Abstract
The development of an effective vaccine against human immunodeficiency virus is considered to be the most practicable means of controlling the advancing global AIDS epidemic. Studies with the domestic cat have demonstrated that vaccinal immunity to infection can be induced against feline immunodeficiency virus (FIV); however, protection is largely restricted to laboratory strains of FIV and does not extend to primary strains of the virus. We compared the pathogenicity of two prototypic vaccine challenge strains of FIV derived from molecular clones; the laboratory strain PETF14 and the primary strain GL8414. PETF14 established a low viral load and had no effect on CD4+- or CD8+-lymphocyte subsets. In contrast, GL8414 established a high viral load and induced a significant reduction in the ratio of CD4+ to CD8+ lymphocytes by 15 weeks postinfection, suggesting that PETF14 may be a low-virulence-challenge virus. However, during long-term monitoring of the PETF14-infected cats, we observed the emergence of variant viruses in two of three cats. Concomitant with the appearance of the variant viruses, designated 627W135 and 628W135, we observed an expansion of CD8+-lymphocyte subpopulations expressing reduced CD8 β-chain, a phenotype consistent with activation. The variant viruses both carried mutations that reduced the net charge of the V3 loop (K409Q and K409E), giving rise to a reduced ability of the Env proteins to both induce fusion and to establish productive infection in CXCR4-expressing cells. Further, following subsequent challenge of naïve cats with the mutant viruses, the viruses established higher viral loads and induced more marked alterations in CD8+-lymphocyte subpopulations than did the parent F14 strain of virus, suggesting that the E409K mutation in the PETF14 strain contributes to the attenuation of the virus.
Infection of the domestic cat with feline immunodeficiency virus (FIV) leads to the development of an immunodeficiency syndrome similar to AIDS in human beings infected with human immunodeficiency virus (HIV). FIV-infected cats are likelier to develop illnesses, including gingivitis, stomatitis, lymphoma, neurological disorders, and wasting (15, 24). Flow cytometry analyses have demonstrated a reduction in the number of circulating CD4+ lymphocytes in FIV-infected cats (1, 22), and longitudinal studies have revealed that the degree of immune impairment correlates with the decline in CD4+-lymphocyte numbers (34), suggesting that, as with HIV infection in humans, the primary lesion in the FIV-infected cat is an impairment of helper T-cell function. During the early, acute phase of infection, the inversion of the CD4/CD8 ratio is compounded by a sharp increase in the number of CD8+ T lymphocytes expressing low levels of CD8 (CD8low) and increased levels of major histocompatibility complex class II antigens (39). This activated T-cell subpopulation persists throughout the course of the infection, suggesting that the early interaction between the virus and the immune system may ultimately determine the outcome of the infection. CD8low cells have since been defined as CD8α+βlow (28, 29), the vpg9 antibody used in previous studies being shown to recognize the CD8αβ heterodimer. An analogous population has since been described in HIV-infected individuals in whom reduced expression of the CD8αβ heterodimer correlates with increased expression of molecules associated with lymphocyte activation, adhesion, and cytotoxic-T-cell activity (27). Further, a significant increase in expression of the CD8αβ heterodimer followed initiation of highly active antiretroviral therapy, suggesting that analysis of discrete CD8+-T-lymphocyte subsets may be of value in assessing the immune status of individuals infected with HIV type 1 (HIV-1) (27).
Previously, it was demonstrated that vaccination with whole-inactivated-virus vaccines or DNA vaccines afforded protection against challenge with the cell culture-adapted PET strain of FIV but not against the primary GL8 strain (10, 12, 13). The induction of protective immunity against primary strains of FIV such as GL8 has proved difficult; outcomes ranged from a suppression of the peripheral blood mononuclear cell (PBMC) viral load and CD4+-T-cell loss with a whole-inactivated-virus vaccine (12) to enhancement of infection following immunization with FIV p24-ISCOMS (14). Encouragingly, protective immunity has been induced following immunization with a fixed- and infected-cell vaccine based on the MBM cell line (19), and in field studies, this vaccine provided immunity to infection following natural exposure to the virus (20). The success of the fixed-cell vaccine may be associated with the observation that the vaccine was based on cells infected with a clade B virus strain (Pisa M2), and the vaccine was tested in a region in which clade B viruses are almost exclusively found (20, 25). Previous studies have suggested that clade B viruses such as Pisa M2 may be more ancient and relatively host adapted, while clade A viruses may have entered their host population more recently and thus may be more virulent (2, 25). As clade A viruses are widespread in the United States and northern Europe, it is important to establish the determinants of virulence in these viruses.
Cell culture-adapted strains of FIV such as PET differ from primary strains of FIV in that they have an expanded cell tropism that permits growth in established cell lines, such as Crandell feline kidney cells (CrFK) or 3201 cells. The underlying mechanism for the expanded cell tropism involves an increase in charge of the V3 loop of the viral envelope glycoprotein (Env) gp120 (31, 36). This increase in charge of the V3 loop facilitates the usage of feline CXCR4 as a sole receptor for infection (11, 37, 41, 42), possibly in conjunction with heparan sulfate (6) and analogous to CD4-independent infection with HIV-2 (8). In contrast, while infection with most primary strains of FIV is CXCR4 dependent (7, 26), the existence of an as-yet-unidentified, high-affinity non-CD4 primary receptor has been suggested (6, 38).
In this study, we compared the virulence of two prototypic clade A vaccine challenge strains of FIV derived from stable molecular clones, PETF14 and GL8414. We demonstrate that the GL8414 molecular clone has virulence characteristics similar to those of the biological isolate of the virus from which it was derived, establishing a high viral load and inducing marked alterations in circulating CD4+- and CD8+-lymphocyte subpopulations. In contrast, the PETF14 virus established a low viral load and had little effect on the CD4+- and CD8+-lymphocyte subpopulations. Further, we show that the emergence of more virulent strains of PETF14 in vivo is associated with alterations in the way that the virus uses CXCR4 as a receptor for infection.
MATERIALS AND METHODS
Viruses and cell lines.
All cell culture media and supplements were obtained from Invitrogen Life Technologies Ltd., Paisley, United Kingdom. Adherent cell lines were maintained in modified Dulbecco's minimal essential medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 0.11 mg of sodium pyruvate/ml, 100 IU of penicillin/ml, and 100 μg of streptomycin/ml. The interleukin 2 (IL-2)-dependent feline T-cell line Mya-1 (21) and PBMCs were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 IU of penicillin/ml, 100 μg of streptomycin/ml, 5 × 10−5 M 2-mercaptoethanol, and 100 IU of recombinant human IL-2/ml.
The challenge viruses were prepared from the GL8414 (12) and PETF14 (33) molecular clones of FIV. Molecular clones were transfected in the murine fibroblast cell line 3T3 using Superfect transfection reagent (Qiagen, Valencia, Calif.). Seventy-two hours posttransfection, supernatants were harvested, treated with a 0.45-μm-pore-size filter, and used to infect the IL-2-dependent feline T-cell line Q201 (40). The infected cultures were monitored visually for cytopathicity and for the production of FIV p24 by enzyme-linked immunosorbent assay (ELISA) (PetCheck FIV antigen ELISA; IDEXX Corp., Portland, Maine). Supernatants were collected at peak cytopathicity and p24 production, treated with a 0.45-μm-pore-size filter, dispensed into 1-ml aliquots, and stored at −70°C.
Quantification of proviral load in PBMCs.
