Abstract
Sheeppox virus (SPPV) and goatpox virus (GTPV), members of the Capripoxvirus genus of the Poxviridae, are etiologic agents of important diseases of sheep and goats in northern and central Africa, southwest and central Asia, and the Indian subcontinent. Here we report the genomic sequence and comparative analysis of five SPPV and GTPV isolates, including three pathogenic field isolates and two attenuated vaccine viruses. SPPV and GTPV genomes are approximately 150 kbp and are strikingly similar to each other, exhibiting 96% nucleotide identity over their entire length. Wild-type genomes share at least 147 putative genes, including conserved poxvirus replicative and structural genes and genes likely involved in virulence and host range. SPPV and GTPV genomes are very similar to that of lumpy skin disease virus (LSDV), sharing 97% nucleotide identity. All SPPV and GTPV genes are present in LSDV. Notably in both SPPV and GTPV genomes, nine LSDV genes with likely virulence and host range functions are disrupted, including a gene unique to LSDV (LSDV132) and genes similar to those coding for interleukin-1 receptor, myxoma virus M003.2 and M004.1 genes (two copies each), and vaccinia virus F11L, N2L, and K7L genes. The absence of these genes in SPPV and GTPV suggests a significant role for them in the bovine host range. SPPV and GTPV genomes contain specific nucleotide differences, suggesting they are phylogenetically distinct. Relatively few genomic changes in SPPV and GTPV vaccine viruses account for viral attenuation, because they contain 71 and 7 genomic changes compared to their respective field strains. Notable genetic changes include mutation or disruption of genes with predicted functions involving virulence and host range, including two ankyrin repeat proteins in SPPV and three kelch-like proteins in GTPV. These comparative genomic data indicate the close genetic relationship among capripoxviruses, and they suggest that SPPV and GTPV are distinct and likely derived from an LSDV-like ancestor.
Capripoxviruses (CaPVs) represent one of eight genera within the chordopoxvirus (ChPV) subfamily of the Poxviridae. The Capripoxvirus genus is currently comprised of sheeppox virus (SPPV), goatpox virus (GTPV), and lumpy skin disease virus (LSDV), causing disease in sheep, goats, or cattle, respectively. These viruses are responsible for some of the most economically significant diseases of domestic ruminants in Africa and Asia (13, 19).
Sheeppox and goatpox are endemic throughout southwest and central Asia, the Indian subcontinent, and northern and central Africa (13, 19). In contrast, LSDV occurs largely in central and southern Africa and is absent in Asia. Sheeppox and goatpox exhibit similar clinical signs that are typical of generalized poxviral diseases, including pyrexia, cutaneous lesions, and notably the development of lung lesions (19, 45). Transmission of sheeppox and goatpox is efficient and suspected to occur via aerosol and insect vector (19, 36, 38).
CaPVs are generally considered to be host specific, because disease outbreaks or virus isolates may preferentially occur or cause disease in one host species (19, 45). This has been shown specifically for Nigerian, Middle Eastern, and Indian strains of SPPV and GTPV and for LSDV (32, 34, 37, 46, 50, 51). However, the ability of SPPV and GTPV strains to naturally or experimentally cross-infect and cause disease in both host species has been described previously (16, 35, 37). This apparent variability in SPPV and GTPV host range, the clinical similarity between sheeppox and goatpox, and the inability to differentiate the two diseases by serological assays have led to the suggestion that sheeppox and goatpox are part of a disease complex caused by a single viral species and that observable host range specificities are the result of regional virus adaptations to sheep or goat hosts (17, 37).
Restriction endonuclease analysis and cross-hybridization studies of SPPV and GTPV indicate that these viruses, although closely related (estimated 96 to 97% nucleotide identity), can be distinguished from one another and may undergo recombination in nature (8, 25-27, 32). These data and limited SPPV and GTPV DNA sequence analysis also indicate a high degree of similarity to LSDV, whose genome sequence contains a conserved ChPV-like complement of replicative genes and a unique complement of virulence and host range genes (11, 23, 25, 26, 29, 59).
Live attenuated SPPV and subunit formulations have been used experimentally and in enzootic and outbreak areas as vaccines against sheeppox, goatpox, and lumpy skin disease (12-14, 33, 47). However, vaccine-induced disease, vaccine failure, and restrictions on the use of live virus vaccines in nonenzootic areas create the need for improved CaPV vaccines (13, 51, 61). An improved understanding of the genetic basis of CaPV virulence and host range will permit rational design of vaccines having greater efficacy and versatility.
Given the economic significance of CaPVs, their potential for spread into nonenzootic regions, and interest in developing more effective CaPV-based vaccines and expression vectors, we have sequenced and analyzed the genomes of three SPPVs and two GTPVs. These data, combined with LSDV sequence data, provide a comparative view of CaPV genomics and describe the genetic basis for CaPV virulence and host range.
MATERIALS AND METHODS
Virus strains.
