Abstract
The hepatitis B virus (HBV) core promoter regulates the transcription of two related RNA products named precore RNA and core RNA. Previous studies indicate that a double-nucleotide mutation that occurs frequently during chronic HBV infection converts a nuclear receptor binding site in the core promoter to the binding site of the transcription factor hepatocyte nuclear factor-1 (HNF-1) and specifically suppresses the transcription of the precore RNA. This mutation also changes two codons in the overlapping X protein coding sequence. In this report, we demonstrate that the X protein and its mutant Xmt can physically bind to HNF-1 both in vitro and in vivo. Further analyses indicate that both X and Xmt can enhance the gene transactivation and the DNA binding activities of HNF-1. This finding demonstrates for the first time that the X protein can stimulate the DNA binding activity of a homeodomain transcription factor. Interestingly, while both X and Xmt can stimulate the HNF-1 activities, they differ in their effects: a smaller amount of Xmt is needed to generate greater transactivation and DNA binding activities of HNF-1. This functional difference between X and Xmt may have important implications in HBV pathogenesis and is apparently why they have different effects on the core promoter bearing the HNF-1 binding site.
Hepatitis B virus (HBV) is a liver-tropic virus. It can cause acute and chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. HBV is a DNA virus with a 3.2-kb, partially double-stranded genome. This genome contains four genes, S, P, C, and X. The S gene codes for the viral envelope proteins commonly known as surface antigens. The P gene codes for the viral DNA polymerase that is also a reverse transcriptase. The C gene codes for the precore protein and the core protein. The precore protein is the precursor of the serum e-antigen, and the core protein is the major protein constituent of the viral core particle. The X gene codes for a 16.5-kDa protein that can regulate the cellular signaling pathways (6, 12, 16, 17, 27), enhance the DNA binding activities of the bZip transcription factors (1, 3, 4, 22), and facilitate hepatocellular oncogenesis in transgenic mice (15, 21). Due to its small genome size, these four HBV genes overlap each other extensively, and every nucleotide in the HBV genome has at least one coding function.
The transcription of the HBV genes is regulated by four promoters and two enhancers (29). One of the promoters, named the core promoter, regulates the transcription of precore RNA and core RNA, which code for the precore protein and the core protein, respectively. Both the precore RNA and the core RNA are larger than the genome size, but only the latter also serves as the mRNA for the synthesis of the viral DNA polymerase (24, 25). In addition, the core RNA also serves as the pregenomic RNA that is packaged by the core protein to form the viral core particle. This pregenomic RNA is subsequently converted to the viral DNA genome by the viral DNA polymerase that is also packaged.
During chronic HBV infection, HBV mutants may be generated. One frequent double mutation that converts nucleotide (nt) 1765 from A to T and nt 1767 from G to A is identified in over 80% of the HBV genomes isolated from patients with chronic hepatitis symptoms (8, 18). This double mutation resides in the core promoter. Previous studies indicate that this double mutation specifically suppresses the precore RNA transcription without affecting the core RNA transcription (9, 10, 14). Further studies indicate that this double mutation converts a nuclear receptor binding site to the binding site of the hepatocyte nuclear factor-1 (HNF-1) (19). HNF-1 is a transcription factor that regulates the expression of a large number of genes in liver, kidney, the digestive tract, and pancreatic β-cells (13). This transcription factor is 628 amino acids in length with a relative molecular mass of approximately 80 to 90 kDa due to posttranslational modification (2, 13). HNF-1 contains an amino-terminal dimerization domain, an atypical homeodomain for DNA binding, and a carboxy-terminal transactivation domain (5, 13).
Due to the sequence overlap, this double mutation also resides in the X protein coding sequence and changes codons 130 and 131 of the X protein from Lys-Val to Met-Ile (19). Our previous studies suggested that the specific suppression of the precore RNA transcription was due to the interaction between HNF-1 and the mutated X protein (19). In this report, we have studied the molecular mechanisms that mediate the interaction between the X protein and HNF-1 and the functional consequences of this interaction. We demonstrate that both the wild-type X protein and the mutated X protein (Xmt) can physically bind to HNF-1 and enhance the DNA binding activity of HNF-1. However, these two X proteins differ in their activities, which results in their differential regulation of the core promoter and the observed suppression of the precore RNA transcription.
MATERIALS AND METHODS
DNA plasmids.
pWTD contains the head-to-tail dimer of the wild-type HBV genome of the adw2 subtype (19). pWTDX− is identical to pWTD with the exception that it contains an A-to-C mutation at nucleotide (nt) 1377, which removes the initiation codon of the X protein coding sequence, and a C-to-T mutation at nt 1398, which creates a premature termination codon in the X protein sequence. These two mutations abolished the expression of the 16.5-kDa X protein (28a). pM1D is identical to pWTD except that it contains the nt 1765 A-to-T and nt 1767 G-to-A double mutation in the core promoter (9, 19). pM1DX− is identical to pM1D with the exception that it contains the two additional mutations at nt 1377 and nt 1398 that prevent the expression of the 16.5-kDa X protein. pCMV-HNF-1 has been described previously (19).
pECE-X is an X protein-expressing plasmid. In this plasmid, the expression of the X protein is under the control of the simian virus 40 (SV40) early promoter (26). pECE-Xmt is identical to pECE-X with the exception that it contains the nt 1765 A-to-T and nt 1767 G-to-A double mutation in its X protein coding sequence. pCMV-HAX contains the hemagglutinin (HA)-tagged X protein coding sequence, which is under the expression control of the immediate-early promoter of cytomegalovirus (CMV). To construct this DNA plasmid, a double-stranded oligonucleotide, 5′-AGCTTCACCATGTACCCATACGACGTCCCAGACTACGCTTC-3′-3′-AGTGGTACATGGGTATGCTGCAGGGTCTGATGCGAAGGTAC-5′, containing the coding sequence of the HA tag was synthesized. This oligonucleotide contained a HindIII site at one end and an NcoI site at the other end. This oligonucleotide was joined in frame to the X protein coding sequence via the NcoI site located at the 5′ end of the X protein coding sequence. The fused HA-X sequence was then inserted into the HindIII and NotI (blunt-ended) sites of the pRc/CMV vector (Invitrogen).
