Abstract
Herpes simplex virus 1 (HSV-1) infection causes the shutoff of host gene transcription and the induction of a transcriptional program of viral gene expression. Cellular RNA polymerase II is responsible for transcription of all the viral genes, but several viral proteins stimulate viral gene transcription. ICP4 is required for all delayed-early and late gene transcription, ICP0 stimulates transcription of viral genes, and ICP27 stimulates expression of some early genes and transcription of at least some late viral genes. The early DNA-binding protein, ICP8, also stimulates late gene transcription. We therefore investigated which HSV proteins interact with RNA polymerase II. Using immunoprecipitation and Western blotting methods, we observed the coprecipitation of ICP27 and ICP8 with RNA polymerase II holoenzyme. The association of ICP27 with RNA polymerase II was detectable as early as 3 h postinfection, while ICP8 association became evident by 5 h postinfection, and the association of both was independent of viral DNA synthesis. Infections with ICP27 gene mutant viruses revealed that ICP27 is required for the association of ICP8 with RNA polymerase II, while studies with ICP8 gene deletion mutants showed no apparent role for ICP8 in the association of ICP27 with RNA polymerase II. The association of ICP27 and ICP8 with RNA polymerase II holoenzyme appeared to be independent of nucleic acids. We hypothesize that the interaction of ICP27 with RNA polymerase II holoenzyme reflects its role in stimulating early and late gene expression and/or its role in inhibiting host transcription and that the interaction of ICP8 with RNA polymerase II holoenzyme reflects its role in stimulating late gene transcription.
Herpes simplex virus type 1 (HSV-1) infection causes a drastic reduction in host cell gene expression while directing a dramatic burst of expression of viral gene products (reviewed in reference 71). During productive infection the approximately 80 viral genes of the 152-kbp double-stranded viral DNA genome are transcribed in the nucleus in a well-regulated, temporal manner. The cascade of viral gene expression is divided into three broad phases: α or immediate-early (IE) genes, β or early (E) genes, and γ or late (L) genes. The α genes are transcribed soon after the release of the viral genome into the nucleus, do not require de novo protein synthesis for expression, and are stimulated by the VP16 virion protein. The expression of β genes is dependent on newly synthesized α gene products, in particular, the ICP4 and ICP27 proteins. Many of the β proteins are involved in viral DNA replication. Viral DNA synthesis, in concert with ICP4, ICP27, and ICP8, triggers the expression of γ gene products, most of which are viral structural proteins that are involved in virion assembly and maturation.
HSV genes are transcribed by the host RNA polymerase II (Pol II) (3, 20), although several viral proteins are involved in the regulation of viral gene expression and modification of host transcription machinery. The virion tegument protein, VP16, binds to several host proteins including the host transcription factor, Oct-1, to form a complex that binds to α gene promoters and stimulates their transcription (37). The α protein ICP4 is required for transcriptional activation of most, if not all, β and γ genes (reviewed in reference 71) but represses α gene expression (24, 94). ICP4 is able to form complexes with the general transcription factors TFIIB, TATA-binding protein (TBP), and TBP-associated factor TAF250 (12, 31, 82). A second α protein, ICP0, stimulates expression of all three temporal classes of HSV genes during lytic infection (11, 73, 88). A third α protein, ICP22, is required for productive infection in some cell types but not in others (62, 63, 80) and for viral modification of host Pol II during productive infection (48, 68, 69). Deletions in the ICP22 gene affect the expression of ICP0 and a subset of γ genes (66). A fourth α protein, ICP27, is also essential for productive virus infection and is the only regulatory protein conserved in all herpesviruses of mammalian and avian origin. ICP27 is required for the accumulation of a subset of viral early and late mRNAs and for the switch from early to late virus gene expression (50, 70, 72). By stimulating early gene expression, ICP27 facilitates viral DNA synthesis (51, 74, 91). The regulatory effects of ICP27 on viral gene expression may be exerted at both the transcriptional and posttranscriptional levels. ICP27 promotes transcription of at least some late genes, e.g., the gC and UL47 genes, in infected Vero cells (39). Evidence for posttranscriptional effects of ICP27 include the observations that ICP27 binds RNA via its RGG motif (7, 38, 53, 76), regulates pre-mRNA 3′ processing (33, 51, 52), stabilizes labile 3′ ends of mRNA (7), interacts with cellular protein p32 (8) and spliceosome-associated protein 145 (9), inhibits the splicing of both viral and cellular transcripts (34), induces retention of intron-containing transcripts in the nucleus (61) but shuttles viral intronless transcripts from the nucleus to the cytoplasm (60, 76, 83), and regulates distribution of host small nuclear ribonucleoproteins (snRNPs) (59, 78). In addition, ICP27 has been found to interact with ICP0 and ICP4 (54, 57), to colocalize with ICP4 within HSV replication compartments in infected cells (23) and transfected cells (102), and to affect the posttranslational modification and DNA-binding ability of ICP4 (57, 97, 98). An additional protein needed for late gene transcription is the single-stranded-DNA-binding protein, ICP8, one of the seven viral β proteins necessary and sufficient for viral DNA replication in transfected cells (13, 96). ICP8 negatively affects transcription from the parental viral genomes (28-30) but stimulates late gene transcription from progeny DNA templates (15, 27).
The shutoff of host gene expression involves inhibition of transcription of host genes, inhibition of RNA splicing, and destabilization of host mRNAs (71). While it is well documented that HSV infection leads to an overall decrease in host cell RNA synthesis (35, 93) and that the transcription by all three RNA polymerases drops to less than 50% of the transcription levels of uninfected cells within 4 h postinfection (p.i.) (64), the molecular mechanism(s) of the inhibition of host transcription has not been defined. ICP27 and ICP4 play major roles in the inhibition of host gene transcription (85).
Although different purification strategies lead to some variation in its protein components, the cellular Pol II holoenzyme is a large, multisubunit complex composed of the core Pol II, general transcription factors such as TFIIB, TFIIE, TFIIF, and TFIIH but not TBP and its associated proteins, the core SRB (for suppressors of RNA polymerase B mutations)-mediator complex, the SRB10 cyclin-dependent kinase complex, and the Swi-Snf complex (36, 55). The RNA polymerase core is a complex of more than 10 protein subunits, ranging in size from 10 to 240 kDa, which include the large subunit and other auxiliary transcription factors (17, 99, 100). The large subunit of the core enzyme—that of human origin has a predicted molecular mass of 217 kDa—contains sites for DNA binding, RNA binding, and catalysis and is expressed in the nuclei of all eukaryotic cells (95). The conserved carboxyl-terminal domain (CTD) of the mammalian Pol II large subunit consists of 52 tandem repeats of heptapeptide consensus sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser, which is required for cell viability (18, 25, 95). Alternative states of phosphorylation of the serine, threonine, and tyrosine residues within the tandem repeats are associated with different steps in the transcription cycle (21, 22). The hypophosphorylated form, Pol IIa, is recruited to the preinitiation complex (49, 58), while the hyperphosphorylated form, Pol IIo, is involved in RNA elongation (10, 49, 58). The transition from Pol IIa to Pol IIo is necessary for the progression of RNA synthesis (58). After the completion of one round of transcription, the Pol IIo form must be dephosphorylated to the Pol IIa form in order to reinitiate another round of transcription (16). By recruiting pre-mRNA processing factors to nascent transcripts, the CTD of the Pol II large subunit may also have a role in pre-mRNA processing (reviewed in references 19, 32, and 87). HSV-1 infection has been reported to induce aberrant phosphorylation of the Pol II large subunit, presumably on the CTD, resulting in the depletion of Pol IIa and Pol IIo and the appearance of a new species, called Pol IIi (69). The subcomplex of the mediator of activation, which contains mainly SRB proteins and other general transcription factors, may exist as separate identities or may tightly associate with the CTD of Pol II large subunit in the core enzyme to form the Pol II holoenzyme (36, 84). The SRB proteins are considered a hallmark of the Pol II holoenzyme (14, 36). The mediator subcomplex can be dissociated from the Pol II holoenzyme by interaction with monoclonal anti-CTD antibodies (41).
The mechanism by which the viral α and β proteins regulate the temporal expression of viral genes and shutoff of cellular gene expression remains to be understood. Because all the viral genes are transcribed by host cell Pol II, we hypothesized that these viral proteins may interact, directly or indirectly, with host Pol II to modify or redirect its function in favor of the expression of viral genes. To address the possible association of the viral transcription regulators with Pol II holoenzyme, we conducted a series of immunoprecipitation experiments with appropriate antibodies. Our results indicated that HSV-1 ICP27 and ICP8 proteins physically associate with the Pol II holoenzyme after viral infection of human HEp-2 cells. At the time of submission of this paper, Jenkins and Spencer (40) reported that ICP27 and ICP22 copurified biochemically with complexes of Pol II that were smaller than the full-size Pol II holoenzyme.
