Abstract
The structure of recombinant virus-like particles of malabaricus grouper nervous necrosis virus (MGNNV), a fish nodavirus isolated from the grouper Epinephelus malabaricus, was determined by electron cryomicroscopy (cryoEM) and three-dimensional reconstruction at 23-Å resolution. The cryoEM structure, sequence comparison, and protein fold recognition analysis indicate that the coat protein of MGNNV has two domains resembling those of tomato bushy stunt virus and Norwalk virus, rather than the expected single-domain coat protein of insect nodaviruses. The analysis implies that residues 83 to 216 fold as a β-sandwich which forms the inner shell of the T=3 capsid and residues 217 to 308 form the trimeric surface protrusions observed in the cryoEM map. The structural similarities between fish nodaviruses and members of the tombusvirus and calicivirus groups provide significant new data for understanding the evolution of the nodavirus family.
Fish nodaviruses are associated with massive mortality in more than 20 species of fish in oceans near Asia, Europe, and Australia (14, 16). They cause nervous necrosis, encephalopathy, and retinopathy, with the appearance of vacuolating lesions in the brain and retina, abnormal swimming behavior, dark color, and high mortality in hatchery-reared larvae and juveniles (1, 2, 4, 7-9, 21). Classification of these viruses into the genus Betanodavirus of the family Nodaviridae is based on similarities to the insect nodaviruses in shape, size, and buoyant density of the particles and the bipartite RNA genome organization. Sequence comparisons, however, show no significant homology between capsid proteins of fish and insect nodaviruses (15). Like the insect nodaviruses, the genome of fish nodaviruses consists of two molecules of messenger sense, single-stranded RNA: a larger RNA1 encoding the RNA-dependent RNA polymerase, and a smaller RNA2 encoding the coat protein. The structure, assembly, and RNA packaging of insect nodaviruses have been studied extensively (5, 18-20, 23). However, the malabaricus grouper nervous necrosis virus (MGNNV) replicates poorly in infected tissue, which causes difficulties in obtaining sufficient material for biophysical analysis.
Recently the capsid protein gene of a fish nodavirus causing nervous necrosis in the grouper Epinephelus malabaricus was expressed in Sf21 insect cells using a recombinant baculovirus vector (12). The virus-like particles that spontaneously assembled were morphologically indistinguishable from authentic MGNNV when examined by negative-stain electron microscopy. RNA1 was not present in the expression system, and the virus-like particles packaged mainly cellular RNA. Here we report the three-dimensional structure of the virus-like particles of MGNNV determined by electron cryomicroscopy (cryoEM), image reconstruction, and a sequence-based domain organization of the protein subunit. The results from the different analyses provide consistent and convincing evidence that the MGNNV subunit organization is different from that of the insect nodavirus.
CryoEM image reconstruction.
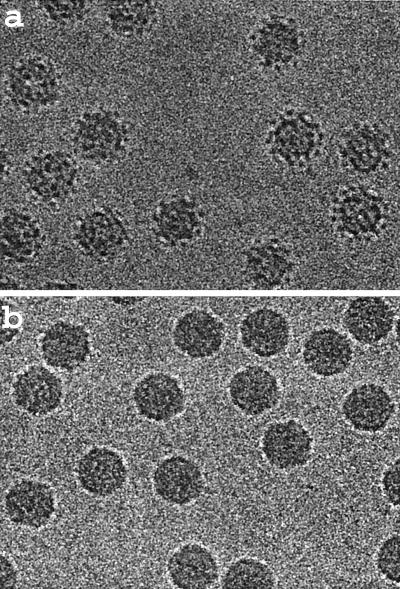
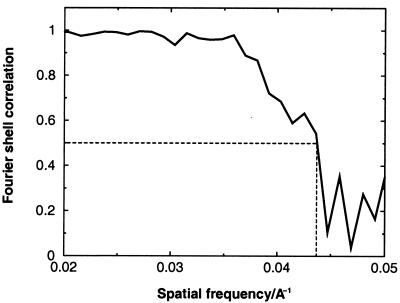
The cryoEM structure of MGNNV was determined as described previously (for an example, see reference 20). Micrographs of frozen hydrated particles were recorded with a Philips CM120 electron microscope at liquid nitrogen temperatures under low-dose conditions (Fig. 1). A total of 263 of 313 boxed particles from a single micrograph recorded at ∼1.2-μm underfocus was used for the final reconstruction. The reconstruction was computed with the program SPIDER (6). The resolution of the reconstruction was ∼23 Å as estimated by the Fourier shell correlation method (Fig. 2) (22).