pCMV-HAXmt is identical to pCMV-HAX with the exception that it contains the nt 1765 A-to-T and nt 1767 G-to-A double mutation. pECE-HAX contains the HA-tagged X protein sequence. The HindIII/XbaI fragment that contains the coding sequence of the HA-tagged X protein was isolated from pCMV-HAX and then inserted into the HindIII and XbaI sites of the pECE-1 vector (26). pECE-HAXmt is identical to pECE-HAX with the exception that it contains the nt 1765 A-to-T and nt 1767 G-to-A double mutation. pET-HAX was constructed by inserting the HA-X coding sequence into the NdeI and BamHI sites of pET-3a (Novagen). The HA-X sequence in this case was isolated by PCR with NdeI site and BglII sites engineered at the 5′ end and the 3′ end, respectively. pET-HAXmt is identical to pET-HAX except that it contains the nt 1765 A-to-T and nt 1767 G-to-A double mutation. pCpHNF1-Luc contains four copies of the HNF-1 binding site derived from nt 1750 to 1779 of the core promoter bearing the nt 1765 A-to-T and nt 1767 G-to-A double mutation. In this plasmid, the minimal thymidine kinase (TK) promoter from pBLCAT-2 (11) was inserted into the SmaI and XhoI sites of pGL3-Luc (Promega) to regulate the expression of the luciferase reporter. The HNF-1 site was inserted upstream of the TK promoter.
Cell culture and DNA transfection.
Huh7 hepatoma cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Cells were plated in a 60-mm petri dish the day before transfection and transfected when they were 80% confluent. Each plate of cells was transfected with 10 μg of DNA with the calcium phosphate precipitation method. In all cases, pXGH5 (Nichols Diagnostics) was included in the transfection procedures to serve as an internal control to monitor the transfection efficiency. pXGH5 contained the human growth hormone reporter under the expression control of the mouse metallothionein promoter. Cells were lysed 48 h after transfection for the luciferase assay. The incubation medium was harvested for analyzing human growth hormone activity with a radioimmunoassay kit (Nichols Diagnostics).
In vitro coimmunoprecipitation of HNF-1 and X proteins.
HNF-1, HA-X, and HA-Xmt RNAs were synthesized in vitro from their respective DNA templates, pCMV-HNF-1, pCMV-HAX, and pCMV-HAXmt, with the T7 RNA polymerase (Promega). The protein translation reaction, which contained 0.5 μg of RNA, 10 μl of rabbit reticulocyte lysates (Promega), 0.5 μl of 1 mM amino acid mixture minus methionine, and 10 μCi of [35S]methionine, was carried out at 30°C for 1 h. For coimmunoprecipitation, 2 μl of HNF-1 translational mixture and 2 μl of either HA-X or HA-Xmt translational mixture were mixed and incubated at room temperature (RT) for 30 min prior to the addition of 0.5 ml of Tris-buffered saline (TBS; 10 mM Tris-HCl, pH 7.0, 150 mM NaCl) containing 0.5% Nonidet P-40 (NP-40). The sample was then mixed with 1 μl of monoclonal anti-HA antibody (Roche Diagnostics) and further incubated at 4°C overnight. The immunocomplex was then precipitated with Pansorbin (Calbiochem, La Jolla, Calif.), subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12.5% gel, and then autoradiographed and analyzed with a PhosphorImager (Molecular Dynamics, Sunnyville, Calif.).
In vivo coimmunoprecipitation of HNF-1 and X proteins.
Huh7 cells in a 10-cm dish were cotransfected with 10 μg of pCMV-HNF-1 and 10 μg of either pECE-HAX or pECE-HAXmt. Forty-eight hours after transfection, cells were lysed by sonication on ice for 2 min in TBS containing 0.5% NP-40. The cell debris was removed by a brief centrifugation in a microcentrifuge. The supernatant was then immunoprecipitated with the mouse anti-HA antibody with the same procedures described above. The immunocomplex precipitated with Pansorbin was then analyzed by Western blotting with the rabbit anti-HNF-1 primary antibody (19) and the horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin secondary antibody. Alternatively, the cell lysates were first immunoprecipitated with 1 μl of rabbit anti-HNF-1 antibody, followed by Western blotting with the anti-HA primary antibody and the horseradish peroxidase-conjugated goat anti-mouse immunoglobulin secondary antibody.
Expression of HA-X and HA-Xmt in E. coli.
Escherichia coli BL21(DE3) cells were transformed with pET-HAX or pET-HAXmt for the expression of HA-X or HA-Xmt, respectively, with our previous procedures (20). After the induction of protein expression with 1 mM isopropylthiogalactopyranoside (IPTG), E. coli cells were pelleted by centrifugation and lysed by sonication on ice. After centrifugation, the pellet, which contained the insoluble HA-X or HA-Xmt, was resuspended in 6 M guanidine hydrochloride. After a brief centrifugation to remove the insoluble cell debris, the protein sample was dialyzed against phosphate-buffered saline (PBS). The dialysates were centrifuged for 2 min in a microcentrifuge, and the pellet was resuspended by sonication in a solution containing 37.5% urea and 5% acetic acid.
The HA-X or HA-Xmt protein was then purified on an acid-urea gel by the published procedures with slight modifications (23). Briefly, methylene blue was added to the sample to a final concentration of 0.1%. The HA-X or HA-Xmt protein was then purified on a 12.5% polyacrylamide gel containing 37.5% urea and 5% glacial acetic acid. The running buffer was 5% acetic acid. Prior to loading the samples, the gel was run for 1.5 to 2 h at 10 V/cm. The samples were then electrophoresed until the methylene blue dye was in the middle of the gel. The gel pieces containing HAX or HAXmt were then isolated and soaked in 6 M urea-5% acetic acid in a dialysis bag at 4°C overnight. The protein sample was then electroeluted. The eluates were collected and first dialyzed against PBS and then against 10 mM NH4HCO3 containing 0.005% NP-40. Protein samples were lyophilized, dissolved in 50 μl of PBS, and stored at −80°C in small aliquots. A blank gel piece was processed simultaneously to serve as a negative control. The purity and the identity of the protein were determined by silver staining (Bio-Rad Silver Stain Plus) and Western blotting.