The FIV proviral load in PBMCs was quantified using real-time PCR measuring PCR product accumulation through a dual-labeled fluorogenic TaqMan probe (9). The primers used were FIV0771f (5′-AGA ACC TGG TGA TAT ACC AGA GAC-3′) and FIV1081r (5′-TTG GGT CAA GTG CTA CAT ATT G-3′). The probe used in this system was FIV1010p (5′-FAM-TAT GCC TGT GGA GGG CCT TCC T-TAMRA-3′). The oligonucleotides were designed to detect a variety of FIV A-subtype isolates and have been previously shown to detect FIV-PET, FIV-GL8, and FIV-AM6 with only minor differences in PCR efficiency (17, 18). The 25-μl PCR mixtures contained 10 mM Tris (pH 8.3); 50 mM KCl; 3 mM MgCl2; 200 nM dATP, dCTP, and dGTP; 400 nM dUTP; a 300 nM concentration of each primer; a 200 nM concentration of the fluorogenic probe; and 2.5 U of Taq DNA polymerase. After the initial denaturation (2 min at 95°C), amplification was performed with 45 cycles of 15 s at 95°C and 60 s at 60°C. The PCR and the online measurement of the emitted fluorescence were performed on a Sequence Detector System ABI 7700 (Applied Biosystems, Foster City, Calif.). The copy number per PCR was calculated by the Sequence Detection Software version 1.6.3 (Applied Biosystems) utilizing a fourfold-dilution series of genomic DNA derived from a CrFK cell line infected with FIV Petaluma. The DNA content per PCR was estimated by a second real-time PCR assay targeting the 18S ribosomal DNA genes (16).
Quantification of viral load in plasma.
The FIV viral load in plasma was quantified by real-time reverse transcriptase PCR (RT-PCR) using the same primers as in the real-time PCR. The 25-μl RT-PCR mixtures contained 12.5 μl of 2× Thermoscript Reaction Mix (Platinum Quantitative RT-PCR Kit; Life Technologies, Karlsruhe, Germany), a 300 nM concentration of each primer, a 200 nM concentration of the fluorogenic probe, 0.5 μl of the Thermoscript Plus/Platinum Taq Enzyme mix, 20 U of RNaseOUT (Life Technologies), and 5 μl of the sample. After a reverse transcription step of 30 min at 60°C followed by a denaturation step (5 min at 95°C), amplification was performed with 45 cycles of 15 s at 95°C and 60 s at 60°C.
Virus isolation.
Virus isolation from PBMCs by bulk cell culture was performed as described previously (12). PBMCs were fractionated from 5 ml of whole blood by centrifugation over a Ficoll-Paque density of 1.077 g/ml. The separated PBMCs were cocultured with Mya-1 cells in the absence of mitogenic stimulation. We have found that this protocol enables the isolation of both primary and cell culture-adapted strains of FIV and does not exert a selective pressure on the virus that is isolated; e.g., Mya-1 cells will support the growth of PETF14 and GL8414 equally, and the viruses harvested from the cells retain their specific cell tropisms.
Antibodies and flow cytometry.
Antibodies were used either un-conjugated or as phycoerythrin (PE) or fluorescein isothiocyanate (FITC) conjugates. Anti-feline CD4-FITC (vpg34) and CD8αβ-PE (vpg9) were obtained from Serotec Ltd., Oxford, United Kingdom; anti-feline CD8α (12A3) from Y. Nishimura, University of Tokyo, Tokyo, Japan; and anti-feline CD8β-FITC and -PE (FT2) from Southern Biotechnology Ltd., Birmingham, Ala. FITC-conjugated anti-feline CD8α (12A3) was prepared using FITC-coupling reagent (Pierce Chemical Co., Rockford, Ill.) according to the manufacturer's instructions. Unconjugated primary antibodies were detected using an FITC-or PE-coupled F(ab′)2 fragment of sheep anti-mouse immunoglobulin G whole molecule (Sigma). EDTA anti-coagulated blood was processed for flow cytometry analyses by whole blood lysis as described previously (39). Samples were analyzed on a Beckman Coulter Epics Elite flow cytometer, and 10,000 events were collected for each sample. Data were analyzed using Expo32 ADC software (Applied Cytometry Systems, Sheffield, United Kingdom).
Molecular cloning and nucleic acid sequencing.
DNAs were prepared from positive viral isolations by column chromatography (QIAamp DNA maxiprep kit; Qiagen). DNAs were prepared as soon as a positive ELISA for FIV p24 was recorded; thus, viruses had undergone minimal passage in vitro. Full-length viral envelope glycoprotein (env) genes were amplified from the replication-competent viruses by using a high-fidelity (proofreading) PCR (High Fidelity PCR system; Roche) using primers corresponding to the 5′ L-SU cleavage site (TAGACGCGTAAGATTTTTAAGGTATTC) and the NdeI site 3′ of the Rev-responsive element (CCCTTTGAGGAAGATGTGTCATATGAATCCATT) incorporating MluI and NdeI restriction sites, respectively. Due to the inherent instability of the full-length env genes from primary isolates of FIV such as GL8, standard high-copy-number PCR product cloning vectors could not be used; thus, all amplified env gene products were digested with MluI/NdeI and were cloned directly into pGL8MYA, a molecular clone of FIV-GL8 in the low-copy-number plasmid pBR328 and in which an MluI site had been introduced at the L-SU junction. The nucleic acid sequence of three independent clones of each env gene was determined using IRD800-labeled oligonucleotides on an automated sequencer (LI-COR Biosciences, Lincoln, Nebr.).
In vitro expression of env genes.
The biological function of each env gene clone was confirmed by the recovery of infectious virus following transfection and by immunofluorescence detection of the Env glycoprotein with an anti-FIV Env monoclonal antibody (vpg71.2). For the recovery of infectious virus, 293T cells were transfected using Superfect (Qiagen) as per the manufacturer's instructions. Supernatants were collected 48 to 72 h posttransfection, treated with a 0.45-μm-pore-size filter, and added to Mya-1 cells. The infected cells were monitored for the production of FIV using the PetCheck FIV antigen ELISA (IDEXX Corp.).
Immunofluorescence was performed on transfected 293T cells following fixation with ice-cold methanol. Fixed cells were rehydrated using phosphate-buffered saline containing 1.0% bovine serum albumin and 0.1% azide (PBA). The cells were then incubated with either 1 μg of vpg71.2 or an isotype-matched control for 30 min on ice, washed twice with PBA by centrifugation, and then incubated with an FITC-coupled F(ab′)2 fragment of sheep anti-mouse immunoglobulin G whole molecule on ice for a further 30 min. Finally, the cells were washed twice with PBA and then analyzed on a Leica UV microscope.
To assess the fusogenicity of the FIV Env proteins, the env genes were subcloned into the mammalian expression vector VR1012 (Vical Inc., San Diego, Calif.). AH927 cells expressing feline CXCR4 were transfected with the constructs using Superfect (Qiagen) and incubated for 48 h at 37°C. The cells were then fixed and stained with 1% methylene blue/0.2% basic fuchsin in methanol. Syncytia were enumerated by light microscopy using an ×12.5 Leitz periplan eyepiece with a 6.5- by 9-mm graticule; three separate fields were counted per well, each well in duplicate. Syncytia were scored as cells with five or more nuclei.
The growth of FIV in vitro was assessed in AH927 cells expressing feline CXCR4. AH927 cells were transduced with retroviral vectors (pDONAI; Takara Biomedicals, Shiga, Japan) expressing feline CXCR4 or CCR5 and were selected in G418. Stable transfectants were seeded in 48-well cell culture plates and were then infected with twofold dilutions of virus grown in IL-2-dependent T cells. Supernatants were collected every 3 days and were assayed for the production of RT using the Lenti-RT nonisotopic RT assay kit (Cavidi Tech, Uppsala, Sweden). RT values were then calculated relative to the purified HIV-1 RT standard.
RESULTS
Early infection. (i) FIV GL8 rapidly establishes a high viral load.
