Abstract
The papillomavirus life cycle is tightly linked to epithelial cell differentiation. Production of virus capsid proteins is restricted to the most terminally differentiated keratinocytes in the upper layers of the epithelium. However, mRNAs encoding the capsid proteins can be detected in less-differentiated cells, suggesting that late gene expression is controlled posttranscriptionally. Short sequence elements (less than 80 nucleotides in length) that inhibit gene expression in undifferentiated epithelial cells have been identified in the late 3′ untranslated regions (UTRs) of several papillomaviruses, including the high-risk mucosal type human papillomavirus type 16 (HPV-16). Here we show that closely related high-risk mucosal type HPV-31 also contains elements that can act to repress gene expression in undifferentiated epithelial cells. However, the HPV-31 negative regulatory element is surprisingly complex, comprising a major inhibitory element of approximately 130 nucleotides upstream of the late polyadenylation site and a minor element of approximately 110 nucleotides mapping downstream. The first 60 nucleotides of the major element have 68% identity to the negative regulatory element of HPV-16, and these elements bind the same cellular proteins, CstF-64, U2AF65, and HuR. The minor inhibitory element binds some cellular proteins in common with the major inhibitory element, though it also binds certain proteins that do not bind the upstream element.
Human papillomaviruses (HPV) are small double-stranded DNA viruses that infect squamous epithelia giving rise to papillomas or warts (16). Papillomaviruses may be divided into types that infect cutaneous or mucosal epithelia and high- and low-risk types depending on whether infection can lead to benign or malignant lesions. HPV type 16 (HPV-16) is the most clinically significant of the high-risk mucosal types. It has been implicated in the development of cervical intraepithelial neoplasias and cervical carcinomas and has been detected in more than 50% of such lesions (52). HPV-31 is another high-risk mucosal type and is very closely related to HPV-16, with 70% sequence identity (11).
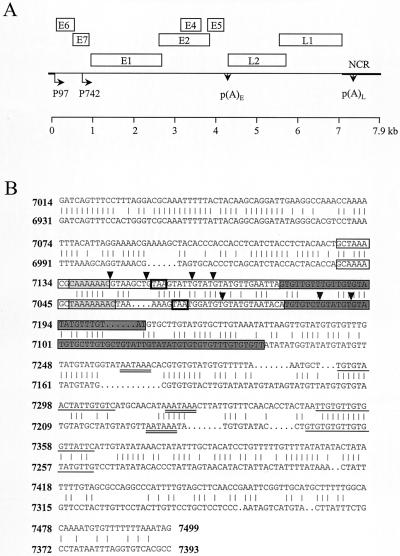
The 8-kb HPV-31 genome, in common with all papillomavirus genomes, comprises an early and a late coding region. The early gene products control viral DNA replication, episomal maintenance of the genome, transcription, and host cell division. The late genes, L1 and L2, encode the major and minor capsid proteins, respectively. A noncoding region lies downstream of the late region and upstream of the early region. This contains sequences controlling the activity of the viral promoter P97 and replication of the genome. At its 5′ end is the late gene 3′ untranslated region (UTR), which contains a single polyadenylation site (16). The P97 promoter is most likely active throughout the viral life cycle (17, 32, 39). The genome comprises a single transcription unit, and individual mRNAs are produced by extensive alternative splicing events and by choice of an early or late polyadenylation site (17, 31). Late gene expression is controlled partly by activation of a cryptic late promoter P742 in the E7 open reading frame (17) and partly by posttranscriptional mechanisms, due to read-through of the early polyadenylation site at early times of infection (47, 48).
Inhibitory elements controlling production of late RNAs have been identified in several papillomaviruses. They include elements in the HPV-16 late gene coding regions (4, 40, 45) and in the bovine papillomavirus type 1 (BPV-1) (8), HPV-1 (46), and HPV-16 (20) late gene 3′ UTRs. These appear to act via diverse posttranscriptional mechanisms. The elements within the L1 and L2 coding regions regulate mRNA stability and translation, the latter through interaction with hnRNPK and poly(rC) binding proteins 1 and 2 (4, 40). The AU-rich 57-nucleotide (nt) HPV-1 3′ UTR inhibitory element binds the elav-like regulator of mRNA stability, HuR (23), suggesting that this element controls decay of late transcripts (41). The 53 nt BPV-1 3′ UTR element acts to inhibit pre-mRNA polyadenylation by binding U1 snRNP (9, 14). The HPV-16 negative regulatory element (NRE) has been mapped to a 79-nt region spanning the 3′ end of the L1 open reading frame and extends into the late gene 3′ UTR (5). The element includes in its 5′ portion four weak 5′ splice sites and a putative stem-loop structure. The 3′ portion comprises a GU-rich region. In vitro UV cross-linking experiments demonstrated the binding of the NRE RNA to several cellular proteins (5, 22). These include auxiliary splicing factor U2AF65 (35), polyadenylation factor CstF-64 (42, 43), and elav-like protein HuR (23), which has roles in mRNA stability (7, 30, 33) and possibly in nucleus-to-cytoplasm transport of RNA (7). The HPV-16 NRE appears to regulate polyadenylation (K. McGuire and S. V. Graham, unpublished results), nuclear export (22), and mRNA stability (21).
In this study we set out to determine whether HPV-31, being closely related to HPV-16, contains a similar NRE. We show that the HPV-31 late gene 3′ UTR indeed contains sequences that inhibit gene expression in undifferentiated epithelial cells. We have identified a region of the HPV-31 late 3′ UTR termed the NRE-like element (NLE) that structurally resembles the HPV-16 NRE. The HPV-31 NLE and the HPV-16 NRE bind many of the same proteins, including RNA-processing factors U2AF65, CstF-64, and HuR. However, precise deletion of the NLE only partially alleviates inhibition of gene expression in undifferentiated epithelial cells. Instead, we have identified a bipartite inhibitory element flanking the late polyadenylation site. The upstream element binds the same proteins as the HPV-16 NRE, and its 5′ portion has high sequence homology to the NRE. The downstream element is quite distinct and binds some different nuclear proteins, suggesting that it may act by a separate mechanism.
MATERIALS AND METHODS
Plasmids.
Plasmid pBSHPV-31 contains the HPV-31 genome (1). Plasmid pLW1 is a chloramphenicol acetyltransferase (CAT) expression vector (10), which lacks a polyadenylation signal. A 445-nt PstI/EcoRI fragment containing the 3′ end of the HPV-16 L1 coding region and late gene 3′ UTR (nt 7008 to 7453) was cloned downstream of the CAT reporter gene to make plasmid pCATPE445. A 227-nt SspI/EcoRI fragment of the HPV-16 late gene 3′ UTR (nt 7226 to 7453) was cloned downstream of the CAT reporter in pLW1 to make plasmid pCATSE227 (20). Plasmid pTer5 is derived from pLW1 and contains the herpes simplex virus type 2 (HSV-2) immediate-early (IE) gene 5 polyadenylation signal downstream of the CAT reporter gene (10). The sequences of all primers used to generate wild-type and mutant HPV-31 late gene 3′ UTR constructs are shown in Tables 1 and 2. A portion of the HPV-31 sequence equivalent to the PstI/EcoRI fragment of HPV-16 was amplified by PCR using primers 1 and 2 and pBSHPV-31 as a template. This fragment was cloned downstream of the CAT reporter gene in pLW1 to generate plasmid pCATPH1 (see Fig. 2A). The deletion constructs used to map inhibitory activity in the HPV-31 late gene 3′ UTR were generated by PCR as follows by using the HPV-31 late gene 3′ UTR cloned into pGEM-T Easy (Promega) as a template. The 101-nt HPV-31 NLE was deleted by using primer 14 with primer 1 and primer 15 with primer 2. The resulting PCR products were ligated together and then cloned downstream of the CAT reporter in pLW1 to make plasmid pCATPH13 (see Fig. 3A). The 5′ end deletion mutants were amplified with reverse primer 2 and forward primers 3 to 10 as required to make plasmids pCATPH2 to pCATPH9 (see Fig. 2A). The sequences with 3′ end deletions were amplified with forward primer 1 and reverse primers 11 to 13 to make plasmids pCATPH10 to pCATPH12 (see Fig. 2A). Internal deletions of the putative major inhibitory element (MIE) and secondary inhibitory element (SIE) were generated by PCR as described previously (27) using primers P6 and T7 with primers 16 and 17 to make plasmid pCATPH14, with primers 18 and 19 to make pCATPH15, with primers 20 and 21 to make pCATPH16, and with primers 22 and 23 to make plasmid pCATPH17 (see Fig. 3A). Primers SP6, T7, 22, and 23 were used to delete the MIE from pCATPH11, which lacks the putative SIE, to make plasmid pCATPH18 (see Fig. 3A). The MIE was amplified by PCR using primers 5 and 24 and then cloned upstream of the polyadenylation signal of pTer5 to make plasmid pTer5+MIE (see Fig. 3A). All clones were checked by sequence analysis.
