Abstract
The 3C-like protease of the Chiba virus, a Norwalk-like virus, is one of the chymotrypsin-like proteases. To identify active-site amino acid residues in this protease, 37 charged amino acid residues and a putative nucleophile, Cys139, within the GDCG sequence were individually replaced with Ala in the 3BC precursor, followed by expression in Escherichia coli, where the active 3C-like protease would cleave 3BC into 3B (VPg) and 3C (protease). Among 38 Ala mutants, 7 mutants (R8A, H30A, K88A, R89A, D138A, C139A, and H157A) completely or nearly completely lost the proteolytic activity. Cys139 was replaceable only with Ser, suggesting that an SH or OH group in the less bulky side chain was required for the side chain of the residue at position 139. His30, Arg89, and Asp138 could not be replaced with any other amino acids. Although Arg8 was also not replaceable for the 3B/3C cleavage and the 3C/3D cleavage, the N-terminal truncated mutant devoid of Arg8 significantly cleaved 3CD into 3C and 3D (polymerase), indicating that Arg8 itself was not directly involved in the proteolytic cleavage. As for position 88, a positively charged residue was required because the Arg mutant showed significant activity. As deduced by the X-ray structure of the hepatitis A virus 3C protease, Arg8, Lys88, and Arg89 are far away from the active site, and the side chain of Asp138 is directed away from the active site. Therefore, these are not catalytic residues. On the other hand, all of the mutants of His157 in the S1 specificity pocket tended to retain very slight activity, suggesting a decreased level of substrate recognition. These results, together with a sequence alignment with the picornavirus 3C proteases, indicate that His30 and Cys139 are active-site residues, forming a catalytic dyad without a carboxylate directly participating in the proteolysis.
Norwalk-like viruses (NLVs) are causative agents of nonbacterial acute gastroenteritis in humans (8, 11, 23, 24). NLV, which is tentatively accepted as the name of the genus, belongs to the family Caliciviridae (16). NLV is well known as a small round-structured virus, the name being derived from the virion structure as observed by electron microscopy. NLVs are serologically and genetically diverse, and hundreds of NLVs have been investigated worldwide (44). However, NLVs cannot be propagated on cell cultures, which restricts collection of vital virus particles containing the genomic RNA. The whole genomes from limited numbers of NLVs have been cloned and sequenced (9, 20, 22, 28, 31, 38, 40).
The NLV genome contains three open reading frames (ORFs). ORF1 encodes a large polyprotein (nonstructural protein), which is processed by the viral protease into six proteins, including putative 2C-like RNA helicase (with NTPase activity, but not RNA helicase activity, proven [35]), 3B VPg, 3C-like protease, and 3D RNA-dependent RNA polymerase. ORF2 encodes a capsid protein (structural protein), and 180 molecules of the protein are organized into a native NLV virion (36). ORF3 encodes a small basic protein, which was recently shown to be a minor structural protein included in a virion (13). When expressed in insect cells by using the baculovirus system, ORF2 alone is capable of producing virus-like particles lacking the genomic RNA (21, 30).
The 3C-like proteases from NLVs and the 3C proteases from picornaviruses are assumed to have chymotrypsin-like folds (2, 10, 14, 32, 39). The X-ray structures of picornaviral 3C proteases from poliovirus (34), rhinovirus (33), and hepatitis A virus (HAV) (1, 3) support this assumption. However, while the active-site nucleophile of chymotrypsin is a serine residue within the GDSG sequence, that of 3C(-like) proteases is a cysteine residue within the GDCG or GMCG sequence. Therefore, 3C(-like) protease is called chymotrypsin-like protease or serine-like protease (10, 14). 3C(-like) proteases constitute a clearly distinct group in the chymotrypsin-like protease family and are different from papain-like cysteine proteases.
Several studies by site-directed mutagenesis have been addressed to identify active-site amino acid residues of picornavirus 3C proteases (7, 15, 17-19, 25, 29), which corresponded to the catalytic triad (His57, Asp102, and Ser195) of chymotrypsin (12). The X-ray crystal structures revealed that amino acid residues His40, Glu71, and Cys147 formed the catalytic triad in the 3C poliovirus protease (34), and His40, Glu71, and Cys146 formed the catalytic triad in the rhinovirus 3C protease (33). However, in the HAV 3C protease, the side chain of Asp84 corresponding to Asp102 of chymotrypsin was directed away from the active site (1, 3). Therefore, it was proposed that the catalytic dyad (His44 and Cys172) was mandatory for the proteolysis.
On the other hand, as for 3C-like protease in the family Caliciviridae, the active-site amino acid residues of 3C-like protease from rabbit hemorrhagic disease virus (RHDV) were determined by site-directed mutagenesis study (5). It was indicated that the catalytic triad composed of His27, Asp44, and Cys104 was critical for the protease activity.
The Chiba virus (Hu/NLV/Chiba407/1987/JP) is one of the NLVs that was isolated in Chiba Prefecture, Japan (43). We have already cloned and sequenced the whole genome of the Chiba virus (40). The Chiba virus 3C-like protease expressed in Escherichia coli has been shown to retain enzymatic activity and substrate specificity (40). In this study, we attempted to identify active-site amino acid residues of the Chiba virus 3C-like protease by using the 3BC fragment derived from Chiba virus ORF1 to assess the protease activity. Thirty-seven charged amino acid residues and putative nucleophile Cys139 (designated by the amino acid number of the 3C-like protease) within the GDCG sequence were subjected to site-directed mutagenesis, and the mutant 3BC construct was tested to see whether or not it was separated into 3B and 3C when expressed in E. coli. It was revealed that the catalytic dyad composed of His30 and Cys139 played a critical role in the proteolysis.
MATERIALS AND METHODS
Reagents.
Pfu Turbo DNA polymerase was purchased from Stratagene (La Jolla, Calif.). Restriction and modifying enzymes were purchased from Takara (Tokyo, Japan), Toyobo (Tokyo, Japan), New England BioLabs (Beverly, Mass.), and Fermentas (Hanover, Md.). The ABI Prism DNA sequencing kit (PE Applied Biosystems, Branchburg, N.J.) was used for DNA sequencing with the ABI Prism 310 genetic analyzer. All other reagents were of reagent grade and were from commercial sources.
Bacterial strains.
E. coli JM109 (45) and TG1 (42) were used for genetic manipulation. E. coli CJ236 (27) was used for the preparation of deoxyuracil-containing single-stranded DNA. E. coli BL21-CodonPlus-RIL (Stratagene) was used for expression of the constructs derived from the Chiba virus ORF1.
Plasmids.