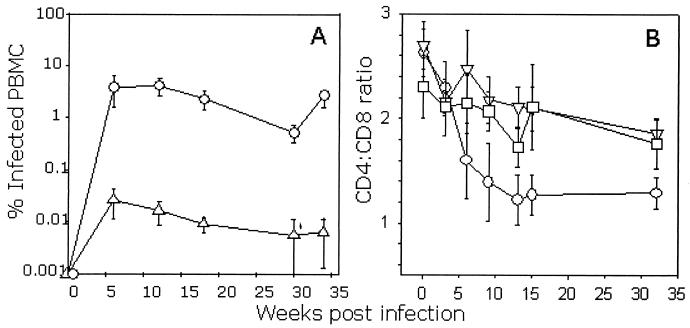
Three groups of three cats were infected with either a matched dose of FIV GL8414 or PETF14 or were mock infected. Samples of peripheral blood were collected at regular intervals after infection, and the PBMC proviral DNA burden was quantified by real-time PCR (Fig. 1A). The proviral loads in the GL8414-infected cats rose sharply, with approximately 1.0% of PBMCs being positive for proviral DNA by 6 weeks postinfection. This high proviral burden was maintained throughout the acute phase of infection and during the first 35 weeks of the study period. In contrast, the PETF14 inoculum established a significantly lower proviral load, with a load of approximately 0.01% being achieved during the acute phase (Fig. 1A). During this period, the effect of viral infection on the CD4/CD8 ratio was monitored by flow cytometry. Coincident with the sharp rise in proviral load in the GL8414-infected cats was a marked decline in the CD4/CD8 ratio manifesting by 6 weeks postinfection (Fig. 1B). In contrast, the CD4/CD8 ratios of the PETF14-infected cats did not differ significantly from those of the controls.
FIG. 1.
Quantification of proviral load and CD4/CD8 ratio in FIV-infected cats during the acute phase of infection. Three cats were inoculated intraperitoneally with either GL8414 (○), PETF14 (▿), or mock infected with PBS (□). (A) Proviral DNA load in PBMCs was estimated by real-time PCR (control cats were consistently negative and thus are not shown). Each point represents the mean proviral load of the three cats in the group (± standard error). (B) The CD4/CD8 ratio was estimated by flow cytometry. Results represent the mean CD4/CD8 ratio of the three study groups (± standard error).
(ii) Expansion of CD8α+βlow T lymphocytes in GL8414-infected cats.
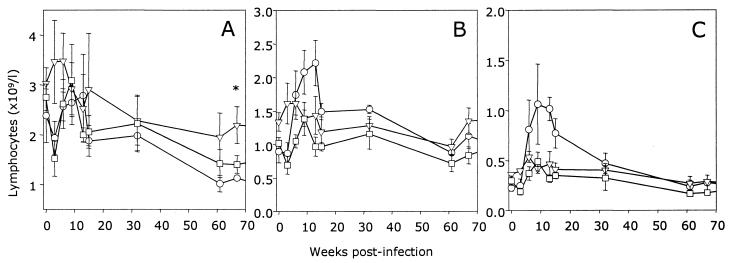
Previously, it was demonstrated that the biological isolate of GL8 induced rapid alterations in the CD8+-lymphocyte subset characterized by reduced expression of CD8 (CD8low) (39). Subsequently, the CD8low population was revealed to consist of CD8α+βlow and CD8αα′ populations (28, 29), analogous to the population described in HIV-infected individuals (27). The absolute numbers of CD4+, CD8+, and CD8α+βlow cells were determined in the three groups of cats (Fig. 2). CD4+-lymphocyte numbers dropped sharply in both the GL8414- and PETF14-infected cats at 3 weeks postinfection. However, this proved to be a transient fall as the levels rebounded by 6 weeks postinfection. Although the data suggest that there was a downward trend in the CD4+-lymphocyte numbers in both the FIV-infected groups, it was not until 67 weeks postinfection that the reduction in the CD4+-lymphocyte number in the GL8414-infected cats reached statistical significance (P = 0.046; t test). The major contributing factor to the depressed CD4/CD8 ratio in the GL8414-infected cats was a transient, sharp increase in the number of CD8+ lymphocytes (Fig. 2B). When the analysis gates for flow cytometry are focused on the CD8α+βlow subset (Fig. 2C), it can be seen that this population expands by 9 weeks postinfection to a maximum of 109/liter, returning to preinfection levels (<0.4 × 109/liter) by 32 weeks postinfection. Due to the small numbers of cats in each group, the elevated numbers of CD8α+βlow lymphocytes did not reach statistical significance. In contrast to results for the GL8414-infected cats, CD8α+βlow lymphocytes could not be detected at any of the time points during the acute phase of infection in the PETF14-infected cats. The data indicate that the virus derived from the GL8414 molecular clone is as pathogenic as the original biological isolate of GL8 and that cats infected with the GL8414 or PETF14 viruses can be differentiated readily on the basis of viral load. Further, at no point during the early acute phase of infection did we observe the CD8α+βlow subset in PETF14-infected cats, consistent with this subset being a marker for infection with a more pathogenic strain of FIV, such as GL8414.
FIG. 2.
Effect of FIV infection on CD4+ (A), CD8+ (B), and CD8α+βlow (C) lymphocyte subsets. The percentage of peripheral blood lymphoid cells expressing CD4 and CD8 was quantified in each of the three study groups, GL8414 infected (○), PETF14 infected (□), or mock infected with PBS (▿), by flow cytometry, and the absolute cell number of each subset was calculated from the results of hematological analysis. Results are expressed as the mean (± standard error) cell number (109/liter); ∗ denotes a statistically significant difference (P = 0.046).
Long-term infection. (i) Emergence of pathogenic variants of FIVF14 during prolonged infection.
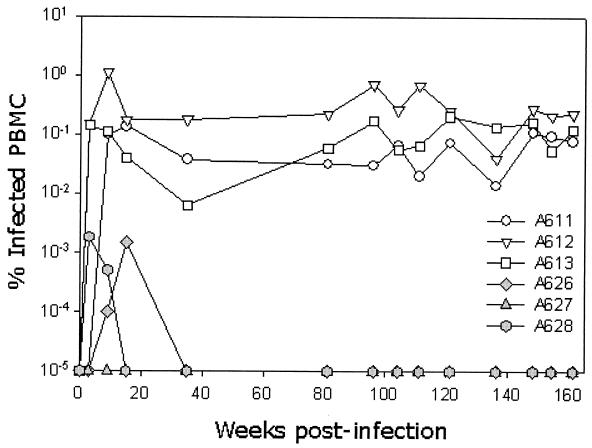
Throughout the acute phase of infection, virus could be isolated readily in vitro from PBMC cultures from both groups of infected cats (data not shown). By 32 weeks postinfection, virus isolation became sporadic from the PETF14-infected cats, consistent with the low viral loads indicated by real-time PCR (Fig. 1). In contrast, virus could be isolated readily from GL8414-infected cats at all time points (data not shown). These findings may indicate that the immune response to infection was limiting the replication of PETF14 but not of GL8414 and are consistent with the contention that the PETF14 virus is less virulent than GL8414. Following the early peak in viral load during the acute phase of infection, the proviral burden in the PBMCs of the PETF14-infected cats fell below the level of assay sensitivity. In contrast, a high proviral burden was maintained in the GL8414-infected cats throughout the study period (Fig. 3).
FIG. 3.
Quantification of proviral load in FIV-infected cats during the acute and chronic phases of infection. Three cats were inoculated intraperitoneally with either GL8414 (open symbols) or PETF14 (closed symbols), and the proviral DNA load in PBMCs was estimated by real-time PCR. The sequential proviral loads of each cat in the two groups are shown as % infected PBMCs.
