Abstract
Reverse transcriptases (RTs) αβ and β from avian Rous sarcoma virus (RSV) harbor an integrase domain which is absent in nonavian retroviral RTs. RSV integrase contains a nuclear localization signal which enables the enzyme to enter the nucleus of the cell in order to perform integration of the proviral DNA into the host genome. In the present study we analyzed the subcellular localization of RSV RT, since previous results indicated that RSV finishes synthesis of the proviral DNA in the nucleus. Our results demonstrate that the heterodimeric RSV RT αβ and the β subunit, when expressed independently, can be detected in the nucleus, whereas the separate α subunit lacking the integrase domain is prevalent in the cytoplasm. These data suggest an involvement of RSV RT in the transport of the preintegration complex into the nucleus. In addition, to analyze whether the integrase domain, located at the carboxyl terminus of β, exhibits integration activities, we investigated the nicking and joining activities of heterodimeric RSV RT αβ with an oligodeoxynucleotide-based assay system and with a donor substrate containing the supF gene flanked by the viral long terminal repeats. Our data show that RSV RT αβ is able to perform the integration reaction in vitro; however, it does so with an estimated 30-fold lower efficiency than the free RSV integrase, indicating that RSV RT is not involved in integration in vivo. Integration with RSV RT αβ could be stimulated in the presence of human immunodeficiency virus type 1 nucleocapsid protein or HMG-I(Y).
After a retrovirus infects a cell, retroviral core particles are released into the cytoplasm, where the viral RNA is reverse transcribed into double-stranded DNA within a nucleoprotein structure, designated the preintegration complex (PIC) (3, 4, 7, 43, 44, 60). In addition to the viral DNA and integrase (IN), PICs include several other viral and cellular proteins, among them the viral enzyme reverse transcriptase (RT), the viral nucleocapsid (NC) protein, and the cellular nonhistone DNA-binding protein HMG-I(Y) (8, 16, 17, 23). For retroviral integration to occur, the newly synthesized viral DNA must associate with the host genome. Human immunodeficiency virus (HIV) is not dependent on host cell division for integration and virus propagation but enters the nucleus via transport through the nuclear pore (46). The viral IN and the Vpr protein carry nuclear localization signals (NLSs) that are thought to be responsible for transport of the HIV PIC into the nucleus (10, 20, 22, 41, 56, 63). A contribution of the matrix protein (MA) has also been suggested (23, 26, 49).
In contrast to HIV, oncogenic retroviruses such as murine leukemia virus (MLV) and the avian sarcoma and leukosis virus (ASLV) are dependent on target cell proliferation for productive replication (27, 35, 37, 61). For MLV it was demonstrated that integration of viral DNA requires passage through mitosis, indicating the necessity of nuclear envelope breakdown (47, 57). For ASLV it was suggested that the viral DNA can be synthesized and integrated into host DNA during the S phase prior to the onset of mitosis (36). It was shown recently that Rous sarcoma virus (RSV) can infect cells arrested by aphidicolin in the G1/S phase of the cell cycle, albeit with moderate efficiency (28). These results indicate that disassembly of the nuclear membrane is not absolutely essential for RSV integration. Moreover, the majority of RSV DNA detected in the cytoplasm is not completely double stranded, suggesting that completion of DNA synthesis is dependent on nuclear entry (44). This model of ASLV DNA synthesis and integration is further supported by the finding that avian sarcoma virus (ASV) IN harbors an NLS in the carboxyl-terminal domain, whereas the IN of MLV does not possess an NLS (41, 42). In addition, when expressed separately in eukaryotic cell lines, RSV IN is found predominantly in the nucleus (41, 51, 54).
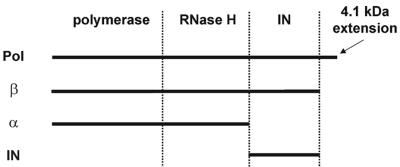
RSV RT, as well as IN, is derived from the pol gene of RSV (Fig. 1). RSV RT is an αβ heterodimer which contains the polymerase, RNase H, and IN domains in the 95-kDa β subunit. A 4.1-kDa polypeptide is cleaved off the carboxyl terminus of the Pol precursor protein to yield β. Deletion of the IN domain from β produces the 63-kDa α subunit and free IN enzyme. In addition, α and β homodimers can be detected, although at much lower concentrations than αβ (31, 32). Since RSV RT contains the IN domain in the β subunit and thus the NLS sequence, we wanted to determine the subcellular localization of RSV RT when expressed independently.
FIG. 1.
Schematic representation of Pol products.
In HIV, the presence of a mature wild-type IN was shown to be required for efficient initiation of reverse transcription in infected cells. A direct physical interaction between IN and RT suggests that the IN protein is directly involved in reverse transcription in vivo (69). In light of these results, we were interested in the structure of the β subunit of RSV RT and, in particular, in whether the IN domain was folded into an active conformation. The correct folding of the IN domain would then offer a significant role for this domain in reverse transcription similar to that observed by Wu et al. (69).
The existence of an RT subunit carrying the IN domain has led to speculations on its possible role in the integration process. Based on this knowledge we wanted to elucidate whether the IN domain of RSV RT possesses catalytic activities similar to those of free IN. Integration requires processing of the viral DNA, nicking of the host DNA, and joining of the processed viral DNA to the host DNA. These biochemically distinguishable activities can be catalyzed by the virally encoded enzyme IN. Through a direct nucleophilic attack the endonuclease activity of free IN has been shown to remove two nucleotides from the 3′OH ends of the viral DNA, thereby creating recessed CA 3′OH ends. (6, 34, 40, 58, 62). The recessed ends are then integrated into the host DNA by IN through coupled joining (1, 18, 39, 64, 65). In this way the processed 3′OH ends of the viral DNA are joined to the host DNA by a second direct nucleophilic attack of the 3′OH ends on an internal phosphate of the host DNA (4, 9, 14, 21, 39).
It was demonstrated previously that the avian retroviral RTs αβ and β harbor a DNA endonuclease activity which can introduce nicks into supercoiled double-stranded DNA (25, 45). Furthermore, these enzymes are able to specifically cleave two or three nucleotides off the long terminal repeat (LTR) ends of proviral DNA (15, 40). However, the joining or strand transfer activity of the IN domain of avian RTs has never been investigated in a highly sensitive in vitro assay.