MATERIALS AND METHODS
Cells and viruses.
Human epidermoid HEp-2 cells were obtained from the American Type Culture Collection (Manassas, Va.) and were grown as monolayer cultures in Dulbecco's modified Eagle's medium (Media Tech, Herndon, Va.) supplemented with 10% heat-inactivated fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) in a humidified 5% CO2 atmosphere at 37°C. The HSV-1 wild type (wt) KOS and KOS1.1 virus strains were propagated and titrated as described previously (43, 46). HSV-1 ICP27 gene mutants d27-1, n59, n263, n406, and n504 (originally called n59R, n263R, n406R, and n504R, but we have dropped the R because this usually refers to a rescued virus) were derived from KOS1.1 and were characterized previously (67). The construction, isolation, and characterization of ICP8 gene mutants HD-2, d101, and d301 are described elsewhere (26).
Antibodies.
The rabbit polyclonal anti-human SRB7 (hSRB7) antibody (36) and mouse monoclonal antibody 8WG16 (90, 95) were kindly provided by Jeffrey Parvin, Brigham and Women's Hospital and Harvard Medical School. Rabbit polyclonal anti-Pol II antibody C21 was purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, Calif.). Rabbit anti-ICP8 antiserum 3-83 was described previously (42). Anti-ICP4 monoclonal antibody 58S (81) was a gift from Neal DeLuca, University of Pittsburgh. Rabbit polyclonal anti-ICP22 antiserum R77 (2) was generously provided by Bernard Roizman, University of Chicago. Monoclonal antibody H1112, specific for ICP0 (1), and monoclonal antibodies H1113 and H1119, specific for ICP27 (1), were purchased from the Goodwin Institute for Cancer Research Inc. (Plantation, Fla.). Rabbit polyclonal anti-RNA helicase A antibody 104 was kindly provided by Jeffrey Parvin. Rabbit polyclonal antibody 367, specific for ICP8, was generously provided by William Ruyechan, State University of New York at Buffalo. The following antibodies were purchased from Santa Cruz Biotechnology: rabbit polyclonal antibodies to protein phosphatase 1 (FL-18) and mitogen-activated protein kinase phosphatase (C-19); goat polyclonal antibodies to SmB (N-18), SC35 (Y-16), hnRNP F (N-15), SMN (N-19), the 70-kDa U1 snRNP protein (C-18), and actin (I-19); and mouse monoclonal antibodies to SH-PTP1 (D-11), SR proteins (1H4), poly(ADP-ribose) polymerase (PARP) (F-2), and actin (C-2).
Viral infections and immunoprecipitations.
HEp-2 cells were plated into 100-mm tissue culture dishes 24 h prior to virus infection in order to get 90% confluence at the time of infection. Cells were infected with either HSV-1 wt strains (KOS or KOS1.1) or various HSV-1 mutants at a multiplicity of infection of 20 PFU per cell in cold phosphate-buffered saline (PBS) containing 0.1% glucose and 1% heat-inactivated newborn calf serum in the presence or absence of sodium phosphonoacetate (PAA; 400 μg/ml). After 1 h of adsorption at 37°C, cells were switched into Dulbecco's modified Eagle's medium containing 1% heat-inactivated bovine calf serum. PAA was added at the time of infection. In experiments where PAA was used, 20 mM HEPES buffer (pH 7.6) was included in all media to maintain the pH of all cultures (15). Caspase inhibitor z-VAD-fmk was purchased from Calbiochem (La Jolla, Calif.) and was added to the media of infected or mock-infected cultures after the adsorption period at a concentration of 100 μM/ml.
Cells were harvested at 8 h p.i. by scraping them into the media. After two washes in cold PBS, cells from each dish were incubated on ice for 30 min in 450 μl of lysis buffer (79) containing 0.5 M potassium acetate-0.25% Nonidet P-40-50 mM Tris-acetate, pH 7.9-10 mM EDTA-10 mM β-glycerophosphate-5 mM sodium fluoride-1 mM phenylmethylsulfonyl fluoride-2 mM Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK; Sigma, St. Louis, Mo.)-1 tablet of Complete protease inhibitor cocktail (Roche Molecular Biochemicals) per 10 ml. The cell lysate was clarified by centrifugation at 10,000 × g at 4°C for 5 min. Immunoprecipitation was carried out on 400 μl of cell lysate with appropriate antibodies and protein A-agarose beads in buffer H (20 mM Tris-acetate [pH 7.9], 1 mM EDTA, 5% glycerol, 1 mM dithiothreitol, 0.12 M potassium acetate, 0.1% Nonidet P-40, 0.2 mg of bovine serum albumin [BSA] per ml, 1 mM phenylmethylsulfonyl fluoride, 2 mM TLCK, Complete protease inhibitor cocktail) at 4°C overnight essentially as described elsewhere (79). After four washes in washing buffer (79) (20 mM Tris-acetate [pH 7.9], 0.12 M potassium acetate, 6 mM magnesium acetate, 0.1% Nonidet P-40, 0.1 mM dithiothreitol, 0.2 mg of BSA per ml, 1 mM phenylmethylsulfonyl fluoride, 2 mM TLCK) the precipitates were dissolved in gel sample buffer (43) for separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
SDS-PAGE and Western blotting.
Proteins in the immunoprecipitates were resolved by SDS-PAGE in diallyltartardiamide cross-linked 9.25% polyacrylamide gels at 14 mA overnight essentially as described previously (43) and then transferred onto a nitrocellulose membrane by electroblotting (75) at 40 V for 2 days. The membranes were blocked in 5% milk in Tris-buffered saline (TBS), probed with appropriate antibodies in TBS containing 0.1% Tween 20, and stained with ECL Western blotting detection reagents (Amersham Pharmacia Biotech Inc., Piscataway, N.J.) in accordance with the manufacturer's procedure.
Nuclease treatment.
All nuclease incubations were performed with cell lysates at 30°C for 30 min, and then the treated cell lysates were directly subjected to immunoprecipitation. When nuclease treatments were performed, EDTA was not included in the lysis buffer. The RNase cocktail (Ambion, Austin, Tex.) used included 25 U of RNase A/ml and 1,000 U of RNase T1/ml as recommended by the manufacturer or 75 U of RNase A/ml and 3,000 U of RNase T1/ml. DNase I (Worthington Diagnostics Corp.) was used at 10 μg/ml in the presence of 10 mM magnesium acetate.
RESULTS
Coprecipitation of HSV proteins with an anti-SRB7 antibody.
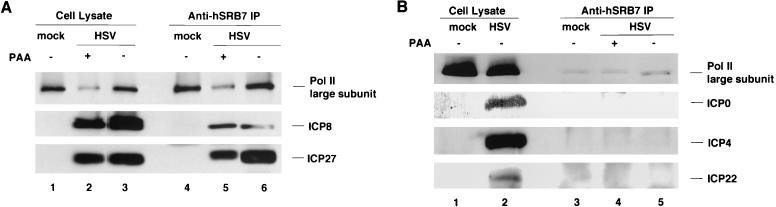
In the first series of experiments, we attempted to determine if any HSV proteins associated with the cellular Pol II holoenzyme in infected cells by immunoprecipitating the holoenzyme and assaying for viral proteins by Western blotting. The conditions for the preparation of the lysate and immunoprecipitation were those used by Scully et al. (79) to show that BRCA1 is a component of the Pol II holoenzyme. We mock infected HEp-2 cells or infected them with wt HSV-1 either with viral DNA synthesis inhibitor PAA to allow only IE and E gene expression or without PAA to allow full viral replication and gene expression. Cells were harvested at 8 h p.i., whole-cell lysates were prepared, and immunoprecipitation was performed with a rabbit polyclonal antibody that recognizes the SRB7 protein, which is a component of the Pol II holoenzyme complex (36). The immunoprecipitated proteins were separated by SDS-PAGE and detected with antibodies against Pol II or HSV-1 proteins.
The anti-SRB7 antibody immunoprecipitated the large subunit of Pol II as a component of the holoenzyme complex from mock-infected cells (Fig. 1A, lane 4) and from virus-infected cells (Fig. 1A, lanes 5 and 6). Thus, the association of SRB7 with Pol II was not affected by HSV-1 infection. Less Pol II large subunit was pulled down by the anti-SRB7 antibody in PAA-treated cultures (Fig. 1A, lane 5), but this was consistent with the decreased level of the large subunit in the PAA-treated cell lysate (lane 2). IE protein ICP27 coprecipitated with SRB7 and Pol II under both infection conditions (Fig. 1A, lanes 5 and 6). In this but not all experiments more ICP27 was coprecipitated at late times of infection. The β protein ICP8 also coprecipitated with SRB7 and Pol II. Approximately equal amounts of ICP8 coprecipitated with Pol II in the absence or presence of viral DNA replication (Fig. 1A, lanes 5 and 6).