Gel shift assay.
The preparation of the Huh7 nuclear extracts and the gel shift assay were described previously (9, 19). The oligonucleotide probe used for the gel shift contained the sequence 5′-GAGGAGATTAGGTTAATGATCTTTGTAT-3′-3′ CTCTAATCCAATTACTAGAAACATAATC-5′. This sequence is derived from the HBV core promoter with the nt 1765 A-to-T and nt 1767 G-to-A double mutation.
Primer extension analyses.
Total cellular RNA was isolated from Huh7 cells transfected with various HBV genomic DNA constructs and used for the primer extension analysis. The sequence of the antisense primer used for primer extension for analyzing the C gene transcripts was 5′-GGTGAGCAATGCTCAGGAGACTCTAAGG-3′, which corresponds to nt 2051 to 2024 of the HBV genome. The antisense primer sequence used for analyzing the human growth hormone transcript was 5′-GCCACTGCAGCTAGGTGAGCGTCC-3′. The primer extension reaction was carried out as previously described (19).
RESULTS
Interaction between HNF-1 and the X protein and its mutant in vitro.
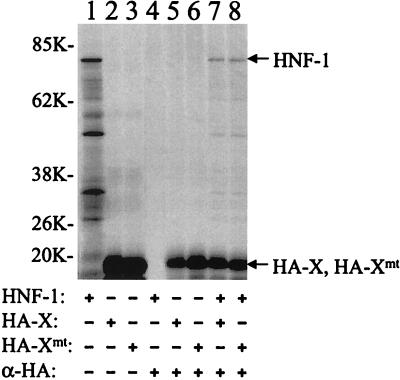
Our previous results suggested that Xmt, the X protein with the Lys-Val to Met-Ile double mutation at amino acids 130 and 131, could interact with HNF-1 to regulate HBV C gene expression (19). To study this interaction, we examined whether HNF-1 and Xmt could physically bind to each other. Since the X protein reacts poorly with its antibodies, we had fused the HA tag, an antigenic epitope derived from influenza virus hemagglutinin, to the amino terminus of the X protein to facilitate the analysis. HNF-1 and HA-tagged X and Xmt proteins were synthesized in vitro with the rabbit reticulocyte lysates and radiolabeled with [35S]methionine. As shown in Fig. 1, although HNF-1 could not be immunoprecipitated by the anti-HA antibody, it could be precipitated by the anti-HA antibody in the presence of either the HA-tagged X protein (HA-X) or the HA-tagged Xmt protein (HA-Xmt). The ability of HNF-1 to be coimmunoprecipitated with HA-X and HA-Xmt indicated that HNF-1 could bind to either of these two proteins.
FIG. 1.
Coimmunoprecipitation of HNF-1 with X and Xmt proteins in vitro. HNF-1, HA-X, and HA-Xmt were labeled with [35S]methionine and synthesized in vitro with rabbit reticulocyte lysates. Details of the procedures are described in Materials and Methods. They were then subjected to gel electrophoresis without immunoprecipitation (lanes 1 to 3) or after immunoprecipitation (lanes 4 to 8) with the anti-HA antibody. Lane 1, HNF-1 without immunoprecipitation; lane 2, HA-X without immunoprecipitation; lane 3, HA-Xmt without immunoprecipitation; lane 4, HNF-1 with immunoprecipitation with the anti-HA antibody; lane 5, HA-X with immunoprecipitation; lane 6, HA-Xmt with immunoprecipitation; lane 7, HA-X and HNF-1 immunoprecipitated with the anti-HA antibody; and lane 8, HA-Xmt and HNF-1 immunoprecipitated with the anti-HA antibody. The arrows mark the locations of the HNF-1, HA-X, and HA-Xmt bands. In lane 1, protein bands smaller than the size of HNF-1 were also detected. The nature of these bands is unclear. They might be degraded HNF-1 or N-terminally truncated HNF-1 synthesized from the internal ATG codons. Sizes are shown on the left in kilodaltons.
Interaction between HNF-1 and the X proteins in vivo.
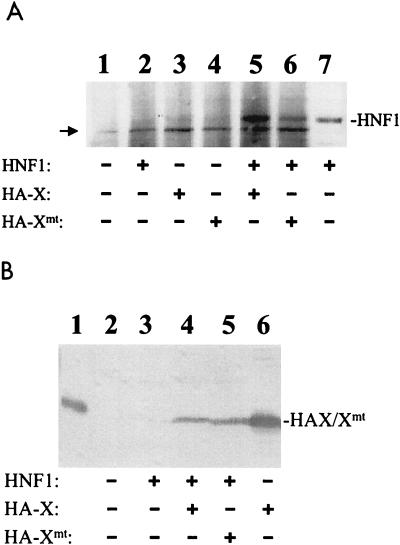
To investigate whether HNF-1 and the two X protein variants could also bind to each other in vivo, we expressed HNF-1, HA-X, and HA-Xmt either separately or together in Huh7 cells, a well-differentiated human hepatoma cell line. The cells were lysed 48 h after transfection, and the cell lysates were immunoprecipitated with the mouse monoclonal anti-HA antibody. The immunoprecipitates were then analyzed by Western blotting with the rabbit anti-HNF-1 antibody. As shown in Fig. 2A, lane 2, the anti-HA antibody could not precipitate HNF-1 if HNF-1 was expressed in the absence of the X protein. However, if HNF-1 was coexpressed with either HA-X or HA-Xmt, its signal could clearly be detected by Western blotting in the coimmunoprecipitation experiment (Fig. 2A, lanes 5 and 6). These results indicated that HNF-1 could also bind to HA-X and HA-Xmt in Huh7 cells.
FIG. 2.