Our results shown here imply that RSV RT is able to perform the integration reaction at low efficiency and that RSV RTs β and αβ harboring the IN domain can be detected in the nucleus, whereas the α subunit, when expressed separately, is found exclusively in the cytoplasm.
MATERIALS AND METHODS
Cloning of β-GFP, α-RFP, and IN-GFP gene fusions.
The DNA fragments containing the genes of the α or the β subunit of RSV RT or RSV IN, respectively, were cloned by a combination of PCR amplification and restriction site cloning by using bacterial expression vectors containing the corresponding genes for amplification. The amplified viral genes possessed a 5′ ATG start codon and a sequence coding for six N-terminal His residues but lacked a 3′ stop codon. The fragments were cloned into the vectors pEGFP-N3 or pDsRed1-N1 (Clontech), expressing a green fluorescent protein (GFP) or a red fluorescent protein (RFP). A multiple cloning site is located between the promoter and the 5′ end of the gene for EGFP or DsRed1, respectively. Thus, the α subunit was expressed as a fusion to the N terminus of DsRed1, and β and IN were expressed as fusions to the N terminus of EGFP.
Growth of NIH 3T3 cells.
Mouse NIH 3T3 fibroblasts (ACC 59; German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) were grown in Dulbecco modified Eagle medium (DMEM; 4.5 g of glucose/liter) containing 10% fetal calf serum (FCS) (vol/vol), 1% l-glutamine (wt/vol), and 1% (each) penicillin, streptomycin, and neomycin (PSN) (DMEM-FCS-l-Glu-PSN) (all supplied by Life Technologies). The cells were cultivated in cell culture flasks at 37°C and 5% CO2 and passaged every 3 to 4 days.
Transfection of NIH 3T3 cells and confocal laser scanning microscopy.
A total of 2.5 × 104 cells/well were seeded on coverslips in 24-well plates 24 h prior to transfection. To transfect the cells by the calcium phosphate procedure (59), the medium was removed, cells were washed once with phosphate-buffered saline (PBS), and 40 μl of the transfection solution containing the desired plasmid was added. After an incubation period of 30 min at room temperature, 400 μl of DMEM-FCS-l-Glu-PSN was added. Expression of RSV RT and RSV IN fusion proteins was determined for 24 h at 37°C under 5% CO2.
Subsequently, the medium was removed and the cells were washed twice with PBS. The cells were fixed with 200 μl of 4% paraformaldehyde in PBS/well for 20 min at room temperature. Afterwards the coverslips were rinsed twice with PBS and once with water and then mounted in a solution of Mowiol 88 (Sigma). For preparation of the Mowiol 88 solution, 6 g of glycerol and 2.4 g of Mowiol 88 were stirred for 2 h at room temperature. Then, 12 ml of 0.3 M Tris-HCl (pH 8.5) was added. After the solution was mixed for 10 min at 50°C, it was centrifuged for 15 min at 5,000 × g, and the supernatant was frozen in aliquots at −20°C.
The fluorescence pattern of the transfected, fixed, and mounted NIH 3T3 cells was imaged and analyzed by confocal laser-scanning microscopy (Zeiss) by using LSM 510 software, version 2.3 (Zeiss).
Purification of RSV RTs.
Growth and infection of Sf21 insect cells with recombinant baculoviruses and the purification of recombinant RSV enzymes were performed as described previously (66, 67). In order to obtain heterodimeric RSV RT αβ, insect cells were infected with two types of recombinant viruses carrying the genes for α or β, respectively.
Purification of RSV IN.
The gene for RSV IN was obtained by PCR amplification of the C-terminal part of the RSV RT β gene and cloned into the bacterial expression vector pdS56RBSII-N6His (13). Expression of a protein with this vector leads to an N-terminal fusion of a histidine hexamer with the recombinant protein. The plasmid was transformed into Escherichia coli M15/pDMI.1. Cultures expressing the His6-tagged RSV IN were grown at 37°C in Luria-Bertani medium (59) containing kanamycin (25 μg/ml) and ampicillin (100 μg/ml) to an optical density at 600 nm of 0.6, induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and harvested after 2 h. The cell pellet was resuspended in a buffer containing 20 mM Tris-HCl (pH 7.5), 1 M NaCl, 25% glycerol, 1 mM MgCl2, 2 mM mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, and 0.1% IGEPAL CA 630 (Sigma) and then sonicated. The cell lysate was centrifuged at 46,000 × g for 60 min at 4°C. The purification procedure was similar to that applied for His6-tagged RSV RTs (66, 67). Briefly, IN was purified from the supernatant by nickel-chelate affinity chromatography, followed by heparin-Sepharose (Pharmacia) chromatography. The eluate was dialyzed and kept at −20°C in a buffer containing 50% glycerol, 20 mM Tris-HCl (pH 7.0), 250 mM NaCl, 0.1% IGEPAL CA 630, and 2 mM dithiothreitol.
Oligodeoxyribonucleotide substrates.
Two complementary oligodeoxynucleotides (ODNs) that correspond to the last 36 bases of the U3 region of the 5′ LTR were used as substrates. The sequence of the RSV U3-sense ODN was 5′-AATGTAGTCTTATGCAATACTCTTGTAGTCTTGCAA-3′. The sequence of the second ODN RSV U3-anti was complementary to RSV U3-sense. The ODNs were purified by electrophoresis on 20% polyacrylamide-7 M urea gels, followed by electroelution in a BioTrap apparatus (Schleicher & Schuell). Eluted ODNs were desalted on NAP10 gel filtration columns (Amersham Pharmacia). After 32P end labeling of RSV U3-anti, the ODN was hybridized to RSV U3-sense in 20 mM Tris-HCl (pH 7.5)-50 mM NaCl by using the unlabeled ODN at a 1.2-fold excess. The mixture was heated for 2 min at 95°C and then slowly (4 h) cooled to room temperature on a heating block.
ODN-based IN activity assay.
The IN activity was measured in a 10-μl reaction volume in a buffer containing 25 mM Tris-HCl (pH 8.0), 80 mM NaCl, 3 mM MnCl2, and 2 mM β-mercaptoethanol. The final concentration of the labeled ODN 5′LTR substrate was 0.1 μM. RSV RT and HIV-1 NC (68) were added at final concentrations of 0.075 and 9.9 μM, respectively. After 90 min at 37°C the reaction was terminated by the addition of 10 μl of formamide buffer (80% formamide in 90 mM Tris-HCl [pH 8.3], 90 mM boric acid, 3 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue). The reaction products were separated on a 10% polyacrylamide-7 M urea gel and analyzed by autoradiography.