The following sequenced viruses were used: SPPV strain TU, isolated during an outbreak of sheeppox in Turkey in the late 1970s, passaged six times in lamb testicle (LT) cells and once in sheep coroid plexus cells, and subsequently reisolated in 2000 from lung lesions of an experimentally infected sheep (Plum Island Animal Disease Center, Animal and Plant Health Inspection Service, U.S. Department of Agriculture, Greenport, N.Y.); SPPV strain A (SA), a field isolate obtained from a sick sheep in the Almatinskaya region, Kazakhstan, and passaged nine times in sheep at the Scientific Research Agricultural Institute (SRAI), Kazakhstan (1987); SPPV strain Niskhi (NK), an attenuated vaccine strain derived from an epizootic strain through 30 passages in lamb kidney (LK) cell cultures at SRAI (1994); GTPV strain Pellor (PL), a pathogenic field isolate passaged three times in LK at SRAI (1996); and GTPV strain G20-LKV (GV), a vaccine strain derived from a field isolate of low pathogenicity through 20 passages in LK at SRAI (2000).
Viral DNA isolation, cloning, sequencing, and sequence analysis.
Viral genomic DNA was extracted from primary LT cells. Random DNA fragments were obtained by incomplete enzymatic digestion with Tsp509I endonuclease (New England Biolabs, Beverly, Mass.), and DNA fragments larger than 1.0 kbp were cloned and used in dideoxy sequencing reactions as previously described (3). Reaction products were analyzed on an ABI PRISM 3700 automated DNA sequencer (Applied Biosystems, Foster City, Calif.). Sequence data were assembled with the Phrap and CAP3 software programs (20, 30), and gaps were closed as described previously (2). The final DNA consensus sequences for each genome represented on average seven to ninefold redundancy at each base position and a Consed estimated error rate of 0.01 to 0.03 per 100 kbp (20, 21, 28).
Genome DNA composition, structure, repeats and restriction enzyme patterns were analyzed as previously described (2) with the Genetics Computer Group GCG v.10 software package (18). Pairwise genomic alignments were done with WABA (Jim Kent; http://www.cse.ucsc.edu/∼kent/) and multiple genomic alignments were done with Dialign (42) and Clustal (58) alignment programs. Open reading frames (ORFs) longer than 30 codons were evaluated for coding potential as previously described (3). All ORFs with coding potential and ORFs greater than 60 codons were subjected to homology searches as previously described (2, 3). Based on these criteria, at least 147 ORFs were annotated as potential genes and numbered to coincide with orthologous ORFs from LSDV (59). Phylogenetic comparisons were done with the PHYLO_WIN software package (22).
Nucleotide sequence accession number.
The genome sequences of SPPV strains TU, SA, and NK, and GTPV strains PL and GV have been deposited in GenBank under accession no. AY077832, AY077833, AY077834, AY077835, and AY077836, respectively.
RESULTS AND DISCUSSION
SPPV and GTPV genomes.
Genome sequences of SPPV field isolate strains TU and SA, SPPV vaccine strain NK, GTPV field isolate PL, and GTPV vaccine strain GV were assembled into contiguous sequences of 149,955, 150,057, 149,662, 149,935, and 149,695 bp, respectively. This agrees with previous restriction enzyme-based size estimates for SPPV and GTPV (SGPV) genomes of approximately 143 to 147 kbp (25). Because poxvirus hairpin loop sequences known to be present at CaPV termini were not sequenced, the leftmost nucleotide of each assembled genome was arbitrarily designated base 1 (8, 43). Nucleotide composition is approximately 75% A+T uniformly distributed for all genomes.
SPPV and GTPV genomes, like those of other poxviruses, contain a central coding region bounded by two identical inverted terminal repeat (ITR) regions. Assembled ITRs of SPPV TU, SA, NK, and GTPV PL and GV contain at least 2,213, 2,349, 2,127, 2,305, and 2,198 bp, respectively, similar to those previously estimated for SPPV (25). Sequences similar to noncoding tandem repeats previously described in SPPV and LSDV terminal regions were identified in SPPV TU, SA, NK, and GTPV PL and GV ITRs (24, 59). Available data from terminal genomic regions indicate that CaPVs differ in the nature and size of tandemly repeated sequence. Notably, all SPPV isolates contain 46 bp of the ORFs closest to the termini (ORFs 001 and 156) in their tandem repeats, compared to LSDV, which contains 5 bp of ORFs 001 and 156 in its tandem repeats (24, 59). Although ORFs 001 and 156 were completely sequenced in each GTPV strain, no terminal tandem repetition was noted. Comparison with experimental and published restriction fragment analysis indicates that additional terminal repeats and hairpin loop sequences of less than 200 bp may be present in each genome (8, 25, 32) (data not shown).