TABLE 1.
Sequences and annealing positions within the HPV-31 genome of PCR primers used to make plasmids with 5′ and 3′ deletions of the 3′ UTR
| Primer no. | Primer name | Restriction site | Genomic location | Sequence (5′-3′)a |
|---|---|---|---|---|
| 1 | pCATPH1 (forward) | PstI | 6931-6951 | GTCACTGCAGGATCAGTTTCCACTGGGTCGC |
| 2 | pCATPH1 (reverse) | HindIII | 7369-7393 | CCAGAAGCTTGGCGTGACACCTAAATTATAGGCAG |
| 3 | pCATPH2 (forward) | PstI | 6981-6995 | GTCACTGCAGACGTCCTAAATTTAA |
| 4 | pCATPH3 (forward) | PstI | 7031-7046 | GTCACTGCAGCTACACCAGCAAAACGT |
| 5 | pCATPH4 (forward) | PstI | 7081-7094 | GTCACTGCAGTACATGTGTCTGTAT |
| 6 | pCATPH5 (forward) | PstI | 7131-7146 | GTCACTGCAGTTTGTGTGTTATATATG |
| 7 | pCATPH6 (forward) | PstI | 7161-7179 | GTCACTGCAGTATGTATGCGTGTGTACTTG |
| 8 | pCATPH7 (forward) | PstI | 7171-7184 | GTCACTGCAGTGTGTACTTGTATAT |
| 9 | pCATPH8 (forward) | PstI | 7211-7224 | GTCACTGCAGTATGCTATGTATGTT |
| 10 | pCATPH9 (forward) | PstI | 7225-7242 | GTCACTGCAGAATAAATATGTGTATACC |
| 11 | pCATPH10 (reverse) | HindIII | 7248-7263 | CCAGAAGCTTCAACATACACAACAC |
| 12 | PCATPH11 (reverse) | HindIII | 7266-7283 | CCAGAAGCTTTAATAGGGTGTATATAAG |
| 13 | pCATPH12 (reverse) | HindIII | 7329-7343 | CCAGAAGCTTAGCAGGAACAAGTAG |
Restriction enzyme sites are underlined.
TABLE 2.
Sequences and annealing positions within the HPV-31 genome of PCR primers used to make internal deletions of plasmids
| Primer no. | Primer name | Restriction site(s) | Genomic location(s) | Sequence (5′-3′)a |
|---|---|---|---|---|
| 14 | pCATPH13 (first half reverse) | EcoRV | 7020-7038 | AGAGATATCTGGTGTAGTGGTAGATGCT |
| 15 | pCATPH13 (second half forward) | EcoRV | 7142-7159 | TGTGATATCTGGTATATGTATGT |
| 16 | PCATPH14 (forward) | None | 7173-7186, 7211-7226 | GTGTACTTGTATAT/TATGCTATGTATGTTA |
| 17 | PCATPH14 (reverse) | None | 7173-7186, 7211-7226 | TAACATACATAGCATA/ATATACAAGTACAC |
| 18 | PCATPH15 (forward) | None | 7142-7161, 7211-7226 | ATATGGTATATGTATGTT/TATGCTATGTATGTTA |
| 19 | PCATPH15 (reverse) | None | 7142-7161, 7211-7226 | TAACATACATAGCATA/AACATACATATACCATAT |
| 20 | PCATPH16 (forward) | None | 7119-7131, 7211-7226 | GTATATGTGTGTG/TATGCTATGTATGTTA |
| 21 | PCATPH16 (reverse) | None | 7119-7131, 7211-7226 | TAACATACATAGCATA/CACACACATATAC |
| 22 | pCATPH17 (forward) | None | 7066-7081, 7212-7225 | ATGGATGTGTATGTAA/TATGCTATGTATGT |
| 23 | pCATPH17 (reverse) | None | 7066-7081, 7212-7225 | ACATACATAGCATA/TTACATACACATCCAT |
| 24 | MIE (reverse) | PstI, XhoI | 7198-7211 | GTCACTGCAGCTCGAGCATACACACATAAC |
Discontinuities in the annealing positions of primers are shown with a shill. Restriction sites are underlined.
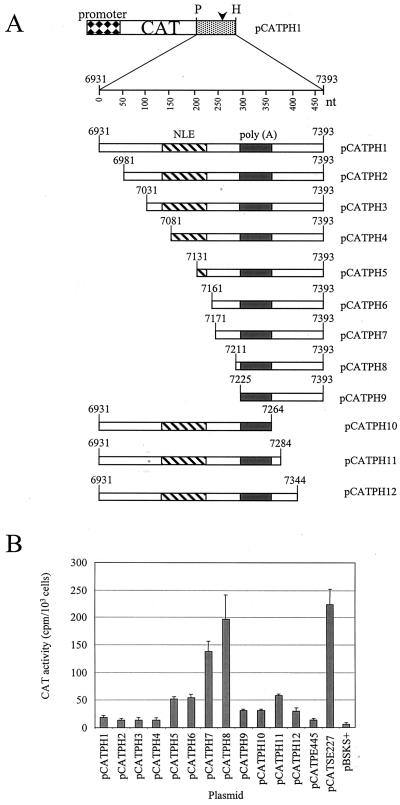
FIG. 3.
Functional assays with HeLa cells to confirm mapping of inhibitory sequences in the HPV-31 late gene 3′ UTR. (A) Diagram of the plasmid constructs used in transfection experiments. Boxes with diamonds, HSV-2 IE gene promoter; open boxes, CAT reporter gene; black boxes, HSV-2 IE gene poly(A) sequences; stippled box; HPV-31 L1/late gene 3′ UTR sequences; arrowheads, poly(A) sites; P, PstI restriction site; H, HindIII restriction site; hatched boxes; NLEs; grey boxes, late poly(A) signals and CstF-64 binding sites. (B) Bar chart of CAT activity in HeLa cells for HPV-31 constructs containing internal deletions assayed in the presence of [3H]chloramphenicol. Values are means plus standard deviations of duplicate transfections from three separate experiments. (C) Bar chart of CAT activity in HeLa cells for constructs containing the HSV-2 IE gene 5 poly(A) sequences. Values are means plus standard deviations of duplicate transfections from three separate experiments.
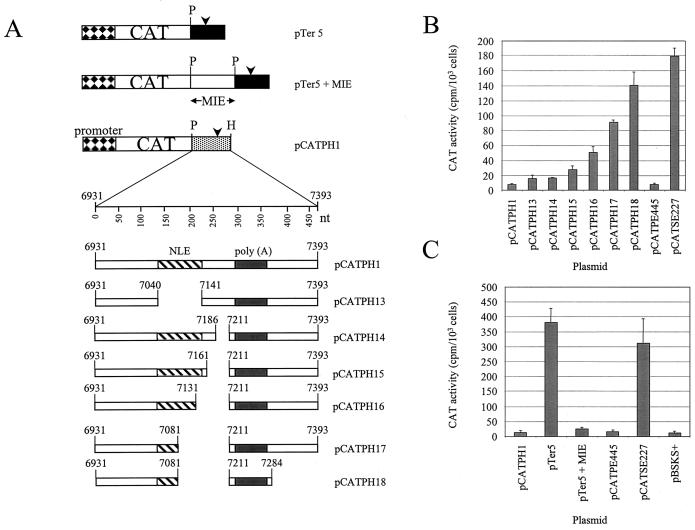
FIG. 2.
Functional assays with HeLa cells to map inhibitory sequences in the HPV-31 late gene 3′ UTR. (A) Diagram of the plasmid constructs used in transfection experiments. Box with diamonds, HSV-2 IE gene promoter; open box, CAT reporter gene; stippled box, HPV-31 L1/late gene 3′ UTR sequences; arrowhead, late poly(A) site; P, PstI restriction site; H, HindIII restriction site; hatched box, NLE; grey box, late poly(A) signal and CstF-64 binding site. (B) Bar chart of CAT activity in HeLa cells for HPV-31 constructs containing 5′ or 3′ deletions assayed in the presence of [3H]chloramphenicol. Values are means plus standard deviations of duplicate transfections from three separate experiments.
Cell culture.
HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 2 mM glutamine (Life Technologies) at 37°C and 5% CO2.
Nuclear extracts.
Nuclear extracts from HeLa cells were prepared as described previously (50) or purchased from the Computer Cell Culture Center (Seneffe, Belgium).