Coimmunoprecipitation of HNF-1 with X and Xmt expressed in Huh7 cells. (A) Coimmunoprecipitation with anti-HA antibody as the primary antibody. Huh7 cells were transfected with the control pRc/CMV vector alone (lane 1), pCMV-HNF-1 (lanes 2 and 7), pCMV-HAX (lane 3), pCMV-HAXmt (lane 4), pCMV-HNF-1 and pCMV-HAX (lane 5), or pCMV-HNF-1 and pCMV-HAXmt (lane 6). Two days after transfection, cells were lysed, immunoprecipitated with the mouse anti-HA antibody, and then analyzed by Western blotting with the rabbit anti-HNF-1 antibody (lanes 1 to 6). In lane 7, the lysates of cells transfected by pCMV-HNF-1 were analyzed directly by Western blotting without immunoprecipitation. This lane served as a positive control for identifying the HNF-1 protein band. The arrow marks the location of a background band. (B) Coimmunoprecipitation with the anti-HNF-1 antibody as the primary antibody. The coimmunoprecipitation procedures were the same as in panel A except that the rabbit anti-HNF-1 antibody was used for immunoprecipitation, followed by Western blotting with the anti-HA antibody. Lane 1, HA-tagged X protein purified from E. coli, which served as a marker; lane 2, Huh7 cells transfected with pRc/CMV control vector; lane 3, Huh7 cells transfected with pCMV-HNF-1; lane 4, Huh7 cells transfected with pCMV-HNF-1 and pCMV-HAX; lane 5, cells transfected with pCMV-HNF-1 and pCMV-HAXmt; lane 6, cell lysates prepared from cells transfected with pCMV-HAX without immunoprecipitation with the anti-HNF-1 antibody.
Note that Huh7 cells also expressed endogenous HNF-1. However, the signal of endogenous HNF-1 was not apparent in the coimmunoprecipitation assay when Huh7 cells were transfected with only the HA-X- or HA-Xmt-expressing plasmid (Fig. 2A, lanes 3 and 4). This lack of signal was likely due to its low expression level and the sensitivity of the coimmunoprecipitation assay. A reciprocal immunoprecipitation with the anti-HNF-1 antibody followed by Western blot analysis with the anti-HA antibody was also conducted to demonstrate the interaction between HNF-1 and the X protein and its mutant Xmt. As shown in Fig. 2B, HA-X and HA-Xmt could also be coprecipitated with HNF-1 by the anti-HNF-1 antibody.
Differential regulation of HNF-1 activity by X and Xmt.
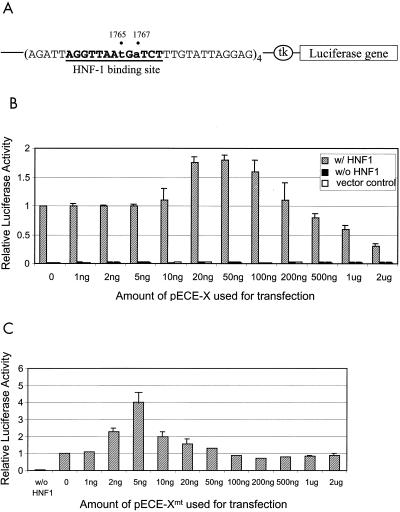
In order to determine the possible effects of X and Xmt on the activities of HNF-1, we created a luciferase reporter plasmid, pCpHNF1-Luc. In this plasmid, the expression of luciferase is under the control of the minimal TK promoter, which was linked to four copies of the HNF-1 binding sites derived from the core promoter that contained the nt 1765 A-to-T and nt 1767 G-to-A double mutation (Fig. 3A). Huh7 cells were cotransfected with pCpHNF1-Luc or its control reporter lacking the HNF-1 binding sites, the HNF-1-expressing plasmid pCMV-HNF-1 or its control vector pR/cCMV, and pECE-X or its control vector pECE-1. As shown in Fig. 3B, cotransfection of the pCpHNF1-Luc reporter with the pR/cCMV control expression vector generated a low luciferase activity similar to that produced by the control reporter lacking the HNF-1 binding site, indicating that the low endogenous HNF-1 level in Huh7 cells was insufficient to enhance the luciferase expression from pCpHNF1-Luc.
FIG. 3.
Effects of X and Xmt on the transactivation activities of HNF-1. (A) Illustration of the HNF-1 reporter construct. The luciferase reporter is boxed, and the HNF-1 binding sequence is shown in boldface letters and underlined. A minimal thymidine kinase (tk) promoter controlled the expression of the luciferase reporter. (B) Histogram of the transactivation activities of X and HNF-1. Shaded box, cotransfection of pCpHNF1-Luc and pCMV-HNF-1; solid box, transfection of pCpHNF1-Luc with pRc/CMV; empty box, transfection of the control reporter lacking the HNF-1 binding site. (C) Histogram of the transactivation activities of Xmt and HNF-1. In both B and C, 3 μg of pCMV-HNF-1 or pRc/CMV was used for the cotransfection with 4 μg of pCpHNF1-Luc or the control reporter ptk-Luc. The amount of pECE-X or pECE-Xmt used for the transfection is indicated at the bottom of the chart. In all cases, 40 ng of pXGH5, a plasmid that expressed the human growth hormone, was also used for cotransfection to monitor the transfection efficiency. All the luciferase activities expressed were normalized against the luciferase activity derived from the cotransfection of pCMV-HNF-1, pCpHNF1-Luc, and pECE-1. The results represent the averages of at least three independent experiments.
In contrast, the cotransfection of pCpHNF1-Luc with the HNF-1-expressing plasmid pCMV-HNF-1 increased the luciferase activity more than 30-fold. This transactivation activity of HNF-1 was further increased by the X protein-expressing plasmid pECE-X in a dose-dependent manner in the cotransfection experiments. The maximal activity was achieved at 50 ng of pECE-X. This activity then gradually decreased when the amount of pECE-X used for the cotransfection was further increased. Since the luciferase activity expressed from the pCpHNF1-Luc reporter in the absence of the HNF-1-expressing plasmid was not affected by pECE-X, the regulatory effect of the X protein on the reporter expression was apparently mediated by HNF-1.