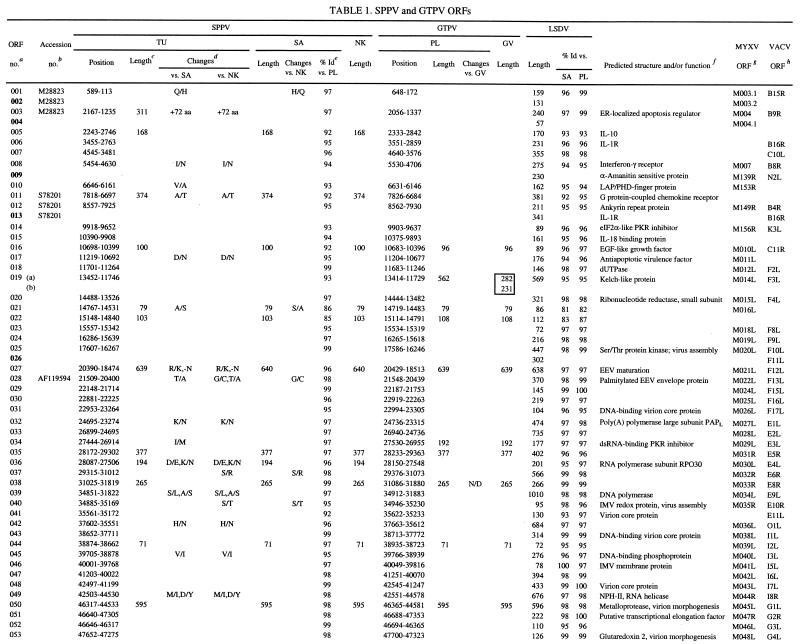
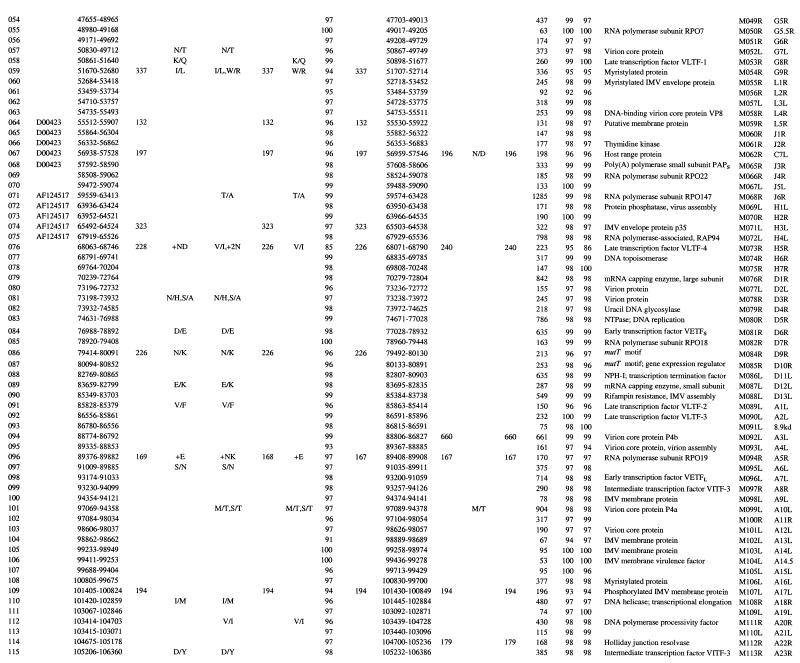
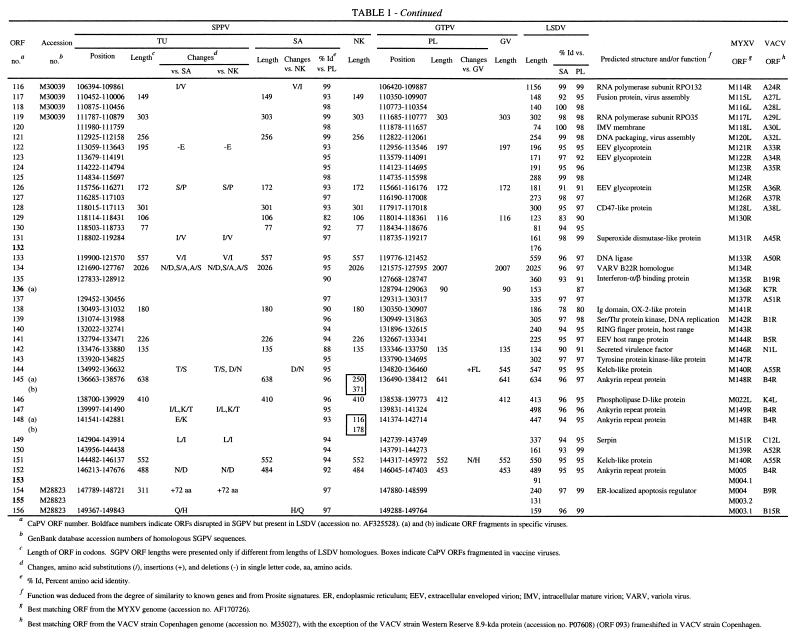
SGPV field isolates contain 147 ORFs, which have been annotated here as putative genes and as orthologues of LSDV genes (Table 1). These genes represent an approximate 93% coding density and encode proteins of 53 to 2,027 amino acids (Table 1). The central genomic region of SGPV (ORFs 024 to 123) contains homologues of conserved poxvirus genes involved in basic replicative mechanisms, including viral transcription and RNA modification, viral DNA replication, and structure and assembly of intracellular mature and extracellular enveloped virions (43) (Table 1). Terminal genomic regions of SGPV (ORFs 001 to 0023 and ORFs 124 to 156) contain genes with putative virulence and host range functions similar to those found in LSDV (59). These include gene families (five genes with ankyrin repeat motifs and three genes with kelch-like repeat motifs) and other genes likely involved in viral modification or evasion of host cellular, apoptotic, and immune responses or processes, including homologues of cytokine binding proteins, interleukin-10 (IL-10), an epidermal growth factor (EGF)-like protein, PKR inhibitors, a serpin, and poxvirus-specific virulence and host range genes (Table 1) (5, 43).