FIG. 1.
CryoEM micrographs of frozen-hydrated samples of MGNNV (a) and PaV (b) recorded at an underfocus of ∼2.7- and ∼2.9-μm, respectively. Note the surface feature of particles in panel a.
FIG. 2.
Fourier shell correlation between two reconstructions, each independently computed by using half of the images. The correlation starts to fall below 0.5 at a spatial frequency of ∼0.043, which corresponds to a resolution of 23 Å.
CryoEM structure of MGNNV.
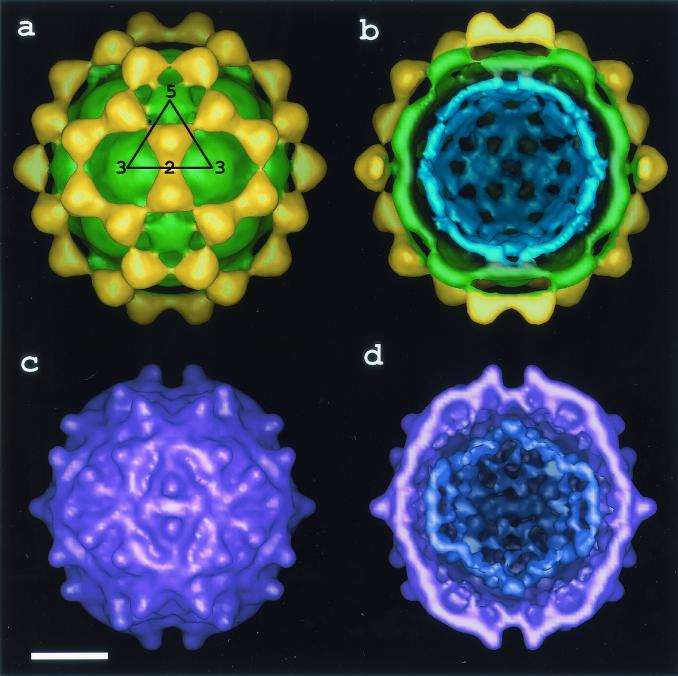
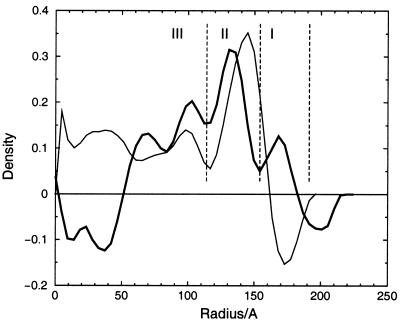
As anticipated from the difference in appearance of the fish and insect nodaviruses in high-contrast, unprocessed images of frozen hydrated particles (Fig. 1), the cryoEM structure of MGNNV differed remarkably from those of insect nodaviruses, such as Pariacoto virus (PaV) (Fig. 3). Like that of the insect nodaviruses, the surface morphology of MGNNV is consistent with a T=3 quasi-equivalent lattice; however, the details of the morphology and the radial density distribution of MGNNV differ dramatically from those of PaV (Fig. 3 and 4). The maximum diameter of MGNNV is approximately 380 Å, significantly larger than the PaV capsid at 360 Å. The density distribution of the MGNNV map shows two shells, at radii corresponding to protein, separated by low density. The outer shell is between radii of 154 and 192 Å with a maximum at 168 Å; the inner shell is between radii of 112 and 154 Å with a maximum at 135 Å. The MGNNV map is contoured at a level such that the resultant volume of these two shells is equal to the expected volume of its protein capsid, which is formed by 180 copies of a 38-kDa subunit. It is noteworthy that the interior boundary of the inner shell (112 Å) of MGNNV and PaV are nearly coincident, and this defines the interior limit of the β-sandwich domain of PaV. Density with a radius of less than 112 Å (Fig. 3b and 4) is probably predominately RNA.
FIG. 3.