FIG. 1.
Coprecipitation of HSV-1 proteins with Pol II holoenzyme. HEp-2 cells were infected with wt HSV-1 KOS or mock infected. When indicated, PAA (400 μg/ml) was added to infected cultures at the beginning of infection. Cells were harvested at 8 h p.i. Immunoprecipitation (IP) was performed on cell lysates using a rabbit polyclonal anti-hSRB7 antibody. Proteins in the cell lysates and immunoprecipitates were resolved by SDS-PAGE, and specific proteins were detected with monoclonal anti-Pol II antibody 8WG16 (1:1,000 dilution), rabbit anti-ICP8 antiserum 3-83 (1:1,000 dilution), monoclonal anti-ICP27 antibody H1113 (1:200 dilution), monoclonal anti-ICP0 antibody H1112 (1:200 dilution), anti-ICP4 monoclonal antibody 58S (1:200 dilution), and rabbit polyclonal anti-ICP22 antiserum R77 (1:500 dilution). (A) Coprecipitation of ICP27 and ICP8 with Pol II. (B) Lack of coprecipitation of ICP0, ICP4, and ICP22 with Pol II. The amount of cell lysate loaded on the gel relative to the amount of lysate used for immunoprecipitation was 1:20.
Because ICP4, ICP0, and ICP22 stimulate viral gene expression in various ways (see the introduction), we expected that these viral proteins might also interact with Pol II holoenzyme. However, we did not detect these three proteins in the SRB7-Pol II immunoprecipitates under our experimental conditions (Fig. 1B, lanes 4 and 5), indicating that they were not stably associated with the Pol II holoenzyme under these conditions. To show that it was possible to immunoprecipitate ICP0 and ICP4 from the lysates, we performed the same assay with anti-ICP0 and anti-ICP4 antibodies. These antibodies immunoprecipitated ICP4 and ICP0 (results not shown), showing that these immunoprecipitation conditions could bring down these proteins with other antibodies.
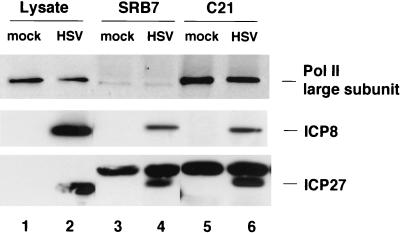
To confirm the interactions of ICP27 and ICP8 with the Pol II holoenzyme, we repeated the immunoprecipitation using other antibodies that directly recognize Pol II. The C21 rabbit polyclonal antibody recognizes epitopes on the tandem repeats within the CTD of the Pol II large subunit. Mouse monoclonal antibody 8WG16 recognizes the highly conserved heptapeptide repeat within the CTD of the Pol II large subunit (90, 95) and reacts with both the hypophosphorylated IIa form and the aberrantly phosphorylated IIi form but not the hyperphosphorylated IIo form of the Pol II CTD (69). When we tested the three antibodies, the anti-hSRB7 antibody (Fig. 2, lane 4), the C21 antibody (lane 6), and the 8WG16 antibody (results not shown), all precipitated ICP27 and ICP8 with the Pol II holoenzyme.
FIG. 2.
Coprecipitation of ICP8, ICP27, and Pol II holoenzyme with different Pol II antibodies. HEp-2 cells were infected with wt HSV-1 KOS or mock infected and harvested 8 h p.i. Immunoprecipitations were carried out on the cell lysate with a rabbit anti-hSRB7 antibody and the C21 rabbit anti-Pol II antibody. Proteins in cell lysates and immunoprecipitates were separated by SDS-PAGE and detected by Western blotting with anti-Pol II antibody 8WG16 (1:1,000 dilution), anti-ICP8 antibody 3-83 (1:1,000 dilution), and monoclonal anti-ICP27 antibody H1113 (1:200 dilution). The ratio of cell lysate loaded on the gel to the amount of lysate used in immunoprecipitation was 1:20. Lanes 1 and 2, cell lysates; lanes 3 and 4, anti-hSRB7 immunoprecipitates; lanes 5 and 6, C21 immunoprecipitates.
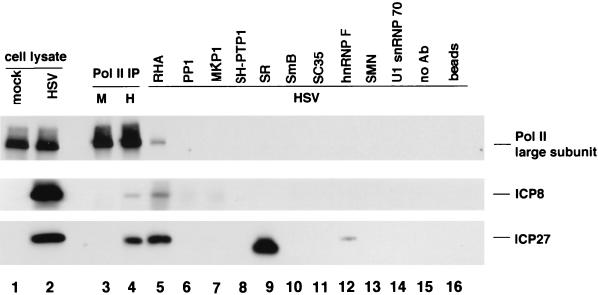
We wished to determine if the coprecipitation of ICP27 and ICP8 with Pol II was specific, so we utilized several antibodies and antisera recognizing other cellular proteins for immunoprecipitation (Fig. 3). As in previous experiments, the C21 antibody coprecipitated ICP27 and ICP8 with the Pol II large subunit (Fig. 3, lane 4). The antibody specific for RNA helicase A coprecipitated Pol II (Fig. 3, lane 5), as observed before (56), confirming its presence in the Pol II holoenzyme complex. In addition, the RHA antibody coprecipitated ICP27 and ICP8 (lane 5). Small amounts of Pol II were precipitated by antibodies specific for MKP, SR, and hnRNP F proteins (lanes 7, 9, and 12). ICP8 was coprecipitated with the RHA antibody (lane 5), and very small amounts of ICP8 were detected in immunoprecipitates with MKP1 and PP1 antibodies (longer exposures are not shown). ICP27 was also coprecipitated with the RHA antibody (lane 5) and the hnRNP F antibody (lane 12), but little or no ICP27 was detected in immunoprecipitates with the other antibodies, except for the SR antibody, which is discussed below. It is notable that little ICP27 was detected in the SmB immunoprecipitate because human autoimmune serum recognizing SmB proteins has been reported to coprecipitate ICP27 (77). The SR protein antibody coprecipitated a small amount of ICP27, which migrated slightly slower than a major mouse immunoglobulin G (IgG) band (lane 9 and results not shown). No ICP27 or ICP8 was precipitated in the absence of the antibody (lane 15) or was detected if protein A-agarose beads were boiled in sample buffer (lane 16). In summary, these control immunoprecipitations demonstrated that the association of ICP27 and ICP8 with Pol II is highly specific.
FIG. 3.
Comparison of immunoprecipitation of ICP27 and ICP8 with Pol II antibody and antibodies specific for other cellular proteins. HEp-2 cells were either mock infected or infected with HSV-1 KOS virus and harvested at 8 h p.i. Lysates were prepared in buffer H in this experiment. Immunoprecipitation (IP) was performed on the mock-infected cell lysate or aliquots of infected-cell lysate with 2 μg of each of the indicated antibodies (Ab). Proteins in the immunoprecipitates were resolved by SDS-PAGE and analyzed by Western blotting with monoclonal antibodies to the Pol II large subunit (8WG16, 1:1,000 dilution) and ICP27 (H1119, 1:300 dilution) and rabbit anti-ICP8 antibody 367 (1:1,500 dilution). The ratio of loaded cell lysate to the input cell lysate for immunoprecipitation was 1:14. Lane 15, wt virus-infected-cell lysate was incubated with the protein A-agarose beads in the absence of an antibody; lane 16, protein A-agarose beads alone. M, mock infection; H, wt HSV-1 virus infection.
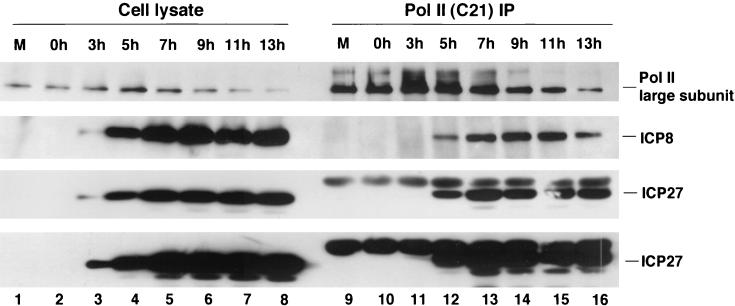
We monitored the kinetics of association of ICP27 and ICP8 with Pol II by performing C21 antibody immunoprecipitation of Pol II from infected-cell lysates harvested at various times after infection. ICP27 was detected in small amounts in the Pol II immunoprecipitate as early as 3 h p.i. (Fig. 4, long exposure in bottom row) and accumulated as viral infection progressed (Fig. 4, third row), while ICP8 was detected in the immunoprecipitates by 5 h p.i. and its association peaked at 9 h p.i. and declined slightly thereafter (Fig. 4, second row). Therefore, ICP27 and ICP8 were associated with Pol II holoenzyme complexes at early and late times p.i.