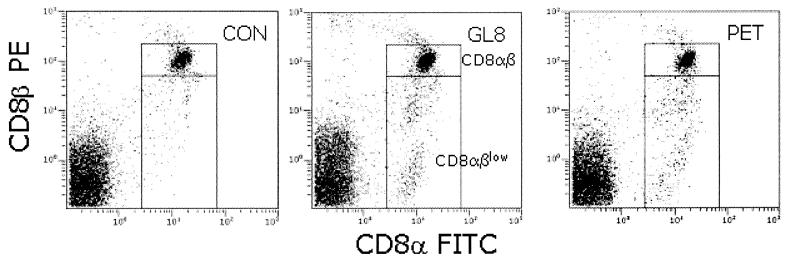
By 144 weeks postinfection, bulk virus isolations from PBMCs from cats 627 and 628 tested positive, despite the PBMC proviral load remaining below assay sensitivity (<10−5% infected PBMCs). Coincident with the positive virus isolations, we observed the expansion of the CD8α+βlow subset in the same cats (Fig. 4). As we had not observed an expansion of the CD8α+βlow subset in the PETF14-infected cats at any time point during the acute phase of infection, we postulated that virulent escape mutants may have evolved with time postinfection.
FIG. 4.
Expansion of CD8α+βlow-lymphocyte subpopulations in FIV-infected cats. Contour plots of representative analyses of one cat from each group at 144 weeks postinfection (CON = control 612, GL8 = GL8414-infected 612, and PET = PETF14-infected 628). Boxed regions illustrate the analysis gates for each population.
(ii) Characterization of the emergent virus strains from PETF14-infected cats.
Virus was isolated from PBMCs from cats 627 and 628, and the env genes were amplified by high-fidelity PCR and cloned. The nucleic acid sequence of the env clones was then determined, and the predicted amino acid sequence translation was established. Figure 5 illustrates the predicted amino acid sequence of the Env region from V3 through V6, a region that has been shown previously to contain determinants of cell tropism (31, 36) and CXCR4 usage (6, 42). The mutation of either of two glutamate residues in V3 (residues 407 and 409 in GL8414 in Fig. 5) to lysine residues is thought to determine the usage of CXCR4 as a sole receptor (analogous to CD4-independent infection with HIV). The PETF14 virus contains a lysine residue at position 409. In the virus reisolated from cat 628 at 135 weeks postinfection (628W135), lysine 409 had been replaced with a glutamate residue (K409E), analogous to GL8414, which reduces the net charge of the V3 loop by replacing a positively charged residue with a negatively charged residue. A second coding mutation (S557N) observed in the reisolate from cat 628 introduced a predicted N-linked glycosylation site in the V5 region. Intriguingly, the week 135 reisolate from cat 627 (627W135) also showed a mutation at position 409 in the V3 loop (K409Q), replacing a positively charged residue with a neutral residue, again reducing the net charge of the V3 loop. A second mutation (N422T) removed a predicted site for N-linked glycosylation from the region at the base of the V3 loop. Given that the glutamate residues 407 and 409 are known to be critical determinants of the interaction between FIV and CXCR4, the data suggest that there may be a direct link between CXCR4 usage and virulence.
FIG. 5.
Predicted amino acid sequence of the V3 -V6 region of the FIV envelope glycoprotein from revertant viruses (627W135 and 628W135) at 135 weeks postinfection. Lines under the sequence delineate the variable regions 3 to 6. Predicted sites for N-linked glycosylation (gray dots) and residues in the V3 loop affecting CXCR4 usage (↓) are marked. Sequences are shown relative to the parent molecular clone PETF14 and to the GL8414 molecular clone for comparison. Sequences were consistent between three independent clones.
Given that changes in the TM protein of FIV affect cell tropism (35), we compared the sequences of the TM protein of the 627W135 and 628W135 clones with that of the parent PETF14. The TM sequence of 628W135 Env showed no changes from the sequence of the parental virus, while the TM sequence of 627W135 showed two coding mutations distinct from those described previously to affect cell tropism. As these changes were not common to the 627W135 and 628W135 viruses, they are unlikely to be responsible for the reversion to a virulent phenotype. Similarly, of the changes observed in the region of SU between the L-SU cleavage site and V3, only a conservative change F208L, was observed in both 627W135 and 628W135. 628W135 displayed additional T154K (neutral hydrophilic to basic) and R252G (basic to amphiphilic) changes, while an additional F388L (conservative) substitution was detected in 627W135.
(iii) In vitro properties of the reverted viruses.
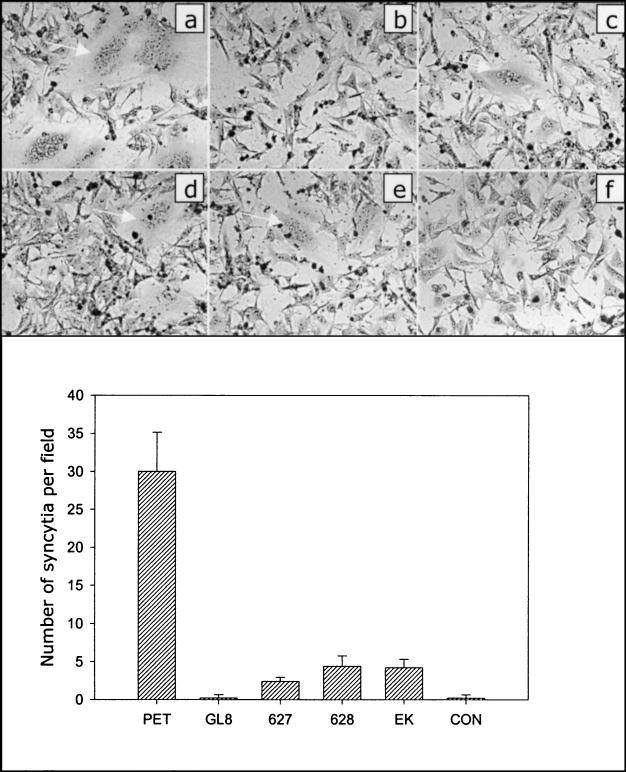
E407K and E409K substitutions in the V3 loop of FIV SU alone are sufficient to permit infection of CrFK cells with FIV, and CrFK-tropic viruses bind CXCR4 with a high affinity (31). Data presented to date suggest that infection of adherent cell lines such as CrFK requires an interaction between the viral envelope glycoprotein and CXCR4 (6, 11, 42) and heparan sulfate (6); indeed, ectopic expression of CXCR4 is sufficient to render cells permissive for syncytium formation and cell-free virus infection with CrFK-adapted strains of FIV (viruses carrying E to K mutations in the V3 loop) (36, 37, 42). In contrast, ectopic expression of feline CXCR4 or CCR5 does not render cells permissive to infection with primary strains of FIV such as GL8, leading to the proposal that the primary strains utilize an as-yet- unidentified, high-affinity binding receptor for infection of primary IL-2-dependent T cells (37). Recent compelling evidence revealed that recombinant FIV envelope glycoproteins bind distinct cell surface molecules on lymphoid cell lines compared with adherent cell lines, further implicating a high-affinity primary binding receptor for attachment to these cell lines (6). The PETF14 virus represents a CrFK-adapted strain of FIV, the viral envelope glycoprotein binds to CXCR4 with a high affinity, and expression of the envelope glycoprotein alone in CXCR4-expressing cells promotes syncytium formation (11, 37). We therefore assessed the in vitro properties of the PETF14 revertant viruses 627W135 and 628W135 in comparison with those of the parental PETF14 virus, the GL8414 virus, and a mutant GL8 envelope glycoprotein bearing an E407K mutation (GL8E407K). DNAs encoding the envelope glycoproteins were subcloned into a mammalian expression vector (VR1012) and were transfected directly into the feline cell line AH927 stably expressing feline CXCR4 (Fig. 6) or CCR5 (not shown). Marked syncytium formation was observed in the cultures transfected with the PETF14 env construct (Fig. 6a), the syncytia often being large (>20 nuclei per syncytium, 30 ± 2 per field). In contrast, the number of syncytia in the GL8414-transfected cultures (0.2 ± 0.2 per field) did not differ from those in the cultures transfected with the vector alone (0.2 ± 0.2 per field). The 627W135 and 628W135 env genes gave an intermediate phenotype, with fewer, smaller syncytia (5 to 10 nuclei per syncytium, 2.4 ± 1.5 per field and 4.4 ± 2.3 per field, respectively). The GL8E407K mutant gave a similar size and number of syncytia to 627W135 and 628W135. No syncytia were observed in AH927 cells stably expressing feline CCR5 following transfection with the envelope constructs (not shown). The data suggest that the observed reduction in net charge of the V3 loop of 627W135 and 628W135 resulted in an impaired ability to induce syncytia in CXCR4-expressing feline cells compared with that of the parental PETF14 strain. Further, despite having a significantly reduced ability to induce syncytium formation in CXCR4-expressing cells, the retention of a degree of syncytium-forming capacity by the 627W135 and 628W135 envelope glycoproteins suggests that other regions in the parental PETF14 envelope glycoprotein contribute to its highly fusogenic phenotype. Accordingly, while the E407K mutation rendered the GL8 envelope glycoprotein fusogenic, the extent of syncytium formation was markedly lower than that of the PETF14 envelope glycoprotein.