The gene fragment encoding the 3B (VPg)-3C (protease) region was amplified by PCR with pUCCVORF1 (40) as a template. The forward primer (5′-ACTAGCGCTGGTAAAAACAAAGGAAAGACCAAG-3′) was designed so as to introduce an Aor51HI site prior to the first codon for the Gly1 of VPg. The reverse primer (5′-TTTGCATGCTTACTCTAGGGTGGTTTCACCTTC-3′) was designed in order to introduce a stop codon (TAA) and a PaeI (SphI) site after the last codon for the Glu181 of protease. After verification by DNA sequencing, the Aor51HI-PaeI fragment containing the entire 3BC region was cloned into the corresponding sites of pUCHisNd (40) to construct pUCHis3BC, on which the expression of the His-tagged 3BC construct was controlled by the lac promoter (Fig. 1B). The SacI-PaeI fragment encoding part of the VPg and the entire protease was subcloned in pUC119 (45), and the resultant plasmid, designated pUCCV3BC3059, was used for site-directed mutagenesis.
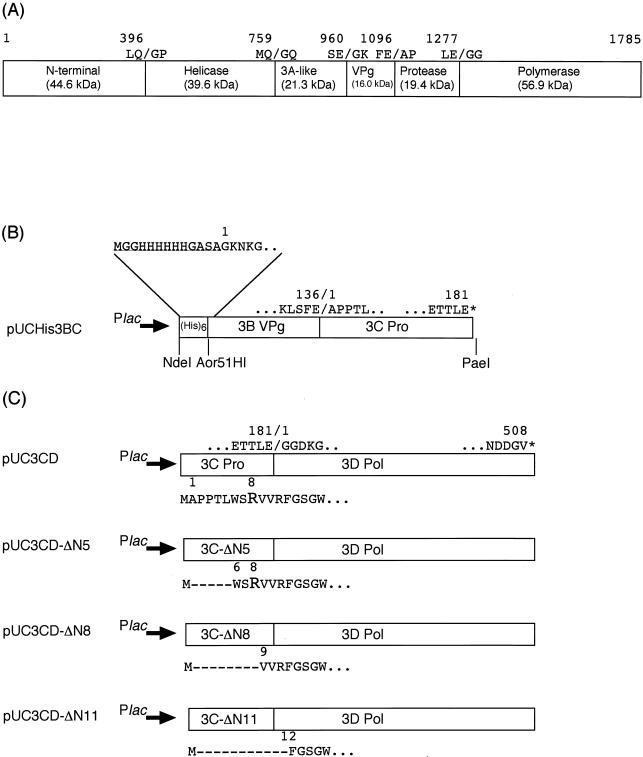
FIG. 1.
Schematic representation of the Chiba virus ORF1 polyprotein and construction of the expression plasmids. (A) Map of the putative proteolytic cleavage sites within ORF1. The numbers indicate the positions of the amino acids. Slashes indicate the cleavage sites. The calculated molecular masses of the cleavage products are also indicated. (B) Plasmid pUCHis3BC expressing the His-3BC fragment. An extra sequence (underlined) containing six consecutive histidine residues was fused to the N terminus of VPg. The His-3BC protein ends with the last residue (Glu181) of the 3C-like protease. (C) Plasmid pUC3CD expressing the 3CD fragment and its related N-terminal truncation mutant plasmids. The 3CD protein starts with the first residues (Ala1) of the 3C-like protease after the initial Met residue and ends with the last residue (Val508) of the 3D RNA-dependent RNA polymerase. The 3CD-ΔN5, 3CD-ΔN8, and 3CD-ΔN11 proteins start with Trp6, Val9, and Phe12, respectively.
The gene fragment encoding the 3C-3D (RNA polymerase) region was amplified by PCR with pUCCVORF1 (40) as a template. The forward primer (5′-GAAAAACTCAGTCATATGGCCCCACCAACCCTC-3′) was designed so as to introduce an NdeI site containing the codon for the initial Met prior to the first codon for the Ala1 of protease. The reverse primer (ORF1-StSp; 5′-GTAGCATGCTTAGACGCCATCATCATTTACGAATTC-3′) was designed in order to introduce a stop codon (TAA) and a PaeI site after the last codon for the Val508 of RNA polymerase. After verification by DNA sequencing, the NdeI-PaeI fragment containing the entire 3CD region was cloned into the corresponding sites of pUC118Nd (40) to construct pUC3CD, in which the expression of the 3CD construct was controlled by the lac promoter (Fig. 1C). The following forward primers were used for the N-terminal truncation of the 3CD precursor: Pro-Nd5 (5′-CCACATATGTGGAGTCGAGTCGTGAGATTT-3′), which substituted Met for Leu5; Pro-Nd8 (5′-TGGCATATGGTCGTGAGATTTGGCTCTGGG-3′), which substituted Met for Arg8; and Pro-Nd11 (5′-GTCCATATGTTTGGCTCTGGGTGGGGCTTT-3′), which substituted Met for Arg11. The gene fragment encoding the mutant 3CD precursor was amplified in combination with the ORF1-StSp primer. Mutant plasmids were constructed in the same manner as pUC3CD and designated pUC3CD-ΔN5, pUC3CD-ΔN8, and pUC3CD-ΔN11, respectively (Fig. 1C).
Site-directed mutagenesis.
Site-directed mutagenesis was based on the method of Kunkel (27) obtained with the Mutan-K mutagenesis kit (Takara). Deoxyuracil-containing single-stranded DNA of pUCCV3BC3059 was prepared with the E. coli CJ236 strain and used as a template. Mutagenic primers are listed in Table 1. Usually, mutations were first detected by the appearance of a new restriction site or the disappearance of the original restriction site. These mutations did not affect the desired amino acid sequences, which were finally verified by DNA sequencing. The SacI-PaeI fragment from mutated pUCCV3BC3059 was cut out and ligated with the SacI-PaeI vector fragment from pUCHis3BC, resulting in mutant pUCHis3BC plasmids.
TABLE 1.