FIG. 4.
Time course of ICP27 and ICP8 association with Pol II in infected cells. HEp-2 cells were infected with wt HSV-1 KOS virus or mock infected and harvested at different times postinfection as indicated. Proteins in the lysates were immunoprecipitated (IP) with C21 anti-Pol II antibody and analyzed by SDS-PAGE and Western blotting with a monoclonal antibody to Pol II (8WG16), polyclonal antiserum to ICP8 (3-83), and monoclonal anti-ICP27 antibody H1113. The ratio of cell lysate loaded on the gel to the amount of lysate used in immunoprecipitation was 1:30. M, mock-infected cells. The bottom two panels are different exposures of the ICP27 antibody blot.
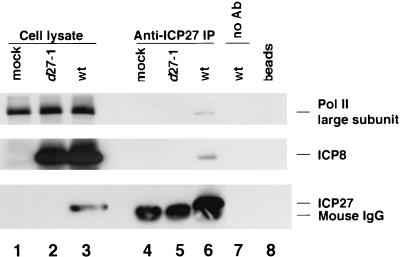
To further verify the association of ICP27 and ICP8 with Pol II, we performed the reverse immunoprecipitation with a monoclonal antibody specific for ICP27 on the HEp-2 cell lysate from cells infected with HSV-1 KOS1.1 wt virus (Fig. 5). We observed that both the Pol II large subunit and ICP8 coprecipitated with ICP27 under normal infection conditions (Fig. 5, lane 6) or in the presence of PAA to inhibit viral DNA synthesis (results not shown). No precipitation of Pol II or ICP8 was observed in the absence of an antibody or in lysates from cells infected with ICP27 null mutant d27-1 (lanes 7 and 5, respectively). Thus, the coprecipitation appeared to involve specific immunoprecipitation of ICP27 and associated proteins.
FIG. 5.
Coprecipitation of Pol II and ICP8 with ICP27. HEp-2 cells were infected with either wt HSV-1 KOS1.1 or the ICP27 null mutant d27-1 virus or were mock infected. Cells were harvested at 8 h p.i. Immunoprecipitation (IP) was performed on the cell lysates with monoclonal anti-ICP27 antibody H1113. Proteins in the immunoprecipitates were resolved by SDS-PAGE and analyzed by Western blotting with monoclonal anti-Pol II antibody 8WG16 (1:1,000), monoclonal anti-ICP27 antibody H1119 (1:300), and rabbit anti-ICP8 antibody 367 (1:1,500). The ratio of loaded cell lysate to the input cell lysate for immunoprecipitation was 1:14. No Ab, wt virus-infected-cell lysate was incubated with the protein A-agarose beads in the absence of an antibody; beads, protein A-agarose beads were boiled in sample buffer.
Apparent lack of a role for ICP8 in association of ICP27 with Pol II.
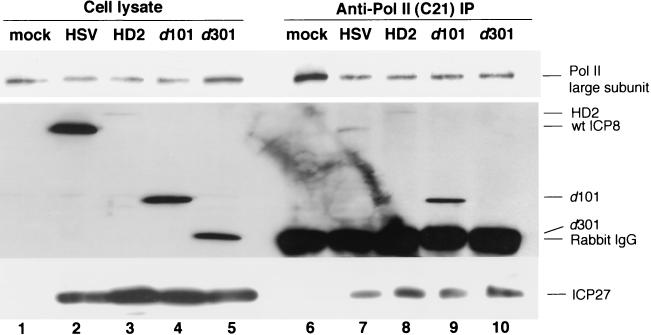
We further investigated if the interactions of ICP27 and ICP8 with Pol II were independent by studying cells infected with mutant viruses defective for ICP8 or ICP27. We performed Pol II immunoprecipitation on lysates from HEp-2 cells infected with ICP8 gene mutants. The HD-2 mutant virus encodes a fusion protein that contains the amino-terminal 281 amino acid residues of ICP8 and the β-galactosidase protein and that is expressed under the control of the ICP8 gene promoter, while d101 and d301 contain ICP8 genes with deletion mutations near the 5′ end and in the middle, respectively (26). Under our experimental conditions, the wt ICP8 and the mutant d101 and HD-2 ICP8 molecules coprecipitated with Pol II (Fig. 6, lanes 7 to 9). The d301 ICP8 comigrated with the rabbit IgG heavy chain (Fig. 6, lane 5 versus 6 to 10), so we were unable to tell whether or not the d301 mutant coprecipitated with Pol II. Comparable amounts of ICP27 coprecipitated with Pol II in both wt- and ICP8 mutant-infected cultures (Fig. 6, lane 7 versus 8 to 10), indicating that the association of ICP27 did not require these portions of ICP8.
FIG. 6.
Association of ICP27 and mutant ICP8 with Pol II. HEp-2 cells were infected with the wt HSV-1 KOS1.1 strain or with ICP8 mutant virus HD-2, d101, or d301 or were mock infected. Cells were harvested at 8 h p.i., and the lysates were subjected to immunoprecipitation (IP) with anti-Pol II antibody C21. The proteins in cell lysates and precipitates were separated by SDS-PAGE and detected with anti-Pol II antibody C21, anti-ICP8 antibody 3-83, and anti-ICP27 antibody H1113. The ratio of cell lysate loaded on the gel to the amount of lysate used in immunoprecipitation was 1:20.
The earlier kinetic association of ICP27 with Pol II (Fig. 4) also suggested that the ICP27 association was independent of ICP8. Because all the ICP8 mutants we used were unable to synthesize viral DNA (26), these results confirmed that viral DNA synthesis was not required for the association of ICP27 and ICP8 with Pol II.
Requirement of ICP27 for association of ICP8 with Pol II.
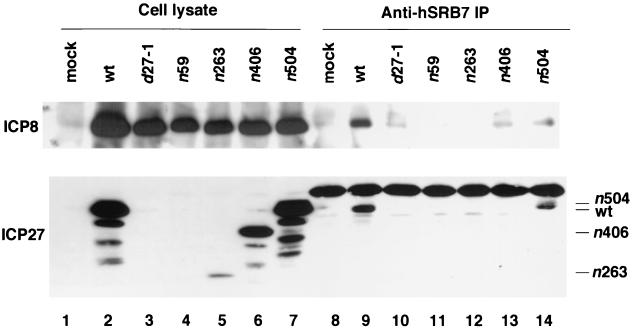
We next investigated if functional ICP27 was required for the interaction between ICP8 and Pol II holoenzyme. We infected HEp-2 cells with ICP27 gene mutant viruses and then carried out immunoprecipitation of Pol II from the cell lysate with an anti-hSRB7 antibody. The d27-1 virus is a null mutant, and n59, n263, n406 and n504 are nonsense mutants that express truncated ICP27 proteins (67). The d27-1, n59, n263, and n406 mutants are partially defective in viral DNA amplification, while n504 manifests a wt phenotype for viral DNA replication (67). From cells infected with these viral strains, only the wt and n504 ICP27 proteins were coprecipitated with Pol II (Fig. 7), indicating that ICP27 residues 407 to 504 and possibly other parts of ICP27 were required for the association of ICP27 with Pol II. Although nearly wt levels of ICP8 were expressed by the ICP27 mutant viruses (Fig. 7, lanes 3 to 7), little ICP8 coprecipitated with Pol II from lysates of cells infected with the d27-1, n59, n263, or n406 virus (Fig. 7, lanes 10 to 13 versus 9). A small amount of ICP8 coprecipitated with Pol II from lysates of cells infected with n504 virus (lane 14).
FIG. 7.
Association of ICP8 and mutant ICP27 proteins with Pol II. HEp-2 cells were infected with wt HSV-1 KOS1.1 strain or with ICP27 mutant virus d27-1, n59, n263, n406, or n504 or were mock infected. The cultures were harvested at 8 h p.i. Immunoprecipitations (IP) were performed with a rabbit anti-hSRB7 antibody. Proteins in cell lysates and immunoprecipitates were analyzed by SDS-PAGE and Western blotting with anti-ICP8 antibody 3-83 and anti-ICP27 antibody H1113. The ratio of cell lysate loaded on the gel to the amount of lysate used for immunoprecipitation was 1:20.