FIG. 6.
Reduced fusogenicity of envelope glycoproteins from 627W135 and 628W135 viruses. env genes from PETF14 (a), GL8414 (b), 627W135 (c), 628W135 (d), and GL8E407K (e) were subcloned into the vector VR1012 and were transfected into CXCR4-expressing AH927 cells. The empty vector VR1012 was transfected as a control (f). Forty-eight hours posttransfection the cells were fixed, stained, and examined by light microscopy. Panels a to f illustrate representative syncytia observed with each construct. The numbers of syncytia per field were enumerated; the data represent the mean number (n = 5) of syncytia per field ± standard error. CON, control.
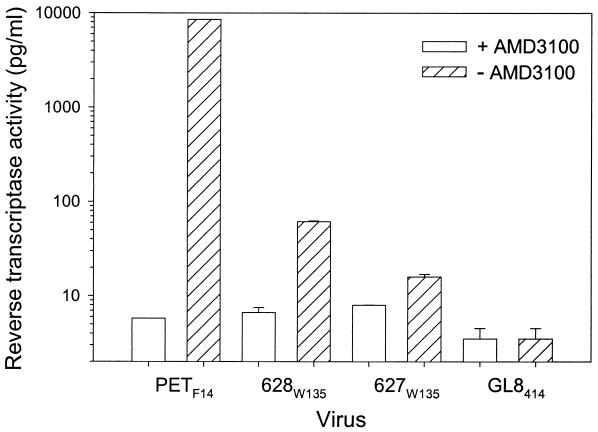
The ability to form syncytia in adherent cell lines appears to be dependent on the expression of CXCR4 (41, 42), and the binding of recombinant envelope glycoprotein involves an interaction with both CXCR4 and heparan sulfate (6). Therefore, we asked whether the reduction in the net charge of the V3 loop of the 627W135 and 628W135 viruses affected the growth of the viruses in vitro in established cell lines. The PETF14, GL8414, 627W135, and 628W135 viruses were used to infect AH927 cells stably expressing feline CXCR4 (AH927-FX4), since these cells are known to support the growth of viruses carrying an Env protein adapted for growth in cell culture (the use of CXCR4 as a sole receptor), while restricting the growth of viruses carrying primary Env proteins (our unpublished observations). In agreement with the studies on the fusogenicity of the viral Env proteins, AH927-FX4 cells supported the growth of PETF14 but not of GL8414. Growth of the 627W135 and 628W135 viruses was severely impaired but was detectable (Fig. 7). Furthermore, the CXCR4 antagonist AMD3100 inhibited the growth of the PETF14, 627W135, and 628W135 viruses (Fig. 7), thus confirming that, while infection was less efficient, it was CXCR4 dependent. Infection of AH927-FX4 with the PETF14, 627W135, and 628W135 viruses was sensitive to pretreatment of the cells with heparinase (data not shown), suggesting that heparan sulfate may play a role in infection of this cell line with these viruses. These findings are in accord with previous findings in which heparan sulfate was found to be required for binding of recombinant 34TF10 FIV SU to adherent cell lines (6). As all four viruses grew readily in IL-2-dependent T cells (data not shown), the data confirm that the mutations detected in the V3 loop of the 627W135 and 628W135 viruses lead to a partial restoration of the characteristics of a primary isolate (loss of the ability to replicate in CXCR4-expressing adherent cell lines). However, the reduced fusogenicity (Fig. 6) and growth (Fig. 7) of the 627W135 and 628W135 viruses suggests that the viruses harbor determinants outside the V3-to-V6 region of Env that are required for efficient replication in these cells.
FIG. 7.
Impaired growth of the 627W135 and 628W135 viruses in CXCR4-expressing feline cells. PETF14, GL8414, 627W135, and 628W135 viruses were used to infect AH927 cells that had been engineered to overexpress feline CXCR4. Infection was performed in the presence (open bars) or absence (hatched bars) of the CXCR4 antagonist AMD3100. Supernatants were collected at 9 days postinfection and were assayed for RT activity. Results represent the mean of duplicate samples and are expressed as picograms per milliliter of RT.
(iv) Virulence of the 627W135 and 628W135 viruses in vivo.
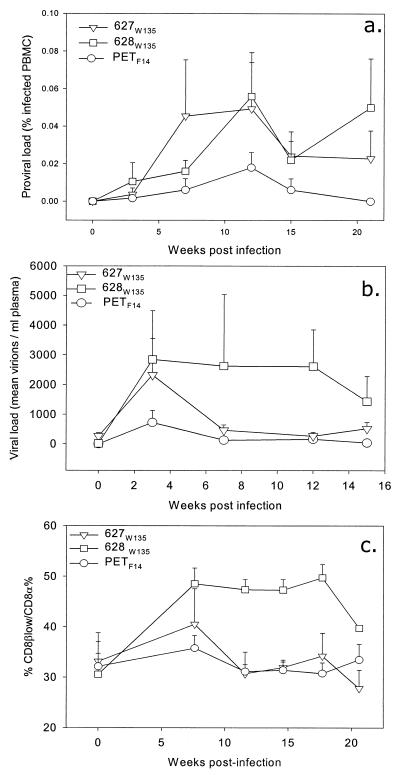
The data presented above suggest that the 627W135 and the 628W135 viruses have reverted to a more pathogenic phenotype than the parental PETF14 molecular clone. In order to test this hypothesis, we infected three groups of cats with matched doses of either the PETF14, 627W135, or 628W135 viruses. Our previous data had indicated that the major differences between the GL8414 and PETF14 viruses were (i) viral load and (ii) expansion of CD8+-lymphocyte subpopulations. GL8414 achieved a high viral load and triggered an expansion of CD8+ lymphocytes expressing reduced CD8β (CD8βlow), while PETF14 achieved a lower viral load and did not affect CD8+-lymphocyte subpopulations. Following infection with the 628W135 virus, marked increases in both proviral load (Fig. 8a) and plasma viral load (Fig. 8b) were detected as early as 3 weeks postinfection. These increased proviral and viral loads were mirrored by an expansion of lymphocytes expressing CD8α+βlow (Fig. 8c). In contrast, while infection with the 627W135 virus achieved a similar initial plasma viremia at 3 weeks postinfection, by week 6 the levels of virus in plasma had dropped markedly. Flow cytometry analysis of peripheral blood lymphocytes from these cats revealed a transient increase in lymphocytes expressing CD8α+βlow at 7 weeks postinfection, returning to control levels by 12 weeks postinfection. Infection with PETF14 resulted in a transient plasma viremia at 3 weeks postinfection, returning to baseline levels by 7 weeks postinfection. While a small increase in lymphocytes expressing CD8α+βlow was observed in the PETF14-infected cats at 7 weeks postinfection, the increase was markedly reduced compared to the number of lymphocytes in the cats infected with 627W135 and in particular with 628W135. Thus, infection of naïve cats with the 627W135 and 628W135 viruses reproduced the alterations observed in the immune systems of cats 627 and 628 with the emergence of the mutant viruses. Further, the infection experiments suggest a correlation between viral load and the expansion of lymphocytes expressing CD8α+βlow. The data confirm that the 627W135 and 628W135 viruses are more pathogenic than the parental PETF14 virus.