CryoEM maps of MGNNV at 23-Å resolution (a) and PaV at 22 Å resolution (c). (b and d) Cutaway views of panels a and c, respectively. Panels a and b are colored radially. The densities for the protrusions (∼154 to 192 Å), the inner shell of the capsid (∼112 to 154 Å), and the RNA (<112 Å) are gold, green, and blue, respectively. In panel d, the RNA density of PaV is dark purple. In panel a, an icosahedral asymmetric unit is outlined by a triangle, and the positions of the icosahedral five-, three-, and two-fold axes are indicated by numbers. The quasi-two-fold axes are centered between the five- and three-fold axes. Bar, 100 Å.
FIG.4.
Radial density distribution of the cryoEM maps of MGNNV (bold line) and PaV (thin line) derived by spherically averaging the density in the maps. The layers of density corresponding to the protrusions, the inner shell of the capsid, and the RNA in MGNNV map are indicated by I, II, and III, respectively.
The outer shell of MGNNV is composed mainly of the large protrusions located at the quasi-three-fold axes. These protrusions are much more prominent than those of PaV (Fig. 3). The inner shell is relatively uniform, and the protrusions are separated from it by nearly a 12-Å void, indicating that the connection between the domains is probably a single, extended polypeptide chain that is not visible in the cryoEM map.
The crystal structures of PaV and Flock house virus showed that the protrusions at highest radius are formed by three two-stranded β-sheets related to each other by quasi-three-fold symmetry (5, 20) (Fig. 5a). These β-sheets are formed by insertions between strands of the canonical viral β-sandwich that forms the contiguous protein shells of the insect nodaviruses, and the strands are twisted together about the quasi-three-fold axes to form the surface protrusions. The inner and outer regions of the insect nodaviruses, however, display continuous density in contrast to the density gap between the outer and inner shells of protein in MGNNV.
FIG. 5.
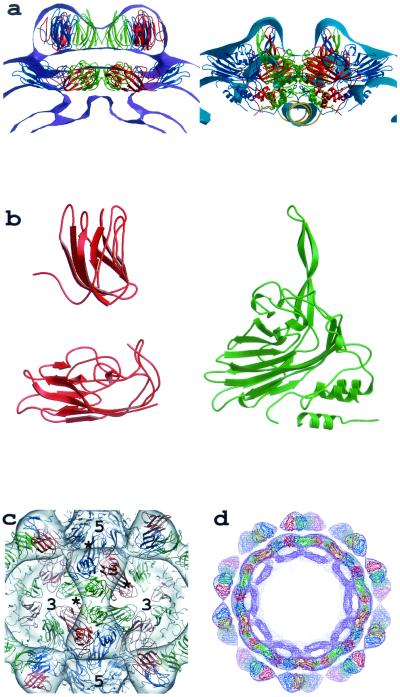
(a) Cross-sectional views of the particle envelopes of MGNNV (left) and PaV (right) defined by the cryoEM density. The refined TBSV model is superimposed with the MGNNV density and the X-ray atomic model of PaV with the PaV density. In both panels, two icosahedral asymmetric units are shown. Subunits A, B, and C are colored blue, red, and green, respectively, and the RNA duplex in the PaV model is colored yellow. (b) Comparison of one subunit of TBSV (left) that was fitted into the MGNNV density in panel a with the equivalent PaV subunit (right). The lack of connectivity between the two domains of TBSV reflects the lack of obvious density connecting the domains in the cryoEM map of MGNNV, suggesting that the connection is made by a single, extended polypeptide. (c) A view down the two-fold axis of the MGNNV density with the TBSV model superimposed. The view emphasizes the strong trimer association of the protruding domains around the quasi-three-fold axes. The C termini of subunits are indicated by stars, and the positions of icosahedral three- and five-fold axes are indicated by 3 and 5, respectively. (d) Cross section of the TBSV model superimposed on the cryoEM density of the full particle of MGNNV viewed down the five-fold axis.