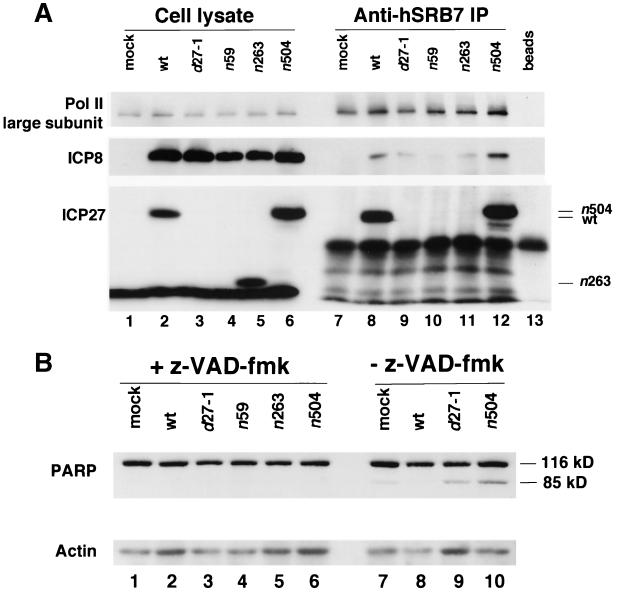
Because ICP27 mutant viruses induce apoptosis in human cells (4), the lack of coprecipitation of ICP8 with Pol II could be due to the induction of apoptosis in these infected cells. We were unable to precipitate Pol II from monkey cell lysates using these antibodies, so we utilized caspase inhibitor z-VAD-fmk to block apoptosis in ICP27 mutant-infected HEp-2 cells (5) and eliminate this as a possible explanation of the results. In the presence of the caspase inhibitor, ICP8 coprecipitated with the SRB7 antibody only in lysates from wt-infected cells (Fig. 8A, lane 8) and n504 mutant-infected cells (lane 12), not in lysates from d27, n59, or n263 mutant virus-infected cells (lanes 9 to 11). Thus, it appeared that ICP27 was needed for association of ICP8 with Pol II, even when apoptosis was blocked. To control for the activity of the drug, we examined PARP, a marker of apoptosis, in these cells (Fig. 8B). Addition of z-VAD-fmk eliminated the appearance of the lower band, indicating that it had blocked caspase activity (Fig. 8B, lanes 1 to 6 versus 7 to 10). Therefore, we concluded that we had inhibited caspase activity and that a functional ICP27 or at least the N-terminal 504 residues of ICP27 were required for efficient association of ICP8 with Pol II.
FIG. 8.
Association of ICP8 and mutant ICP27 proteins with Pol II in the presence of an apoptosis inhibitor. HEp-2 cells were infected with wt HSV-1 KOS1.1 strain or with ICP27 mutant virus d27-1, n59, n263, or n504 or were mock infected. Caspase inhibitor I (z-VAD-fmk) was added to 100 μM after the adsorption period. The cultures were harvested at 8 h p.i. (A) Immunoprecipitation (IP) was carried out on the cell lysates with the rabbit anti-SRB7 antibody. Proteins in the cell lysates and immunoprecipitates were analyzed by SDS-PAGE and Western blotting with antibodies specific for Pol II large subunit (8WG16), ICP8 (367), or ICP27 (H1113). The ratio of loaded cell lysate to the amount used in immunoprecipitation was 1:20. Beads, protein A-Sepharose beads alone. (B) Proteins in equal volumes of the cell lysates were separated by SDS-PAGE and analyzed by Western blotting with antibodies to PARP (F-2) and actin (C-2).
Does RNA or DNA play a role in the association of ICP27 and ICP8 with Pol II?
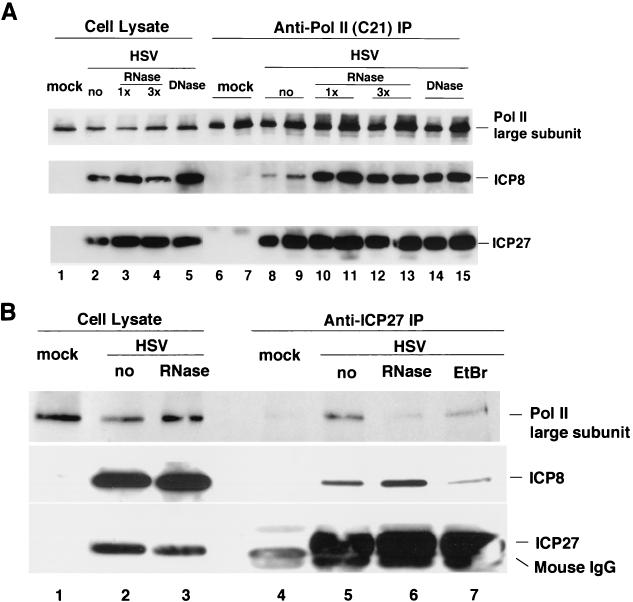
ICP8, the major DNA-binding protein encoded by HSV, interacts with DNA both in vitro and in vivo (46). Likewise, ICP27 is known to interact with RNA in vitro (7, 38, 53, 76). The RNA-binding properties of ICP27 and the DNA-binding property of ICP8 raised the possibility that their associations with Pol II might be mediated by RNA or DNA. To investigate this hypothesis, we treated lysates of wt HSV-1 KOS strain-infected HEp-2 cells with a cocktail of RNase A and RNase T1 or with DNase I prior to anti-Pol immunoprecipitation. In several repetitions of this experiment, we consistently observed that equal or possibly increased amounts of ICP27 and ICP8 coprecipitated with Pol II after RNase or DNase I treatment (Fig. 9A, lanes 10 to 13 versus 8 and 9 or 14 and 15 versus 8 and 9). From these results, it appeared that nucleic acids did not play a role in the association of these two proteins with Pol II. In contrast, immunoprecipitation of ICP27 from RNase-treated lysates showed reduced levels of Pol II being coprecipitated (Fig. 9B, lane 6), compared to levels precipitated from control lysates (lane 5). The coprecipitation of ICP8 with ICP27 may be partially dependent on double-stranded DNA (dsDNA) because ethidium bromide treatment, which dissociates proteins from dsDNA (86), reduced the amount of ICP8 coprecipitating with ICP27 in some experiments (Fig. 9B, lane 7).
FIG. 9.
Role of nucleic acids in association of HSV proteins with the Pol II holoenzyme. HEp-2 cells were infected with wt HSV-1 or mock-infected and harvested at 8 h p.i. (A) Cell lysates were untreated or treated with an various amounts of RNases or with DNase I and then were subjected to immunoprecipitation (IP) with rabbit anti-Pol II antibody C21. Proteins in cell lysates and the Pol II immunoprecipitates were resolved by SDS-PAGE and detected by Western blotting with anti-Pol II antibody 8WG16 (1:1,000 dilution), anti-ICP8 antiserum 3-83 (1:1,000 dilution), and anti-ICP27 antibody H1113 (1:200 dilution). The ratio of cell lysate loaded on the gel to the amount of lysate used in immunoprecipitation was 1:13 for lanes 6, 8, 10, 12, and 14 and 1:26 for lanes 7, 9, 11, 13, and 15. 1x, cell lysate was treated with 25 U of RNase A/ml and 1,000 U of RNase T1/ml at 30°C for 30 min; 3x, cell lysate was treated with 75 U of RNase A/ml and 3,000 U of RNase T1/ml at 30°C for 30 min; DNase, cell lysate was treated with 10 μg of DNase I/ml. No, no nuclease during incubation. (B) Cell lysates were untreated or treated with 25 U of RNase A/ml and 1,000 U of RNase T1/ml at 30°C for 30 min. Immunoprecipitations were performed with monoclonal anti-ICP27 antibody H1113 on RNase-treated and untreated cell lysates in the absence or presence of ethidium bromide. Proteins in cell lysates and immunoprecipitates were resolved by SDS-PAGE and detected with anti-Pol II antibody 8WG16 (1:1,000 dilution), anti-ICP8 antiserum 3-83 (1:1,000 dilution), and anti-ICP27 antibody H1113 (1:200 dilution). The ratio of cell lysate loaded on the gel to the amount of lysate used in immunoprecipitation was 1:40. EtBr, immunoprecipitation was done on untreated HSV-infected cell lysate in the presence of 50 μg of ethidium bromide/ml.
We are uncertain as to why the reciprocal immunoprecipitations gave somewhat different results, but it is possible that, under the nuclease and immunoprecipitation conditions, some of the large Pol II subunit protein was degraded, making it appear that it was lost under the ICP27 immunoprecipitation conditions. Because, at least under certain conditions for coprecipitation, the association of ICP27 and ICP8 with Pol II was resistant to nuclease treatment, we conclude that at least a portion of the interactions do not involve a nucleic acid and may be due to direct protein-protein interactions.