The recombinant NC protein of HIV-1 was expressed and purified as described previously (68). Lyophilized HIV-1 NC protein was resuspended in RT buffer, frozen in small aliquots, and used only once after thawing.
Integration reaction mixtures with a 294-bp donor DNA substrate.
ASV IN used for this assay was prepared by G. Merkel (Fox Chase Cancer Center, Philadelphia, Pa.) as described by Jones et al. (38). HMG-I(Y) was purified as described by Nissen et al. (55). ODNs were purified by denaturing polyacrylamide gel electrophoresis, followed by reversed-phase chromatography as previously described (1). The following ODNs were used to prepare the donor substrate: U5 (5′-AATGAAGCCTTCTGCTGGGCGGAGCCTATG-3′) and U3 (5′-AATGTAGTCTTATGCGTTGCCCGGATCCGG-3′).
Radioactively labeled integration donors harboring the supF gene flanked by U3 and U5 were obtained by PCR as described previously (29).
The integration reaction conditions were similar to those described previously (1, 29). Briefly, 15 ng (0.15 pmol of the ends) of donor was mixed with 50 ng of acceptor (0.02 pmol) and 115 nM dimeric ASV IN in a 26-μl preincubation reaction mixture containing 20 mM Tris-HCl (pH 7.5), 166 mM NaCl, 5 mM dithiothreitol, 10% dimethyl sulfoxide, 0.05% Nonidet P-40, 1% glycerol, 1.6 mM HEPES (pH 8.0), and 3.3 mM EDTA. The preincubation reaction mixtures were placed on ice overnight. The volume of each preincubation mixture was then increased to 30 μl with the addition of MgCl2 to a final concentration of 6.7 mM, and the integration assay mixture was incubated at 37°C for 90 min. Further treatment of the reaction products was as described previously (29).
Integration assay conditions with avian RT were identical to those of IN, with a final RT concentration of 500 nM. Where specified, HMG-I(Y) was added to the IN or RT integration reaction mixtures at a concentration of 154 nM.
RESULTS
Subcellular localization of the α and β subunits of RSV RT.
It is not known whether the IN domain of RSV RT adopts the same conformation as free IN or whether the NLS sequence is masked by an alternative folding of this domain. A functional NLS in RT might contribute to the transfer of the PIC and might enable RSV RT to traverse the nuclear envelope independently from other proteins. Thus, completion of reverse transcription in the nucleus could be ensured. To analyze whether the NLS present in RSV RT is active, we determined the subcellular localization of the separately expressed RT subunits and of heterodimeric RT αβ.
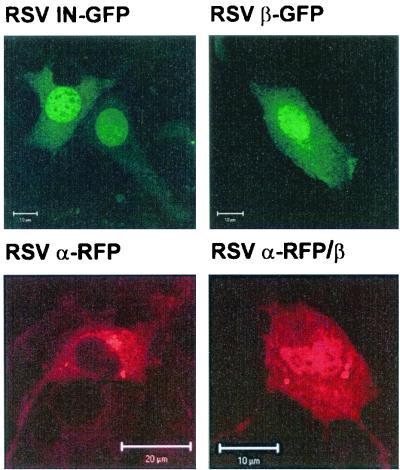
Fluorescent fusion proteins of RSV IN and of the α and β subunits of RSV RT were transiently expressed in NIH 3T3 mouse fibroblasts and investigated by confocal laser scanning microscopy. RSV IN and RSV RT β were fused to the N terminus of the GFP, and RSV RT α was expressed as an amino-terminal fusion to the RFP DsRed1. Figure 2 shows representative images of NIH 3T3 cells expressing the different fluorescent fusion proteins. Our data demonstrate clearly that RSV IN-GFP is localized in the nucleus. This confirms the results shown previously by Kukolj et al. (41).
FIG. 2.
Subcellular localization of RSV RTs and of RSV IN by confocal laser scanning microscopy. Representative confocal micrographs of transiently transfected NIH 3T3 fibroblasts expressing GFP or RFP fusion proteins are shown. GFP fusion proteins were expressed with RSV IN (RSV IN-GFP) or the β subunit of RSV RT (RSV RT β-GFP). The α subunit of RSV RT was expressed as a fusion to RFP (RSV RT α-RFP). Localization of the heterodimer (RSV RT α-RFP/β) was detected by coexpression of RSV RT α-RFP and β.
When the α subunit of RSV RT was fused to GFP, Western blots with anti-GFP antibodies showed that GFP was partially cleaved off the fusion protein (data not shown). Since free GFP is only ca. 28 kDa in size, it might be transferred to the nucleus by diffusion. To avoid misinterpretation, we chose to use the α-RFP fusion protein for our analyses since in this case the Western blots did not show any cleavage of the fusion protein. α-RFP, which lacks the IN domain and thus the NLS, is restricted to the cytoplasm, whereas β-GFP, similar to IN-GFP, accumulates in the nucleus (Fig. 2).
To analyze the subcellular localization of the heterodimer, we expressed α-RFP, together with β-GFP. Colocalization of the two proteins should have resulted in yellow fluorescence. However, since no yellow nuclear fluorescence could be detected, we assumed that due to the bulky RFP or GFP fusion domains the two proteins were unable to form dimers. To investigate this hypothesis further, dimerization experiments were performed by coexpressing β-GFP containing a histidine extension at the N terminus (His6-β-GFP) and α-RFP lacking the histidine extension. Heterodimeric RT can be purified via the His6 tag of β-GFP if dimerization with α-RFP takes place. Purification by nickel-chelate affinity chromatography yielded His6-β-GFP but not the heterodimer, indicating that dimer formation is hampered by the fluorescent fusion proteins (data not shown).