TABLE 1.
SPPV and GTPV ORFs
CaPV ORF number. Boldface numbers indicate ORFs disrupted in SGPV but prsent in LSDV (accession no. AF325528). (a) and (b) indicate ORF fragments in specific viruses.
GenBank database accession numbers of homologous SGPV sequences.
Length of ORF in codons. SGPV ORF lengths were presented only if different from lengths of LSDV homologous. Boxes indicate CaPV ORFs fragmented in vaccine viruses.
Changes, amino acid substitutions (/), insertions (+), and deletions (−) in single letter code, aa, amino acids.
% Id, Percent amino acid identity.
Function was deduced from the degree of similarity to known genes and from Prosite signatures. ER, endoplasmic reticulum; EEV, extracellular enveloped virion; IMV, intracellular mature virion; VARV, variola virus.
Best matching ORF from the MYXF genome (accession no. AF170726).
Comparison of SPPV and GTPV.
SPPV and GTPV are highly similar to each other at the genomic level, sharing colinearity (147 orthologous genes) and average nucleotide identity of 96% over the length of their genomes (Tables 1 and 2). Intraspecies nucleotide identity was greater than 99%, because SPPV TU and SA contained only 192 genomic changes, including 131 single-nucleotide substitutions (Table 2). Analysis of entire genome sequences suggests that SPPV and GTPV, although highly similar, are phylogenetically distinct (Fig. 1). This phylogenetic grouping of CaPVs isolated from a given host species supports a genetic basis for CaPV species-specific host range, and it agrees with data suggesting that viruses of sheep and goat origins can be differentiated via genomic restriction fragment pattern analysis (8, 25, 32).
TABLE 2.
Comparisons between CaPV genomesa
| Genome | Identity
|
||||
|---|---|---|---|---|---|
| SPPV
|
GTPV
|
||||
| TU | SA | NK | PL | GV | |
| SA | 193-131 (99.7) | ||||
| NK | 188-137 (99.8) | 71-36 (99.9) | |||
| PL | 2939-1972 (96.2) | 2949-1979 (96.2) | 2960-1986 (96.2) | ||
| GV | 2945-1974 (96.2) | 2955-1980 (96.2) | 2964-1987 (96.2) | 7-4 (100.0) | |
| LSDV | 2603-1816 (96.8) | 2617-1822 (96.9) | 2615-1825 (97.0) | 2373-1676 (97.2) | 2380-1676 (97.2) |
Pairwise comparisons of CaPV genomes presented as total differences-single-nucleotide substitutions (percent nucleotide identity rounded to the nearest 0.1%). Boldface numbers reflect intraspecies comparisons. Changes were calculated with the Diffseq program (ftp:uk.embnet.org/pub/EMBOSS).
FIG. 1.
Phylogenetic comparison of CaPVs. Genomic nucleotide sequences excluding terminal repetition were aligned by using Dialign (42) to generate the unrooted tree. The maximum-likelihood algorithm with HKY correction for multiple substitutions was used as implemented by the Phylip package (22). Branch length values indicate changes per nucleotide. Similar results were obtained by using the maximum-parsimony algorithm and the neighbor-joining algorithm, which maintained 100% support for species-specific groupings after 1,000 bootstrap replicates (data not shown).
SPPV and GTPV field isolates demonstrate average amino acid differences of 4% (Table 1). SPPV SA and GTPV PL share 101, 39, and 7 genes with 96 to 100%, 91 to 95%, and 80 to 90% amino acid identity, respectively (Table 1). Twenty-six SPPV and GPV ORFs differ in size (1 to 18 amino acids) due to insertion or deletion of amino acids within the ORF or alterations in start or stop codons. SPPV TU ORFs 003 and 154 are 73 amino acids longer than in other SPPVs due to two upstreat insertions resulting in frameshifts. Additionally, GTPV contains a 90-amino-acid ORF (ORF 136a) that is homologous to the carboxyl terminus of LSDV136. This ORF is absent in SPPV.