Mutagenic primers used for site-directed mutagenesis
| Namea | Nucleotide sequenceb | Codon change | Restriction site |
|---|---|---|---|
| R8A | 5′-CCCTCTGGTCTGCAGTCGTGAG-3′ | CGA→GCA | PstI |
| R8K | 5′-CACCAACCCTCTGGTCCAAGGTCGTGAGATTTGG-3′ | CGA→AAG | EcoT14I |
| R8Q | 5′-CACCAACCCTCTGGTCGCAGGTCGTGAGATTTGG-3′ | CGA→CAG | BspMIc |
| R8L | 5′-CACCAACCCTCTGGTCCTTGGTCGTGAGATTTGG-3′ | GCA→TTG | EcoT14I |
| R11A | 5′-GAGTCGTGGCATTCGGCTCTGG-3′ | AGA→GCA | Mva1269I |
| H30A | 5′-TCACCACCACTGCAGTTATACCAA-3′ | CAT→GCA | PstI |
| H30Y | 5′-TTTATCACCACTACGTATGTTATACCA-3′ | CAT→TAT | BsaAI |
| H30Q | 5′-TCACCACCACGCAGGTTATACCAA-3′ | CAT→CAG | BspMIc |
| H30N | 5′-TTTATCACCACGACCAATGTCATACCAACTGG-3′ | CAT→AAT | PshAIc |
| H30E | 5′-TTTATCACCACGACCGAAGTCATACCAACTGG-3′ | CAT→GAA | PshAIc |
| H30D | 5′-CACCACCACCGACGTCATACCAACTG-3′ | CAT→GAC | AatII |
| H30R | 5′-TCACCACCACGCGTGTTATACCA-3′ | CAT→CGT | MluI |
| R37A | 5′-CTGGGGTGGCTGAGTTCTTTGG-3′ | CGT→GCT | (EcoRI)d |
| E38A | 5′-GGGTGCGTGCATTCTTTGG-3′ | GAA→GCA | (EcoRI)d |
| E42A | 5′-TCTTTGGGGCGCCCATTGA-3′ | GAG→GCG | EheI |
| E45A | 5′-GGAGCCCATTGCTAGCATAGCAATCCA-3′ | GAA→GCT | NheI |
| H50A | 5′-TAGCAATCGCTCGAGCTGGTGA-3′ | CAT→GCT | XhoI |
| R51A | 5′-CAATCCATGCTGCAGGTGAATT-3′ | CGT→GCT | PstI |
| E54A | 5′-CGTGCTGGCGCCTTTACACA-3′ | GAA→GCC | EheI |
| R59A | 5′-CACAATTCGCATTCTCACGCAA-3′ | AGG→GCA | Mva1269I |
| R62A | 5′-GGTTTTCAGCCAAGGTCCGCCC-3′ | CGC→GCC | EcoT14I |
| K63A | 5′-AGGTTTTCCCGGGCAGTCCGCC-3′ | AAA→GCA | SmaI |
| R65A | 5′-ACGCAAAGTGGCGCCGGATCTG-3′ | CGC→GCG | EheI |
| D67A | 5′-AAAGTCCGGCCGGCTCTGACTG-3′ | GAT→GCT | NaeI |
| E74A | 5′-GAATGGTGCTAGCGGAAGGCT-3′ | GAG→GCG | NheI |
| E75A | 5′-TGTTGGAGGCCGGCTGTCCT-3′ | GAA→GCC | NaeI |
| E79A | 5′-GCTGTCCTGCAGGTGTCGTG-3′ | GAG→GCA | PstI |
| K88A | 5′-TTCTTATCGCTCGAGACTCTGG-3′ | AAG→GCT | XhoI |
| K88R | 5′-CTATTCTTATCCGTCGCGACTCTGGTGA-3′ | AAG→CGT | NruI |
| K88Q | 5′-TATTCTTATCCAGCGGGACTCTGGTG-3′ | AAG→CAG | MspA1I |
| K88L | 5′-CTATTCTTATCCTTCGCGACTCTGGTGA-3′ | AAG→CTT | NruI |
| R89A | 5′-TTATCAAGGCAGACTCTGGT-3′ | CGT→GCA | AlwNI |
| R89K | 5′-TTCTTATCAAGAAAGACTCGGGTGAGCTGCT-3′ | CGT→AAA | AvaI |
| R89Q | 5′-TCTTATCAAGCAGGACTCGGGTGAGCTGCT-3′ | CGT→CAG | AvaI |
| R89L | 5′-CTTATCAAGCTTGACTCTGGT-3′ | CGT→CTT | HindIII |
| D90A | 5′-TATCAAGCGTGCTAGCGGTGAGCTGCT-3′ | GAC→GCT | NheI |
| E93A | 5′-TGACTCTGGTGCACTGCTACCCCT-3′ | GAG→GCA | ApaLI |
| R100A | 5′-CCCTTGCTGTCGCGATGGGCGCTAT-3′ | CGA→GCG | NruI |
| K108A | 5′-GCGTCCATGGCGATCCAGGG-3′ | AAG→GCG | NcoI |
| R112A | 5′-GATCCAGGGCGCCCTAGTGCAT-3′ | AGG→GCC | EheI |
| H115A | 5′-GGCTAGTGGCCGGCCAATCTGG-3′ | CAT→GCC | NaeI |
| K128A | 5′-GCTAACGCTGCAGGGATGGAT-3′ | AAG→GCA | PstI |
| D131A | 5′-GGGGATGGCCTTGGGTACCCT-3′ | GAT→GCC | EcoT14I |
| D138A | 5′-CTACCAGGCGCCTGTGGTGC-3′ | GAT→GCC | EheI |
| D138E | 5′-TACCAGGTGAATGCGGTGCCCCTT-3′ | GAT→GAA | Mva1269I |
| D138N | 5′-CCCTACCAGGCAATTGTGGTGC-3′ | GAT→AAT | MunI |
| D138M | 5′-AGGTACCCTACCCGGGATGTGTGTGGTGCCCCT-3′ | GAT→ATG | SmaI |
| C139Ae | 5′-CTACCAGGTGATGCCGGCGCCCCTTATGTG-3′ | TGT→GCC | EheI |
| C139S | 5′-CCAGGTGATTCCGGAGCCCCTTATG-3′ | TGT→TCC | AccIII |
| C139T | 5′-TACCAGGTGATACCGGTGCCCCTTA-3′ | TGT→ACC | BshTI |
| C139Y | 5′-CCAGGTGATTATGGCGCCCCTTATG-3′ | TGT→TAT | EheI |
| C139M | 5′-TACCAGGTGATATGGGGCGCCCCTTATGT-3′ | TGT→ATG | EheI |
| K146A | 5′-ATGTGTACGCACGTAACAATGA-3′ | AAA→GCA | BsaAI |
| R147A | 5′-CCTTATGTATACAAAGCAAACAATG-3′ | AGA→GCA | AccI |
| D150A | 5′-GAAACAATGCATGGGTGGTT-3′ | GAC→GCA | EcoT22I |
| H157Af | 5′-TGTGGTGTAGCTGCAGCTGC-3′ | CAT→GCT | PstI |
| H157A-2f | 5′-TTGTGGTGTAGCCGCGGCTGCCACGA-3′ | CAT→GCC | SacII |
| H157Y | 5′-TTGTGGTGTATACGCAGCTGCCA-3′ | CAT→TAC | AccI |
| H157Q | 5′-GTGGTGTACAGGCAGCTGCCA-3′ | CAT→CAG | (NspI)d |
| H157R | 5′-TGTGGTGTACGTGCAGCTGCC-3′ | CAT→CGT | BsaAI |
| K162A | 5′-GCAGCTGCCACGGCTAGCGGTAACACTGTA-3′ | AAG→GCT | NheI |
| E175A | 5′-GTTCAGGCTGGCGCCGGTGAAACCAC-3′ | GAA→GCC | EheI |
| E177A | 5′-GGGGAAGGCGCCACCACCCT-3′ | GAA→GCC | EheI |
| E181A | 5′-GAAACCACGCTAGCGTAAGCAT-3′ | GAG→GCG | NheI |
The name of the primer represents the amino acid change. Amino acids are shown in the one-letter code. Letters before the number indicate the original amino acid residues, and letters after the number indicate the introduced amino acid residues.
Restriction sites used for detection of mutations are underlined.
Since the BspMI and PshAI sites were not cleaved, R8Q, H30Q, H30N, and H30E mutations were detected by DNA sequencing.