FIG. 8.
Quantification of proviral load (a), viral load (b), and CD8αβlow (c) in FIV-infected cats during the acute phase of infection. Three cats were inoculated intraperitoneally with either PETF14 (○), 627W135 (▿), or 628W135 (□). (a) Proviral DNA load in PBMC was estimated by real-time PCR. Each point represents the mean % infected PBMCs of the three cats in the group (+ standard error). (b) Viral load in plasma was estimated by real-time PCR. Each point represents the mean number of virions per milliliter of plasma from the three cats in the group (+ standard error). (c) Expansion of CD8α+βlow-lymphocyte subpopulations following infection with the three isolates, expressed as the ratio of CD8α+βlow cells to CD8α cells. Each point represents the mean ratio of CD8α+βlow to CD8α cells for the three cats in each group (+ standard error).
DISCUSSION
In this study we compared the pathogenicity of two strains of FIV used routinely as challenge strains for vaccine studies. As the viruses were derived from molecular clones, they represent a reproducible source of virus for interstudy comparison. While it is possible to induce protective immunity to infection with the PET isolate (10, 43), the GL8414 strain represents a more stringent challenge. To date, protective immunity has not been observed against the GL8 isolate of FIV, although a reduction in viral load has been observed following immunization with a whole-inactivated-virus vaccine (12). The data presented here suggest that the GL8 strain of FIV is intrinsically more pathogenic than the PET strain of FIV, a strain of virus that has been suggested to be of low pathogenicity (5, 12). Using minimally passaged virus derived from the GL8414 molecular clone of FIV, we have established that GL8 achieves a significantly greater viral load in infected cats than PET (approximately 100-fold higher). Further, the GL8414-derived virus reproduces the sharp drop in the CD4/CD8 ratio described previously for the biological isolate of GL8 (39) and representative of pathogenic strains of FIV(1, 22, 44). Having established that the PETF14-derived virus was less virulent than the GL8414-derived virus, we monitored the two groups of infected cats and noted the emergence of pathogenic variants of PETF14 in two of the infected cats 135 weeks after infection (627W135 and 628 W135). The Env sequences of these viruses displayed characteristic changes in the V3 loop residues that are known to govern the ability of FIV to infect CXCR4-expressing adherent cell lines. Subsequently, we showed that the mutant viruses had a greatly reduced ability to trigger syncytium formation, were less able to establish a productive infection in adherent cells expressing CXCR4, and were more pathogenic than the parental PETF14 virus when inoculated in vivo.
The reversion from an avirulent (PETF14) to a virulent phenotype (627W135 and 628W135) at approximately 2 years postinfection is of particular significance, given that “attenuated” laboratory strains of FIV have been suggested as potential vaccines for protection against virulent strains of the virus. Many laboratory strains of FIV display an increase in the net charge of the V3 loop (e.g., PETF14 [23], PET34TF10 [33], UT113 [36], AM6 [31], and GL8CrFK [37]), analogous to results for CD4-independent strains of HIV. The increase in charge of the V3 loop correlates with enhanced replication in CXCR4-expressing feline cell lines, such as CrFK (31, 36). It is thought that such mutations enable the virus to infect cells in the absence of an as-yet-unidentified primary receptor (37, 38), perhaps by facilitating a direct interaction with CXCR4. Alternatively, it is possible that the decreased net charge of the V3 loop in 627W135 and 628W135 might result in a decreased ability of either of these viruses to adsorb efficiently to the cell surface via heparan sulfate (6). Indeed, it is conceivable that the limited replication of PETF14 in vivo may result from the virus being trapped on endothelial cells and other tissues expressing large amounts of heparan sulfate on their surface.
Using the PETF14 as a typical CXCR4-tropic strain of FIV, we show here that, while this phenotype is positively selected in vitro, it is not stable in vivo. Further, given that PETF14 neither established a high viral load nor induced the expansion of activated CD8+-T-lymphocyte subpopulations and that the emergence of the variants 627W135 and 628W135 coincided with the expansion of CD8+-lymphocyte populations and an increased frequency of successful virus isolations, the data suggest that the V3 loop mutation associated with high-affinity CXCR4 usage in vitro leads to attenuation in vivo.
The sequence changes that were detected in the V3 loop of the envelope glycoprotein may have arisen as a result of selection from virus-neutralizing antibodies, since this region represents a major neutralizing determinant for FIV. It is possible that the PETF14 is more readily neutralizable than the 627W135 and 628W135 variants, thus accounting for the lower viral load in PETF14-infected cats. In previous studies, escape from neutralizing antibodies was correlated with mutations in the V4 or V5 region of the virus, the emergence of neutralization-resistant strains having been examined both in vitro and in vivo (3, 4, 30, 32). By comparison, however, we did not observe consistent changes in this region of the 627W135 and 628W135 variants. Neither reisolate contained mutations in the V4 region, and, while 628W135 had acquired a mutation that would give rise to a predicted N-linked glycosylation site (S557N), this was not present in the 627W135 reisolate. While the additional glycosylation site in V5 of 628W135 may confer a replicative advantage to the virus by conferring resistance to neutralizing antibodies, this alone was clearly not the main determinant of virulence. In contrast, the K409Q and K409E mutations in the 627W135 and 628W135 variants are consistent with previous in vivo-selected variants based on the PET34TF10 clone of FIV (3, 4). The PET34TF10 clone of FIV has lysine residues at positions 407 and 409 in the V3 loop. However, following passage in vivo, the reisolated virus showed K407E and K409G mutations (4).
The emergence of the 627W135 and 628W135 variants in the PETF14-infected cats correlated well with the appearance of activated CD8+-lymphocyte subpopulations, characterized by reduced expression of CD8β. One of the first indications that more virulent viruses had evolved in cats 627 and 628 was an increased frequency with which virus could be isolated from bulk PBMC cultures. As real-time PCR for proviral DNA load in PBMC during this period remained consistently negative, the data suggest that, although the viral load had increased in the cats, the levels of virus were still very low, below the assay sensitivity of 10−5% infected PBMCs. In contrast, the proviral loads in the GL8414-infected cats remained high throughout the study. If the variant viruses in cats 627 and 628 were more virulent than the parental PETF14 virus, we would have expected an increased proviral load following the emergence of the variant viruses. The finding that the proviral load remained below the sensitivity of the real-time PCR assay may indicate suppression of viral replication by the host immune response. We addressed this possibility by challenging three groups of naïve cats with either the variant 627W135 and 628W135 or the parental virus PETF14. Following challenge, the variant viruses achieved higher viral loads than the parental strain, indicating that, in the absence of a preexisting immune response, the viruses were more virulent than the parental strain. Further, infection with the 627W135 and 628W135 viruses induced a concomitant expansion of CD8α+βlow lymphocytes.