Superposition of the cryoEM map of MGNNV with the atomic model of PaV derived from the crystal structure showed relatively good agreement for the region occupied by the contiguous β-sandwich shell in PaV. However, there was poor agreement in the outer radial region (Fig. 5a). The protrusions at the quasi-three-fold axes in MGNNV have much larger volume than could be accounted for by the three twisted β-sheets at the surface of the PaV structure (Fig. 5a). Thus, these protrusions must contain more protein that may form individual domains. Moreover, the PaV model could not account for differences in density observed in the MGNNV reconstruction at the icosahedral and quasi-two-fold symmetry axes. Significant density exists at the icosahedral two-fold axes between protrusions. However, there is only weak density at the corresponding regions between protrusions related by the quasi-two-fold symmetry (Fig. 3a). This density difference implies different patterns of contacts between protrusions at icosahedral and quasi-two-fold axes.
Both the cryoEM reconstruction and the crystal structure of PaV revealed that an ordered portion of the viral RNA forms a dodecahedral cage composed of 30 segments of RNA duplex closely associated with the capsid (20). In the reconstruction of MGNNV, there is no significant density adjacent to the capsid that can be interpreted as duplex RNA. However, the density at lower radius (<112 Å) may be attributed to the cellular RNA randomly packaged inside the capsid and/or portions of the coat protein, e.g., the N-terminal basic-residue-rich segments which are likely to interact with RNA (Fig. 3b).
Fold recognition of MGNNV capsid protein.
The MGNNV capsid protein shows no significant sequence homology with any known insect nodavirus, although sequence identity among fish nodaviruses varies from 71 to 88% (15, 16). We used 3D-PSSM (three-dimensional position-specific scoring matrix) to investigate the folding motif of the MGNNV coat protein. 3D-PSSM combines knowledge of three-dimensional structures with secondary-structure matching and solvation potentials to recognize protein folds with remote sequence homologues (11). The amino acid sequence of the coat protein of MGNNV was entered into the web server of 3D-PSSM (http://www.bmm.icnet.uk/∼3dpssm/) to search for the homologous folds in the database of known structures. Four structures were identified as comparable with MGNNV with a confidence level of ≥95%, while the rest of the candidates had a confidence level of <50% (Table 1). Strikingly, there were no insect nodaviruses with known atomic structure in the top 20 comparable structures. As a control, the primary sequence of PaV was entered, and all known structures of insect nodaviruses were identified as the top candidates for similar folding (data not shown).
TABLE 1.
The top five proteins with folds comparable to that of MGNNV coat protein by 3D-PSSM searching
| Templatea | Template length (no. of residues) | Sequence identity (%) | Confidenceb (%) |
|---|---|---|---|
| 1smv_c | 222 | 14 | >95 |
| 1f2n_a | 189 | 17 | >95 |
| 1c8n_c | 219 | 15 | >95 |
| 2tbv_c | 321 | 16 | >95 |
| 2mev_3 | 231 | 12 | <50 |
1smv, Sesbania mosaic virus; 1f2n, rice yellow mottle virus; 1c8n, tobacco necrosis virus; 2tbv, tomato bushy stunt virus; 2mev, Mengo encephalomyocarditis virus. The first four letters of the template name are the Protein Data Bank code of the structure, and the last letter is the chain name.
Confidence is defined as 100 × probability (p) of a match being correct. For a definition of p, see reference 11.