Therefore, we expressed RSV RT α-RFP, together with an untagged β subunit. Since RSV RT α-RFP does not enter the nucleus when expressed separately (Fig. 2), detection of α-RFP in the nucleus indicates dimerization with the β subunit. Only in the heterodimer complex can the NLS sequence of the β subunit mediate transport of RSV RT α-RFP to the nucleus. Transfection rates in these assays were low (<5%), and only a few cotransfected cells were present. This was probably due to the toxicity of DsRed1. In addition, we observed that expression of the β subunit of RSV RT, together with α-RFP, led to aggregates in the transfected cells. This might also have contributed to cell toxicity. We did not observe transfected cells where the red fluorescence was exclusively located in the nucleus. Thus, to guide more of the α-RFP into the nucleus, we attempted to increase the amount of the RSV RT β-expressing plasmid in the transfection assay. However, we were not able to obtain cotransfected cells that showed exclusive nuclear fluorescence. Higher expression levels of β are probably toxic for the cells, since we observed previously that cells overexpressing β are impaired in cell growth.
The cotransfected cells show a distribution of red fluorescence in the cytoplasm and the nucleus, indicating dimerization of the RT subunits and the ability of the heterodimer to pass the nuclear membrane. Western blot analysis with a polyclonal antibody directed against IN (2) demonstrated that β was expressed in the cells transfected with the two plasmids carrying the gene for α and β, respectively (data not shown).
Our data indicate that RSV RTs β and αβ can transfer to the nucleus without the need for other viral proteins.
Integration activities with an ODN-based assay system.
To further elucidate the function of the IN domain in RSV RT, we analyzed the integration activities of RT. Two different assays were performed. First, we used an in vitro ODN assay similar to the one published by Müller et al. (53). This assay was designed for a quick and simultaneous analysis of the specific removal of the last two nucleotides of the 3′ end of the LTR and of the joining activities of free IN. On account of the larger size of the RSV RT, we used 36-mer ODNs to ensure that the entire enzyme was able to bind to the DNA properly (53). To optimize the IN activity of RT, different reaction conditions were tested, e.g., by changing the enzyme-to-substrate ratios, the Mg2+ or Mn2+ concentrations, the ionic strengths, and the buffer pH.
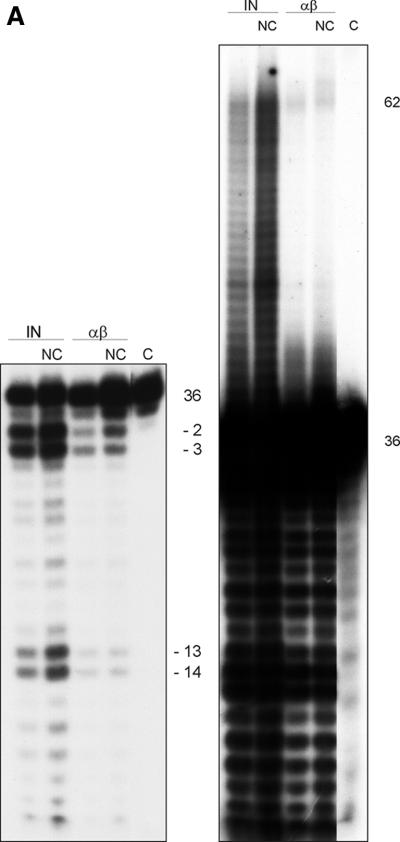
Previous experiments by Carteau et al. (11) with HIV-1 IN indicated a stimulation of the integration reaction of HIV-1 by NC protein in vitro (12). Figure 3A shows the IN activities of free IN and of RSV RT αβ in the absence or presence of the HIV-1 NC protein. The molarities of RT and IN were calculated by assuming that RT αβ is present as a heterodimer and that IN is present as a homodimer. The amount of RT used in the assay shown is the concentration that worked best after optimization of the assay conditions. It has been suggested that activation of integration by NC in the presence of Mn2+ is through competition of the NC protein with IN for binding to nonspecific sequences (11). This implies that the effect is not enzyme specific and allows the use of HIV NCp7 in our assay. The short exposure of the autoradiograph (left panel) shows the products of the specific processing reaction. The result demonstrates that, like RSV IN, RSV RT αβ cuts the ODN substrate predominantly at positions −2 and −3. The positions of the cleavage sites are the same as with free IN. In addition to these cleavage sites there is another major site at position −12/−13. These sites are also present with free IN. The long exposure of the autoradiograph (right panel) shows the strand transfer products. The result reveals that RSV RT αβ possesses joining activity; however, it is much weaker than the one found with free RSV IN (see also below). Similar results were obtained when homodimeric RSV RT β was used (data not shown). Upon addition of HIV-1 NC protein, integration by RSV RT αβ can be slightly stimulated. A stimulation is also visible with free IN.
FIG. 3.
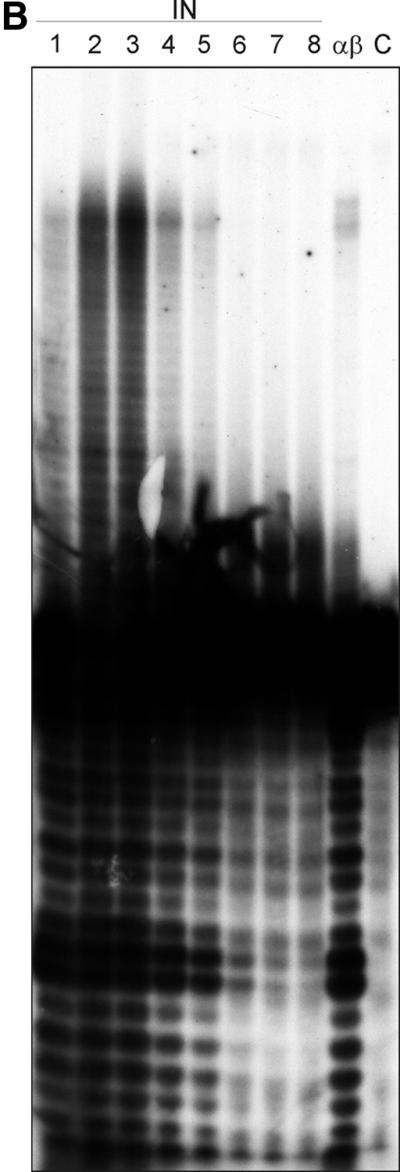
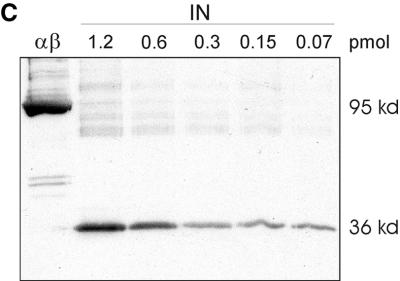
(A) Integration activity of RSV RT αβ and RSV IN in the absence or presence of HIV-1 NC protein. A double-stranded 36-mer ODN substrate (100 nM) corresponding to the 3′ LTR of the viral DNA was used. The ODN harboring the 3′-terminal CATT sequence was 5′ 32P end labeled. The concentration of the enzymes was 75 nM; HIV-1 NC was added at a concentration of 9.9 μM. Samples were incubated for 90 min at 37°C. The left panel of the figure shows a short exposure of an autoradiograph of the lower part of the gel to visualize the 3′OH processing activities of IN and RSV RT αβ. The right panel shows a long exposure of an autoradiograph of the same gel to visualize the integration products in the upper region. (B) Determination of the lowest IN concentration exhibiting integration activities. The ODN-based IN assay was performed in a 10-μl assay by using decreasing amounts of IN in order to determine the lowest IN concentration still exhibiting activity. Lane 1, 0.75 pmol of IN; lane 2, 0.5 pmol of IN; lane 3, 0.25 pmol of IN; lane 4, 0.05 pmol of IN; lane 5, 0.025 pmol of IN; lane 6, 0.005 pmol of IN; lane 7, 0.0025 pmol of IN; lane 8, 0.001 pmol of IN; lane αβ, 0.75 pmol of RT αβ. (C) Western blot with RT αβ and with different concentrations of IN to detect a possible contamination with free IN in our RT preparation. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then blotted onto a nitrocellulose membrane. The concentration of RSV RT αβ was 48 pmol. Detection of free IN was achieved with an antiserum directed against the C terminus of RSV IN (2). The second antibody was an anti-rabbit immunoglobulin G antibody conjugated with alkaline phosphatase.