Restriction enzymes in parentheses indicate the enzymes that disappeared by mutagenesis.
C139A was described as C1235A in reference 40.
With the H157A primer, the Ser (CAT→TCT) and Pro (CAT→CCT) mutants were obtained instead of the Ala mutant. Therefore, the H157A-2 primer was used for the Ala mutation.
Construction of the Arg8 substitution mutant pUC3CD plasmids.
The XcmI-KpnI restriction fragment containing the Arg8 substitution from the respective mutant pUCHis3BC plasmids was ligated with the equivalent position of pUC3CD.
Expression of the 3BC and 3CD constructs.
E. coli BL21-CodonPlus-RIL was transformed with each of the wild-type pUCHis3BC plasmids, mutant pUCHis3BC plasmids, or pUC3CD derivatives. E. coli cells were grown at 37°C. The transformant was first grown in 3 ml of 2×YT broth (3ba). One hundred microliters of the culture was inoculated into 30 ml of fresh 2×YT broth, and the culture was further grown overnight (about 16 h.). Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM and incubated for 3 h. Cells were harvested, washed with 20 mM Tris-HCl (pH 7.4), and resuspended with 3 ml of the same buffer. After sonication, the cell lysate was centrifuged to remove unbroken cells, and the supernatant was used as a sample. The protein concentration was determined by the Bradford method (6), with bovine serum albumin as a standard.
Western blotting.
The supernatant containing 10 μg of proteins was used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by electroblotting onto a nitrocellulose membrane. The His-tagged proteins were detected by mouse anti-His monoclonal antibody (Sigma, St. Louis, Mo.) and alkaline phosphatase-linked goat anti-mouse immunoglobulin G (IgG) (Jackson ImmunoResearch Laboratories, West Grove, Pa.). The Chiba virus 3C-like protease was detected by rabbit antiserum raised against the purified His-tagged 3C-like protease (40) and alkaline phosphatase-linked goat anti-rabbit IgG (Vector Laboratories, Burlingame, Calif.). Rabbit antiserum raised against keyhole limpet hemocyanin conjugates of the synthetic peptide derived from RNA polymerase (40) was used for the detection of the Chiba virus RNA polymerase.
RESULTS AND DISCUSSION
Alanine-scanning mutagenesis -
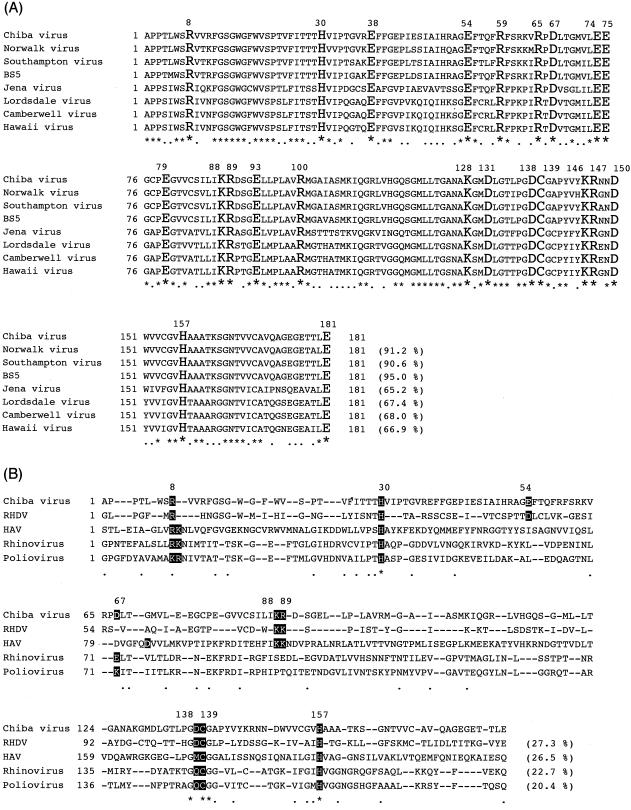
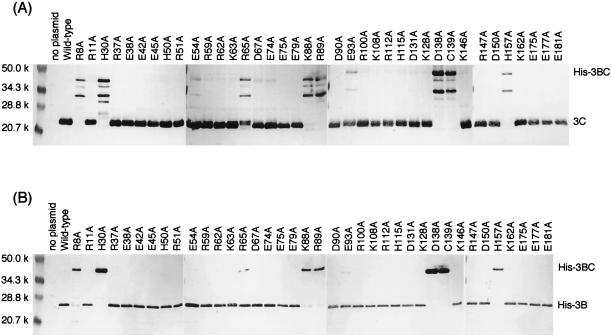
By analogy with chymotrypsin and the picornavirus 3C proteases, it was likely that some of charged amino acid residues played critical roles in the proteolysis in the NLV 3C-like proteases. In this study, we focused on 37 charged amino acid residues as well as putative nucleophile Cys139 of the Chiba virus 3C-like protease to identify active-site amino acid residues of the 3C-like proteases. As shown in Fig. 2A, charged residues are distributed throughout amino acid sequence of the 3C-like protease. Twenty-two charged residues are conserved in all of the known NLV 3C-like proteases, which are highly homologous to each other. These 38 residues were independently replaced with Ala. Both mutant His-tagged 3BC (His-3BC) constructs and wild-type His-3BC were expressed in E. coli cells and were investigated by Western blot analyses to determine whether or not the specific proteolytic cleavage between 3B (VPg) and 3C (protease) took place (Fig. 1B). If cleavage occurs, the 20-kDa 3C and the 23-kDa His-3B (the calculated mass of His-3B is 17.5 kDa [see reference 40]) can be detected by the antiprotease antiserum and anti-His monoclonal antibody, respectively. If not, the 45-kDa His-3BC precursor will be detected by both antibodies. Figure 3 shows the results of Western blotting analyses. The wild-type His-3BC was cleaved into His-3B and His-3C, as detected by the respective antibodies. We previously showed that the C139A mutation (described as C1235A in reference 40) rendered the enzyme inactive. Here, the C139A His-3BC was not cleaved at all, and the 45-kDa band was detected by both antibodies. This result reconfirmed that the Cys139 residue played a critical role in the proteolysis. Out of 37 charged amino acid residues found in the Chiba virus 3C-like protease, 6 residues (Arg8, His30, Lys88, Arg89, Asp138, and His157) seemed to be critical for the proteolysis, since their Ala mutants were not cleaved. The Ala mutation of Arg65 seemed to affect protease activity, since the uncleaved His-3BC precursor, as well as the cleaved His-3B and His-3C, was detected. Although the Arg65 residue might be important for efficiency of the proteolysis, further analysis was not done in this study. As shown in Fig. 3A, the 34-kDa band and some minor bands were detected by antiprotease antiserum. These bands might be nonspecific proteolytic products by E. coli proteases, but the details were not investigated in this study. Alanine-scanning mutagenesis revealed the six critical charged amino acid residues (Arg8, His30, Lys88, Arg89, Asp138, and His157) in addition to Cys139 (Fig. 2A). These seven residues were further mutated to investigate their role in the proteolysis.