DISCUSSION
HSV infection leads to the shutoff of host transcription and the selective activation of viral gene transcription by cellular Pol II. Because several viral proteins have been observed to play crucial roles in the repression of host expression and the regulation of viral gene expression, we hypothesized that at least some of those viral proteins might physically interact, either directly or indirectly, with host Pol II holoenzyme. Using immunoprecipitation conditions similar to those used to demonstrate that the BRCA1 protein was a component of the Pol II holoenzyme (79), we demonstrated that the HSV α ICP27 protein and β ICP8 protein coimmunoprecipitated with Pol II. Despite the reports that ICP4 can interact with general transcription factors TFIIB, TBP, and TAF250 (12, 31, 82), that ICP22 is required for the aberrant modification of Pol II (48, 68, 69), and that ICP4 and ICP22 colocalize with Pol II in discrete nuclear structures after the onset of viral DNA replication (47), we were unable to detect these proteins in immunoprecipitates with Pol II in the anti-hSRB7 immunoprecipitates under our experiment conditions. The lack of evidence for the coprecipitation of these proteins with Pol II under these conditions does not exclude their presence in the Pol II holoenzyme complex; it could mean that they are not present in the complex, but it could also mean that they do not interact in a way that allows their recovery in the immunoprecipitate. Using biochemical purification of Pol II, Spencer and Jenkins have also observed ICP27 associating with Pol II, although in complexes smaller than the full-size holoenzyme (40). They observed ICP22 copurifying with Pol II complexes, so different approaches may identify different types of complexes.
Interaction of ICP27 with Pol II.
The nature of the association of ICP27 with Pol II remains to be defined in molecular detail. The reciprocal coprecipitation of Pol II by an antibody directed against ICP27 further argued for the existence of a bona fide complex. Studies of nonsense mutants showed that ICP27 residues 407 to 504 and possibly other regions of ICP27 are required for its association with Pol II. The question of a role for RNA in the association of ICP27 with Pol II has not been fully resolved. RNase digestion of the lysates prior to immunoprecipitation with a Pol II antibody did not affect the amount of ICP27 coprecipitated, indicating that the bulk of the interaction was due to protein-protein interactions. However, immunoprecipitation with the ICP27 antibody showed reduced coprecipitation of Pol II following RNase treatment. Thus, part of the association may involve RNA. We believe that the reduction in Pol II in ICP27 antibody immunoprecipitations after RNase treatment may be due to degradation of the large Pol II subunit. Therefore, because under some immunoprecipitation conditions the association appears to be resistant to RNase digestion, we conclude that at least a portion of the interaction is due to direct protein-protein interactions. RNA-binding ability is not likely to be sufficient for ICP27 to associate with Pol II because n406 ICP27 does not bind to Pol II even though a mutant ICP27 comprising residues 1 to 405 can bind to RNA (53). As other RNA-binding and -processing proteins such as mRNA-processing factors (65, 87), guanyl transferase and methylase for mRNA capping, SR-like CTD-associated factor (SCAF) for intronic splicing, cleavage polyadenylation specificity factor (CPSF), and cleavage stimulation factor (CstF) for 3′ cleavage and polyadenylation (6, 65, 87) bind to the CTD on the large Pol II subunit, it will be of considerable interest to determine if ICP27 can bind directly to the CTD.
The specificity of the coprecipitation of ICP27 with Pol II could be questioned because ICP27 has been reported previously to interact with HSV ICP4 (57), and cellular proteins p32 (8), casein kinase 2 (92), hnRNP (92), Sm small ribonucleoprotein (77), spliceosome-associated protein 145 (9), and RNA and export factor binding proteins (REFs) (44). We therefore compared the abilities of a number of control antibodies to coimmunoprecipitate ICP27. Many of the antibodies brought down little to no ICP27, and only a small amount of ICP27 was brought down by an anti-Sm antibody, while much larger amounts of ICP27 were brought down by an anti-Pol II antibody. While no quantitative comparisons can be made from these data, the relatively efficient coprecipitation of ICP27 with the anti-Pol II antibody is consistent with this being a real interaction.
Interaction of ICP8 with Pol II.
Similarly, the molecular basis for the interaction of ICP8 with RNA Pol II remains to be defined. While we were able to coprecipitate ICP8 when Pol II antibodies were used, we were not able to perform a clean reciprocal coprecipitation because anti-ICP8 antibodies appeared to cross-react with some component of the Pol II complex in mock-infected cells (results not shown). Our genetic studies demonstrated that ICP27 is needed for the association of ICP8 with Pol II, but this could be due to direct interactions of ICP8 with ICP27 and/or associated proteins or due to an alteration of Pol II by ICP27 that allows ICP8 to bind. Some additional studies suggest that ICP8 and ICP27 may interact directly (E. McNamee and D. Knipe, unpublished results). Although ICP8 binds single-stranded DNA and dsDNA tightly (46), DNase treatment of the lysates did not affect the coprecipitation of ICP8 with Pol II. Thus, ICP8 seems to interact with Pol II by protein-protein interactions.
Implications for viral transcriptional regulation.
ICP27 has been shown to stimulate transcription of true late genes such as gC and UL47 (39) and the expression of certain β genes (51, 74, 91), although it has not been determined if the latter effect is transcriptional or posttranscriptional. In addition, ICP27 has been proposed to play posttranscriptional roles such as promoting the use of either weak polyadenylation sites (51, 52) or downstream polyadenylation sites (33) or shuttling mRNA to the cytoplasm (60, 76, 83). The interaction of ICP27 with the Pol II enzyme could promote its initiation or elongation on viral late gene promoters. Late promoters have only a TATA box, an initiator element, and a downstream activating sequence (reviewed in reference 71). The HSV ICP4 protein has been shown to bind to general transcription factors and associated factors (12, 31, 82), and evidence for interactions between ICP4 and ICP27 has been reported (57). Thus, it is conceivable that ICP4 associates with general transcription factors and viral DNA while ICP27 associates with the Pol II complex and that the interaction between ICP4 and ICP27 brings Pol II to the viral genome for efficient transcription of late viral promoters. In this way, the interaction of ICP27 with Pol II could serve to promote transcription of the viral genome.
ICP8 plays a role in stimulating transcription of late genes, which is independent of viral DNA replication (15, 27). Its interaction with Pol II is likely a reflection of that function. As ICP8 does not activate transcription directly, it is possible that on binding to the Pol II complex it interacts with another protein that stimulates transcription. Alternatively, ICP8 bound to viral DNA could recruit the Pol II complexes containing ICP27 onto viral DNA and thereby promote the transcription of viral genes.
Potential role for ICP27 in linking viral transcription and RNA transport.
In addition to promoting transcription of the viral genome, ICP27 may promote the binding of the RNA transport apparatus to the transcription complex by binding to a component of the Pol II complex and to a component of the RNA transport apparatus, such as hnRNP K (92), REF (44), and other proteins. Combining all of these potential interactions, ICP27 could thereby serve to couple transcription and RNA transport. This is likely to be a critical role in HSV-infected cells where RNA splicing is inhibited.
Functional homology between ICP27 and RNA helicase A.
Many of the properties associated with ICP27 parallel those of cellular protein RNA helicase A. RNA helicase A has both RNA and DNA helicase activities (45, 101), is associated with the Pol II holoenzyme, and plays a role in bridging the CREB protein (CBP or P300) to Pol II (56). Furthermore, RHA shuttles between the nucleus and cytoplasm and serves as a cofactor for the constitutive transport element of the type D retrovirus (89). Thus, it has been proposed that RHA plays roles in both transcriptional and posttranscriptional processes. Although ICP27 has not been shown to have any enzymatic activities, it promotes transcription of HSV late genes, associates with Pol II, binds to RNA, shuttles between the nucleus and cytoplasm, and may play a role in transporting viral RNA to the cytoplasm. It will be of considerable interest to determine how ICP27 coordinates its activities with those of the functionally similar cellular RHA protein.
This study raises many potential areas for further investigation. First, how do the associations of ICP27 and ICP8 with Pol II affect the transcription of viral and host genes? Second, how do these associations affect the multitude of cellular proteins that associate with the Pol II holoenzyme? Is Pol II tethered to the viral late genes by a new set of protein-protein interactions? Is the splicing apparatus displaced from the complex? Is a new RNA transport apparatus assembled at sites of viral transcription? Answers to these questions will tell us much about how HSV takes control of the host cell nucleus.
Acknowledgments
This research was supported by grants AI20530 and CA26345 from the National Institutes of Health.
We thank Jeff Parvin for providing antibodies and advice on immunoprecipitation and Neal DeLuca, Bernard Roizman, and William Ruyechan for providing antibodies. We thank Karen Hart for technical assistance and Lisa Holik for assistance in preparation of the manuscript.