To exclude that a contamination with free IN is the actual cause for the integration activities of RSV RT αβ, we determined the lowest concentration of free IN still exhibiting catalytic activity in the ODN integration assay (Fig. 3B). For our calculations we assumed that IN is present as a dimer. Weak IN activity is already visible with 0.025 pmol (2.5 nM) of IN (lane 5), whereas IN activity is not detectable anymore at 0.005 pmol in a 10-μl assay (0.5 nM). RSV RT αβ shows noticeable integration activity at a 30-fold-higher concentration (0.75 pmol or 75 nM). This implies that there should be at least 0.005 pmol of contaminating IN in 0.75 pmol of RT to obtain the integration activity observed. The Western blot with RSV IN in Fig. 3C demonstrates that 70 fmol of free IN is still detectable and leads to a distinctly visible band (2). We loaded 48 pmol of RT αβ onto the gel which, on account of the result above, should contain at least 320 fmol of free IN in order to exhibit strand transfer activity. This amount should be clearly visible on the blot. However, no band of the size of IN is traceable, indicating that the IN activity detectable with RSV RT αβ can be ascribed to the RT itself.
Moreover, the result presented in Fig. 3B gives us a good estimate of the differences in strand transfer efficiencies between RSV IN and RSV RT αβ. The data show that the integration activity of RT is ca. 30-fold lower than that of IN.
Analysis of the integration reaction with a 294-bp donor DNA substrate.
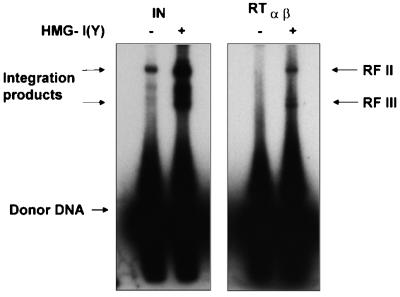
The standard integration assay used above examines the action of IN on only a single viral terminus, in our case U3. However, during integration in the cell both viral DNA ends are recognized by IN and coordinately integrated into the host DNA (18). To analyze concerted integration, a double-stranded blunt-ended DNA donor substrate was used that contained the supF gene flanked by the terminal 15 bp of the RSV U3 and U5 ends. A 3.4-kb plasmid DNA was used as the acceptor (Fig. 4) (30). Since it has been shown that the efficiency of the integration reaction catalyzed by free IN can be increased by a host protein from the high-mobility group (HMG), we analyzed the integration reaction of RSV RT in the presence of HMG-I(Y) (1, 16, 30). HMG proteins fulfill several functions in the cell. For example, they bind to the minor groove of double-stranded DNA and preferentially interact with bent or supercoiled DNA and are involved in the unwinding of DNA.
FIG. 4.
Integration reaction with a supF containing substrate to investigate coupled joining. In vitro integration products with IN and RT αβ are shown in the absence or presence of HMG-I(Y) protein. The upper band (RF II) of the integrated products represents a mixture of plasmids, with one molecule of donor integrated either in a concerted (both ends) or nonconcerted (one end or two ends integrated independently) way. The lower band (RF III) of products represents plasmids where two donor molecules are integrated in a concerted way, thus leading to a linear product (29).
By using this minidonor DNA, we could show integration into the acceptor plasmid with both enzymes RSV IN and RSV RT αβ. The integration products were separated on agarose gels as described previously (29) (Fig. 4). The fastest-migrating band corresponds to the labeled donor DNA. Again, the strand transfer efficiency of RSV RT is much lower than that of RSV IN. In the absence of HMG-I(Y), integrated products by RSV RT are barely visible. However, in the presence of HMG-I(Y) the integration activity of RSV RT αβ can be stimulated remarkably. Two major product bands are detectable with RSV RT or RSV IN, respectably. The upper band (RF II) represents a mixture of concerted integration products (i.e., both ends of the substrate were integrated into the plasmid in a concerted event), single-ended nonconcerted integration (i.e., only one end of the donor was integrated into the acceptor plasmid) and two-ended nonconcerted integration (i.e., both ends of a single donor molecule were integrated, but by two independent integration events). The lower band (RF III) represents concerted integration, where one end of two different donor DNA molecules integrates at the same site in the acceptor DNA, thus creating a linear DNA molecule (29). Due to the low integration activity of RT and the significant amount of RF III product, we have been unable to obtain transformants after supF biological selection that carry the plasmid with the integrated donor DNA.
DISCUSSION
In this study we analyzed possible functions of the IN domain of RSV RT for the viral replication cycle by determining its subcellular localization and integration activities. We were able to demonstrate that RSV RTs αβ and β can cross the nuclear membrane and that RSV RT αβ exhibits integration activities, although at a 30-fold-lower level than free IN, suggesting that this domain is folded in an active form of IN.