Differences between SPPV and GTPV in amino acid identity and ORF size are greater in terminal genomic regions and in genes with likely virulence and host range functions. Seventy-six percent of the most variable ORFs (<96% amino acid identity) and 54% of the ORFs differing in size between SPPV and GTPV occur in the terminal 35% of the genome (ORFs 001 to 023 and ORFs 122 to 156) (Table 1). Of the seven least similar ORFs (82 to 90% amino acid identity), three are similar to myxoma virus (MYXV) M016L (ORF 021), vaccinia virus (VACV) interferon-α/β binding protein (ORF 135), and VACV N1L-secreted virulence factor (ORF 142), and four differ in size (4 to 14 amino acids) and include a CaPV-specific ORF (ORF 022) and homologues of late transcription factor 4 (ORF 076), MYXV M130R (ORF 129) and an OX-2-like immunoglobulin (Ig) domain protein (ORF 138) (Table 1). Size differences (4 to 19 amino acids) are also present between SPPV and GTPV homologues of CC chemokine receptor (ORF 011), epidermal growth factor (EGF)-like growth factor (ORF 016), kelch-like protein (ORF 019), double-stranded RNA (dsRNA)-binding PKR inhibitor (ORF 035), RPO30 (ORF 036), VACV D9R mutT motif protein (ORF 086), Holliday junction resolvase (ORF 114), variola virus B22R (ORF 134), and ankyrin repeat protein (ORF 152). These differences in proteins located in terminal genomic regions likely affect aspects of viral virulence and host range.
Comparison of SGPV and LSDV.
Nine LSDV genes with likely virulence and host range functions are disrupted in SPPV and GTPV (Table 1). Genes affected by insertions, deletions, and substitutions include an LSDV-specific gene (LSDV132) and those similar to IL-1 receptor (IL-1R) (LSDV013), MYXV M003.2 (LSDV002 and LSDV155), MYXV M004.1 (LSDV004 and LSDV153), VACV N2L (LSDV009), VACV F11L (LSDV026), and VACV K7L (LSDV136) genes (24) (Table 1). Affected SGPV genes are highly fragmented, with 4 to 23 potentially frameshifting nucleotide changes per ORF compared to LSDV. The number of genomic changes relative to LSDV in these nine SGPV genomic regions is relatively high (average of 2.6 nucleotide insertion/deletion sites per 100 bases) compared to changes throughout the remainder of the genome (average of 0.09 nucleotide insertion/deletion sites per 100 bases). Although ORF 136 is highly fragmented in SGPV, two-thirds of the predicted protein remains as a carboxyl-terminal fragment in GTPV (Table 1). Given the likely ancestral nature of several LSDV genes disrupted in SGPV (present in other poxvirus genera), available data suggest that gene fragmentation and sequence divergence occurred during adaptation of an LSDV-like ancestor to sheep and/or goats.
Extensive fragmentation of these nine ORFs in SGPV likely results in functional inactivation. Of the nine genes disrupted, only the IL-1R gene (ORF 013) is orthologous to genes of known function, including VACV WR B15R homologues, secreted proteins that bind and inactivate host IL-1β to affect viral virulence (6, 56). LSDV013 contains the three Ig domains common to IL-1R and likely functions as an IL-1 binding protein (59). Disruption of ORF 013 in SGPV likely affects the ability of the virus to modulate host IL-1-mediated responses. ORF 009 is similar to VACV N2L, an ORF associated with viral sensitivity to α-amanitin (57). ORF 136 has limited similarity to a previously described poxvirus gene family, which includes VACV A52R, an antagonist for host cell IL-1 and Toll-like receptor-mediated intracellular signaling (9, 55, 59). The absence of these genes in SGPV likely affects aspects of CaPV virulence and host range and suggests a specific role for them in bovine host range.
ORFs 002 and 155 and ORF 013, disrupted in all SGPVs described here and in an Indian strain of SPPV, are intact in both the Kenyan “sheep and goat poxvirus” 0240 strain (KS-1) and LSDV (Table 1) (11, 16, 17, 24). Strain 0240 yields restriction patterns very similar to LSDV, and it causes mild disease in sheep and goats (8, 32, 33). Notably, published sequences from strain 0240 are more similar to LSDV (99.6 to 99.8% nucleotide identity) than to SPPV (92.0 to 97.1% nucleotide identity). Although considered of low virulence in cattle, strain 0240 induced more severe reactions than other SPPV and GTPV strains when administered experimentally, and it caused LSDV-like disease when used as a vaccine (8, 32, 60). These data suggest that strain 0240 may in fact be LSDV (32).
Comparison of SGPV field and vaccine strains.
SPPV NK and GTPV GV vaccine strains have greater than 99.9% amino acid identity to their respective field strains (Table 2). SPPV NK contains only 71 genomic differences compared to SPPV SA, including 36 single-nucleotide substitutions, 15 insertions of 1 to 29 nucleotides, and 20 deletions of 1 to 4 nucleotides and affecting 17 proteins (Table 1). GTPV GV is extremely similar to the PL field isolate; differences occur in only seven genomic locations and include four single-nucleotide substitutions, two single-nucleotide insertions, and one deletion of 28 nucleotides and affect six proteins. Comparative genomic data suggest that these genomic changes account for viral attenuation and that the changes affect genes with predicted virulence and host range functions (Table 1).