REFERENCES
- 1.Ackermann, M., D. K. Braun, L. Pereira, and B. Roizman. 1984. Characterization of herpes simplex virus 1 alpha proteins 0, 4, and 27 with monoclonal antibodies. J. Virol. 52:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann, M., M. Sarmiento, and B. Roizman. 1985. Application of antibody to synthetic peptides for characterization of the intact and truncated α22 protein specified by herpes simplex virus 1 and the R325 α22− deletion mutant. J. Virol. 56:207-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alwine, J. C., W. L. Steinhart, and C. W. Hill. 1974. Transcription of herpes simplex type 1 DNA in nuclei isolated from infected HEp-2 and KB cells. Virology 60:302-307. [DOI] [PubMed] [Google Scholar]
- 4.Aubert, M., and J. A. Blaho. 1999. The herpes simplex virus type 1 regulatory protein ICP27 is required for the prevention of apoptosis in infected human cells. J. Virol. 73:2803-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aubert, M., S. A. Rice, and J. A. Blaho. 2001. Accumulation of herpes simplex virus type 1 early and leaky-late proteins correlates with apoptosis prevention in infected human HEp-2 cells. J. Virol. 75:1013-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley, D. 1999. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 11:347-351. [DOI] [PubMed] [Google Scholar]
- 7.Brown, C. R., M. S. Nakamura, J. D. Mosca, G. S. Hayward, S. E. Straus, and L. P. Perera. 1995. Herpes simplex virus trans-regulatory protein ICP27 stabilizes and binds to 3′ ends of labile mRNA. J. Virol. 69:7187-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant, H. E., D. A. Matthews, S. Wadd, J. E. Scott, J. Kean, S. Graham, W. C. Russell, and J. B. Clements. 2000. Interaction between herpes simplex virus type 1 IE63 protein and cellular protein p32. J. Virol. 74:11322-11328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant, H. E., S. E. Wadd, A. I. Lamond, S. J. Silverstein, and J. B. Clements. 2001. Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J. Virol. 75:4376-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cadena, D. L., and M. E. Dahmus. 1987. Messenger RNA synthesis in mammalian cells is catalyzed by the phosphorylated form of RNA polymerase II. J. Biol. Chem. 262:12468-12474. [PubMed] [Google Scholar]
- 11.Cai, W., and P. A. Schaffer. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66:2904-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrozza, M., and N. DeLuca. 1996. Interactions of the viral activator protein ICP4 with TFIID through TAF250. Mol. Cell. Biol. 16:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Challberg, M. D. 1986. A method for identifying the viral genes required for herpesvirus DNA replication. Proc. Natl. Acad. Sci. USA 83:9094-9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chao, D. M., E. L. Gadbois, P. J. Murray, S. F. Anderson, M. S. Sonu, J. D. Parvin, and R. A. Young. 1996. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature 380:82-85. [DOI] [PubMed] [Google Scholar]
- 15.Chen, Y. M., and D. M. Knipe. 1996. A dominant mutant form of the herpes simplex virus ICP8 protein decreases viral late gene transcription. Virology 221:281-290. [DOI] [PubMed] [Google Scholar]
- 16.Chesnut, J. D., J. H. Stephens, and M. E. Dahmus. 1992. The interaction of RNA polymerase II with the adenovirus-2 major late promoter is precluded by phosphorylation of the C-terminal domain of subunit IIa. J. Biol. Chem. 267:10500-10506. [PubMed] [Google Scholar]
- 17.Conaway, R. C., and J. W. Conaway. 1993. General initiation factors for RNA polymerase II. Annu. Rev. Biochem. 62:161-190. [DOI] [PubMed] [Google Scholar]
- 18.Corden, J. L. 1990. Tails of RNA polymerase II. Trends Biochem. Sci. 15:383-387. [DOI] [PubMed] [Google Scholar]
- 19.Corden, J. L., and M. Patturajan. 1997. A CTD function linking transcription to splicing. Trends Biochem. Sci. 22:413-416. [DOI] [PubMed] [Google Scholar]
- 20.Costanzo, F., G. Campadelli-Fiume, L. Foa-Tomasi, and E. Cassai. 1977. Evidence that herpes simplex virus DNA is transcribed by cellular RNA polymerase B. J. Virol. 21:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahmus, M. E. 1996. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 271:19009-19012. [DOI] [PubMed] [Google Scholar]
- 22.Dahmus, M. E. 1994. The role of multisite phosphorylation in the regulation of RNA polymerase II activity. Prog. Nucleic Acid Res. Mol. Biol. 48:143-179. [DOI] [PubMed] [Google Scholar]
- 23.de Bruyn Kops, A., S. L. Uprichard, M. Chen, and D. M. Knipe. 1998. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in different viral functions. Virology 252:162-178. [DOI] [PubMed] [Google Scholar]
- 24.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emili, A., and C. J. Ingles. 1995. The RNA polymerase II carboxy-terminal domain: links to a bigger and better “holoenzyme”? Curr. Opin. Genet. Dev. 5:204-209. [DOI] [PubMed] [Google Scholar]
- 26.Gao, M., and D. M. Knipe. 1989. Genetic evidence for multiple nuclear functions of the herpes simplex virus ICP8 DNA-binding protein. J. Virol. 63:5258-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao, M., and D. M. Knipe. 1991. Potential role for herpes simplex virus ICP8 DNA replication protein in stimulation of late gene expression. J. Virol. 65:2666-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Godowski, P. J., and D. M. Knipe. 1985. Identification of a herpes simplex virus function that represses late gene expression from parental viral genomes. J. Virol. 55:357-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godowski, P. J., and D. M. Knipe. 1983. Mutations in the major DNA-binding protein gene of herpes simplex virus type 1 result in increased levels of viral gene expression. J. Virol. 47:478-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Godowski, P. J., and D. M. Knipe. 1986. Transcriptional control of herpesvirus gene expression: gene functions required for positive and negative regulation. Proc. Natl. Acad. Sci. USA 83:256-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu, B., R. Kuddus, and N. A. DeLuca. 1995. Repression of activator-mediated transcription by herpes simplex virus ICP4 via a mechanism involving interactions with the basal transcription factors TATA-binding protein and TFIIB. Mol. Cell. Biol. 15:3618-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hampsey, M. 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62:465-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hann, L. E., W. J. Cook, S. L. Uprichard, D. M. Knipe, and D. M. Coen. 1998. The role of herpes simplex virus ICP27 in the regulation of UL24 gene expression by differential polyadenylation. J. Virol. 72:7709-7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardy, W. R., and R. M. Sandri-Goldin. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68:7790-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hay, J., G. J. Koteles, H. M. Keir, and H. Subak-Sharpe. 1966. Herpes virus specified ribonucleic acids. Nature 210:387-390. [DOI] [PubMed] [Google Scholar]
- 36.Hengartner, C. J., C. M. Thompson, J. Zhang, D. M. Chao, S. M. Liao, A. J. Koleske, S. Okamura, and R. A. Young. 1995. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 9:897-910. [DOI] [PubMed] [Google Scholar]
- 37.Herr, W. 1998. The herpes simplex virus VP16-induced complex: mechanisms of combinatorial transcriptional regulation. Cold Spring Harbor Symp. Quant. Biol. 63:599-607. [DOI] [PubMed] [Google Scholar]
- 38.Ingram, A., A. Phelan, J. Dunlop, and J. B. Clements. 1996. Immediate early protein IE63 of herpes simplex virus type 1 binds RNA directly. J. Gen. Virol. 77:1847-1851. [DOI] [PubMed] [Google Scholar]
- 39.Jean, S., K. M. LeVan, B. Song, M. Levine, and D. M. Knipe. 2001. Herpes simplex virus 1 ICP27 is required for transcription of two viral late (gamma2) genes in infected cells. Virology 283:273-284. [DOI] [PubMed] [Google Scholar]
- 40.Jenkins, H. L., and C. A. Spencer. 2001. RNA polymerase II holoenzyme modifications accompany transcription reprogramming in herpes simplex virus type 1-infected cells. J. Virol. 75:9872-9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim, Y. J., S. Bjorklund, Y. Li, M. H. Sayre, and R. D. Kornberg. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599-608. [DOI] [PubMed] [Google Scholar]
- 42.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knipe, D. M., and A. E. Spang. 1982. Definition of a series of stages in the association of two herpesviral proteins with the cell nucleus. J. Virol. 43:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20:5769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee, C. G., and J. Hurwitz. 1992. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J. Biol. Chem. 267:4398-4407. [PubMed] [Google Scholar]