FIG. 2.
Amino acid alignments of the Chiba virus 3C-like protease. (A) Alignment of the 3C-like proteases of NLVs for which whole genomic sequences had been determined. Chiba virus has 37 charged amino acid residues; 22 out of 37 residues indicated by large letters are conserved in all NLV 3C-like proteases. The putative nucleophile Cys139 is also indicated by a large letter. Asterisks indicate conserved residues. Dots indicate similar amino acid residues. The numbers in parentheses indicate the amino acid identities with the Chiba virus 3C-like proteases. Accession numbers: Chiba virus, AB042808; Norwalk virus, M87661; Southampton virus, L07418; BS5 strain, AF093797; Jena virus, AJ011099; Lordsdale virus, X86557; Camberwell virus, AF145896; Hawaii virus, U07611. (B) Alignment with the RHDV 3C-like protease and the picornaviral 3C proteases whose structures had been determined. The Chiba virus 3C-like protease was aligned with the RHDV (M67473), the HAV (P14553), rhinovirus (P03303), and poliovirus (P03300) 3C(-like) proteases. The roles of the residues whose background color is black or gray were discussed in the text.
FIG. 3.
Western blot analyses of alanine-scanning mutants of the His-3BC constructs. Ten micrograms of the proteins from E. coli BL21-CodonPlus-RIL cells harboring the wild-type or mutant pUCHis3BC plasmid were separated by SDS-PAGE, followed by electroblotting. Relevant proteins were detected by antiprotease antiserum (A), or anti-His monoclonal antibody (B). Mutations were represented by using one-letter codes of amino acids. Numbers indicate the positions of amino acid residues. Letters preceding the number and following the number indicate the wild-type amino acid and the introduced amino acid, respectively.
Cys139 as a nucleophile of the active site.
Cys139 of the Chiba virus 3C-like protease is thought to correspond to the active-site nucleophile Ser195 of chymotrypsin, because these amino acid residues are found within the similar amino acid sequence motifs GDCG and GDSG, respectively. Cys104 of the RHDV 3C-like protease, Cys146 of the poliovirus 3C protease, Cys147 of the rhinovirus 3C protease, Cys144 of the HAV 3C protease, and Cys163 of the foot-and-mouth disease virus 3C protease are also found in the GDCG, GDCG, GDCG, GMCG, and GYCG sequences, respectively (Fig. 2B). The Chiba virus 3C-like protease did not have overall sequence similarity (less than 30% amino acid identity) to the picornaviral 3C proteases, whereas similarity was relatively high around this sequence motif (Fig. 2B). These findings suggest that Cys139 is a nucleophile of the active site of the Chiba virus 3C-like protease. The X-ray structures of the chymotrypsin and picornavirus 3C proteases indicated that these Ser or Cys residues were nucleophiles of their active site (1, 3, 33, 34, 41).
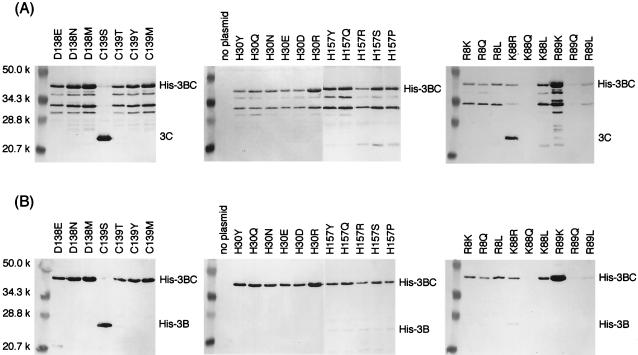
To investigate the role of Cys139, this residue was replaced with Ser, Thr, Tyr, and Met. As shown in Fig. 4, only the Ser mutant had protease activity comparable to that of the wild-type enzyme. Although Thr and Tyr, like Ser, have the OH group and Met has a lone pair of electrons on the sulfur atom, these mutants exhibited no activity. Since the Thr residue has an additional methyl group bound to the β carbon compared with the Ser residue, rotation of the side chain is assumed to be limited in the folded molecule. The side chain of the Tyr residue is much bulkier than those of the Cys and Ser residues. Therefore, the OH group of Thr or Tyr would not be able to work as a nucleophile. Even if a lone pair of electrons on the side chain of the Met residue might work as a nucleophile under certain conditions and a nucleophilic attack against a peptide carbonyl happened, a methyl cation, compared with a proton, would not be stabilized by its environment and would not be suitable as a leaving group. Thus, the presence of the flexible SH or OH group is required for the side chain of the amino acid residue at position 139, supporting the idea that Cys139 is a nucleophile of the active site.
FIG. 4.
Western blot analyses of the Arg8, His30, Lys88, Arg89, Asp138, Cys139, and His157 mutants of the His-3BC constructs. Experiments were performed as described in the legend to Fig. 3.
His30 as the general base of the active site.
Alanine-scanning mutagenesis revealed that two histidine residues, His30 and His157, were critical for the proteolysis. According to amino acid sequence alignment (Fig. 2B), it is likely that His30 of the Chiba virus 3C-like protease corresponds to His57 of the chymotrypsin, His27 of the RHDV 3C-like protease, His40 of the poliovirus and rhinovirus 3C proteases, and His44 of the HAV 3C protease. These His residues are close to a nucleophile Ser or Cys residue in the respective enzymes (1, 3, 33, 34, 41) and are likely to function as the general base of the active site. On the other hand, His157 of the Chiba virus 3C-like protease corresponds to His117 of the RHDV 3C-like protease, His161 of the poliovirus 3C protease, His160 of the rhinovirus 3C protease, and His191 of the HAV 3C protease (Fig. 2B), and seems to be one of the residues forming the S1 specificity pocket (4, 7, 17, 19, 29).
His30 was replaced with Tyr, Gln, Asn, Glu, Asp, and Arg. All of the mutants exhibited no protease activity (Fig. 4). The presence of the imidazole ring was essential for the proteolysis, since any chemical nature (aromaticity, hydrophilicity, the ability to accept a proton, or having a positive charge) of the side chain could not be substituted for a tautomeric property of imidazole. It was thought that both the basic nature of an imidazole ring contributed to accepting a proton released from the nucleophile after an attack against the peptide carbonyl, and the aromaticity of a protonated imidazole ring contributed to delocalization of a positive charge. These results were consistent with the idea that His30 was the general base of the active site.
His157 in the S1 specificity pocket.
As described above, His157 was thought to be one of the residues forming the S1 specificity pocket. The corresponding histidine residues in poliovirus and rhinovirus 3C proteases were shown to be involved in recognition of the side chain of the P1-position Gln residue at the cleavage sites (4, 7, 17, 19, 29).