In SPPV NK, single in-frame stop and frameshift mutations are present in ankyrin repeat-containing genes ORF 145 and ORF 148, respectively. Each gene is represented by two smaller ORFs in NK (ORFs 145a and 145b and ORFs 148a and 148b). Poxvirus ankyrin repeat genes have been associated with host range functions in orthopoxviruses and leporipoxviruses, and they may inhibit virally induced apoptosis (31, 44, 48). It has also been suggested that specific complements of ankyrin repeat genes may affect poxvirus host range (7, 54). The attenuated phenotype of the VACV Ankara strain may be due in part to mutations in ankyrin repeat genes (7). Ankyrin repeat motifs in other proteins are clearly involved in mediating protein-protein interactions, and the VACV K1L ankyrin repeat host range protein interacts with at least one other viral protein (41, 52). Disruption of two of the five ankyrin repeat genes in SPPV vaccine strain NK further suggests a significant role for them in viral virulence and/or host range.
GTPV vaccine strain GV contains mutations in all genes encoding kelch-like family proteins (ORFs 019, 144, and 151). ORF 019 contains a single frameshift that results in two smaller ORFs (ORFs 019a and 019b), ORF 144 contains a 28-nucleotide deletion in the carboxyl and 3′-noncoding region, and ORF 151 contains a single nonconservative amino acid substitution (N to H). These three ORFs in CaPV field isolates contain four to five imperfect carboxyl-terminal repeats similar to those found in the Drosophila kelch protein and to kelch-like proteins present in several ChPV genera (4, 10, 40, 53). Kelch and poxviral kelch-like proteins also contain an amino-terminal domain (BTB/POZ domain) known to mediate oligomerization and protein complex formation in numerous cellular actin-binding and transcriptional regulatory proteins (15, 53). Kelch repeat motifs are found in functionally diverse proteins known to affect cellular transcription, development, and organization, and they are involved in protein-protein interactions (1). Notably, proteins from influenza virus and herpes simplex virus bind cellular kelch repeat proteins and affect viral pathogenesis (1). The function of poxvirus kelch-like proteins is unknown; however, they are nonessential for replication of VACV in cell culture and have been speculated to mediate viral interaction with specific cellular components (39, 49, 54). The changes observed in all kelch-like proteins of the GTPV vaccine strain GV may be highly significant for viral attenuation. Overall, data from attenuated CaPVs suggest that further elucidation of functions of ankyrin repeat and kelch-like proteins may be helpful in design of more effective and versatile CaPV vaccines.
Conclusions.
Genome sequences of SPPV and GTPV described here, together with their comparison to the complete genomic sequence of LSDV, provides a comprehensive view of CaPV genomics. CaPV genomes are very similar to each other, averaging no less than 96% nucleotide identity over their entire length. SPPV, GTPV, and LSDV contain the same repertoire of orthologous genes, with the exception that SPPV and GTPV lack nine LSDV genes with likely CaPV virulence and host range functions. SPPV and GTPV genomes sequenced here are phylogenetically distinct from each other and from LSDV, and they contain species-specific nucleotide differences that may be associated with aspects of host range. Relatively few genomic changes in SPPV and GTPV vaccine viruses account for viral attenuation.
Acknowledgments
We thank J. Lubroth for providing the TU strain of SPPV and A. Zsak and A. Lakowitz for providing excellent technical assistance.
REFERENCES
- 1.Adams, J., R. Kelso, and L. Cooley. 2000. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell. Biol. 10:17-24. [DOI] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, E. Oma, G. F. Kutish, and D. L. Rock. 1999. The genome of Melanoplus sanguinipes entomopoxvirus. J. Virol. 73:533-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2000. The genome of fowlpox virus. J. Virol. 74:3815-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, F. A. Osorio, C. Balinsky, G. F. Kutish, and D. L. Rock. 2002. The genome of swinepox virus. J. Virol. 76:783-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Trends Microbiol. 8:410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcami, A., and G. L. Smith. 1992. A soluble receptor for interleukin-1 beta encoded by vaccinia virus: a novel mechanism of virus modulation of the host response to infection. Cell 71:153-167. [DOI] [PubMed] [Google Scholar]
- 7.Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365-396. [DOI] [PubMed] [Google Scholar]
- 8.Black, D. N., J. M. Hammond, and R. P. Kitching. 1986. Genomic relationship between capripoxviruses. Virus Res. 5:277-292. [DOI] [PubMed] [Google Scholar]