- 46.Lee, C. K., and D. M. Knipe. 1983. Thermolabile in vivo DNA-binding activity associated with a protein encoded by mutants of herpes simplex virus type 1. J. Virol. 46:909-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leopardi, R., P. L. Ward, W. O. Ogle, and B. Roizman. 1997. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the UL13 protein kinase. J. Virol. 71:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Long, M. C., V. Leong, P. A. Schaffer, C. A. Spencer, and S. A. Rice. 1999. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 73:5593-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu, H., W. Flores, R. Weinmann, and D. Reinberg. 1991. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc. Natl. Acad. Sci. USA 88:10004-10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCarthy, A. M., L. McMahan, and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J. Virol. 63:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGregor, F., A. Phelan, J. Dunlop, and J. B. Clements. 1996. Regulation of herpes simplex virus poly(A) site usage and the action of immediate-early protein IE63 in the early-late switch. J. Virol. 70:1931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLauchlan, J., A. Phelan, C. Loney, R. M. Sandri-Goldin, and J. B. Clements. 1992. Herpes simplex virus IE63 acts at the posttranscriptional level to stimulate viral mRNA 3′ processing. J. Virol. 66:6939-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 70:7445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mullen, M. A., S. Gerstberger, D. M. Ciufo, J. D. Mosca, and G. S. Hayward. 1995. Evaluation of colocalization interactions between the IE110, IE175, and IE63 transactivator proteins of herpes simplex virus within subcellular punctate structures. J. Virol. 69:476-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Myer, V. E., and R. A. Young. 1998. RNA polymerase II holoenzymes and subcomplexes. J. Biol. Chem. 273:27757-27760. [DOI] [PubMed] [Google Scholar]
- 56.Nakajima, T., C. Uchida, S. F. Anderson, C.-G. Lee, J. Hurwitz, J. D. Parvin, and M. Montminy. 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90:1107-1112. [DOI] [PubMed] [Google Scholar]
- 57.Panagiotidis, C. A., E. K. Lium, and S. J. Silverstein. 1997. Physical and functional interactions between herpes simplex virus immediate-early proteins ICP4 and ICP27. J. Virol. 71:1547-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Payne, J. M., P. J. Laybourn, and M. E. Dahmus. 1989. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J. Biol. Chem. 264:19621-19629. [PubMed] [Google Scholar]
- 59.Phelan, A., M. Carmo-Fonseca, J. McLaughlan, A. I. Lamond, and J. B. Clements. 1993. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc. Natl. Acad. Sci. USA 90:9056-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phelan, A., and J. B. Clements. 1997. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J. Gen. Virol. 78:3327-3331. [DOI] [PubMed] [Google Scholar]
- 61.Phelan, A., J. Dunlop, and J. B. Clements. 1996. Herpes simplex virus type 1 protein IE63 affects the nuclear export of virus intron-containing transcripts. J. Virol. 70:5255-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poffenberger, K. L., E. Tabares, and B. Roizman. 1983. Characterization of a viable, noninverting herpes simplex virus 1 genome derived by insertion and deletion of sequences at the junction of components L and S. Proc. Natl. Acad. Sci. USA 80:2690-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Post, L. E., and B. Roizman. 1981. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell 25:227-232. [DOI] [PubMed] [Google Scholar]
- 64.Preston, C. M., and A. A. Newton. 1976. The effects of herpes simplex virus type 1 on cellular DNA-dependent RNA polymerase activities. J. Gen. Virol. 33:471-482. [DOI] [PubMed] [Google Scholar]
- 65.Proudfoot, N. 2000. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 25:290-293. [DOI] [PubMed] [Google Scholar]
- 66.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 90:6701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 64:1704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rice, S. A., M. C. Long, V. Lam, P. A. Schaffer, and C. A. Spencer. 1995. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J. Virol. 69:5550-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rice, S. A., M. C. Long, V. Lam, and C. A. Spencer. 1994. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J. Virol. 68:988-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rice, S. A., L. S. Su, and D. M. Knipe. 1989. Herpes simplex virus alpha protein ICP27 possesses separable positive and negative regulatory activities. J. Virol. 63:3399-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2460. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott, Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 72.Sacks, W. R., C. C. Greene, D. P. Aschman, and P. A. Schaffer. 1985. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 55:796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sacks, W. R., and P. A. Schaffer. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61:829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Samaniego, L. A., A. L. Webb, and N. A. DeLuca. 1995. Functional interactions between herpes simplex virus immediate early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J. Virol. 69:5705-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 76.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sandri-Goldin, R. M., and M. K. Hibbard. 1996. The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum, and the C terminus appears to be required for this interaction. J. Virol. 70:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sandri-Goldin, R. M., M. K. Hibbard, and M. A. Hardwicke. 1995. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J. Virol. 69:6063-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Scully, R., S. F. Anderson, D. M. Chao, W. Wei, L. Ye, R. A. Young, D. M. Livingston, and J. D. Parvin. 1997. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 94:5605-5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sears, A. E., I. W. Halliburton, B. Meignier, S. Silver, and B. Roizman. 1985. Herpes simplex virus 1 mutant deleted in the alpha 22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J. Virol. 55:338-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith, C. A., P. Bates, R. Rivera-Gonzalez, B. Gu, and N. A. DeLuca. 1993. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J. Virol. 67:4676-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Soliman, T. M., R. M. Sandri-Goldin, and S. J. Silverstein. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 71:9188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spahr, H., J. Beve, T. Larsson, J. Bergstrom, K. A. Karlsson, and C. M. Gustafsson. 2000. Purification and characterization of RNA polymerase II holoenzyme from Schizosaccharomyces pombe. J. Biol. Chem. 275:1351-1356. [DOI] [PubMed] [Google Scholar]
- 85.Spencer, C. A., M. E. Dahmus, and S. A. Rice. 1997. Repression of host RNA polymerase II transcription by herpes simplex virus type 1. J. Virol. 71:2031-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stahl, H., M. Bauer, and R. Knippers. 1983. The simian-virus-40 large-tumor antigen in replicating viral chromatin: a salt-resistant protein-DNA interaction. Eur. J. Biochem. 134:55-61. [DOI] [PubMed] [Google Scholar]
- 87.Steinmetz, E. J. 1997. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell 89:491-494. [DOI] [PubMed] [Google Scholar]
- 88.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67:2571-2585. [DOI] [PubMed] [Google Scholar]
- 89.Tang, H., G. M. Gaietta, W. H. Fischer, M. H. Ellisman, and F. Wong-Staal. 1997. A cellular cofactor for the constitutive transport element of type D retrovirus. Science 276:1412-1415. [DOI] [PubMed] [Google Scholar]
- 90.Thompson, N. E., D. B. Aronson, and R. R. Burgess. 1990. Purification of eukaryotic RNA polymerase II by immunoaffinity chromatography. Elution of active enzyme with protein stabilizing agents from a polyol-responsive monoclonal antibody. J. Biol. Chem. 265:7069-7077. [PubMed] [Google Scholar]
- 91.Uprichard, S. L., and D. M. Knipe. 1996. Herpes simplex virus ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J. Virol. 70:1969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wadd, S., H. Bryant, O. Filhol, J. E. Scott, T. Y. Hsieh, R. D. Everett, and J. B. Clements. 1999. The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J. Biol. Chem. 274:28991-28998. [DOI] [PubMed] [Google Scholar]
- 93.Wagner, E. K., and B. Roizman. 1969. Ribonucleic acid synthesis in cells infected with herpes simplex virus. I. Patterns of ribonucleic acid synthesis in productively infected cells. J. Virol. 4:36-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Watson, R. J., and J. B. Clements. 1980. A herpes simplex virus type 1 function continuously required for early and late virus RNA synthesis. Nature 285:329-330. [DOI] [PubMed] [Google Scholar]
- 95.Wintzerith, M., J. Acker, S. Vicaire, M. Vigneron, and C. Kedinger. 1992. Complete sequence of the human RNA polymerase II largest subunit. Nucleic Acids Res. 20:910.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu, K. C., and M. Pfahl. 1988. Variable responsiveness of hormone-inducible hybrid genes in different cell lines. Mol. Endocrinol. 2:1294-1301. [DOI] [PubMed] [Google Scholar]
- 97.Xia, K., N. A. DeLuca, and D. M. Knipe. 1996. Analysis of phosphorylation sites of the herpes simplex virus type 1 ICP4. J. Virol. 70:1061-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xia, K., D. M. Knipe, and N. A. DeLuca. 1996. Role of protein kinase A and the serine-rich domain of herpes simplex virus type 1 ICP4 in viral replication. J. Virol. 70:1050-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Young, R. A. 1991. RNA polymerase II. Annu. Rev. Biochem. 60:689-715. [DOI] [PubMed] [Google Scholar]
- 100.Zawel, L., and D. Reinberg. 1993. Initiation of transcription by RNA polymerase II: a multi-step process. Prog. Nucleic Acid Res. Mol. Biol. 44:67-108. [DOI] [PubMed] [Google Scholar]
- 101.Zhang, S. S., and F. Grosse. 1991. Purification and characterization of two DNA helicases from calf thymus nuclei. J. Biol. Chem. 266:20483-20490. [PubMed] [Google Scholar]
- 102.Zhong, L., and G. S. Hayward. 1997. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient contransfection assays. J. Virol. 71:3146-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]