The H157A mutation (CAT→GCT), performed with the H157A primer (Table 1), was not successful. Instead, the Ser and Pro mutants of His157, which had undesired nucleotide changes (CAT→TCT and CAT→CCT, respectively), were spontaneously obtained and characterized here. Furthermore, His157 was replaced with Tyr, Gln, and Arg. All of the His157 mutants, as well as H157A, produced a very slight amount of His-3B (Fig. 4B), although the cleaved protease fragment was not clearly detected by the antiprotease antiserum (Fig. 4A). These indicated that His157 was not essential for the protease activity. Severely reduced protease activity might be due to impaired substrate recognition. The relationship between the substrate specificity of His157 and the amino acid sequence at the cleavage site will be the subject of a future study.
Asp138.
Asp138 was the only negatively charged residue found to be essential for the proteolysis by alanine-scanning mutagenesis. When Asp138 was replaced with Asn, Glu, and Met, all mutants had no protease activity (Fig. 4), indicating that the Asp residue is essential at this position in the Chiba virus 3C-like protease. Asp138 is the amino acid residue next to a nucleophile, Cys139. In the X-ray structures of chymotrypsin and picornavirus 3C proteases, the side chain of the residue preceding a nucleophile was oriented away from the catalytic residues in the active site (1, 3, 33, 34, 41) and therefore may not be able to participate in the catalysis. This observation is consistent with the fact that the Asp residue was not conserved at the corresponding position. Asp appears at this position in the chymotrypsin, RHDV 3C-like protease, and poliovirus and rhinovirus 3C proteases, while Met occupies the HAV 3C protease and Tyr occupies the FMDV 3C protease (Fig. 2B). Asp138 may therefore be involved in stabilizing the protein structure through the ionic interaction or hydrogen bonding, although the details of this will not be clear until information on the high-order structure is obtained.
Site-directed mutagenesis performed here did not find the third member carboxylate of the active site corresponding to Asp102 in chymotrypsin. Amino acid alignment shown in Fig. 2B indicated that either of the following two residues of the Chiba virus 3C-like protease might work as the third member carboxylate: Glu54, which corresponded to the third member Asp44 of the RHDV 3C-like protease; or Asp67, which corresponded to Glu71 of poliovirus and rhinovirus 3C proteases. As shown by the crystal structure of the HAV 3C protease (1, 3), its side chain might point away from the active site and might not interact with the general base histidine (His30) in the Chiba virus 3C-like protease. This feature of these two proteases is in marked contrast with those of the chymotrypsin, poliovirus, and rhinovirus 3C proteases. However, in the HAV 3C protease, since Tyr143 is almost perpendicular to the plane of the imidazole of His44 and seems to be deprotonated and negatively charged, it might contribute to the electrostatic stabilization and the delocalization of a positive charge on the imidazole ring (3). Therefore, we cannot rule out the possibility that an anion, as served by the side chain of a tyrosine residue, may stabilize a positive charge on the imidazole ring of His30 or withdraw a proton from the imidazole ring as the third member of the active site in the Chiba virus 3C-like protease.
Lys88 and Arg89.
Lys88 was replaced with Arg, Gln, and Leu, and Arg89 was replaced with Lys, Gln, and Leu. As shown in Fig. 4, both the Gln88 and Gln89 mutants were not expressed substantially in E. coli. The Arg88 mutant had significant protease activity, although uncleaved His-3BC remained. In contrast, the Leu88 mutant had no activity. Therefore, it was likely that a positively charged residue at position 88 was important for the protease activity. The Arg residue was likely to be essential at position 89, since the Lys and Leu mutants of Arg89 did not show any protease activity.
When amino acid sequences of the Chiba virus 3C-like protease are aligned with that of the HAV 3C protease, Lys88 and Arg89 of the Chiba virus are likely to correspond to Lys69 and Lys70 of RHDV and Lys105 and Lys106 of HAV (Fig. 2B), which were located on the surface of the enzyme distant from the active site according to the crystal structure of the HAV 3C protease and were implicated in RNA recognition and/or the maintenance of the integrity of the RNA recognition site (3). Although these two Lys residues are not found in the poliovirus and rhinovirus 3C proteases, this may be explained by the difference in local structures (α-helix in HAV [3] versus nonstructured in poliovirus [34] and rhinovirus [33]). In any case, Lys88 and Arg89 of the Chiba virus 3C-like protease are likely to be far from the active site and are not involved in the catalysis.
Effects of Arg8 mutations on 3B/3C and 3C/3D cleavage.
Arg8 was replaced with Lys, Gln, and Leu. None of the mutants showed protease activity (Fig. 4). Therefore, Arg was essential for the proteolysis. According to the amino acid sequence alignment with the picornavirus 3C proteases, Arg8 is likely to correspond to the first or second positively charged residue in the highly conserved sequence motif K(/R)-R(/K)-N-I(/L) (Lys12 or Arg13 in poliovirus 3C, Arg12 or Lys13 in rhinovirus 3C, and Arg10 or Lys11 in HAV 3C), although the motif was not conserved in NLVs and RHDV (Fig. 2B). The crystal structures revealed that this sequence motif formed the last turn of the N-terminal α-helix. Khan et al. (26) previously suggested the role of the motif and proposed the intriguing model of autocatalytic (intramolecular) cleavage at the 3B/3C junction by using the structure of the poliovirus 3C protease. In this model, before the intramolecular or cis cleavage at the 3B/3C site occurs, the region including the C terminus of the 3B VPg and the N terminus of the 3C protease is in an extended conformation, so as to reach into the active site of the protease. After the cleavage, the N terminus dissociates out of the active site and then forms a final conformation identical to the crystal structure. The idea was derived from the crystal structure of the precursor of the α-lytic protease, a serine protease, which underwent autocatalytic cleavage during maturation (37). This model implies that the Arg8 residue is important for the 3B/3C cleavage, but is not necessary for cleavage at the other junctions.
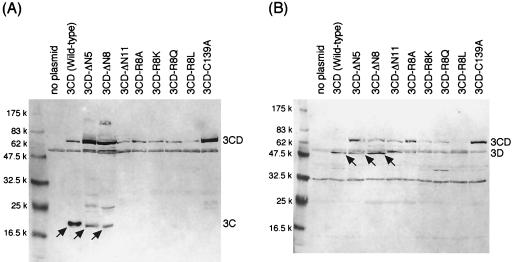
To examine whether or not Arg8 is important for the 3C/3D cleavage, first, the R8A, R8K, R8Q, and R8L mutants were constructed on the basis of the 3CD precursor (Fig. 1C). E. coli cells expressing the wild-type 3CD precursor generated the 20-kDa 3C and the 56-kDa 3D fragments, as detected by the respective antisera (Fig. 5), indicating that the 3C-like protease moiety was enzymatically active. The 60-kDa band in Fig. 5A and B and the 33-kDa band in Fig. 5B were nonspecific ones because they also appeared in the lysate of plasmidless host E. coli cells. As for four Arg8 substitution mutant 3CD precursors, not any 3C-like protease-specific proteolytic events were observed, since only the 75-kDa bands were detected by both antiprotease and antipolymerase antisera (Fig. 5). This result seemed to suggest that the Arg8 residue was also involved in the 3C/3D cleavage.
FIG. 5.