The ability of Xmt to stimulate HNF-1-mediated transcription was also tested. A similar dose-dependent activation of the luciferase activity by the Xmt protein in the presence of pCMV-HNF-1 was also observed, with the luciferase activity reaching the peak of an approximately fourfold stimulation when 5 ng of pECE-Xmt was used for cotransfection. The luciferase activity then gradually decreased when the pECE-Xmt amount was further increased. As with X, Xmt was not able to stimulate reporter expression in the absence of the HNF-1-expressing plasmid pCMV-HNF-1 (data not shown). Thus, both X and Xmt could stimulate the activity of HNF-1, although they differed in their effects: a smaller amount of Xmt was needed to generate a greater stimulatory effect on the HNF-1 activity.
The difference between the twofold stimulation and the fourfold stimulation of HNF-1 activity by X and Xmt (Fig. 3B and 3C, respectively) was reproducible and significant (P < 0.01). This difference was not due to the different stabilities of X and Xmt in Huh7 cells, as the Western blot analysis of the Huh7 cell lysates with the anti-HA antibody revealed no difference in their expression levels (data not shown).
Enhancement of the DNA binding activity of HNF-1 by X and Xmt.
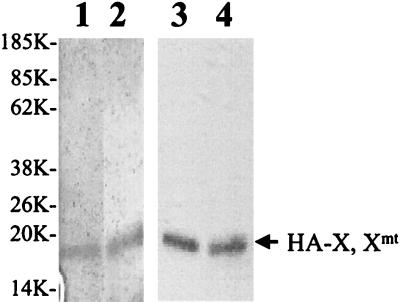
To further understand how X and Xmt might regulate the HNF-1 activity, we also performed gel shift assays to examine whether the DNA binding activity of HNF-1 was affected by X and Xmt. An oligonucleotide containing the HNF-1 binding sequence derived from the core promoter with the nt 1765 A-to-T and nt 1767 G-to-A double mutation was used as the probe. The HA-X and HA-Xmt proteins were expressed in E. coli and subsequently purified. The purity of these two proteins was analyzed by silver staining, and the identity was verified by Western blotting (Fig. 4). Based on silver staining, the purity of these two X proteins was estimated to be at least 80%.
FIG. 4.
HA-X and HA-Xmt proteins expressed in E. coli. The procedures for the expression of HA-X and HA-Xmt in E. coli and their subsequent purifications are described in Materials and Methods. Lanes 1 and 2, silver staining of HA-X (lane 1) and HA-Xmt (lane 2) purified from E. coli. Lanes 3 and 4, Western blot analysis of HA-X (lane 3) and HA-Xmt (lane 4) with the anti-HA antibody. The arrow marks the locations of HA-X and HA-Xmt. Sizes are shown in kilodaltons.
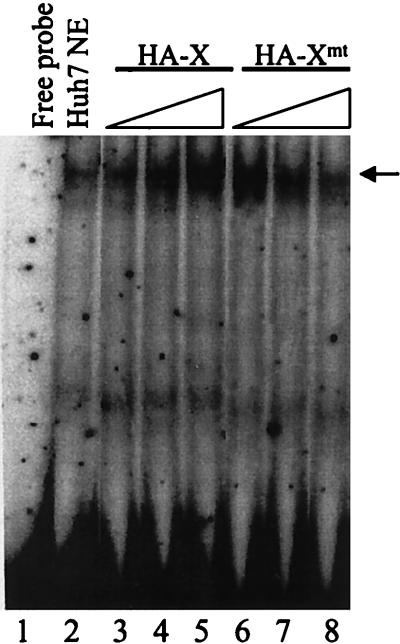
As shown in Fig. 5, in agreement with our previous results, the addition of Huh7 nuclear extracts resulted in the generation of the HNF-1 band shift (19) (also see below). The signal of this band shift could be enhanced by the X protein in a dose-dependent manner. The addition of 0.2 ng, 20 ng, and 100 ng of HA-X protein resulted in successive increases in the DNA binding activity of HNF-1. The HA-Xmt protein was also able to stimulate the DNA binding activity of HNF-1 in the same assay. However, 0.2 ng of HA-Xmt was sufficient to stimulate the DNA binding activity of HNF-1 to about the same degree as 100 ng of HA-X did. Further increase of the HA-Xmt amount in the gel shift assay resulted in reduction of the HNF-1 DNA binding activity, and when 100 ng of HA-Xmt was used in the binding reaction, the HNF-1 DNA binding activity was reduced to a level similar to that when no HA-Xmt was used. Thus, as in the luciferase reporter assay, although both HA-Xmt and HA-X could stimulate the DNA binding activity of HNF-1, they differed in their effects.
FIG. 5.
Enhancement of the DNA binding activity of HNF-1 by HA-X and HA-Xmt. The procedures for the gel shift experiment are described in Materials and Methods. Lane 1, free DNA probe; lane 2, free DNA probe mixed with the Huh7 nuclear extracts; lanes 3 to 5, the addition of 0.2 ng, 20 ng, and 100 ng of HA-X, respectively, into the binding reaction mixture; lanes 6 to 8, the addition of 0.2 ng, 20 ng, and 100 ng of HA-Xmt, respectively, into the binding reaction mixture. The location of the HNF-1 band shift is indicated by an arrow.