- 9.Bowie, A., E. Kiss-Toth, J. A. Symons, G. L. Smith, S. K. Dower, and L. A. O'Neill. 2000. A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proc. Natl. Acad. Sci. USA 97:10162-10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cameron, C., S. Hota-Mitchell, L. Chen, J. Barrett, J. X. Cao, C. Macaulay, D. Willer, D. Evans, and G. McFadden. 1999. The complete DNA sequence of myxoma virus. Virology 264:298-318. [DOI] [PubMed] [Google Scholar]
- 11.Cao, J. X., P. D. Gershon, and D. N. Black. 1995. Sequence analysis of HindIII Q2 fragment of capripoxvirus reveals a putative gene encoding a G-protein-coupled chemokine receptor homologue. Virology 209:207-212. [DOI] [PubMed] [Google Scholar]
- 12.Capstick, P. B., and W. Coackley. 1961. Protection of cattle against lumpy skin disease. Res. Vet. Sci. 2:362-375. [Google Scholar]
- 13.Carn, V. M. 1993. Control of capripoxvirus infections. Vaccine 11:1275-1279. [DOI] [PubMed] [Google Scholar]
- 14.Carn, V. M., C. P. Timms, P. Chand, D. N. Black, and R. P. Kitching. 1994. Protection of goats against capripox using a subunit vaccine. Vet. Rec. 135:434-436. [DOI] [PubMed] [Google Scholar]
- 15.Collins, T., J. R. Stone, and A. J. Williams. 2001. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 21:3609-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies, F. G. 1976. Characteristics of a virus causing a pox disease in sheep and goats in Kenya, with observation on the epidemiology and control. J. Hyg. 76:163-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies, F. G., and C. Otema. 1981. Relationships of capripox viruses found in Kenya with two Middle Eastern strains and some orthopox viruses. Res. Vet. Sci. 31:253-255. [PubMed] [Google Scholar]
- 18.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esposito, J. J., and F. Fenner. 2001. Poxviruses, p. 2885-2921. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monathy, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 20.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 21.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 22.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biol. Sci. 12:543-554. [DOI] [PubMed] [Google Scholar]
- 23.Gershon, P. D., D. M. Ansell, and D. N. Black. 1989. A comparison of the genome organization of capripoxvirus with that of the orthopoxviruses. J. Virol. 63:4703-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gershon, P. D., and D. N. Black. 1989. A capripoxvirus pseudogene whose only intact homologs are in other poxvirus genomes. Virology 172:350-354. [DOI] [PubMed] [Google Scholar]
- 25.Gershon, P. D., and D. N. Black. 1988. A comparison of the genomes of capripoxvirus isolates of sheep, goats, and cattle. Virology 164:341-349. [DOI] [PubMed] [Google Scholar]
- 26.Gershon, P. D., and D. N. Black. 1989. The nucleotide sequence around the capripoxvirus thymidine kinase gene reveals a gene shared specifically with leporipoxvirus. J. Gen. Virol. 70:525-533. [DOI] [PubMed] [Google Scholar]
- 27.Gershon, P. D., R. P. Kitching, J. M. Hammond, and D. N. Black. 1989. Poxvirus genetic recombination during natural virus transmission. J. Gen. Virol. 70:485-489. [DOI] [PubMed] [Google Scholar]
- 28.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:192-202. [DOI] [PubMed] [Google Scholar]
- 29.Heine, H. G., M. P. Stevens, A. J. Foord, and D. B. Boyle. 1999. A capripoxvirus detection PCR and antibody ELISA based on the major antigen P32, the homolog of the vaccinia virus H3L gene. J. Immunol. Methods 227:187-196. [DOI] [PubMed] [Google Scholar]
- 30.Huang, X., and A. Madan. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ink, B. S., C. S. Gilbert, and G. I. Evan. 1995. Delay of vaccinia virus-induced apoptosis in nonpermissive Chinese hamster ovary cells by the cowpox virus CHOhr and adenovirus E1B 19K genes. J. Virol. 69:661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitching, R. P., P. P. Bhat, and D. N. Black. 1989. The characterization of African strains of capripoxvirus. Epidemiol. Infect. 102:335-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitching, R. P., J. M. Hammond, and W. P. Taylor. 1987. A single vaccine for the control of capripox infection in sheep and goats. Res. Vet. Sci. 42:53-60. [PubMed] [Google Scholar]
- 34.Kitching, R. P., J. J. McGrane, J. M. Hammond, A. H. Miah, A. H. Mustafa, and J. R. Majumder. 1987. Capripox in Bangladesh. Trop. Anim. Health Prod. 19:203-208. [DOI] [PubMed] [Google Scholar]
- 35.Kitching, R. P., J. J. McGrane, and W. P. Taylor. 1986. Capripox in the Yemen Arab Republic and the Sultanate of Oman. Trop. Anim. Health Prod. 18:115-122. [DOI] [PubMed] [Google Scholar]
- 36.Kitching, R. P., and P. S. Mellor. 1986. Insect transmission of capripoxvirus. Res. Vet. Sci. 40:255-258. [PubMed] [Google Scholar]
- 37.Kitching, R. P., and W. P. Taylor. 1985. Clinical and antigenic relationship between isolates of sheep and goat pox viruses. Trop. Anim. Health Prod. 17:64-74. [DOI] [PubMed] [Google Scholar]