Further issues to address in the FIV system are the relationship between determinants of virulence in vitro and pathogenesis in vivo. This study focused on a comparison of two isolates of FIV that have been used extensively as challenge viruses in vaccine studies. It is not known whether viruses isolated from cats infected in the field are predominantly virulent and pathogenic, similar to GL8414, or whether a spectrum of isolates with various degrees of virulence and pathogenicity exists. It is possible that the virulence characteristics of field isolates depend on the stage of infection. Proviral loads measured in field cats by real-time PCR were comparable to those of cats infected with the GL8414 isolate (D. Klein, unpublished data) rather than the low loads detected in cats infected with the PETF14 isolate. In future studies, it will be important to examine the virulence characteristics and pathogenicity of field isolates to establish whether particular virulence determinants exist that characterize the behavior of FIV isolates in vivo. This will enable the selection of vaccine challenge strains that are most representative of field isolates and thus will impact upon future lentiviral vaccine strategies.
Acknowledgments
This work was supported by The Wellcome Trust.
We are grateful to G. Law, D. Graham, and P. McGowan, University of Glasgow, and Elzbieta Knapp, University of Veterinary Sciences, Vienna, Austria, for technical assistance and to P. Johnson, National Institute of Allergy and Infectious Diseases, National Institutes of Health, for kindly providing the F14 molecular clone.
REFERENCES
- 1.Ackley, C. D., J. K. Yamamoto, N. Levy, N. C. Pedersen, and M. D. Cooper. 1990. Immunologic abnormalities in pathogen-free cats experimentally infected with feline immunodeficiency virus. J. Virol. 64:5652-5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann, M. H., C. Mathiason-Dubard, G. H. Learn, A. G. Rodrigo, D. L. Sodora, P. Mazzetti, E. A. Hoover, and J. I. Mullins. 1997. Genetic diversity of feline immunodeficiency virus: dual infection, recombination, and distinct evolutionary rates among envelope sequence clades. J. Virol. 71:4241-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendinelli, M., M. Pistello, D. Del Mauro, G. Cammarota, F. Maggi, A. Leonildi, S. Giannecchini, C. Bergamini, and D. Matteucci. 2001. During readaptation in vivo, a tissue culture-adapted strain of feline immunodeficiency virus reverts to broad neutralization resistance at different times in individual hosts but through changes at the same position of the surface glycoprotein. J. Virol. 75:4584-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cammarota, G., D. Matteucci, M. Pistello, E. Nicoletti, S. Giannecchini, and M. Bendinelli. 1996. Reduced sensitivity to strain-specific neutralization of laboratory-adapted feline immunodeficiency virus after one passage in vivo: association with amino acid substitutions in the V4 region of the surface glycoprotein. AIDS Res. Hum. Retrovir. 12:173-175. [DOI] [PubMed] [Google Scholar]
- 5.Dean, G. A., S. Himathongkham, and E. E. Sparger. 1999. Differential cell tropism of feline immunodeficiency virus molecular clones in vivo. J. Virol. 73:2596-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Parseval, A., and J. H. Elder. 2001. Binding of recombinant feline immunodeficiency virus surface glycoprotein to feline cells: role of CXCR4, cell-surface heparans, and an unidentified non-CXCR4 receptor. J. Virol. 75:4528-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egberink, H. F., E. De Clerq, A. L. Van Vliet, J. Balzarini, G. J. Bridger, G. Henson, M. C. Horzinek, and D. Schols. 1999. Bicyclams, selective antagonists of the human chemokine receptor CXCR4, potently inhibit feline immunodeficiency virus replication. J. Virol. 73:6346-6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Endres, M. J., P. R. Clapham, M. Marsh, M. Ahuja, J. D. Turner, A. McKnight, J. F. Thomas, B. Stoebenau-Haggarty, S. Choe, P. J. Vance, T. N. C. Wells, C. A. Power, S. S. Sutterwala, R. W. Doms, N. R. Landau, and J. A. Hoxie. 1996. CD4-independent infection by HIV-2 is mediated by fusin. Cell 87:745-756. [DOI] [PubMed] [Google Scholar]
- 9.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 10.Hosie, M., R. Osborne, J. K. Yamamoto, J. C. Neil, and O. Jarrett. 1995. Protection against homologous but not heterologous challenge induced by inactivated feline immunodeficiency virus vaccines. J. Virol. 69:1253-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosie, M. J., N. Broere, J. Hesselgesser, J. D. Turner, J. A. Hoxie, J. C. Neil, and B. J. Willett. 1998. Modulation of feline immunodeficiency virus infection by stromal cell-derived factor (SDF-1). J. Virol. 72:2097-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosie, M. J., T. Dunsford, D. Klein, B. J. Willett, C. Cannon, R. Osborne, J. Macdonald, N. Spibey, N. Mackay, O. Jarrett, and J. C. Neil. 2000. Vaccination with inactivated virus but not viral DNA reduces virus load following challenge with a heterologous and virulent isolate of feline immunodeficiency virus. J. Virol. 74:9403-9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosie, M. J., J. N. Flynn, M. A. Rigby, C. Cannon, T. H. Dunsford, N. A. Mackay, D. Argyle, B. J. Willett, T. Miyazawa, D. E. Onions, O. Jarrett, and J. C. Neil. 1998. DNA vaccination affords significant protection against feline immunodeficiency virus infection without inducing detectable antiviral antibodies. J. Virol. 72:7310-7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosie, M. J., R. Osborne, G. Reid, J. C. Neil, and O. Jarrett. 1992. Enhancement after feline immunodeficiency virus vaccination. Vet. Immunol. Immunopathol. 35:191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosie, M. J., C. Robertson, and O. Jarrett. 1989. Prevalence of feline leukaemia virus and antibodies to feline immunodeficiency virus in cats in the United Kingdom. Vet. Rec. 128:293-297. [DOI] [PubMed] [Google Scholar]
- 16.Klein, D., B. Bugl, W. H. Gunzburg, and B. Salmons. 2000. Accurate estimation of transduction efficiency necessitates a multiplex real-time PCR. Gene Ther. 7:458-463. [DOI] [PubMed] [Google Scholar]
- 17.Klein, D., P. Janda, R. Steinborn, M. Muller, B. Salmons, and W. H. Gunzburg. 1999. Proviral load determination of different feline immunodeficiency virus isolates using real-time polymerase chain reaction: influence of mismatches on quantification. Electrophoresis 20:291-299. [DOI] [PubMed] [Google Scholar]
- 18.Klein, D., C. M. Leutenegger, C. Bahula, P. Gold, R. Hofmann-Lehmann, B. Salmons, H. Lutz, and W. H. Guenzburg. 2001. Influence of preassay and sequence variations on viral load determination by a multiplex real-time reverse transcriptase-polymerase chain reaction for feline immunodeficiency virus. J. Acquir. Immune Defic. Syndr. 26:8-20. [DOI] [PubMed] [Google Scholar]