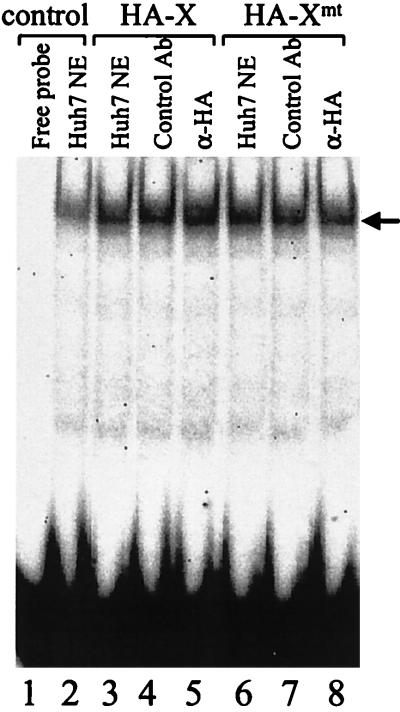
Since the X protein could bind to HNF-1, we decided to examine whether HA-X and HA-Xmt were part of the HNF-1-DNA complex by performing the supershift assay with the anti-HA antibody. As shown in Fig. 6, addition of the anti-HNF-1 antibody in the binding reaction resulted in the removal of the band shift signal (lane 3), confirming that the band shift was indeed caused by HNF-1. The inclusion of 1 ng of HA-X or HA-Xmt in the binding reaction stimulated the DNA binding activity of HNF-1 roughly twofold (lanes 4 to 9), indicating that HA-X and HA-Xmt at this amount had similar stimulatory effects on HNF-1. Since the addition of the anti-HA antibody could not supershift or reduce the band shift signal of HNF-1, HA-X or HA-Xmt was apparently not part of the HNF-1-DNA complex in this assay.
FIG. 6.
Absence of HA-X and HA-Xmt in the HNF-1 band shift. The supershift assay was conducted as described in Materials and Methods. Lane 1, free DNA probe; lane 2, DNA probe incubated with Huh7 nuclear extract (NE); lane 3, Huh7 nuclear extract plus 1 ng of X protein; lane 4, the same as lane 3 but with the addition of a control mouse antibody; lane 5, the same as lane 4 but with the addition of the anti-HA antibody; lane 6, nuclear extracts plus 1 ng of HA-Xmt protein; lane 7, the same as lane 6 but with the addition of a control mouse antibody; lanes 8, the same as lane 7 but with the addition of the anti-HA antibody. The location of the HNF-1 band shifts is indicated by an arrow.
Regulation of HBV C gene transcription by HNF-1, X, and Xmt.
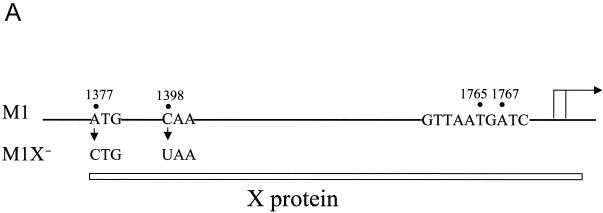
To further investigate how HNF-1, X, and Xmt might interact with each other to regulate the expression of the HBV C gene, we introduced an A-to-C mutation at nt 1377 and a C-to-T mutation at nt 1398 in the HBV genome (Fig. 7A). The former removed the initiation codon for the translation of the X protein, and the latter created a premature termination codon in the X protein coding sequence. Both mutations would prevent the expression of the 16.5-kDa X protein. These two mutations were introduced into both the wild-type HBV genome and the HBV genome that contained the nt 1765 A-to-T and nt 1767 G-to-A double mutation (the M1 mutant). HBV genomic dimers with or without the ability to express the X protein were then transfected into Huh7 cells. The cellular RNA was then isolated and analyzed for transcription of HBV precore RNA and core RNA by primer extension.
FIG. 7.
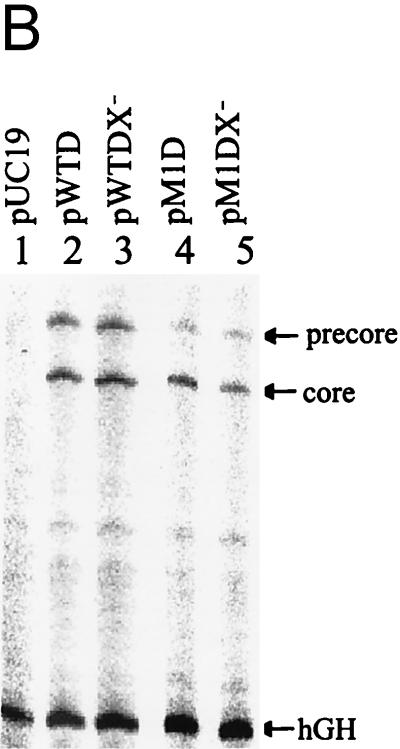
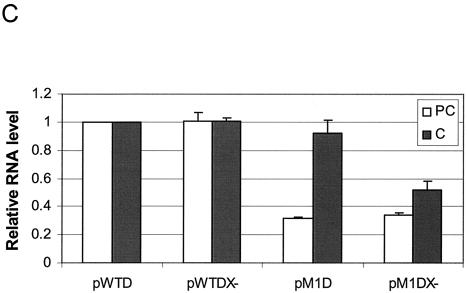
Effects of X and Xmt on transcription of precore and core RNAs. (A) Illustration of the HBV genome with the mutations that abolished the expression of the Xmt protein. The locations of the nt 1376 A to C and nt 1397 C to U double mutations that abolished the expression of the Xmt protein are indicated. The HNF-1 binding sequence in the M1 genome is also shown. The rightward arrow marks the locations of precore RNA and core RNA transcription initiation sites. (B) Primer extension analysis of precore RNA and core RNA transcribed from various HBV genomic DNA constructs. Huh7 cells transfected with various HBV genomic dimers were lysed 2 days after transfection. The total cellular RNA was then extracted and used for the primer extension analysis. Lane 1, cells transfected with the control pUC19 vector; lane 2, cells transfected with the wild-type HBV genome (pWTD); lane 3, cells transfected with the wild-type HBV genome with mutations that abolished the expression of the X protein (pWTDX−); lane 4, the M1 HBV genome (pM1D); lane 5, the M1 genome with mutations that abolished Xmt protein expression (pM1DX−). In all cases, pXGH5, a plasmid that expresses the human growth hormone (hGH), was used for cotransfection to serve as the internal transfection control. The locations of the precore RNA and the core RNA as well as the internal control human growth hormone RNA are indicated by arrows. The procedures for the primer extension analysis are described in Materials and Methods. (C) Quantitation analysis of the results shown in panel B. The signals of the precore (PC) RNA and the core (C) RNA bands were measured by densitometry and calculated based on the equation [(Xa − B)/(XWTD − B)]/[TEa/TEWTD], where Xa is the precore RNA (or the core RNA) signal of sample a, XWTD is the precore RNA (or the core RNA) signal expressed by pWTD, B is the background signal from a randomly selected area of the gel, TEa is the transfection efficiency of sample a as determined by the human growth hormone RNA internal control, and TEWTD is the transfection efficiency of pWTD. The results represent the averages of three independent experiments.