Transient transfections and CAT assays.
HeLa cells (105 cells/35-mm-diameter well) were transfected with 2 μg of CsCl-purified plasmid DNA by using LipofectAce reagent (Life Technologies) in accordance with the manufacturer's instructions. Cells were harvested and assayed after 48 h. Transfected cells were harvested by scraping and then lysed by freezing and thawing. CAT assays were for 2 h at 37°C in the presence of [3H]chloramphenicol (NEN), and then CAT activity was determined as described previously (37).
UV cross-linking and (EMSAs).
Primer sequences used to prepare riboprobe templates are shown in Table 3. HPV-31 probe templates were made by PCR amplification of pBSHPV-31 using forward primers with a 5′ extension encoding the bacteriophage T7 promoter. HPV-16 NRE probe templates were amplified by PCR using plasmid pCATPE445 as a template and included T3 RNA polymerase promoter sequences. PCR products were purified by fractionation on a 6% acrylamide gel. Riboprobes were synthesized by in vitro transcription with the Stratagene RNA transcription kit in the presence of 25 μCi of [α-32P]UTP (800 mCi/mmol; NEN) and 0.5 μg of DNA as the template, according to the manufacturer's protocol. Full-length transcripts were purified from a 5% denaturing acrylamide gel for use in electrophoretic mobility shift assays (EMSAs) or were purified with Mini Quick Spin Sephadex columns (Roche) for UV cross-linking experiments. UV cross-linking experiments were carried out as described previously (29); a Stratalinker (Stratagene) at a setting of 250 mJ was used to cross-link the samples. EMSAs were carried out as described previously (2). For EMSA competition experiments, specific competitor RNAs were transcribed in vitro with 0.5 μg of DNA as the template as described above except that unlabeled UTP was substituted for [α-32P]UTP. Nonspecific competitor RNA was Escherichia coli tRNA or a 65-nt RNA homologous to the pBluescript KS(+) polylinker region transcribed from a plasmid template linearized with EcoRI.
TABLE 3.
Sequences and annealing positions within the HPV-31 genome (primers 25 to 32) and the HPV-16 genome (primers 33 to 36) of PCR primers used to make riboprobes
| Primer no. | Primer name | Promoter | Genomic location | Sequence (5′-3′)a |
|---|---|---|---|---|
| 25 | NLE (forward) | T7 (sense) | 7040-7062 | CGACTTAATACGACTCACTATAGCAAAACGTAAAAAAACTAAAAAG |
| 26 | 3′ NLE (forward) | T7 (sense) | 7085-7101 | TGTAATACGACTCACTATAGGGTGTGTCTGTATGTGTAT |
| 27 | NLE (reverse) | SP6 (antisense) | 7122-7140 | GATCGATTTAGGTGACACTATAGAACACACAAACACACACAT |
| 28 | 5′ NLE (reverse) | SP6 (antisense) | 7065-7084 | GATCGATTTAGGTGACACTATAGTGTATTACATACACATCCAT |
| 29 | M (forward) | T3 (sense) | 7161-7179 | CAGAGATGCAATTAACCCTCACTAAAGGGATATGTATGCGTGTGTACTT |
| 30 | M (reverse) | None | 7191-7210 | CATACACACATAACATACTA |
| 31 | S (forward) | T3 (sense) | 7283-7301 | CAGAGATGCAATTAACCCTCACTAAAGGGAGTAACATACTATTACTAT |
| 32 | S (reverse) | None | 7324-7343 | AGCAGGAACAAGTAGGAACA |
| 33 | NRE (forward) | T3 (sense) | 7128-7145 | CAGAGATGCAATTAACCCTCACTAAAGGGAGAATTCGCTAAACGCAAAAAACGT |
| 34 | 3′ NRE (forward) | T3 (sense) | 7177-7192 | CAGAGATGCAATTAACCCTCACTAAAGGGAGAATTCGTGTTGTTTGTTGTGT |
| 35 | 5′ NRE (reverse) | None | 7155-7176 | TAGAGCTCTAATTCAACATACATACAATAC |
| 36 | NRE (reverse) | None | 7186-7207 | CTGAGCTCACATACAAACATATACACAAC |
Bacteriophage RNA polymerase promoter sequences are underlined.
Expression and purification of GST-tagged CstF-64 RBD.
Plasmid pGEX-2T plus the CstF-64 RNA binding domain (RBD) (44) was transformed into E. coli BL-21 (DE3) (Novagen), which was grown at 37°C for 3 h to an optical density at 600 nm of 0.5, and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h at 25°C. Purification of glutathione S-transferase (GST) or the GST-tagged CstF-64 RBD was carried out as described previously (44). The protein concentration was determined by using a Bradford assay (Bio-Rad), and protein integrity was checked on a Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. Full-length protein was estimated to constitute >95% of the sample.
Purification of RNA binding proteins on agarose beads.
Proteins binding to NLE RNA were purified as described previously (3). Briefly, 500 pmol of RNA was prepared by in vitro transcription, treated with sodium m-periodate, and then incubated with 400 μl of adipic acid dihydrazide agarose beads for 12 h at 4°C to cross-link the RNA to the beads. The beads were washed three times with 2 M NaCl and then equilibrated with buffer D (20 mM HEPES-KOH [pH 7.9], 5% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol). HeLa nuclear extract (250 μl; 1.25 mg) in buffer D was applied, and the mixture was incubated at 30°C for 20 min. The beads were washed four times with buffer D containing 4 mM MgCl2. The bound proteins were then eluted in protein loading buffer by heating to 90°C for 5 min. The affinity-selected proteins were electrophoresed on a SDS-12% PAGE gel.
Western blotting.
SDS-PAGE gels were electroblotted onto a nitrocellulose membrane. The blots were blocked overnight at 4°C in 5% (wt/vol) dried milk powder in phosphate-buffered saline (PBS). Primary antibodies were diluted in a solution containing PBS and 0.05% Tween plus 5% (wt/vol) dried milk powder. Anti-HuR antibody 19F12 was used at a dilution of 1 in 250, and anti-U2AF65 antibody MC3 was used at 1 in 100. Blots were incubated in the primary antibody for 1 h at room temperature with shaking. After being washed in PBS-0.05% Tween, the membrane was incubated in the secondary antibody for 1 h at room temperature with shaking. The secondary antibody was antimouse horseradish peroxidase (Sigma) diluted 1 in 1,000 in PBS-0.05% Tween plus 5% (wt/vol) dried milk powder. After being washed, the membranes were visualized with ECL reagents (Amersham Pharmacia Biotech) in accordance with the manufacturer's instructions.
RESULTS
HPV-31 contains an NLE-like element (NLE).
HPV-16 and HPV-31 have 70% overall sequence identity (11). For HPV-16, a 79-nt element overlapping the 3′ end of the L1 coding region and extending into the late gene 3′ UTR is known to inhibit gene expression in undifferentiated epithelial cells (20, 21). To determine whether such a regulatory sequence was also present in the HPV-31 late region, we aligned the sequence of the region of HPV-16 containing the NRE and late poly(A) sites (nt 7014 to 7499) (20, 38) with the equivalent region of HPV-31 (nt 6931 to 7393) (Fig. 1B). Sequence identity in this region was 68%. We identified an element, in a position equivalent to that of the HPV-16 NRE, which strongly resembles the HPV-16 NRE structurally. It contains the L1 stop codon, three weak consensus 5′ splice site sequences (compared with four in the HPV-16 NRE), and a highly GU-rich 3′ portion (Fig. 1B). A computer prediction of the secondary structure of the HPV-31 late gene 3′ UTR RNA made using the Zuker MFOLD algorithm (26) suggests that a series of A residues in the 5′ portion (nt 7047 to 7055) of the NLE, like those in the HPV-16 NRE, form the loop of a stem-loop structure. We have defined the HPV-31 NLE as 101 nt in length; this is 32 nt longer than the HPV-16 NRE to encompass the more extensive 3′ GU-rich portion of the NLE.
FIG. 1.
Diagram of the HPV-31 genome, and alignment of HPV-31 (nt 6931 to 7393) and HPV-16 (nt 7014 to 7499) sequences. (A) Linearized genomic structure of HPV-31, showing early (E) and late (L) gene coding regions (boxes). P97 is a constitutively active promoter; P742 is a promoter activated in differentiated cells. p(A)E and p(A)L, positions of the early and late poly(A) sites, respectively; heavy line, noncoding region (NCR) of the virus. (B) HPV-31 L1/late gene 3′ UTR sequences contain an NLE. Shown is an alignment of HPV-16 L1/late gene 3′ UTR sequences (upper sequences) and HPV-31 L1/late gene 3′ UTR sequences (lower sequences). Boxed regions, positions of the HPV-16 NRE and the HPV-31 NLE; light grey boxes, loops of predicted stem-loop structures; boldface boxes, L1 stop codons (TAA); dark grey boxes, GU-rich 3′ portions of the NRE and NLE; inverted triangles, intron-exon boundaries of weak consensus 5′ splice sites; double-underlined sequences; poly(A) hexanucleotides (AAUAAA); single-underlined sequences, GU/U-rich CstF-binding sites.