The subcellular localization of RSV RT also suggests an involvement in nuclear transport of the PIC. Since most of the viral DNA detected in the cytoplasm of ALV-infected cells is incomplete, a functional NLS ensures nuclear localization of RT and hence the ability to complete reverse transcription of the viral genome. A scenario where RT α lacking an NLS is responsible for reverse transcription in the cytoplasm and the αβ heterodimer ensures reverse transcription in the nucleus appears conceivable and could explain the presence of the different RT types in virions.
There is evidence that ASLV and HIV are able to enter the nucleus before synthesis of the viral DNA is finished (43, 50). Mutations in the NLS lead to a significant delay in replication in cell culture assays, indicating that efficient propagation of the virus is dependent on the presence of a functional NLS even in cycling cells (42). However, nothing is known about the mechanism of transport of the PIC of avian retroviruses on a molecular level. Furthermore, the composition of the ASV PIC is not very well characterized. The only NLS identified for ASV so far is the one at the carboxyl terminus of IN. However, other viral proteins in addition to IN and RT might contribute to nuclear transport. Garbitt et al. (24) were able to show recently that wild-type and mutant MA proteins of RSV exhibit distinctly different patterns of subcellular localization. The wild-type MA-GFP fusion protein was present in the cytoplasm and the nucleus, but there was an enhanced nuclear concentration, indicating the presence of an unidentified signal sequence on MA that could contribute to localization of the PIC.
Transport of RSV RT into the nucleus raised the question whether RSV RT is also involved in the integration process or involved in other events related to integration. Recently, two groups demonstrated that repair of the DNA damage incurred during integration can be mediated by RT and IN or a combination of host and viral enzymes and may represent a potential role for RT in the PIC (5, 70). We report here for the first time that RSV RT αβ exhibits not only IN nicking but also joining activities (Fig. 3 and 4). End processing by RT in the ODN assay is highly active at cleaving the 3′ strand, predominantly releasing 2 and 3 bases, in addition to releasing 13 and 14 bases. Similar cleavage patterns have been identified in reconstituted concerted ASV integration assays where IN releases 2 and 12 bases (1, 29). In addition, RSV RT αβ creates a significant amount of RF III product (concerted integration of one end of two different donor DNA molecules) compared to RF II. These results suggest that the majority of RF II products may be single-ended integration events. Similar results leading to single-ended integration events have been identified in ASV reconstituted integration assays when substitutions have been introduced into the IN recognition sequences. Single-ended integration events would not be rescued in a biological selection (29). This might explain why we have been unable so far to obtain transformants. We cannot yet answer the question whether these activities are biologically relevant, especially because the strand transfer activity of RSV RT αβ is at least 30-fold lower than that of the free IN (Fig. 3).
The results presented here suggest that the IN domain of RSV RT αβ is folded in an active conformation and is capable of many of the activities related to integration. IN functions as a dimer or higher-order multimer (38). It would be expected that in the context of RSV RT αβ the IN domain is a monomer. So how then is a monomer capable of IN activity? IN activity may require only one active IN subunit within a dimer (19). In the case of the heterodimer of RT, the α subunit could function as a platform to stabilize the viral DNA for processing and joining while the IN domain in the β subunit is functionally active. Alternatively, the RSV RT αβ heterodimer could form tetramers (αβ)2. It has been shown previously that, under certain conditions, the formation of RSV RT tetramers (αβ)2 can be observed (48). In such a complex the IN domain could form the required dimer necessary for activity (38).
The low integration activity of RSV RT suggests that it might not be biologically relevant; however, we cannot yet exclude activation of RT by other proteins in vivo. Although it has been shown that the free IN protein is sufficient to catalyze the various steps necessary for integration in vitro, the catalytic activities of retroviral IN proteins are rather poor. In vivo, several viral and cellular proteins are associated with the viral double-stranded DNA in the PIC. It is assumed that these proteins contribute to increase the efficiency of integration. Even though IN is present only as a free enzyme in nonavian retroviruses, it has been shown that RT and IN are in intimate contact in virus particles of MLV and HIV and that wild-type IN is required for reverse transcription of the HIV genome (33, 69). These results indicate that a close proximity of IN and RT might be important for viral replication. Since the integration process directly succeeds reverse transcription, close contact of the two enzymes is desirable. Thus, one possible role of RSV RT in the integration process might be to associate with free IN via its IN domain to direct it to the viral DNA ends after reverse transcription has been finished.
Acknowledgments
We thank R. Katz, Anna Marie Skalka, and George Merkel (Fox Chase Cancer Center, Philadelphia, Pa.) for the kind gift of a peptide antiserum directed against the C terminus of RSV IN. We thank Steve Hughes, National Cancer Institute (NCI), Frederick, Md., for providing a plasmid carrying the RSV pol gene; Robert Gorelick, NCI, Frederick, Md., for the generous gift of HIV-1 NC protein; and Raymond Reeves, University of Washington at Pullman, for the kind gift of HMG-I(Y). The help of Martina Wischnewski with enzyme purifications and assays is also greatly appreciated. We thank Roger Goody (Max-Planck-Institut für Molekulare Physiolgie, Dortmund, Germany) and Jonathan Leis (Northwestern University School of Medicine, Chicago, Ill.) for support.
This work was supported by the Max-Planck-Gesellschaft and by a grant from the Deutsche Forschungsgemeinschaft to B.M.W.