As shown in Fig. 7B, the two mutations that abolished the expression of the X protein from the wild-type genome did not affect the transcription of the precore RNA and the core RNA in Huh7 cells. This was consistent with the previous reports (7, 30), which indicated that the X protein did not play a role in HBV replication in Huh7 cells. Also in agreement with the previous reports, the nt 1765 A-to-T and nt 1767 G-to-A double mutation, which created the M1 genome, reduced the precore RNA level by 70% without significantly affecting the core RNA level. The additional double mutation that abolished the expression of the Xmt protein from the M1 genome reduced the transcription level of the core RNA by nearly 50% without further reducing the precore RNA level. The quantitation results are shown in Fig. 7C. These results indicated that Xmt was a weak transactivator and could increase the transcriptional efficiency of the core RNA.
DISCUSSION
Our previous studies indicated that a frequent double nucleotide mutation in the HBV core promoter converted a nuclear receptor binding site to an HNF-1 binding site and mutated two codons in the X protein sequence (9, 19). Further analysis indicated a suppression of the precore RNA transcription, possibly due to the interaction between HNF-1 and the mutated X protein (9, 19).
In this report, we have investigated this possible interaction between HNF-1 and the X protein. Our coimmunoprecipitation studies indicated that HNF-1 could physically bind to the X protein and its mutant Xmt both in vitro and in vivo (Fig. 1 and 2). This interaction is likely the reason why the activities of HNF-1 could be stimulated by both X and Xmt in a dose-dependent manner (Fig. 3). Although X and Xmt could both stimulate the gene transactivation activity of HNF-1, they were different in their activities. A smaller amount of Xmt than of X was needed to stimulate greater HNF-1 activity. The ability of X and Xmt to stimulate the gene transactivation activity of HNF-1 suggested that these two proteins might interact with HNF-1 in the nucleus to enhance its activity. Indeed, linking the nuclear localization signal of the SV40 T antigen to the X protein to suppress its cytoplasmic localization did not impair its ability to stimulate the gene transactivation activity of HNF-1 (data not shown).
The molecular mechanisms by which the X protein might regulate the HNF-1 activity were also studied. The results shown in Fig. 5 indicated that X and Xmt could stimulate the DNA binding activity of HNF-1. However, in spite of their abilities to bind to HNF-1 directly, X and Xmt did not appear to stably associate with HNF-1 under our assay conditions (Fig. 6). Previous studies indicated that the X protein could stimulate the DNA binding activities of bZip transcription factors (1, 3, 4, 22, 28). Similarly, X was not observed to be part of the protein-DNA complex in those studies (3, 4, 22, 28). Thus, our finding was consistent with the previous observations. Although both X and Xmt could stimulate the DNA binding activity of HNF-1, similar to the reporter studies shown in Fig. 3, they again differed in their activities: a smaller amount of Xmt than of X could generate a higher DNA binding activity of HNF-1 (Fig. 5).
The studies of the roles of X and Xmt in HBV C gene regulation in the context of the entire HBV genome also suggested that these proteins were functionally different. While abolishing X protein expression had no effect on transcription of the precore RNA and the core RNA from the wild-type HBV genome (Fig. 7) (7, 30), abolishing the expression of Xmt reduced the expression level of the core RNA from the M1 genome (Fig. 7). It is interesting that the double codon mutation in Xmt is located in a sequence that is highly conserved among mammalian hepadnavirus X proteins (16). Thus, it is conceivable that this double mutation affects the activity of the Xmt protein and/or its interaction with the transcriptional cofactors.
In conclusion, we have demonstrated that X and Xmt physically interact with HNF-1 and stimulate its DNA binding activity. Our finding thus expanded the transcription factors activated by X beyond the bZip family to include the homeodomain protein HNF-1. We have addressed the relevance of the interaction on core RNA and precore RNA transcription in the context of the entire HBV genome. The double mutation, which converted nt 1765 from A to T and nt 1767 from G to A in the HBV genome, is highly specific. Based on our previous results and the current studies, it is clear that this specificity is due in part to the need to create the HNF-1 binding site and in part to the need to create Xmt that is functionally similar to the wild-type X protein but yet distinct from it. As the creation of the HNF-1 site in the core promoter by the frequent double mutation would inevitably lead to the production of Xmt, it is unlikely that X would interact with HNF-1 during natural HBV infection to regulate the transcription of the C gene.
The HBV X protein has been shown to enhance hepatocellular oncogenesis in transgenic mice (15, 21). It will be interesting to determine whether Xmt also possesses the same or enhanced activity in similar studies.
Acknowledgments
We thank Ben Yen for critical reading of the manuscript and members of J. Ou's laboratory for helpful discussions during this project.
This work was supported by a research grant from the National Institutes of Health.
