, C. Liesnard, C.-M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Foueille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Coms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV infection in Caucasian individuals bearing mutant alleles of the CCR5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 50.Seman, A. L., W. F. Pewen, L. F. Fresh, L. N. Martin, and M. Murphey-Corb. 2000. The replicative capacity of rhesus macaque peripheral blood mononuclear cells for simian immunodeficiency virus in vitro is predictive of the rate of progression to AIDS in vivo. J. Gen. Virol. 81:2441-2449. [DOI] [PubMed] [Google Scholar]
- 51.Sharma, D. P., M. C. Zink, M. G. Anderson, R. Adams, J. E. Clements, S. V. Joag, and O. Narayan. 1992. Derivation of neurotropic SIV from exclusively lymphocyte-tropic parental virus: pathogenesis of infection in macaques. J. Virol. 66:3550-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shin, H. D., C. Winkler, J. C. Stephens, J. Bream, H. Young, J. J. Goedert, T. R. O'Brien, D. Vlahov, S. Buchbinder, J. Giorgi, C. Rinaldo, S. Donfield, A. Willoughby, S. J. O'Brien, and M. W. Smith. 2000. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL-10. Proc. Natl. Acad. Sci. USA 97:14467-14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, M. W., M. Dean, M. Carrington, C. Winkler, G. A. Huttley, D. A. Lomb, J. J. Goedert, T. R. O'Brien, L. P. Jacobsen, R. Kaslow, S. Buchbinder, E. Vittinghoff, D. Vlahov, K. Hoots, M. W. Hilgartner, S. J. O'Brien, et al. 1997. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Science 277:959-965. [DOI] [PubMed] [Google Scholar]
- 54.Suryanarayana, K., T. A. Wiltrout, G. M. Vasquez, V. M. Hirsch, and J. D. Lifson. 1998. Plasma SIV RNA viral load determination by real-time quantification of product generation in reverse transcriptase-polymerase chain reaction. AIDS Res. Hum. Retrovir. 14:183-189. [DOI] [PubMed] [Google Scholar]
- 55.Telesnitsky, A., and S. P. Goff. 1997. Reverse transcriptase and the generation of retroviral DNA, p. 121-160. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 56.Thali, M., A. A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Specific association of cyclophilin A with human immunodeficiency virus type 1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 57.van Rij, R. P., S. Broersen, J. Goudsmit, R. A. Coutinho, and H. Schitemaker. 1998. The role of a stromal cell-derived factor-1 chemokine gene variant in the clinical course of HIV-1 infection. AIDS 12:F85-F90. [PubMed] [Google Scholar]
- 58.Winkler, C., W. Modi, M. W. Smith, G. W. Nelson, X. Wu, M. Carrington, M. Dean, T. Honjo, K. Tashiro, D. Yabe, S. Buchbinder, E. Vittinghoff, J. J. Goedert, T. R. O'Brien, L. P. Jacobson, R. Detels, S. Donfield, A. Willoughby, E. Gomperts, D. Vlahov, J. Phair, S. J. O'Brien, et al. 1998. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. Science 279:389-397. [DOI] [PubMed] [Google Scholar]
- 59.Wu, L., W. A. Paxton, N. Kassam, N. Ruffing, J. B. Rottman, N. Sullivan, H. Choe, J. Sodroski, W. Newman, R. A. Koup, and C. R. Mackay. 1997. CCR5 levels and expression pattern correlate with infectibility by macrophage-tropic HIV-1 in vitro. J. Exp. Med. 185:1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, L., T. Ho, A. Talal, G. Wang, S. S. Frankel, and D. D. Ho. 1998. In vivo distribution of the human immunodeficiency virus/simian immunodeficiency virus co-receptors: CXCR4, CCR3, and CCR5. J. Virol. 72:5035-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]