- 38.Kitching, R. P., and W. P. Taylor. 1985. Transmission of capripoxvirus. Res. Vet. Sci. 39:196-199. [PubMed] [Google Scholar]
- 39.Kotwal, G. J., and B. Moss. 1988. Analysis of a large cluster of nonessential genes deleted from a vaccinia virus terminal transposition mutant. Virology 167:524-537. [PubMed] [Google Scholar]
- 40.Lee, H. J., K. Essani, G. L. Smith, F. Jeanmougin, and D. G. Higgins. 2001. The genome sequence of Yaba-like disease virus, a yatapoxvirus. Virology 281:170-192. [DOI] [PubMed] [Google Scholar]
- 41.McCraith, S., T. Holtzman, B. Moss, and S. Fields. 2000. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc. Natl. Acad. Sci. USA 97:4879-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgenstern, B., K. Frech, A. Dress, and T. Werner. 1998. DIALIGN: finding local similarities by multiple sequence alignment. Bioinformatics 14:290-294. [DOI] [PubMed] [Google Scholar]
- 43.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monathy, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 44.Mossman, K., S. F. Lee, M. Barry, L. Boshkov, and G. McFadden. 1996. Disruption of M-T5, a novel myxoma virus gene member of the poxvirus host range superfamily, results in dramatic attenuation of myxomatosis in infected European rabbits. J. Virol. 70:4394-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munz, E., and K. Dumbell. 1994. Sheeppox and goatpox, p. 613-615. In J. A. W. Coetzer, G. R. Thomson, and R. C. Tustin (ed.), Infectious diseases of livestock, vol. 1. Oxford University Press, Cape Town, South Africa. [Google Scholar]
- 46.Murty, D. K., and P. P. Singh. 1971. Epidemiological studies on an outbreak of sheep-pox in a mixed flock in Uttar Pradesh. Indian J. Anim. Sci. 41:1072-1079. [Google Scholar]
- 47.Ngichabe, C. K., H. M. Wamwayi, T. Barrett, E. K. Ndungu, D. N. Black, and C. J. Bostock. 1997. Trial of a capripoxvirus-rinderpest recombinant vaccine in African cattle. Epidemiol. Infect. 118:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perkus, M. E., S. J. Goebel, S. W. Davis, G. P. Johnson, K. Limbach, E. K. Norton, and E. Paoletti. 1990. Vaccinia virus host range genes. Virology 179:276-286. [DOI] [PubMed] [Google Scholar]
- 49.Perkus, M. E., S. J. Goebel, S. W. Davis, G. P. Johnson, E. K. Norton, and E. Paoletti. 1991. Deletion of 55 open reading frames from the termini of vaccinia virus. Virology 180:406-410. [DOI] [PubMed] [Google Scholar]
- 50.Rafyi, A., and H. Ramyar. 1959. Goat pox in Iran. J. Comp. Pathol. 69:141-147. [PubMed] [Google Scholar]
- 51.Rao, T. V., and S. K. Bandyopadhyay. 2000. A comprehensive review of goat pox and sheep pox and their diagnosis. Anim. Health Res. Rev. 1:127-136. [DOI] [PubMed] [Google Scholar]
- 52.Sedgwick, S. G., and S. J. Smerdon. 1999. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24:311-316. [DOI] [PubMed] [Google Scholar]
- 53.Senkevich, T. G., G. L. Muravnik, S. G. Pozdnyakov, V. E. Chizhikov, O. I. Ryazankina, S. N. Shchelkunov, E. V. Koonin, and V. I. Chernos. 1993. Nucleotide sequence of XhoI O fragment of ectromelia virus DNA reveals significant differences from vaccinia virus. Virus Res. 30:73-88. [DOI] [PubMed] [Google Scholar]
- 54.Shchelkunov, S. N., P. F. Safronov, A. V. Totmenin, N. A. Petrov, O. I. Ryazankina, V. V. Gutorov, and G. J. Kotwal. 1998. The genome sequence analysis of the left and right species-specific terminal region of a cowpox virus strain reveals unique sequences and a cluster of intact ORFs for immunomodulatory and host range proteins. Virology 243:432-460. [DOI] [PubMed] [Google Scholar]
- 55.Smith, G. L., Y. S. Chan, and S. T. Howard. 1991. Nucleotide sequence of 42 kbp of vaccinia virus strain WR from near the right inverted terminal repeat. J. Gen. Virol. 72:1349-1376. [DOI] [PubMed] [Google Scholar]
- 56.Spriggs, M. K., D. E. Hruby, C. R. Maliszewski, D. J. Pickup, J. E. Sims, R. M. Buller, and J. VanSlyke. 1992. Vaccinia and cowpox viruses encode a novel secreted interleukin-1-binding protein. Cell 71:145-152. [DOI] [PubMed] [Google Scholar]
- 57.Tamin, A., J. Esposito, and D. Hruby. 1991. A single nucleotide substitution in the 5′-untranslated region of the vaccinia N2L gene is responsible for both alpha-amanitin-resistant and temperature-sensitive phenotypes. Virology 182:393-396. [DOI] [PubMed] [Google Scholar]
- 58.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2001. Genome of lumpy skin disease virus. J. Virol. 75:7122-7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeruham, I., O. Nir, Y. Braverman, M. Davidson, H. Grinstein, M. Haymovitch, and O. Zamir. 1995. Spread of lumpy skin disease in Israeli dairy herds. Vet. Rec. 137:91-93. [DOI] [PubMed] [Google Scholar]
- 61.Yeruham, I., S. Perl, A. Nyska, A. Abraham, M. Davidson, M. Haymovitch, O. Zamir, and H. Grinstein. 1994. Adverse reactions in cattle to a capripox vaccine. Vet. Rec. 135:330-332. [DOI] [PubMed] [Google Scholar]