- 19.Matteucci, D., M. Pistello, P. Mazzetti, S. Giannecchini, D. Del Mauro, L. Zaccaro, P. Bandecchi, F. Tozzini, and M. Bendinelli. 1996. Vaccination protects against in vivo-grown feline immunodeficiency virus even in the absence of detectable neutralizing antibodies. J. Virol. 70:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matteucci, D., A. Poli, P. Mazzetti, S. Sozzi, F. Bonci, P. Isola, L. Zaccaro, S. Giannecchini, M. Calandrella, M. Pistello, S. Specter, and M. Bendinelli. 2000. Immunogenicity of an anti-clade B feline immunodeficiency fixed-cell virus vaccine in field cats. J. Virol. 74:10911-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyazawa, T. M., T. Furuya, S. Itagaki, Y. Tohya, E. Takahashi, and T. Mikami. 1989. Establishment of a feline T-lymphoblastoid cell line highly sensitive for replication of feline immunodeficiency virus. Arch. Virol. 108:131-135. [DOI] [PubMed] [Google Scholar]
- 22.Novotney, C., R. V. English, J. Housman, M. G. Davidson, M. P. Nasisse, C. R. Jeng, W. C. Davis, and M. B. Tompkins. 1990. Lymphocyte population changes in cats naturally infected with feline immunodeficiency virus. AIDS 4:1213-1218. [DOI] [PubMed] [Google Scholar]
- 23.Olmsted, R. A., A. K. Barnes, J. K. Yamamoto, V. M. Hirsch, R. H. Purcell, and P. R. Johnson. 1989. Molecular cloning of feline immunodeficiency virus. Proc. Natl. Acad. Sci. USA 86:2448-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen, N. C., E. W. Ho, M. L. Brown, and J. K. Yamamoto. 1987. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency syndrome. Science 235:790-793. [DOI] [PubMed] [Google Scholar]
- 25.Pistello, M., G. Cammarota, E. Nicoletti, D. Matteucci, M. Curcio, D. Del Mauro, and M. Bendinelli. 1997. Analysis of the genetic diversity and phylogenetic relationship of Italian isolates of feline immunodeficiency virus indicates a high prevalence and heterogeneity of subtype B. J. Gen. Virol. 78:2247-2257. [DOI] [PubMed] [Google Scholar]
- 26.Richardson, J., G. Pancino, T. Leste-Lasserre, J. Schneider-Mergener, M. Alizon, P. Sonigo, and N. Heveker. 1999. Shared usage of the chemokine receptor CXCR4 by primary and laboratory-adapted strains of feline immunodeficiency virus. J. Virol. 73:3661-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitz, J. E., M. A. Forman, M. A. Lifton, O. Concepcion, K. A. Reimann, Jr., C. S. Crumpacker, J. F. Daley, R. S. Gelman, and N. L. Letvin. 1998. Expression of the CD8alpha beta-heterodimer on CD8(+) T lymphocytes in peripheral blood lymphocytes of human immunodeficiency virus− and human immunodeficiency virus+ individuals. Blood 92:198-206. [PubMed] [Google Scholar]
- 28.Shimojima, M., T. Miyazawa, M. Kohmoto, Y. Ikeda, Y. Nishimura, K. Maeda, Y. Tohya, and T. Mikami. 1998. Expansion of CD8alpha+beta- cells in cats infected with feline immunodeficiency virus. J. Gen. Virol. 79:91-94. [DOI] [PubMed] [Google Scholar]
- 29.Shimojima, M., M. R. Pecoraro, K. Maeda, Y. Tohya, T. Miyazawa, and T. Mikami. 1998. Characterization of anti-feline CD8 monoclonal antibodies. Vet. Immunol. Immunopathol. 61:17-23. [DOI] [PubMed] [Google Scholar]
- 30.Siebelink, K. H. J., W. Huisman, J. A. Karlas, G. F. Rimmelzwaan, M. L. Bosch, and A. D. M. E. Osterhaus. 1996. Neutralization of feline immunodeficiency virus by polyclonal feline antibody: simultaneous involvement of hypervariable regions 4 and 5 of the surface glycoprotein. J. Virol. 69:5124-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siebelink, K. H. J., J. A. Karlas, G. F. Rimmelzwaan, A. D. M. E. Osterhaus, and M. L. Bosch. 1995. A determinant of feline immunodeficiency virus involved in CrFK tropism. Vet. Immunol. Immunopathol. 46:61-69. [DOI] [PubMed] [Google Scholar]
- 32.Siebelink, K. H. J., G. F. Rimmelzwaan, M. L. Bosch, R. H. Meloen, and A. D. M. E. Osterhaus. 1993. A single amino acid substitution in hypervariable region 5 of the envelope protein of feline immunodeficiency virus allows escape from virus neutralization. J. Virol. 67:2202-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talbott, R. L., E. E. Sparger, K. M. Lovelace, W. M. Fitch, N. C. Pedersen, P. A. Luciw, and J. H. Elder. 1989. Nucleotide-sequence and genomic organization of feline immunodeficiency virus. Proc. Natl. Acad. Sci. USA 86:5743-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torten, M., M. Franchini, J. E. Barlough, J. W. George, E. Mozes, H. Lutz, and N. C. Pedersen. 1991. Progressive immune dysfunction in cats experimentally infected with feline immunodeficiency virus. J. Virol. 65:2225-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vahlenkamp, T. W., E. J. Verschoor, N. N. M. P. Schuurman, A.L.W. van Vliet, M. C. Horzinek, H. F. Egberink, and A. de Ronde. 1997. A single amino acid substitution in the transmembrane envelope glycoprotein of feline immunodeficiency virus alters cellular tropism. J. Virol. 71:7132-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verschoor, E. J., L. A. Boven, H. Blaak, A.R.W. van Vliet, M. C. Horzinek, and A. de Ronde. 1995. A single mutation within the V3 envelope neutralization domain of feline immunodeficiency virus determines its tropism for CRFK cells. J. Virol. 69:4752-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willett, B. J., K. Adema, N. Heveker, A. Brelot, L. Picard, M. Alizon, J. D. Turner, J. A. Hoxie, S. Peiper, J. C. Neil, and M. J. Hosie. 1998. The second extracellular loop of CXCR4 determines its function as a receptor for feline immunodeficiency virus. J. Virol. 72:6475-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willett, B. J., and M. J. Hosie. 1999. The role of the chemokine receptor CXCR4 in infection with feline immunodeficiency virus. Mol. Membr. Biol. 16:67-72. [DOI] [PubMed] [Google Scholar]
- 39.Willett, B. J., M. J. Hosie, J. J. Callanan, J. C. Neil, and O. Jarrett. 1993. Infection with feline immunodeficiency virus is followed by the rapid expansion of a CD8+ lymphocyte subset. Immunology 78:1-6. [PMC free article] [PubMed] [Google Scholar]
- 40.Willett, B. J., M. J. Hosie, T. H. Dunsford, J. C. Neil, and O. Jarrett. 1991. Productive infection of T-helper lymphocytes with feline immunodeficiency virus is accompanied by reduced expression of CD4. AIDS 5:1469-1475. [DOI] [PubMed] [Google Scholar]
- 41.Willett, B. J., M. J. Hosie, J. C. Neil, J. D. Turner, and J. A. Hoxie. 1997. Common mechanism of infection by lentiviruses. Nature 385:587.. [DOI] [PubMed] [Google Scholar]
- 42.Willett, B. J., L. Picard, M. J. Hosie, J. D. Turner, K. Adema, and P. R. Clapham. 1997. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J. Virol. 71:6407-6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto, J. K., T. Okuda, C. D. Ackley, H. Louie, E. Pembroke, H. Zochlinski, R. J. Munn, and M. B. Gardner. 1991. Experimental vaccine protection against feline immunodeficiency virus. AIDS Res. Hum. Retrovir. 7:911-921. [DOI] [PubMed] [Google Scholar]
- 44.Yang, J. S., R. V. English, J. W. Ritchey, M. G. Davidson, T. Wasmoen, J. K. Levy, D. H. Gebhard, M. B. Tompkins, and W. A. Tompkins. 1996. Molecularly cloned feline immunodeficiency virus NCSU1 JSY3 induces immunodeficiency in specific-pathogen-free cats. J. Virol. 70:3011-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]