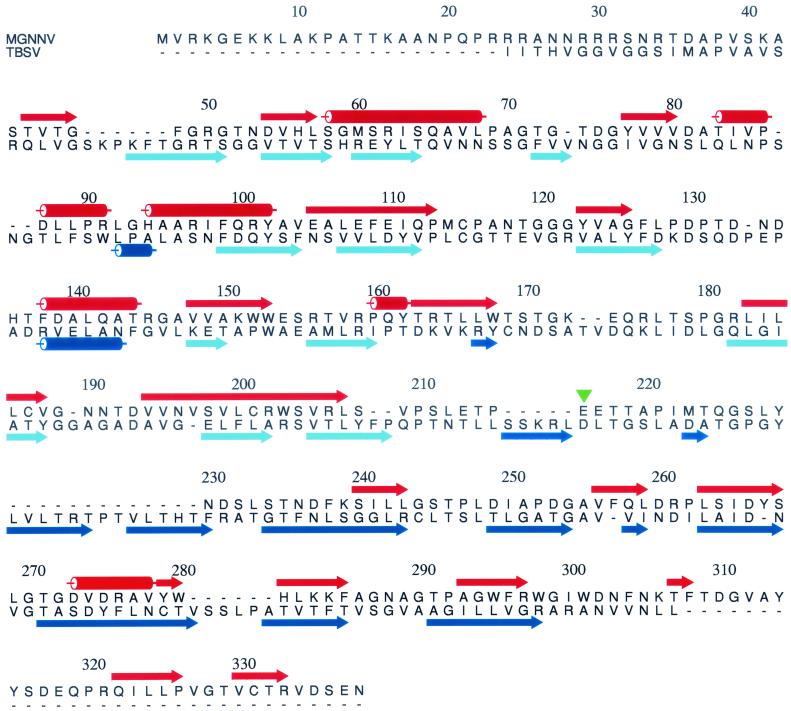
All four structures identified as likely homologues of MGNNV were virus coat proteins that have canonical, uninterrupted viral β-sandwich folds. The first three varied from 189 to 222 aligned residues, and they predicted the fold of MGNNV residues 31 to 235, 49 to 214, and 27 to 213, respectively, as a β-sandwich (Table 1). The fourth structure provided prediction for almost the entire MGNNV sequence (residues 24 to 319) (Fig. 6). The fourth structure was the coat protein of tomato bushy stunt virus (TBSV), which contains two domains: the N-terminal domain (residues 103 to 271) forming a β-sandwich and the contiguous protein capsid, and the C-terminal domain (residues 272 to 387) forming a protrusion at the capsid surface (10). Sequence comparisons of the coat proteins of several fish nodaviruses showed that the protein could be divided into a conserved region and a variable region (15). The conserved region comprises residues 83 to 216 and shows pairwise sequence identity of 86 to 96%; the variable region comprises residues 235 to 315, with pairwise sequence identity of 66 to 84%. These results suggested that residues 83 to 216 of MGNNV might form a conserved β-sandwich domain that is similar to that in TBSV. Residues 217 to the C-terminal residue 338 would then form a protruding domain that corresponds to the surface protrusions observed in the cryoEM map. The 122 residues in the protruding domain correspond to 48% of the 256 residues in the two domains. This percentage is close to the ratio of the volume of the protrusions to the total capsid volume (45%) in the cryoEM map. No homologous structure was identified with a confidence factor above 50% when only the variable region of MGNNV (residues 217 to 338) was entered into 3D-PSSM, indicating that this protruding domain may have a novel fold. The N-terminal segment from residues 1 to 82 shares more than 79% sequence identity among the four fish nodaviruses MGNNV, Dicentrarchus labrax encephalitis virus, striped jack nervous necrosis virus, and Atlantic halibut virus. It is rich in basic residues and may contribute to neutralization of the negative charge of the viral nucleic acid in virus assembly and RNA packaging as demonstrated for insect nodaviruses (13, 20).
FIG. 6.
Alignment of MGNNV and TBSV coat protein sequences by 3D-PSSM. Although there is no significant sequence identity between the coat proteins of the two viruses, the pattern of secondary structures of MGNNV is rich in β-elements and matches that of TBSV to a reasonable extent. The secondary structures of MGNNV (red) are predicted by 3D-PSSM, and those of TBSV (blue and light blue) are assigned based on the crystal structure. The α-helices and β-strands are shown as rods and arrows, respectively. In TBSV, the β-strands forming the N-terminal β-sandwich domain are colored light blue. The green triangle near residue 216 indicates the boundary between the putative two domains of MGNNV.
Modeling the domain structure of MGNNV capsid protein.