Western blot analyses of the 3CD precursor and its derivatives. Ten micrograms of the proteins from E. coli BL21-CodonPlus-RIL cells harboring the wild-type or mutant pUC3CD plasmid were separated by SDS-PAGE, except that 50 μg of proteins was used for the 3CD-ΔN8 and 3CD-ΔN11 mutants, followed by electroblotting. Relevant proteins were detected by antiprotease antiserum (A) or antipolymerase antiserum (B). Arrows in panel A indicate the 3C, 3C-ΔN5, or 3C-ΔN8 fragments, and those in panel B indicate the 3D RNA polymerase fragments.
On the contrary, the effect of the N-terminal truncation of the 3C-like protease was quite different. Next, the truncation mutants of the 3C-like protease, which lacked the N-terminal 5 (3CD-ΔN5), 8 (3CD-ΔN8), or 11 (3CD-ΔN11) amino acids, were constructed and examined (Fig. 1C). As shown in Fig. 5, the 3CD-ΔN5 truncation mutant and even the 3CD-ΔN8 mutant lacking Arg8 could significantly generate about 18.5-kDa N-terminal-truncated 3C and the 56-kDa 3D peptides, although both mutants had less proteolytic activity than the wild-type 3CD, because a portion of the mutant 3CD precursors remained uncut (Fig. 5). In the 3CD-ΔN8 mutant, the amount of the truncated 3C fragment was quite low, consistent with the low level of expression of the 3CD-ΔN8 precursor in E. coli cells. It was clear that the cleavage of the 3CD-ΔN5 and 3CD-ΔN8 mutant proteins was exerted by the proteolytic activity of the respective truncated 3C moiety, since the truncated mutants carrying an active-site mutation, C139A, produced only the 75-kDa species, exhibiting no proteolytic activity (data not shown). The 3CD-ΔN11 truncation mutant was inactive as to the protease activity, since only the 75-kDa precursor band was observed (Fig. 5). These results indicated that the N-terminal region containing 8 amino acids rather than the Arg8 residue itself was not necessary for the 3C/3D cleavage and that the N-terminal portion itself of the 3C-like protease was not involved in the proteolysis. The effect of the Arg8 substitution on the protein structure might be more detrimental than that of the N-terminal truncation, and/or the Arg8 mutation might alter the conformation of the N terminus, resulting in the fact that the N-terminal region containing the Arg8 mutation hindered access of the cleavage site to the active center of the protease. However, whether the model proposed by Khan et al. (26) is valid and is the explanation for the NLV 3C-like proteases and picornavirus 3C proteases cannot be judged from these results. To judge if this is true or not, we will have to await information on the highly ordered structure of the protease.
Conclusion.
First, in analogy to chymotrypsin and picornavirus 3C proteases, the catalytic dyad composed of His30 and Cys139 functions as active-site residues in the Chiba virus 3C-like protease. While the catalytic triad works in the proteolysis in chymotrypsin (12) and 3C proteases from poliovirus (34) and rhinovirus (33), it was found that the HAV 3C protease was the first chymotrypsin-like protease in which the catalytic dyad was likely to work (1, 3). Therefore, it is very likely that the dyad composed of a nucleophile (Ser or Cys) and a general base (His) is mandatory for the protease activity. In the HAV 3C protease and the Chiba virus 3C-like protease, the protonated imidazole ring carrying a positive charge might have been strongly stabilized by its environment through an effect such as a stack of aromatic rings. However, we cannot rule out the possibility that an anion such as a tyrosine phenolate rather than a carboxylate might participate in the proteolysis as the third member of the active site.
Second, the substitution of Arg8, which seemed to be located on the N-terminal α-helix, affected both the cleavage at the 3B/3C junction and the cleavage at the 3C/3D junction, whereas the deletion of at least eight residues of the N terminus, including Arg8, did not affect the cleavage at the 3C/3D junction. These results indicated that the Arg8 residue itself was not directly involved in the proteolysis. It was also suggested that the Arg8 substitution, but not the deletion of the N-terminus region containing Arg8, altered a conformation of the protease, leading to inactivation of the protease as to both the cleavage at the 3B/3C junction and the cleavage at the 3C/3D junction.
This study was the first evaluation of the active site of the NLVs at the molecular level. The information obtained in this study is expected to provide the basis for drug design in diarrheal therapy for humans. Determination of the high-order structures of the protease and its related precursors will not only elucidate the accuracy of results described here, but also greatly contribute to and accelerate computer-based drug design.
Acknowledgments
We thank T. Suzuki for helpful discussions and suggestions. We also thank T. Mizoguchi for secretarial work.
This work was supported by a grant for Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Labour and Welfare and a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
REFERENCES
- 1.Allaire, M., M. M. Chernaia, B. A. Malcom, and M. N. G. James. 1994. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature (London) 369:72-76. [DOI] [PubMed] [Google Scholar]
- 2.Bazan, J. F., and R. J. Fletterick. 1988. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc. Natl. Acad. Sci. USA 85:7872-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann, E. M., S. C. Mosimann, M. M. Chernaia, B. A. Malcom, and M. N. G. James. 1997. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J. Virol. 71:2436-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blair, W. S., J. H. C. Nguyen, T. B. Parsley, and B. L. Semler. 1996. Mutations in the poliovirus 3CD proteinase S1-specificity pocket affect substrate recognition and RNA binding. Virology 218:1-13. [DOI] [PubMed] [Google Scholar]
- 5.Boniotti, B., C. Wirblich, M. Sibilia, G. Meyers, H.-J. Thiel, and C. Rossi. 1994. Identification and characterization of a 3C-like protease from rabbit hemorrhagic disease virus, a calicivirus. J. Virol. 68:6487-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Cheah, K.-C., L. E.-C. Leong, and A. G. Porter. 1990. Site-directed mutagenesis suggests close functional relationship between a human rhinovirus 3C cysteine protease and cellular trypsin-like serine protease. J. Biol. Chem. 265:7180-7187. [PubMed] [Google Scholar]
- 8.Clarke, I. N., P. R. Lambden, and E. O. Caul. 1998. Human enteric RNA viruses: caliciviruses and astroviruses, p. 511-535. In B. W. Mahy and L. Collier (ed.), Topley & Wilson's microbiology and microbial infections, vol. 1. Arnold, London, United Kingdom. [Google Scholar]
- 9.Dingle, K. E., P. R. Lambden, E. O. Caul, and I. N. Clarke. 1995. Human enteric Caliciviridae: the complete genome sequence and expression of virus-like particles from a genetic group II small round structured virus. J. Gen. Virol. 76:2349-2355. [DOI] [PubMed] [Google Scholar]
- 10.Dougherty, W. G., and B. L. Semler. 1993. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 57:781-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estes, M. K., R. L. Atmar, and M. E. Hardy. 1997. Norwalk and related diarrhea viruses, p. 1073-1095. In D. D. Richmann, R. J. Whitley, and F. G. Hayden (ed.), Clinical virology. Churchill Livingstone, Inc., New York, N.Y.