REFERENCES
- 1.Aiyar, A., P. Hindmarsh, A. M. Skalka, and J. Leis. 1996. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J. Virol. 70:3571-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, F., J. Leis, D. A. Soltis, R. M. Crowl, W. Danho, M. S. Poonian, Y. C. Pan, and A. M. Skalka. 1987. Proteolytic processing of avian sarcoma and leukosis viruses pol-endo recombinant proteins reveals another pol gene domain. J. Virol. 61:534-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltimore, D. 1970. RNA-dependent DNA polymerase in virions of RNA tumor viruses. Nature 226:1209-1211. [DOI] [PubMed] [Google Scholar]
- 4.Bowerman, B., P. O. Brown, J. M. Bishop, and H. E. Varmus. 1989. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 3:469-478. [DOI] [PubMed] [Google Scholar]
- 5.Brin, E., J. Yi, A. M. Skalka, and J. Leis. 2000. Modeling the late steps in HIV-1 retroviral integrase-catalyzed DNA integration. J. Biol. Chem. 275:39287-39295. [DOI] [PubMed] [Google Scholar]
- 6.Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1989. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc. Natl. Acad. Sci. USA 86:2525-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukrinsky, M. I., N. Sharova, M. P. Dempsey, T. L. Stanwick, A. G. Bukrinskaya, S. Haggerty, and M. Stevenson. 1992. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. USA 89:6580-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushman, F. D., T. Fujiwara, and R. Craigie. 1990. Retroviral DNA integration directed by HIV integration protein in vitro. Science 249:1555-1558. [DOI] [PubMed] [Google Scholar]
- 10.Cannon, P. M., E. D. Byles, S. M. Kingsman, and A. J. Kingsman. 1996. Conserved sequences in the carboxyl terminus of integrase that are essential for human immunodeficiency virus type 1 replication. J. Virol. 70:651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carteau, S., S. C. Batson, L. Poljak, J. F. Mouscadet, H. De Rocquigny, J. L. Darlix, B. P. Roques, E. Kas, and C. Auclair. 1997. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J. Virol. 71:6225-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carteau, S., R. J. Gorelick, and F. D. Bushman. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 73:6670-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Certa, U., W. Bannwarth, D. Stüber, R. Gentz, M. Lanzer, S. Le Grice, F. Guillot, I. Wendler, G. Hunsmann, H. Bujard, and J. Mous. 1986. Subregions of a conserved part of the HIV gp41 transmembrane protein are differentially recognized by antibodies of infected individuals. EMBO J. 5:3051-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craigie, R., T. Fujiwara, and F. Bushman. 1990. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62:829-837. [DOI] [PubMed] [Google Scholar]
- 15.Duyk, G., J. Leis, M. Longiaru, and A. M. Skalka. 1983. Selective cleavage in the avian retroviral long terminal repeat sequence by the endonuclease associated with the αβ form of avian reverse transcriptase. Proc. Natl. Acad. Sci. USA 80:6745-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farnet, C. M., and F. D. Bushman. 1997. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell 88:483-492. [DOI] [PubMed] [Google Scholar]
- 17.Farnet, C. M., and W. A. Haseltine. 1991. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 65:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald, M. L., A. C. Vora, W. G. Zeh, and D. P. Grandgenett. 1992. Concerted integration of viral DNA termini by purified avian myeloblastosis virus integrase. J. Virol. 66:6257-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fletcher, T. M., M. A. Soares, S. McPhearson, H. Hui, M. Wiskerchen, M. A. Muesing, G. M. Shaw, A. D. Leavitt, J. D. Boeke, and B. H. Hahn. 1997. Complementation of integrase function in HIV-1 virions. EMBO J. 16:5123-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, A. V. Albright, F. Gonzalez-Scarano, and M. H. Malim. 1998. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J. Virol. 72:6004-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujiwara, T., and K. Mizuuchi. 1988. Retroviral DNA integration: structure of an integration intermediate. Cell 54:497-504. [DOI] [PubMed] [Google Scholar]
- 22.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallay, P., S. Swingler, J. Song, F. Bushman, and D. Trono. 1995. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell 83:569-576. [DOI] [PubMed] [Google Scholar]
- 24.Garbitt, R. A., J. A. Albert, M. D. Kessler, and L. J. Parent. 2001. trans-Acting inhibition of genomic RNA dimerization by Rous sarcoma virus matrix mutants. J. Virol. 75:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golomb, M., and D. Grandgenett. 1979. Endonuclease activity of purified RNA-directed DNA polymerase from avian myeloblastosis virus. J. Biol. Chem. 254:1606-1613. [PubMed] [Google Scholar]
- 26.Haffar, O. K., S. Popov, L. Dubrovsky, I. Agostini, H. Tang, T. Pushkarsky, S. G. Nadler, and M. Bukrinsky. 2000. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. J. Mol. Biol. 299:359-368. [DOI] [PubMed] [Google Scholar]
- 27.Harel, J., E. Rassart, and P. Jolicoeur. 1981. Cell cycle dependence of synthesis of unintegrated viral DNA in mouse cells newly infected with murine leukemia virus. Virology 110:202-207. [DOI] [PubMed] [Google Scholar]
- 28.Hatziioannou, T., and S. P. Goff. 2001. Infection of nondividing cells by Rous sarcoma virus. J. Virol. 75:9526-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hindmarsh, P., M. Johnson, R. Reeves, and J. Leis. 2001. Base-pair substitutions in avian sarcoma virus U5 and U3 long terminal repeat sequences alter the process of DNA integration in vitro. J. Virol. 75:1132-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hindmarsh, P., T. Ridky, R. Reeves, M. Andrake, A. M. Skalka, and J. Leis. 1999. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J. Virol. 73:2994-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hizi, A., and W. K. Joklik. 1977. RNA-dependent DNA polymerase of avian sarcoma virus B77. I. Isolation and partial characterization of the α, β2, and αβ forms of the enzyme. J. Biol. Chem. 252:2281-2289. [PubMed] [Google Scholar]
- 32.Hizi, A., J. P. Leis, and W. K. Joklik. 1977. The RNA-dependent DNA polymerase of avian sarcoma virus B77. Binding of viral and nonviral ribonucleic acids to the α, β2, and αβ forms of the enzyme. J. Biol. Chem. 252:6878-6884. [PubMed] [Google Scholar]