The β-sandwich domain (residues 103 to 271) and the protruding domain (residues 272 to 387) of TBSV (Protein Data Bank entry code 1tbv) were manually fitted into the inner shell and protruding density of the cryoEM map of MGNNV, respectively. The resultant model was subjected to cycles of rigid body refinement against a set of structure amplitudes generated from the cryoEM map. For this calculation, the map was modified in such a way that density above or equal to 1.4 standard deviation was set to 1, and lower density was set to 0. This adjustment ensured that the refinement was carried out against a molecular volume and shape as shown in Fig. 3a and b. During the refinement, each domain was defined as an independent rigid group. The crystallographic R-factor dropped from 0.56 to 0.48. The final model agreed well with the cryoEM map (Fig. 5). Compared to the original TBSV coordinates, the β-sandwich domain was shifted slightly towards the center of the virus particle (the root mean square difference in Cα positions is ∼3 Å) whereas the protruding domain moved by ∼24 Å with a rotation of ∼87°. Such a large change reflects an improved fit to the density but also suggests, like the 3D-PSSM searches, that there may be no relationship between the folds of the outer domains in TBSV and MGNNV and that the TBSV domain is only shifted to optimally occupy the MGNNV density. Nevertheless, the pattern of interactions between the outer domains confirms the T=3 lattice of the capsid (Fig. 5a and c). These domains make more extensive contacts at the icosahedral two-fold axes than at the quasi-two-fold axes, which is consistent with the observation of more-significant density at icosahedral two-fold axes than at quasi-two-fold axes in the cryoEM map.
The model suggests that the C terminus of the coat protein of MGNNV is probably located at the outer surface of the capsid. In contrast, the X-ray structures of insect nodaviruses showed that the C-terminal regions are situated inside the capsid (5, 20, 23). In all known insect nodaviruses, a short, C-terminal segment of the coat protein is cleaved via postassembly autocatalysis to yield mature particles with full infectivity, and it is proposed to play a role in the delivery of viral RNA into host cells (3, 5, 20, 23). However, there is no biochemical evidence for such autoproteolysis in the fish nodaviridae, indicating that RNA translocation in fish nodaviruses upon infection may involve a different molecular mechanism.
The cryoEM structure and fold analysis demonstrated that the MGNNV coat protein bears a closer resemblance to TBSV than to the insect nodavirus capsid proteins. The conserved region from residues 83 to 216 probably folds into a β-sandwich that forms the inner shell of the capsid. The variable region from residues 217 to the C terminus probably forms the surface-protruding domain and may have a novel fold as suggested by results with 3D-PSSM. Presumably, residues around 216 form a polypeptide hinge to connect the two domains. It is likely that the relative position and orientation of the outer domain in MGNNV are different from those in TBSV. The protruding domains in MGNNV are held together around the quasi-three-fold axes to form 60 protrusions, whereas the protruding domains in TBSV join at the icosahedral and quasi-two-fold axes to form 90 protrusions. The coat protein of Norwalk virus also consists of a β-sandwich domain, which forms the inner shell of the capsid, and a protruding domain, which forms dimers at the icosahedral and quasi-two-fold axes as in TBSV (17). In all these viruses, the β-sandwich domain serves to form the structural scaffold of the virus capsid, while the protruding domain may be involved in specific biological processes such as host cell recognition.
The structural similarity between fish nodaviruses and members of the tombusvirus and calicivirus groups suggests that fish and insect nodaviruses have converged to bipartite viruses, with their structures and some functions reflecting different origins.
Acknowledgments
This work was supported by NIH grants to J.E.J., A.S., and M.Y. M.Y. is the recipient of a Clinical Scientist Award in Translation Research from the Burroughs Wellcome Fund. N.K.K. was supported by an NIH fellowship award.