HPV-31 late gene 3′ UTR sequences inhibit reporter gene expression.
In HPV-16, a 445-nt PstI/EcoRI region of the HPV-16 genome (nt 7008 to 7453) was originally shown to exert a repressive effect on gene expression in undifferentiated epithelial cells (20). We asked whether the equivalent region of the HPV-31 genome (nt 6931 to 7393) also contains inhibitory sequences. Therefore this 462-nt fragment was cloned downstream of the CAT reporter gene in plasmid pLW1 to make plasmid pCATPH1 (Fig. 2A). When transiently transfected into HeLa cells, as a model for basal epithelial cells, this construct gave low levels of CAT activity, similar to levels obtained with pCATPE445, which contains the equivalent region of the HPV-16 genome. Plasmid pCATSE227, where the HPV-16 NRE has been deleted, gave high levels of reporter gene expression as expected (see Table 5; Fig. 2B). This result suggests that the HPV-31 late gene 3′ UTR also contains inhibitory sequences.
TABLE 5.
Result of CAT assay with HeLa cells transiently transfected with 5′ and 3′ end deletion constructs
| Plasmid | Genomic location | Mean CAT activity (cpm/103 cells) ± SD | Fold increase over pCATPH1 |
|---|---|---|---|
| pCATPH1 | 6931-7393 | 18.03 ± 3.66 | |
| pCATPH2 | 6981-7393 | 13.89 ± 1.56 | 0.8 |
| pCATPH3 | 7031-7393 | 13.19 ± 3.52 | 0.7 |
| pCATPH4 | 7081-7393 | 13.48 ± 3.62 | 0.7 |
| pCATPH5 | 7131-7393 | 51.99 ± 3.29 | 2.9 |
| pCATPH6 | 7161-7393 | 53.85 ± 6.88 | 3.0 |
| pCATPH7 | 7171-7393 | 137.25 ± 19.31 | 7.6 |
| pCATPH8 | 7211-7393 | 196.18 ± 44.92 | 10.9 |
| pCATPH9 | 7226-7393 | 30.7 ± 2.5 | 1.7 |
| pCATPH10 | 6931-7264 | 30.4 ± 2.97 | 1.7 |
| pCATPH11 | 6931-7284 | 57.55 ± 2.78 | 3.2 |
| pCATPH12 | 6931-7344 | 30.05 ± 6.19 | 1.7 |
| pCATPE445 (HPV-16) | 7008-7453 | 13.29 ± 3.05 | |
| pCATSE227 (HPV-16) | 7226-7453 | 217.01 ± 24.87 |
Inhibitory sequences in the HPV-31 late gene 3′ UTR map to regions flanking the late poly(A) signal.
We deleted the 101-nt HPV-31 NLE from pCATPH1 to make plasmid pCATPH13 (Fig. 3A). In transient transfection experiments with HeLa cells, pCATPH13 consistently gave higher levels of CAT activity than pCATPH1 (Table 4; Fig. 3B), but reporter expression increased only 2- to 3-fold, compared to the 20- to 100-fold increase observed with HPV-16 constructs lacking the NRE (20) (Fig. 3B). This suggests that, for HPV-31, sequence elements within the late 3′ UTR apart from the NLE could also contribute to the inhibitory activity. To test this, we made a series of 5′ and 3′ deletions of the wild-type sequence in the plasmid pCATPH1 (Fig. 2A), in each case maintaining the poly(A) signal and downstream GU/U-rich CstF-64 binding site. We found that, in transient transfection experiments with HeLa cells, 5′ deletions of between 200 and 280 nt (constructs pCATPH5 to pCATPH8) significantly increased reporter gene expression (Table 5; Fig. 2B). Construct pCATPH8, which has a 5′ deletion of 280 nt, gave a level of CAT activity very similar to that obtained with HPV-16 plasmid pCATSE227, which lacks the NRE. Thus further inhibitory sequences lie downstream of the NLE but upstream of the HPV-31 late poly(A) signal. A further 5′ end deletion of 295 nt (pCATPH9) gave low CAT activity, perhaps because the deleted sequence abuts the polyadenylation signal and may inhibit efficient polyadenylation (Table 5; Fig. 2B). Interestingly, a 3′ deletion of 110 nt (construct pCATPH11) increased CAT activity threefold above that obtained with pCATPH1 (Table 5; Fig. 2B), suggesting that there is a subsidiary inhibitory element (SIE) downstream of the late poly(A) signal and GU/U-rich downstream sequence element. However, a 3′ end deletion of 130 nt (pCATPH10) gave low CAT activity (Table 5; Fig. 2B), again most likely because the deletion is close to essential polyadenylation sequences. Due to this technical difficulty, we cannot exclude the possibility that in our mapping we have omitted small portions of the 3′ end of the major inhibitory element (MIE), and the 5′ end of the SIE, in the region of the poly(A) signal.
TABLE 4.
Result of CAT assay of HeLa cells transiently transfected with constructs with internal deletions
| Plasmid | Genomic location(s) of deletion(s) | Mean CAT activity (cpm/103 cells) ± SD | Fold increase over pCATPH1 |
|---|---|---|---|
| pCATPH1 | None | 7.64 ± 0.73 | |
| pCATPH13 | 7040-7141 | 16.26 ± 4.72 | 2.1 |
| pCATPH14 | 7186-7211 | 16.28 ± 1.53 | 2.1 |
| pCATPH15 | 7171-7211 | 27.6 ± 5.2 | 3.6 |
| pCATPH16 | 7141-7211 | 50.52 ± 8.32 | 6.6 |
| pCATPH17 | 7081-7211 | 91.09 ± 3.74 | 11.9 |
| pCATPH18 | 7081-7211, 7284-7393 | 140.62 ± 17.41 | 18.4 |
| pCATPE445 (HPV-16) | 7008-7453 | 7.78 ± 1.98 | |
| pCATSE227 (HPV-16) | 7226-7453 | 179.68 ± 10.68 |
Delineating the inhibitory elements.
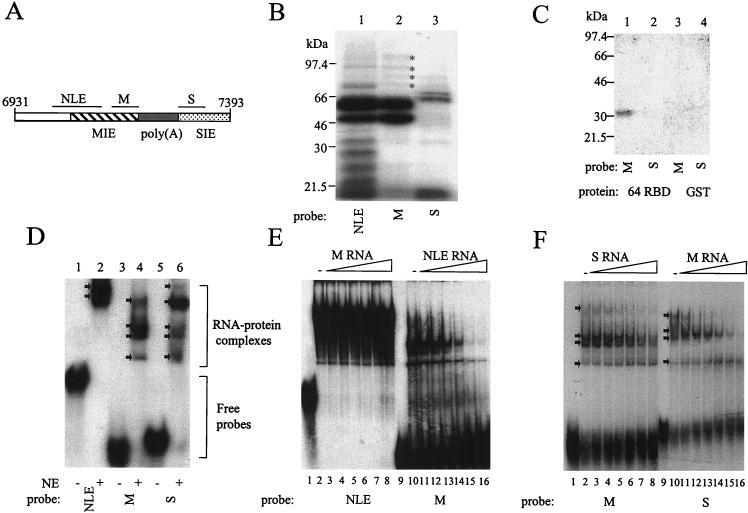
On the basis of our 5′-to-3′ deletion constructs we reasoned that the sequences lying between nt 7081 and 7210, which we termed the MIE, comprised the most significantly inhibitory portion of the late gene 3′ UTR. To confirm this, we carried out 3′-to-5′ deletion analysis of the element. We deleted 25 (pCATPH14), 50 (pCATPH15), and 80 nt (pCATPH16) from the 3′ end of the proposed MIE, retaining the polyadenylation signal and downstream sequences (Fig. 3A). These deletions gave CAT activities twofold (pCATPH14), fourfold (pCATPH15), and sevenfold (pCATPH16) higher than that for pCATPH1 (Fig. 3B; Table 4), suggesting that the 3′ end of the MIE does indeed lie near nt 7210. Next, we deleted the MIE from CAT expression plasmid pCATPH1 to make pCATPH17 (Fig. 3A). We also deleted this region from CAT expression plasmid pCATPH11 to make pCATPH18, which lacks both the MIE and the SIE which lies downstream of the late poly(A) signal (Fig. 3A). Plasmid pCATPH17 gave CAT activity 12-fold higher than that obtained with pCATPH1 (Table 4; Fig. 3B). These results support our initial conclusion that the 130-nt region comprising the 3′ NLE and downstream sequences contains the MIE in the HPV-31 late gene 3′ UTR. In this experiment, the CAT activity for the HPV-16 construct lacking the NRE, pCATSE227, was twofold higher than that for pCATPH17. In the previous experiment (Fig. 2B), the CAT activity for pCATSE227 was only 10% higher than that for 5′ deletion construct pCATPH8. Plasmid pCATPH18 gave a CAT activity 18-fold higher than that obtained with control plasmid pCATPH1 and 1.5-fold higher than that obtained with pCATPH17 (Table 4; Fig. 3B). This is consistent with the small, though significant, increase over pCATPH1 obtained with 3′ deletion construct pCATPH11 (Fig. 2B). Results with these plasmids, having internal deletions of the predicted inhibitory elements, confirm that the two inhibitory elements in the HPV-31 late gene 3′ UTR are located within the sequences from nt 7081 to 7210 and from nt 7284 to 7393 (Fig. 4).