- 12.Fersht, A. R., and J. Sperling. 1973. The charge relay system in chymotrypsin and chymotrypsinogen. J. Mol. Biol. 74:137-149. [DOI] [PubMed] [Google Scholar]
- 13.Glass, P. J., L. J. White, J. M. Ball, I. Leparc-Goffart, M. E. Hardy, and M. K. Estes. 2000. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 74:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorbalenya, A. E., A. P. Donchenko, V. M. Blinov, and E. V. Koonin. 1989. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. FEBS Lett. 243:103-114. [DOI] [PubMed] [Google Scholar]
- 15.Gosert, R., G. Dollenmaier, and M. Weitz. 1997. Identification of active-site residues in protease 3C of hepatitis A virus by site-directed mutagenesis. J. Virol. 71:3062-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H.-J. Thiel. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181(Suppl. 2):S322-S330. [DOI] [PubMed] [Google Scholar]
- 17.Grubman, M. J., M. Zellner, G. Bablanian, P. W. Mason, and M. E. Piccone. 1995. Identification of the active-site residues of the 3C proteinase of foot-and-mouth disease virus. Virology 213:581-589. [DOI] [PubMed] [Google Scholar]
- 18.Hämmerle, T., C. U. T. Hellen, and E. Wimmer. 1991. Site-directed mutagenesis of the putative catalytic triad of poliovirus 3C proteinase. J. Biol. Chem. 266:5412-5416. [PubMed] [Google Scholar]
- 19.Ivanoff, L. A., T. Towatari, J. Ray, B. D. Korant, and S. R. Petteway, Jr. 1986. Expression and site-specific mutagenesis of the poliovirus 3C protease in Escherichia coli. Proc. Natl. Acad. Sci. USA 83:5392-5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, X., D. Y. Graham, M. Wang, and M. K. Estes. 1990. Norwalk virus genome cloning and characterization. Science 250:1580-1583. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, X., M. Wang, D. Y. Graham, and M. K. Estes. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 23.Kapikian, A. Z. 1994. Norwalk and Norwalk-like viruses, p. 471-518. In A. Z. Kapikian (ed.), Viral infections of the gastrointestinal tract. Marcel Dekker, Inc., New York, N.Y.
- 24.Kapikian, A. Z., M. K. Estes, and R. M. Chanock. 1996. Norwalk group of viruses, p. 783-810. In B. N. Fields, D. M. Knipe, P. M. Howly, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Strauss (ed.), Fields virology, 3rd ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 25.Kean, K. M., N. L. Teterina, D. Marc, and M. Girard. 1991. Analysis of putative active site residues of the poliovirus 3C protease. Virology 181:609-619. [DOI] [PubMed] [Google Scholar]
- 26.Khan, A. R., N. Khazanovich-Bernstein, E. M. Bergmann, and M. N. G. James. 1999. Structural aspects of activation pathways of aspartic protease zymogens and viral 3C protease precursor. Proc. Natl. Acad. Sci. USA 96:10968-10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunkel, T. A., J. D. Roberts, and A. Z. Richard. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 28.Lambden, P. R., E. O. Caul, C. R. Ashley, and I. N. Clarke. 1993. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science 259:516-519. [DOI] [PubMed] [Google Scholar]
- 29.Lawson, M. A., and B. L. Semler. 1991. Poliovirus thiol proteinase 3C can utilize a serine nucleophile within the putative catalytic triad. Proc. Natl. Acad. Sci. USA 88:9919-9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leite, J. P. G., T. Ando, J. S. Noel, B. Jiang, C. D. Humphrey, J. F. Lew, K. Y. Green, R. I. Glass, and S. S. Monroe. 1996. Characterization of Toronto virus capsid protein expressed in baculovirus. Arch. Virol. 141:865-875. [DOI] [PubMed] [Google Scholar]
- 31.Liu, B. L., P. R. Lambden, H. Gunther, P. Otto, M. Elschner, and I. N. Clarke. 1999. Molecular characterization of a bovine enteric calicivirus: relationship to the Norwalk-like viruses. J. Virol. 73:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malcom, B. A. 1995. The picornaviral 3C proteinases: cysteine nucleophiles in serine proteinase fold. Protein Sci. 4:1439-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matthews, D. A., W. W. Smith, R. A. Ferre, B. Condon, G. Budahazi, W. Sisson, J. E. Villafranca, C. A. Janson, H. E. McElroy, C. L. Gribskov, and S. Worland. 1994. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell 77:761-771. [DOI] [PubMed] [Google Scholar]
- 34.Mosimann, S. C., M. M. Cherney, S. Sia, S. Plotch, and M. N. G. James. 1997. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 273:1032-1047. [DOI] [PubMed] [Google Scholar]
- 35.Pfister, T., and E. Wimmer. 2001. Polypeptide p41 of a Norwalk-like virus is a nucleic acid-independent nucleoside triphosphatase. J. Virol. 75:1611-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prasad, B. V. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus particle. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 36a.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Sauter, N. K., T. Mau, S. D. Rader, and D. A. Agard. 1998. Structure of α-lytic protease complexed with its pro region. Nat. Struct. Biol. 5:945-950. [DOI] [PubMed] [Google Scholar]
- 38.Seah, E. L., J. A. Marshall, and P. J. Wright. 1999. Open reading frame 1 of the Norwalk-like virus Camberwell: completion of sequence and expression in mammalian cells. J. Virol. 73:10531-10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seipelt, J., A. Guarné, E. Bergmann, M. James, W. Sommergruber, I. Fita, and T. Skern. 1999. The structures of picornaviral proteinases. Virus Res. 62:159-168. [DOI] [PubMed] [Google Scholar]
- 40.Someya, Y., N. Takeda, and T. Miyamura. 2000. Complete nucleotide sequence of the Chiba virus genome and functional expression of the 3C-like protease in Escherichia coli. Virology 278:490-500. [DOI] [PubMed] [Google Scholar]
- 41.Sprang, S., T. Standing, R. J. Fletterick, R. M. Stroud, J. Finer-Moore, N.-H. Xuong, R. Hamlin, W. J. Rutter, and C. S. Craik. 1987. The three-dimensional structure of Asn102 mutant of trypsin: role of Asp102 in serine protease catalysis. Science 237:905-909. [DOI] [PubMed] [Google Scholar]
- 42.Taylor, J. W., J. Ott, and F. Eckstein. 1985. The rapid generation of oligonucleotide-directed mutagenesis at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 13:8764-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Utagawa, E. T., N. Takeda, S. Inouye, K. Kasuga, and S. Yamazaki. 1994. 3′-Terminal sequence of a small round structured virus (SRSV) in Japan. Arch. Virol. 135:185-192. [DOI] [PubMed] [Google Scholar]
- 44.Vinje, J., J. Green, D. C. Lewis, C. I. Gallimore, D. W. G. Brown, and M. P. G. Koopmans. 2000. Genetic polymorphism across regions of the three open reading frames of “Norwalk-like viruses.” Arch. Virol. 145:223-241. [DOI] [PubMed] [Google Scholar]
- 45.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vector and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]