REFERENCES
- 1.Bovo, G., T. Nishizawa, C. Maltese, F. Borghesan, F. Mutinelli, F. Montesi, and S. De Mas. 1999. Viral encephalopathy and retinopathy of farmed marine fish species in Italy. Virus Res. 63:143-146. [DOI] [PubMed] [Google Scholar]
- 2.Breuil, G., J. R. Bonami, J. F. Pepin, and Y. Pichot. 1991. Viral infection (picorna-like virus) associated with mass mortalities in hatchery-reared sea bass (Dicentrarchus labrax) larvae and juveniles. Aquaculture 97:109-116. [Google Scholar]
- 3.Cheng, R. H., V. S. Reddy, N. H. Olson, A. J. Fisher, T. S. Baker, and J. E. Johnson. 1994. Functional implications of quasi-equivalence in a T=3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure 2:271-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chua, F. H. C., M. L. Ng, K. L. Ng, J. J. Loo, and J. Y. Wee. 1994. Investigation of outbreaks of a novel disease, ‘sleepy grouper disease,’ affecting the brown-spotted grouper, Epinephelus tauvina Forskal. J. Fish Dis. 17:417-427. [Google Scholar]
- 5.Fisher, A. J., and J. E. Johnson. 1993. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature 361:176-179. [DOI] [PubMed] [Google Scholar]
- 6.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 7.Glazebrook, J. S., M. P. Heasman, and S. W. de Beer. 1990. Picorna-like viral particles associated with mass mortalities in larval barramundi, Lates calcarifer Bloch. J. Fish Dis. 13:245-249. [Google Scholar]
- 8.Grotmol, S., G. K. Totland, K. Thorud, and B. K. Hjeltnes. 1997. Vacuolating encephalopathy and retinopathy with a nodavirus-like agent: a probable cause of mass mortality of cultured larval and juvenile Atlantic halibut Hippoglossus hippoglossus. Dis. Aquat. Organ. 29:85-97. [Google Scholar]
- 9.Grotmol, S., O. Bergh, and G. K. Totland. 1999. Transmission of viral encephalopathy and retinopathy (VER) to yolk-sac larvae of the Atlantic halibut Hippoglossus hippoglossus: occurrence of nodavirus in various organs and possible route of infection. Dis. Aquat. Organ. 36:95-106. [DOI] [PubMed] [Google Scholar]
- 10.Harrison, S., A. Olson, C. Schutt, F. Winkler, and G. Bricogne. 1978. Tomato bushy stunt virus at 2.9 Å resolution. Nature 276:368-373. [DOI] [PubMed] [Google Scholar]
- 11.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 12.Lin, C.-S., M.-W. Lu, L. Tang, W. Liu, C.-B. Chao, C. J. Lin, N. K. Krishna, J. E. Johnson, and A. Schneemann. 2001. Characterization of virus-like-particles assembled in a recombinant baculovirus system expressing the capsid protein of a fish nodavirus. Virology 290:50-58. [DOI] [PubMed] [Google Scholar]
- 13.Marshall, D., A. Schneemann. 2001. Specific packaging of nodaviral RNA2 requires the N-terminus of the capsid protein. Virology. 285:165-175. [DOI] [PubMed] [Google Scholar]
- 14.Munday, B. L., and T. Nakai. 1997. Special topic review: nodaviruses as pathogens in larval and juvenile marine finfish. World J. Microbiol. Biotechnol. 13:375-381. [Google Scholar]
- 15.Nishizawa, T., K. Mori, M. Furuhashi, T. Nakai, I. Furusawa, and K. Muroga. 1995. Comparison of the coat protein genes of five fish nodaviruses, the causative agents of viral nervous necrosis in marine fish. J. Gen. Virol. 76:1563-1569. [DOI] [PubMed] [Google Scholar]
- 16.Nishizawa, T., M. Furuhashi, T. Nagai, T. Nakai, and K. Muroga. 1997. Genomic classification of fish nodaviruses by molecular phylogenetic analysis of the coat protein gene. Appl. Environ. Microbiol. 63:1633-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 18.Schneemann, A., and D. Marshall. 1998. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J. Virol. 72:8738-8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneemann, A., V. S. Reddy, and J. E. Johnson. 1998. The structure and function of nodavirus particles: a paradigm for understanding chemical biology. Adv. Virus Res. 50:381-446. [DOI] [PubMed] [Google Scholar]
- 20.Tang, L., K. N. Johnson, L. A. Ball, T. Lin, M. Yeager, and J. E. Johnson. 2001. The structure of Pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat. Struct. Biol. 8:77-83. [DOI] [PubMed] [Google Scholar]
- 21.Thiery, R., J. C. Raymond, and J. Castric. 1999. Natural outbreak of viral encephalopathy and retinopathy in juvenile sea bass, Dicentrarchus labrax: study by reverse transcriptase-polymerase chain reaction. Virus Res. 63:11-17. [DOI] [PubMed] [Google Scholar]
- 22.van Heel, M. 1987. Similarity measures between images. Ultramicroscopy 21:95-100. [Google Scholar]
- 23.Wery, J. P., V. S. Reddy, M. V. Hosur, and J. E. Johnson. 1994. The refined three-dimensional structure of an insect virus at 2.8 Å resolution. J. Mol. Biol. 235:565-586. [DOI] [PubMed] [Google Scholar]