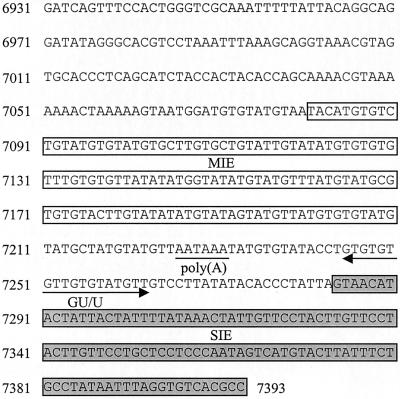
FIG. 4.
HPV-31 L1/late gene 3′ UTR sequences (nt 6931 to 7393), showing the position of inhibitory elements and polyadenylation signals. Open boxes, MIE (nt 7081 to 7210); underlined region, poly(A) hexanucleotide; arrow, CstF binding site (GU/U); Grey box, SIE (nt 7284 to 7393).
The MIE functions as an inhibitory element in a heterologous system.
We next asked whether the inhibition of gene expression by the MIE was specific to the HPV-31 late poly(A) signal. To test this, we cloned the MIE upstream of the HSV-2 IE gene 5 polyadenylation signal in CAT expression plasmid pTer5 to make plasmid pTer5+MIE (Fig. 3A). Like pCATSE227, pTer5 gave very high CAT reporter gene expression following transient transfection into HeLa cells (Table 6; Fig. 3C). pTer5+MIE gave much lower gene expression, equivalent to a 15-fold inhibition in comparison to expression with pTer5, which was broadly comparable to that with pCATPH1 (Table 6; Fig. 3C). The inhibitory effect brought about by the MIE is not, therefore, specific to the HPV-31 poly(A) signal.
TABLE 6.
Result of CAT assay using constructs containing heterologous (HSV-2 IE gene 5) poly(A) sequences
| Plasmid | Genomic location of HPV sequences | Mean CAT activity (cpm/103 cells) ± SD | Fold inhibition by MIE |
|---|---|---|---|
| pCATPH1 | 6931-7393 | 13.30 ± 1.25 | |
| pTer5 | 380.38 ± 48.74 | ||
| pTer5+MIE | 7081-7211 | 25.16 ± 3.73 | 15.1 |
| pCATPE445 (HPV-16) | 7008-7453 | 11.17 ± 2.56 | |
| pCATSE227 (HPV-16) | 7226-7453 | 311.20 ± 81.40 |
The HPV-31 NLE binds the same proteins as the HPV-16 NRE.
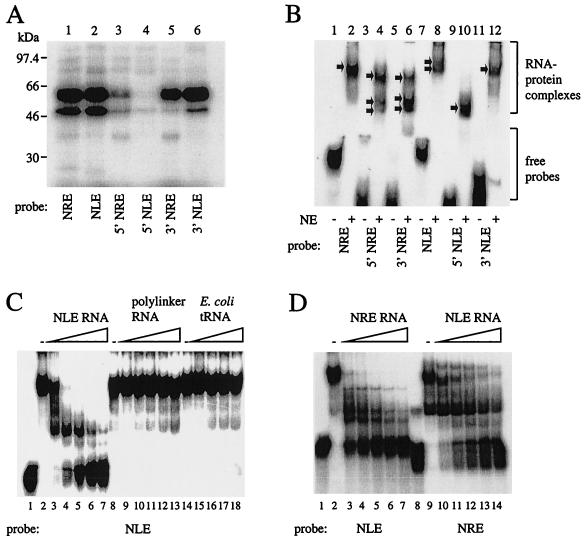
The HPV-16 NRE has previously been shown to interact with several cellular factors (5, 22). We compared the binding of the HPV-16 NRE and the HPV-31 NLE to HeLa cell nuclear extracts by UV cross-linking (Fig. 5A). The full-length NRE and NLE probes UV cross-linked to proteins of similar sizes, including proteins that comprise an abundant 65-kDa band (Fig. 5A, lanes 1 and 2). Another abundant protein of 50 kDa (Fig. 5A, lanes 1 and 2), as well as several less abundant proteins, was also bound by both the NRE and NLE. Probes corresponding to the 3′ GU-rich portion of each element (Fig. 5A, lanes 5 and 6) bound more protein than the 5′ NRE or 5′ NLE probes (Fig. 5A, lanes 3 and 4). The greater extent of protein binding to the 3′ NLE (lane 6) than to the 3′ NRE (lane 5) may be partly a function of its length (56 nt, compared with 30 nt for the 3′ NRE).
FIG. 5.
UV cross-linking and EMSA experiments to compare protein binding to the HPV-16 NRE and HPV-31 NLE. (A) UV cross-linking of 32P-labeled NRE and NLE probes to HeLa cell nuclear extracts. Lane 1, full-length (HPV-16) NRE; lane 2, full-length (HPV-31) NLE; lane 3, 5′ NRE (49 nt); lane 4, 5′ NLE (46 nt); lane 5, 3′ NRE (30 nt); lane 6, 3′ NLE (56 nt). (B) EMSA using 32P-labeled RNA probes and HeLa cell nuclear extracts very similar to those used for panel A run on a nondenaturing polyacrylamide gel. RNA-protein complexes and free probes are bracketed. Arrows, RNA-protein complexes. NE, HeLa cell nuclear extracts. (C) EMSA competition assay using a 32P-labeled NLE probe (1.5 pmol) and HeLa nuclear extracts. Lane 1, no extracts; lane 2, no competitor RNA; lanes 3 to 7, 1- to 16-fold molar excess of specific competitor, i.e., 1.5 to 24 pmol of in vitro-transcribed, unlabeled NLE RNA; lane 8, no competitor RNA; lanes 9 to 13, 1- to 16-fold molar excess of nonspecific competitor, i.e., 1.5 to 24 pmol of in vitro-transcribed pBluescript KS(+) polylinker RNA; lane 14, no competitor RNA; lanes 15 to 18, nonspecific competitor, i.e., 500 ng to 4 μg of E. coli tRNA. (D) EMSA competition assay using 32P-labeled NLE and NRE probes and HeLa nuclear extracts. Lanes 1 to 7, NLE probe (1.5 pmol); lane 1, no extracts; lane 2, no competitor; lanes 3 to 7, 1- to 16-fold molar excess, i.e., 1.5 to 24 pmol of in vitro-transcribed, unlabeled NRE RNA; lanes 8 to 14, NRE probe (1.5 pmol); lane 8, no extracts; lane 9, no competitor; lanes 10 to 14, 1- to 16-fold molar excess, i.e., 1.5 to 24 pmol of in vitro-transcribed, unlabeled NLE RNA.
Next, we carried out an EMSA using half- and full-length NRE and NLE probes and HeLa cell nuclear extracts (Fig. 5B). The full-length NRE probe (lane 2) generated a single shifted complex. The 5′ NRE (Fig. 5B, lane 4) and 3′ NRE (Fig. 5B, lane 6) each gave rise to three complexes. The patterns obtained differ slightly: the upper band predominates for the 5′ NRE probe (lane 4), and the lower two predominate for the 3′ NRE probe (lane 6). The full-length NLE probe generated two highly retarded complexes (lane 8). The 5′ NLE probe gave rise to a single complex (lane 10). The 3′ NLE generated a single complex (lane 12) resembling in mobility the lower complex formed on the full-length NLE probe. This is consistent with the UV cross-linking experiment (Fig. 5A), in which the full-length and 3′ NLEs gave similar patterns of bands.