- 33.Hu, S. C., D. L. Court, M. Zweig, and J. G. Levin. 1986. Murine leukemia virus pol gene products: analysis with antisera generated against reverse transcriptase and endonuclease fusion proteins expressed in Escherichia coli. J. Virol. 60:267-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes, S. H., A. Mutschler, J. M. Bishop, and H. E. Varmus. 1981. A Rous sarcoma virus provirus is flanked by short direct repeats of a cellular DNA sequence present in only one copy prior to integration. Proc. Natl. Acad. Sci. USA 78:4299-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphries, E. H., and J. M. Coffin. 1976. Rate of virus-specific RNA synthesis in synchronized chicken embryo fibroblasts infected with avian leukosis virus. J. Virol. 17:393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Humphries, E. H., C. Glover, and M. E. Reichmann. 1981. Rous sarcoma virus infection of synchronized cells establishes provirus integration during S-phase DNA synthesis prior to cellular division. Proc. Natl. Acad. Sci. USA 78:2601-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Humphries, E. H., and H. M. Temin. 1974. Requirement for cell division for initiation of transcription of Rous sarcoma virus RNA. J. Virol. 14:531-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones, K. S., J. Coleman, G. W. Merkel, T. M. Laue, and A. M. Skalka. 1992. Retroviral integrase functions as a multimer and can turn over catalytically. J. Biol. Chem. 267:16037-16040. [PubMed] [Google Scholar]
- 39.Katz, R. A., G. Merkel, J. Kulkosky, J. Leis, and A. M. Skalka. 1990. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell 63:87-95. [DOI] [PubMed] [Google Scholar]
- 40.Katzman, M., R. A. Katz, A. M. Skalka, and J. Leis. 1989. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol. 63:5319-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kukolj, G., K. S. Jones, and A. M. Skalka. 1997. Subcellular localization of avian sarcoma virus and human immunodeficiency virus type 1 integrases. J. Virol. 71:843-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kukolj, G., R. A. Katz, and A. M. Skalka. 1998. Characterization of the nuclear localization signal in the avian sarcoma virus integrase. Gene 223:157-163. [DOI] [PubMed] [Google Scholar]
- 43.Lee, Y. M., and J. M. Coffin. 1990. Efficient autointegration of avian retrovirus DNA in vitro. J. Virol. 64:5958-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, Y. M., and J. M. Coffin. 1991. Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol. Cell. Biol. 11:1419-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leis, J., G. Duyk, S. Johnson, M. Longiaru, and A. Skalka. 1983. Mechanism of action of the endonuclease associated with the αβ and ββ forms of avian RNA tumor virus reverse transcriptase. J. Virol. 45:727-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewis, P. F., and M. Emerman. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin, T. H., T. Quinn, M. Walsh, D. Grandgenett, and J. C. Lee. 1991. Avian myeloblastosis virus reverse transcriptase: effect of glycerol on its hydrodynamic properties. J. Biol. Chem. 266:1635-1640. [PubMed] [Google Scholar]
- 49.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller, M. D., B. Wang, and F. D. Bushman. 1995. Human immunodeficiency virus type 1 preintegration complexes containing discontinuous plus strands are competent to integrate in vitro. J. Virol. 69:3938-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris-Vasios, C., J. P. Kochan, and A. M. Skalka. 1988. Avian sarcoma-leukosis virus pol-endo proteins expressed independently in mammalian cells accumulate in the nucleus but can be directed to other cellular compartments. J. Virol. 62:349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Müller, B., D. Bizub-Bender, M. D. Andrake, K. S. Jones, and A. M. Skalka. 1995. Monoclonal antibodies against Rous sarcoma virus integrase protein exert differential effects on integrase function in vitro. J. Virol. 69:5631-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Müller, B., K. S. Jones, G. W. Merkel, and A. M. Skalka. 1993. Rapid solution assays for retroviral integration reactions and their use in kinetic analyses of wild-type and mutant Rous sarcoma virus integrases. Proc. Natl. Acad. Sci. USA 90:11633-11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mumm, S. R., P. J. Hippenmeyer, and D. P. Grandgenett. 1992. Characterization of a stable eukaryotic cell line expressing the Rous sarcoma virus integrase. Virology 189:500-510. [DOI] [PubMed] [Google Scholar]
- 55.Nissen, M. S., T. A. Langan, and R. Reeves. 1991. Phosphorylation by cdc2 kinase modulates DNA binding activity of high mobility group I nonhistone chromatin protein. J. Biol. Chem. 266:19945-19952. [PubMed] [Google Scholar]
- 56.Pluymers, W., P. Cherepanov, D. Schols, C. De, and Z. Debyser. 1999. Nuclear localization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology 258:327-332. [DOI] [PubMed] [Google Scholar]
- 57.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roth, M. J., P. L. Schwartzberg, and S. P. Goff. 1989. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell 58:47-54. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1994. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Temin, H., and S. Mizutani. 1970. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226:1211-1213. [DOI] [PubMed] [Google Scholar]
- 61.Temin, H. M. 1967. Studies on carcinogenesis by avian sarcoma viruses. VI. Differential multiplication of uninfected and of converted cells in response to insulin. J. Cell Physiol. 69:377-384. [DOI] [PubMed] [Google Scholar]
- 62.Vink, C., E. Yeheskiely, G. A. van der Marel, J. H. van Boom, and R. H. Plasterk. 1991. Site-specific hydrolysis and alcoholysis of human immunodeficiency virus DNA termini mediated by the viral integrase protein. Nucleic Acids Res. 19:6691-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vodicka, M. A., D. M. Koepp, P. A. Silver, and M. Emerman. 1998. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 12:175-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vora, A. C., R. Chiu, M. McCord, G. Goodarzi, S. J. Stahl, T. C. Mueser, C. C. Hyde, and D. P. Grandgenett. 1997. Avian retrovirus U3 and U5 DNA inverted repeats: role of nonsymmetrical nucleotides in promoting full-site integration by purified virion and bacterial recombinant integrases. J. Biol. Chem. 272:23938-23945. [DOI] [PubMed] [Google Scholar]
- 65.Vora, A. C., and D. P. Grandgenett. 1995. Assembly and catalytic properties of retrovirus integrase-DNA complexes capable of efficiently performing concerted integration. J. Virol. 69:7483-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Werner, S., and B. M. Wöhrl. 1999. Soluble Rous sarcoma virus reverse transcriptases α, αβ and β purified from insect cells are processive DNA polymerases that lack an RNase H 3′ > 5′ directed processing activity. J. Biol. Chem. 274:26329-26336. [DOI] [PubMed] [Google Scholar]
- 67.Werner, S., and B. M. Wöhrl. 2000. Asymmetric subunit organization of heterodimeric Rous sarcoma virus reverse transcriptase αβ: localization of the polymerase and RNase H active sites in the α subunit. J. Virol. 74:3245-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu, W. X., L. E. Henderson, T. D. Copeland, R. J. Gorelick, W. J. Bosche, A. Rein, and J. G. Levin. 1996. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J. Virol. 70:7132-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu, X. Y., H. M. Liu, H. L. Xiao, J. A. Conway, E. Hehl, G. V. Kalpana, V. Prasad, and J. C. Kappes. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 73:2126-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoder, K. E., and F. D. Bushman. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74:11191-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]