REFERENCES
- 1.Andrisani, O. M., and S. Barnabas. 1999. The transcriptional function of the hepatitis B virus X protein and its role in hepatocarcinogenesis. Int. J. Oncol. 15:373-379. [DOI] [PubMed] [Google Scholar]
- 2.Bach, I., M. Pontoglio, and M. Yaniv. 1992. Structure of the gene encoding hepatocyte nuclear factor 1 (HNF-1). Nucleic Acids Res. 20:4199-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnabas, S., and O. M. Andrisani. 2000. Different regions of hepatitis B virus X protein are required for enhancement of bZip-mediated transactivation versus transrepression. J. Virol. 74:83-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnabas, S., T. Hai, and O. M. Andrisani. 1997. The hepatitis B virus X protein enhances the DNA binding potential and transcription efficacy of bZip transcription factors. J. Biol. Chem. 272:20684-20690. [DOI] [PubMed] [Google Scholar]
- 5.Baumhueter, S., D. B. Mendel, P. B. Conley, C. J. Kuo, C. Turk, M. K. Graves, C. A. Edwards, G. Courtois, and G. R. Crabtree. 1990. HNF-1 shares three sequence motifs with the POU domain proteins and is identical to LF-B1 and APF. Genes Dev. 4:372-379. [DOI] [PubMed] [Google Scholar]
- 6.Benn, J., F. Su, M. Doria, and R. J. Schneider. 1996. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J. Virol. 70:4978-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blum, H. E., Z. S. Zhang, and E. Galun. 1992. Hepatitis B virus X protein is not essential to the viral life cycle in vitro. J. Virol. 12:4229-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckwold, V. E., and J. Ou. 1999. Hepatitis B virus C-gene expression and function: the lessons learned from viral mutants. Curr. Top. Virol. 1:71-81. [Google Scholar]
- 9.Buckwold, V. E., Z. Xu, M. Chen, T. S. Yen, and J. H. Ou. 1996. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J. Virol. 70:5845-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckwold, V. E., Z. Xu, T. S. Yen, and J. H. Ou. 1997. Effects of a frequent double-nucleotide basal core promoter mutation and its putative single-nucleotide precursor mutations on hepatitis B virus gene expression and replication. J. Gen. Virol. 78:2055-2065. [DOI] [PubMed] [Google Scholar]
- 11.Chen, M., and J. H. Ou. 1995. Cell type-dependent regulation of the activity of the negative regulatory element of the hepatitis B virus core promoter. Virology 214:198-206. [DOI] [PubMed] [Google Scholar]
- 12.Doria, M., N. Klein, R. Lucito, and R. J. Schneider. 1995. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 14:4747-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frain, M., G. Swart, P. Monaci, A. Nicosia, S. Stampfli, R. Frank, and R. Cortese. 1989. The liver-specific transcription factor LF-B1 contains a highly diverged homeobox DNA binding domain. Cell 59:145-157. [DOI] [PubMed] [Google Scholar]
- 14.Gunther, S., N. Piwon, and H. Will. 1998. Wild-type levels of pregenomic RNA and replication but reduced pre-C RNA and e-antigen synthesis of hepatitis B virus with C(1653)-> T, A(1762)-> T and G(1764)-> A mutations in the core promoter. J. Gen. Virol. 79:375-380. [DOI] [PubMed] [Google Scholar]
- 15.Kim, C. M., K. Koike, I. Saito, T. Miyamura, and G. Jay. 1991. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature 351:317-320. [DOI] [PubMed] [Google Scholar]
- 16.Klein, N. P., M. J. Bouchard, L. H. Wang, C. Kobarg, and R. J. Schneider. 1999. Src kinases involved in hepatitis B virus replication. EMBO J. 18:5019-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein, N. P., and R. J. Schneider. 1997. Activation of Src family kinases by hepatitis B virus HBx protein and coupled signaling to Ras. Mol. Cell. Biol. 17:6427-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurosaki, M., N. Enomoto, Y. Asahina, I. Sakuma, T. Ikeda, S. Tozuka, N. Izumi, F. Marumo, and C. Sato. 1996. Mutations in the core promoter region of hepatitis B virus in patients with chronic hepatitis B. J. Med. Virol. 49:115-123. [DOI] [PubMed] [Google Scholar]
- 19.Li, J., V. E. Buckwold, M. W. Hon, and J. H. Ou. 1999. Mechanism of suppression of hepatitis B virus precore RNA transcription by a frequent double mutation. J. Virol. 73:1239-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo, S.-Y., and J.-H. Ou. 1998. Expression and dimerization of hepatitis C virus core protein in E. coli, p. 325-330. In J. N. Y. Lau (ed.), Hepatitis C protocol. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 21.Madden, C. R., M. J. Finegold, and, B. L. Slagle. 2001. Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J. Virol. 75:3851-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maguire, H. F., J. P. Hoeffler, and A. Siddiqui. 1991. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science 252:842-844. [DOI] [PubMed] [Google Scholar]
- 23.McNabb, D. S., and R. J. Courtney. 1992. Posttranslational modification and subcellular localization of the p12 capsid protein of herpes simplex virus type 1. J. Virol. 66:4839-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nassal, M., M. Junker-Niepmann, and H. Schaller. 1990. Translational inactivation of RNA function: discrimination against a subset of genomic transcripts during HBV nucleocapsid assembly. Cell 63:1357-1363. [DOI] [PubMed] [Google Scholar]
- 25.Ou, J. H., H. Bao, C. Shih, and S. M. Tahara. 1990. Preferred translation of human hepatitis B virus polymerase from core protein- but not from precore protein-specific transcript. J. Virol. 64:4578-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seto, E., T. S. Yen, B. M. Peterlin, and J. H. Ou. 1988. Trans-activation of the human immunodeficiency virus long terminal repeat by the hepatitis B virus X protein. Proc. Natl. Acad. Sci. USA 85:8286-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su, F., C. N. Theodosis, and, R. J. Schneider. 2001. Role of NF-κB and Myc proteins in apoptosis induced by hepatitis B virus HBx protein. J. Virol. 75:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams, J. S., and O. M. Andrisani. 1995. The hepatitis B virus X protein targets the basic region-leucine zipper domain of CREB. Proc. Natl. Acad. Sci. USA 92:3819-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Xu, Z., T. S. B. Yen, L. Wu, C. R. Madden, W. Tan, B. L. Slagle, and J. H. Ou. 2002. Enhancement of hepatitis B virus replication by its X protein in transgenic mice. J. Virol. 76:2579-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yen, T. S. B. 1993. Regulation of hepatitis B virus gene expression. Semin. Virol. 4:33-42. [Google Scholar]
- 30.Zoulim, F., J. Saputelli, and C. Seeger. 1994. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 68:2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]