NLE-protein interactions are specific, since complex formation was abolished by adding increasing amounts of specific (unlabeled NLE RNA) (Fig. 5C, lanes 3 to 7), but not nonspecific (in vitro-transcribed pBluescript polylinker RNA or E. coli tRNA) competitor RNA (Fig. 5C, lanes 9 to 13 and 15 to 18). As the binding patterns generated by the NRE and NLE in UV cross-linking experiments appeared similar, we performed competition experiments for protein binding to the NLE using excess unlabeled NRE RNA, and vice versa. In an EMSA, unlabeled NRE RNA competed with an NLE probe for binding HeLa nuclear extract proteins (Fig. 5D, lanes 3 to 7) and, conversely, unlabeled NLE RNA competed for binding with an NRE probe (Fig. 5D, lanes 10 to 14). NRE RNA competed efficiently with the labeled NLE: complexes were disrupted by a one- to twofold or greater molar excess of unlabeled RNA (Fig. 5D, lanes 3 to 7). NLE RNA competed less efficiently with the labeled NRE: complexes were disrupted by a fourfold or greater molar excess of unlabeled RNA (Fig. 5D, lanes 12 to 14). The two elements therefore bind substantially the same proteins present in HeLa nuclear extracts, but the affinities of the complexes are different.
The NLE binds the known NRE-binding proteins CstF-64, U2AF65, and HuR.
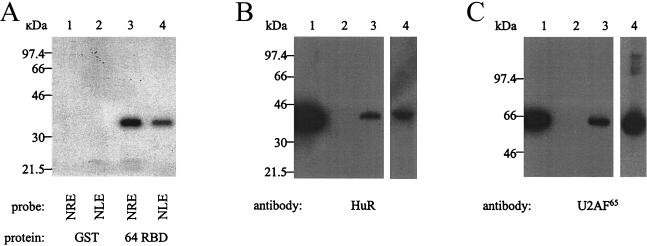
Next, we asked whether the NLE could bind RNA-processing factors CstF-64, U2AF65, and HuR, all of which have previously been shown to bind the HPV-16 NRE (22). First, we UV cross-linked NLE or NRE probes to a GST-tagged CstF-64 RBD (44). We found that the HPV-31 NLE probe UV cross-linked to the CstF-64 RBD (Fig. 6A, lane 4). The HPV-16 NRE, used as a positive control for binding, produced a slightly stronger band than the NLE probe (lane 3), suggesting that CstF-64 has a greater affinity for the NRE than for the NLE. Neither probe cross-linked to a GST control protein (lanes 1 and 2). In the absence of expression constructs for U2AF65 or HuR, we adopted an alternative approach to test the binding of these proteins to the NLE. NLE RNA was covalently cross-linked to agarose beads and then was used to purify binding proteins present in HeLa nuclear extracts. The affinity-purified proteins were subjected to SDS-PAGE and then Western blotted. We detected HuR (Fig. 6B, lane 3) and U2AF65 (Fig. 6C, lane 3) in the proteins that bound the NLE. We used poly(U) RNA linked to beads as a positive control for binding and detected HuR (Fig. 6B, lane 4) and U2AF65 (Fig. 6C, lane 4). The beads alone did not bind HuR (Fig. 6B, lane 2) or U2AF65 (Fig. 6C, lane 2). The NLE, like the NRE, therefore binds 3′ end-processing factor CstF-64, splicing factor U2AF65, and nucleus-to-cytoplasm shuttling protein HuR.
FIG. 6.
UV cross-linking and Western blotting to identify specific RNA-processing factors that bind to the NLE. (A) UV cross-linking of 32P-labeled NRE and NLE probes to bacterially expressed GST-tagged CstF-64 RBD protein. Lane 1, GST protein and NRE probe; lane 2, GST protein and NLE probe; lane 3, GST-tagged CstF-64 RBD and NRE probe; lane 4, GST-tagged CstF-64 RBD and NLE probe. (B) Western blot with the 19F12 anti-HuR monoclonal antibody. Lane 1, 20 μg of HeLa nuclear extracts; lane 2, 20 μl of proteins purified with beads alone; lane 3, 20 μl of proteins purified with NLE RNA; lane 4, 20 μl of proteins purified with poly(U) RNA. (C) Western blot with the MC3 anti-U2AF65 monoclonal antibody. Lane 1, 20 μg of HeLa nuclear extracts; lane 2, 20 μl of proteins purified with beads alone; lane 3, 20 μl of proteins purified using NLE RNA; lane 4, 20 μl of proteins purified with poly(U) RNA.
The sequences flanking the late poly(A) signal also bind nuclear proteins.
Next we asked whether the inhibitory sequences upstream and downstream of the late poly(A) signal bind cellular proteins. We used as probes the 50-nt sequences upstream (nt 7161 to 7211; probe M), and 60-nt sequences downstream (nt 7284 to 7343, probe S), of the late poly(A) signal (Fig. 7A). These probes derive from the MIE and SIE, respectively, and do not include any signal sequences involved in polyadenylation. In a UV cross-linking experiment using HeLa cell nuclear extracts (Fig. 7B), probe M (lane 2) bound proteins of approximately 65 and 50 kDa, which are similar in size to the most abundant proteins bound by the NLE (lane 1). The M probe also bound at least four proteins between 70 and 100 kDa that have mobilities similar to those of proteins that UV cross-link to the NLE. The NLE probe, however, UV cross-links to many proteins, including several between 25 and 50 kDa that do not UV cross-link to the M probe. Probe S (lane 3) binds proteins that do not resemble those binding to the NLE or M probes, the most abundant of which are two proteins around 70 kDa. The M probe, like the NLE and NRE, UV cross-links to the CstF-64 RBD (Fig. 7C, lane 1). The S probe, however, does not (Fig. 7C, lane 2). Neither probe binds GST (lanes 3 and 4). This is consistent with the M and S probes UV cross-linking to different sets of proteins present in HeLa nuclear extracts and with the similar sets of proteins bound by NLE and M probes (Fig. 7B). In an EMSA using HeLa cell nuclear extracts, the NLE probe (Fig. 7D, lane 2) generated two major retarded complexes, as before (Fig. 5B). Probe M (Fig. 7D, lane 4) gave rise to four shifted complexes, whereas probe S (Fig. 7D, lane 6) gave rise to five complexes.
FIG. 7.
UV cross-linking and EMSA of protein binding to sequences surrounding the late poly(A) signal. (A) Diagram of the HPV-31 late gene 3′ UTR sequences showing the positions of the probes used for protein binding studies. Hatched box, MIE; grey box, poly(A) and CstF binding sites; stippled box, SIE; lines, positions of NLE probe (nt 7041 to 7141), M probe (nt 7161 to 7211), and S probe (nt 7284 to 7343). (B) UV cross-linking of 32P-labeled NLE, M, and S probes to HeLa nuclear extracts. Lane 1, NLE (101 nt); lane 2, M (50 nt); lane 3, S (60 nt). Asterisks, minor proteins of similar sizes that bind both NLE and M probes. (C) UV cross-linking of 32P-labeled M and S probes to bacterially expressed CstF-64 RBD. Lane 1, GST-tagged CstF-64 RBD protein with an M probe; lane 2, GST-tagged CstF-64 RBD protein with an S probe; lane 3, GST protein with an M probe; lane 4, GST protein with an S probe. 64 RBD, GST-tagged CstF-64 RBD. (D) EMSA of 32P-labeled NLE, M, and S probes using HeLa cell nuclear extracts. RNA-protein complexes and free probes are bracketed. Arrows, RNA-protein complexes; NE, HeLa nuclear extracts. (E) EMSA competition assay. Lanes 1 to 8, 32P-labeled NLE probe (1.5 pmol); lane 1, no extracts; lane 2, no competitor RNA; lanes 3 to 8, 1- to 32-fold molar excess, i.e., 1.5 to 48 pmol of in vitro-transcribed unlabeled M RNA; lanes 9 to 16, 32P-labeled M probe (1.5 pmol); lane 9, no extracts; lane 10, no competitor RNA; lanes 11 to 16, 1- to 32-fold molar excess, i.e., 1.5 to 48 pmol of in vitro-transcribed unlabeled NLE RNA. (F) EMSA competition assay. Lanes 1 to 8, 32P-labeled M probe (1.5 pmol); lane 1, no extracts; lane 2, no competitor RNA; lanes 3 to 6, 1- to 32-fold molar excess, i.e., 1.5 to 48 pmol of in vitro-transcribed unlabeled S RNA; lanes 9 to 16, 32P-labeled S probe (1.5 pmol); lane 9, no extracts; lane 10, no competitor RNA; lanes 11 to 16, 1- to 32-fold molar excess, i.e., 1.5 to 48 pmol of in vitro-transcribed, unlabeled M RNA.
The upstream probe M binds a subset of the proteins bound by the NLE.
Since NLE and M probes UV cross-link to several proteins of similar sizes (Fig. 7B, lanes 1 and 2) and since both bind CstF (Fig. 6A and 7C), we asked whether these RNAs compete for binding proteins present in HeLa cell nuclear extracts. In an EMSA experiment, unlabeled M RNA could not compete for binding of HeLa nuclear extract proteins with an NLE probe (Fig. 7E, lanes 3 to 8). However, a fourfold or greater molar excess of unlabeled NLE RNA disrupted RNA-protein complexes formed on an M probe (Fig. 7E, lanes 13 to 16), suggesting that the proteins bound by M are a subset of those binding the NLE. Since NLE RNA can displace proteins from M RNA, but not vice versa, the affinity of protein binding to the NLE may be higher than the affinity of binding to M.
Upstream and downstream probes M and S can compete for binding cellular proteins.
Although the M and S probes UV cross-link to proteins of different sizes, in an EMSA experiment they both generated certain retarded complexes of similar mobilities (Fig. 7D). We therefore asked whether these two probes could compete for binding of proteins present in HeLa cell nuclear extracts. In an EMSA experiment, unlabeled S RNA could compete to a limited extent with an M probe for binding HeLa nuclear extract proteins, but only when present in an eightfold or greater molar excess (Fig. 7F, lanes 6 to 8). Unlabeled M RNA could compete efficiently with an S probe for binding proteins in HeLa cell nuclear extracts when present in a twofold or greater molar excess (Fig. 7F, lanes 12 to 16). Taken together, the UV cross-linking and EMSA experiments may suggest that M and S bind some, but not all, proteins in common. They also suggest that the affinity of protein binding to M is higher than that of binding to S.
DISCUSSION
We have shown that HPV-31 L1/late gene 3′ UTR sequences inhibit gene expression in undifferentiated epithelial cells. A short element, the NLE, has high sequence similarity to the NRE previously described for HPV-16. The inhibitory sequences in the late gene 3′ UTR, however, are not limited to the NLE. Instead, there is an MIE, approximately 130 nt in length, which overlaps the NLE by 60 nt and which lies upstream of the late poly(A) signal. The MIE also functions as an inhibitory element when placed upstream of a heterologous poly(A) signal. An SIE, approximately 110 nt in length, lies downstream of the poly(A) signal. The 3′ and 5′ borders of the MIE and SIE, respectively, have not been precisely delineated, since deletion constructs that terminate close to the poly(A) signal give low gene expression, most likely because polyadenylation is disrupted. The NLE and NRE compete for binding proteins present in HeLa cell nuclear extracts. The NLE binds RNA-processing factors CstF-64, U2AF65, and HuR, previously shown to bind the NRE (5, 22). The downstream portion of the MIE also binds proteins resembling those bound by the NLE and NRE, and the entire MIE may be required for optimum efficiency of protein binding. The SIE, however, binds some different, and some similar, proteins present in HeLa nuclear extracts.
The elements we have identified differ from those previously identified in the late gene 3′ UTRs of HPV-16 (20), HPV-1 (46), and BPV-1 (8). The control sequences present in HPV-31 appear more complex: the upstream element is substantially larger than those previously described, and inhibitory sequences present downstream of the poly(A) signal are novel. Further, since the two HPV-31 elements bind some different cellular proteins, they may act by distinct mechanisms.
It is perhaps surprising that the 101-nt NLE does not substantially inhibit gene expression in undifferentiated epithelial cells, since it closely resembles the NRE, both structurally and in terms of protein binding. The 5′ portion of the NLE, unlike that of the NRE (data not shown), does not inhibit gene expression on its own. This could be because it contains only a single weak consensus 5′ splice site, a feature known to be an important determinant of the ability of the NRE to inhibit gene expression (9; S. A. Cumming and S. V. Graham, unpublished observations).
RNA probes derived from the MIE bind many cellular proteins, including RNA-processing factors HuR, CstF-64, and U2AF65. HuR has roles in mRNA stability and RNA transport (7, 30, 33). It binds the HPV-16 NRE (22), as well as the HPV-1 AU-rich element (41), and may act by stabilizing late mRNAs in the cytoplasm of differentiated cells (36). The HPV-31 element may act in a similar way.
The GU-rich MIE also binds CstF-64 in vitro. During 3′ end processing, CstF-64 binds a GU/U-rich element downstream of the cleavage site (24, 43). The cleavage-polyadenylation specificity factor (CPSF) binds to the AAUAAA hexanucleotide, which directs cleavage downstream. CPSF also binds to CstF, allowing cleavage of the transcript between the two protein binding sites and addition of poly(A) to the cleaved 3′ end (19, 34). The carboxy-terminal domain of RNA polymerase II is found in association with CstF and CPSF (28) and is essential for 3′ end processing (15). During transcription of the late gene 3′ UTR, CstF comes into contact with the GU-rich tracts in the MIE before reaching the proper CstF binding site downstream of the polyadenylation site. Thus the MIE could compete for binding of CstF with the downstream GU/U-rich element. This may reduce the rate of polyadenylation at the late site by reducing the availability of free CstF subunits to bind the correct GU/U-rich site, leading to inhibition of late gene expression in undifferentiated epithelial cells.
Terhune et al. have demonstrated that there is a higher concentration of all three CstF subunits in undifferentiated than in differentiated epithelial cells (47). If the MIE indeed binds CstF-64 in vivo, then this would be expected to compromise efficient polyadenylation in differentiated cells. However, the MIE binds many cellular proteins, and formation of a differentiation-specific complex may block the binding of CstF in differentiated cells, resulting in a higher concentration of free CstF-64 subunits. These would then be available to bind to the downstream GU/U-rich element, thus increasing the efficiency of polyadenylation.
The MIE also binds U2AF65. U2AF65 is one subunit of an auxiliary splicing factor that interacts with the U2 snRNP at intronic branch points and 3′ splice sites (35). The binding of splicing factors to transcripts may prevent nuclear export (18). For example, the Caenorhabditis elegans U2AF65 protein was shown to bind reporter gene transcripts containing U2AF65 binding sequences in their 3′ UTRs, causing nuclear retention and preventing gene expression (25). Some splicing factors can disrupt polyadenylation. For example, the U1 snRNP components U1A (12, 13) and the U1 70-kDa subunit (U1 70K) (14) interact with and inhibit poly(A) polymerase. U2AF65, which is closely related to U1 70K, also binds poly(A) polymerase (49) and could perhaps also inhibit polyadenylation.
The inhibitory element located downstream of the late poly(A) site is probably rather short-lived in the RNA. Transcription proceeds for up to 500 nt beyond the poly(A) site before cleavage and polyadenylation occur (6); however the 3′ product is rapidly degraded following cleavage (51). The downstream element binds two proteins of around 70 kDa in vitro. These have yet to be identified. If these were also polyadenylation factors, their binding could reduce the efficiency of upstream cleavage and hence the polyadenylation of transcripts.
We have shown that HPV-31, like certain other papillomaviruses, contains inhibitory RNA sequence elements in its late gene 3′ UTR. Unlike those other papillomaviruses, HPV-31 contains complex inhibitory sequences located downstream, as well as upstream, of the late poly(A) site. In light of the interactions with RNA-processing factors we have shown, we propose that these elements most likely act by disrupting polyadenylation at the late site in undifferentiated cells. It seems likely, however, that the HPV-31 inhibitory elements, like the HPV-16 NRE, are multifunctional, operating via more than one posttranscriptional mechanism.
Acknowledgments
This work was supported by the Wellcome Trust and the Biotechnology and Biological Sciences Research Council.
We thank Richard Elliott for critical reading of the manuscript. We also thank Craig Meyers for the pBSHPV-31 plasmid, Yoshio Takagaki and James Manley for the CstF-64 RBD expression construct, Henry Furneaux for the 19F12 HuR antibody, and Juan Valcárcel for the MC3 U2AF65 antibody.
REFERENCES
- 1.Bedell, M. A., J. B. Hudson, T. R. Golub, M. E. Turyk, M. Hosken, G. D. Wilbanks, and L. A. Laimins. 1991. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J. Virol. 65:2254-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, D. J., R. Chan, H. Min, J. Wang, and L. Bell. 1998. The electrophoretic mobility shift assay for RNA binding proteins, p. 109-136. In C. W. J. Smith (ed.), RNA:protein interactions: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 3.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collier, B., L. Goobar-Larsson, M. Sokolowski, and S. Schwartz. 1998. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogeneous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J. Biol. Chem. 273:22648-22656. [DOI] [PubMed] [Google Scholar]
- 5.Dietrich-Goetz, W., I. M. Kennedy, B. Levins, M. A. Stanley, and J. B. Clements. 1997. A cellular 65-kDa protein recognizes the negative regulatory element of human papillomavirus late mRNA. Proc. Natl. Acad. Sci. USA 94:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dye, M. J., and N. J. Proudfoot. 1999. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell 3:371-378. [DOI] [PubMed] [Google Scholar]
- 7.Fan, X. C., and J. A. Steitz. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17:3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furth, P. A., and C. C. Baker. 1991. An element in the bovine papillomavirus late 3′ untranslated region reduces polyadenylated cytoplasmic RNA levels. J. Virol. 65:5806-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furth, P. A., W.-T. Choe, J. H. Rex, J. C. Byrne, and C. C. Baker. 1994. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol. Cell. Biol. 14:5278-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaffney, D., J. L. Whitton, M. Lynch, J. McLauchlan, and J. B. Clements. 1985. A modular system for the assay of transcriptional regulatory signals: the sequence TAATGARAT is required for herpes simplex virus immediate early gene activation. Nucleic Acids Res. 13:7847-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldsborough, M. D., D. DiSilvestre, G. F. Temple, and A. T. Lorincz. 1989. Nucleotide sequence of human papillomavirus type 31: a cervical neoplasia-associated virus. Virology 171:306-311. [DOI] [PubMed] [Google Scholar]
- 12.Gunderson, S. I., K. Beyer, G. Martin, W. Keller, W. C. Boelens, and I. W. Mattaj. 1994. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell 76:531-541. [DOI] [PubMed] [Google Scholar]
- 13.Gunderson, S. I., S. Vagner, M. Polycarpou-Schwartz, and I. W. Mattaj. 1997. Involvement of the carboxy terminus of vertebrate poly(A) polymerase in U1A autoregulation and in coupling of splicing and polyadenylation. Genes Dev. 11:761-773. [DOI] [PubMed] [Google Scholar]
- 14.Gunderson, S. I., M. Polycarpou-Schwartz, and I. W. Mattaj. 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell 1:255-264. [DOI] [PubMed] [Google Scholar]
- 15.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential polyadenylation factor. Nature 395:93-96. [DOI] [PubMed] [Google Scholar]
- 16.Howley, P. M. 1996. Papillomavirinae: the viruses and their replication, p. 2045-2076. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 17.Hummel, M., J. B. Hudson, and L. A. Laimins. 1992. Differentiation-induced and constitutive expression of human papillomavirus type 31b in cell lines containing viral episomes. J. Virol. 66:6070-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izaurralde, E., and S. Adam. 1998. Transport of macromolecules between the nucleus and the cytoplasm. RNA 4:351-364. [PMC free article] [PubMed] [Google Scholar]
- 19.Keller, W. 1995. No end yet to messenger RNA 3′ processing! Cell 81:829-832. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy, I. M., J. K. Haddow, and J. B. Clements. 1990. Analysis of human papillomavirus type 16 late mRNA 3′ processing signals in vitro and in vivo. J. Virol. 64:1825-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennedy, I. M., J. K. Haddow, and J. B. Clements. 1991. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J. Virol. 65:2093-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koffa, M. D., S. V. Graham, Y. Takagaki, J. L. Manley, and J. B. Clements. 2000. The human papillomavirus type 16 negative regulatory element interacts with three proteins that act at different posttranscriptional levels. Proc. Natl. Acad. Sci. USA 97:4677-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma, W.-J., S. Cheng, C. Campbell, A. Wright, and H. Furneaux. 1996. Cloning and characterisation of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271:8144-8151. [DOI] [PubMed] [Google Scholar]
- 24.MacDonald, C. C., J. Wilusz, and T. Shenk. 1994. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol. 14:6647-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacMorris, M. A., D. A. R. Zorio, and T. Blumenthal. 1999. An exon that prevents export of a mature mRNA. Proc. Natl. Acad. Sci. USA 96:3813-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 27.Matthews, K. R., C. Tschudi, and E. Ullu. 1994. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 8:491-501. [DOI] [PubMed] [Google Scholar]
- 28.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 29.Moore, M. J., and C. C. Query. 1998. Use of site-specifically modified RNAs constructed by RNA ligation, p. 75-108. In C. W. J. Smith (ed.), RNA:protein interactions: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 30.Myer, V. E., X. C. Fan, and J. A. Steitz. 1997. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 16:2130-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozbun, M. A., and C. Meyers. 1997. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J. Virol. 71:5161-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozbun, M. A., and C. Meyers. 1998. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. J. Virol. 72:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng, S. S.-Y., C.-Y. A. Chen, N. Xu, and A.-B. Shyu. 1998. RNA stabilization by the AU-rich element binding protein HuR, an ELAV protein. EMBO J. 17:3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proudfoot, N. 1996. Ending the message is not so simple. Cell 87:779-781. [DOI] [PubMed] [Google Scholar]
- 35.Ruskin, B., P. D. Zamore, and M. R. Green. 1988. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52:207-219. [DOI] [PubMed] [Google Scholar]
- 36.Schwartz, S., M. Sokolowski, B. Collier, A. Carlsson, and L. Goobar-Larsson. 1999. CIS acting regulatory sequences on human papillomavirus late mRNAs. Recent Res. Dev. Virol. 1:53-74. [Google Scholar]
- 37.Seed, B., and S.-Y. Sheen. 1988. A simple phase-extraction assay for chloramphenicol acetyltransferase activity. Gene 67:271-277. [DOI] [PubMed] [Google Scholar]
- 38.Seedorf, K., G. Krämmer, M. Dürst, S. Suhai, and W. G. Röwekamp. 1985. Human papillomavirus type 16 DNA sequence. Virology 145:181-185. [DOI] [PubMed] [Google Scholar]
- 39.Smotkin, D., and F. O. Wettstein. 1986. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc. Natl. Acad. Sci. USA 83:4680-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokolowski, M., W. Tan, M. Jellne, and S. Schwartz. 1998. mRNA instability elements in the human papillomavirus type 16 L2 coding region. J. Virol. 72:1504-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sokolowski, M., H. Furneaux, and S. Schwartz. 1999. The inhibitory activity of the AU-rich RNA element in the human papillomavirus type 1 late 3′ untranslated region correlates with its affinity for the elav-like HuR protein. J. Virol. 73:1080-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takagaki, Y., L. C. Ryner, and J. L. Manley. 1989. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev. 3:1711-1724. [DOI] [PubMed] [Google Scholar]
- 43.Takagaki, Y., J. L. Manley, C. C. MacDonald, J. Wilusz, and T. Schenk. 1990. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 4:2112-2120. [DOI] [PubMed] [Google Scholar]
- 44.Takagaki, Y., and J. L. Manley. 1997. RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol. 17:3907-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan, W., B. K. Felber, A. S. Zolotukhin, G. N. Pavlakis, and S. Schwartz. 1995. Efficient expression of the human papillomavirus type 16 L1 protein in epithelial cells by using Rev and the Rev-responsive element of human immunodeficiency virus or the cis-acting transactivation element of simian retrovirus type 1. J. Virol 69:5607-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan, W., and S. Schwartz. 1995. The Rev protein of human immunodeficiency virus type 1 counteracts the effect of an AU-rich negative element in the human papillomavirus type 1 late 3′ untranslated region. J. Virol. 69:2932-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terhune, S. S., C. Milcarek, and L. A. Laimins. 1999. Regulation of human papillomavirus type 31 polyadenylation during the differentiation-dependent life cycle. J. Virol. 73:7185-7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terhune, S. S., W. G. Hubert, J. T. Thomas, and L. A. Laimins. 2001. Early polyadenylation signals of human papillomavirus type 31 negatively regulate capsid gene expression. J. Virol. 75:8147-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vagner, S., C. Vagner, and I. W. Mattaj. 2000. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 14:403-413. [PMC free article] [PubMed] [Google Scholar]
- 50.Wahle, E., and W. Keller. 1994. 3′ end-processing of mRNA, p. 1-34. In S. J. Higgins and B. D. Hames (ed.), RNA processing—a practical approach. Oxford University Press, New York, N.Y.
- 51.Yonaha, M., and N. J. Proudfoot. 2000. Transcriptional termination and coupled polyadenylation in vitro. EMBO J. 19:3770-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.zur Hausen, H. 1989. Papillomaviruses in anogenital cancer as a model to understand the role of viruses in human cancers. Cancer Res. 49:4677-4681. [PubMed] [Google Scholar]