Abstract
The invasion strategy of many viruses involves the synthesis of viral gene products that mimic the functions of the cellular proteins and thus interfere with the key cellular processes. Here we show that adenovirus infection is accompanied by an increased ubiquitin-cleaving (deubiquitinating) activity in the host cells. Affinity chromatography on ubiquitin aldehyde (Ubal), which was designed to identify the deubiquitinating proteases, revealed the presence of adenovirus L3 23K proteinase (Avp) in the eluate from adenovirus-infected cells. This proteinase is known to be necessary for the processing of viral precursor proteins during virion maturation. We show here that in vivo Avp deubiquitinates a number of cellular proteins. Analysis of the substrate specificity of Avp in vitro demonstrated that the protein deubiquitination by this enzyme could be as efficient as proteolytic processing of viral proteins. The structural model of the Ubal-Avp interaction revealed some similarity between S1-S4 substrate binding sites of Avp and ubiquitin hydrolases. These results may reflect the acquisition of an advantageous property by adenovirus and may indicate the importance of ubiquitin pathways in viral infection.
Selective protein degradation plays an essential role in a number of key cellular processes, including cell cycle progression, transcription, and intracellular signaling. The major cytoplasmic pathway used by eukaryotic cells is ubiquitin-dependent proteolysis, which involves the covalent addition of a polyubiquitin chain to target protein in a cascade of enzymatic reactions, catalyzed by the E1 ubiquitin-activating enzyme, the E2 ubiquitin-conjugating enzymes, and the E3 ubiquitin-protein ligases (13). Conjugation of ubiquitin to specific proteins is regulated by a complex interplay between the E2s, the E3s, and deubiquitinating enzymes (DUBs). While the substrate specificity is conferred by the E3s, the reversibility of ubiquitination is provided by the DUBs (7, 42, 43).
DUBs have been shown to be involved in various biological processes including cell growth, differentiation, and oncogenesis (7, 42, 43). Although specific targets for DUBs in these processes are often unknown, the data suggest that some DUBs can regulate gene expression by acting on the regulators of transcription or on chromatin structure. The DUBs, which are positive regulators of proteolysis, like Doa4/tre-2 and Ubp14/IsoT subfamilies, function downstream of the proteasome by processing poly-ubiquitinated products. This protects the inhibition of the 26S proteasome by free polyubiquitin chains. The negative regulators such as Uch2p/UCH37 act upstream of the proteasome by trimming ubiquitin from ubiquitinated substrates and thus preventing their degradation.
Here we show that adenovirus (Ad) infection is accompanied by an increased deubiquitinating activity in infected cells. Affinity chromatography on ubiquitin aldehyde (Ubal), which was designed to purify the deubiquitinating proteases, revealed the presence of Ad L3 23K proteinase (Avp) in the eluate from Ad type 2 (Ad2)-infected cells. We demonstrate further that Avp can function as a DUB in vitro and in vivo.
MATERIALS AND METHODS
Cell infection and transfection.
HeLa cells were cultured in Eagle’s minimal essential medium (EMEM) with Earle's balanced salt solution and 25 mM HEPES supplemented with 10% fetal calf serum (FCS), 2 mM glutamine, streptomycin (100 μg/ml), and penicillin (100 U/ml) in a humidified atmosphere at 37°C with 5% CO2. The cells were grown on 100-mm-diameter dishes (Falcon) to 70 to 80% confluence and infected with wild-type Ad2 (multiplicity of infection = 100) in 1 ml of EMEM without FCS. After 1 h at 37°C the medium was exchanged on EMEM with 10% FCS (supplemented with 100 μM LaggH [z-Leu-Ala-Gly-NH-CH2-CHO] where indicated). Then, at different times postinfection (p.i.), cells were washed with phosphate-buffered saline plus 1 mM MgCl2, scraped, and analyzed. The same conditions were used for the infection with the Ad2ts1 mutant (a gift from M. Rosa) except that the temperature was 39.5°C throughout. The transfection of HeLa cells was performed on six-well plates (Falcon) with FuGENE 6 according to manufacturer instructions (Roche). Generally, 1 μg of expression vector (0.5 μg plus 0.5 μg for double transfection) per well was used, and the cells were analyzed 24 h after transfection.
Cell fractionation and Western blotting.
Harvested HeLa cells (∼5 × 106 cells) were pelleted by centrifugation at 600 × g for 10 min at 4°C and subjected to one of the three different treatments. First, the cell pellet was extracted at 4°C with 200 μl of cell lysis buffer (CLB) (20 mM EDTA, 0.8% sodium dodecyl sulfate [SDS], 20 mM N-ethylmalcimide (NEM), 100 mM Tris [pH 7.4], supplemented with a protease inhibitor cocktail [PIC; Roche]) and centrifuged at 13,000 × g for 10 min, and supernatant was used as a whole-cell extract. Second, the nuclei were prepared by osmotic swelling of the cells in 0.5 ml of RSB buffer (10 mM NaCl, 1.5 mM MgCl2, 10 mM Tris [pH 7.4], supplemented with 1 mM NEM and PIC) for 10 min at 4°C. Swollen cells were disrupted with Dounce-type homogenizer, and the nuclei were recovered in the pellet by centrifugation at 1,000 × g for 3 min. Nuclei were lysed in 100 μl of CLB to give nuclear extract. Third, for the preparation of acid-soluble nuclear extract (AS), the pellet of nuclei obtained as described above was resuspended in 50 μl of RSB plus 0.1% NP-40, mixed with 50 μl of 0.8 N H2SO4, and incubated on ice for 30 min. The AS fraction (∼100 μl) was collected following centrifugation at 13,000 × g for 15 min, and the pH was adjusted to 8 by addition of 4 N NaOH (∼9 μl). Protein concentrations were determined using the Micro BCA Protein Assay (Bio-Rad).
Transfected cells were lysed in urea buffer (UB) (150 μl/well; 8 M urea, 20 mM NEM, 100 mM Tris [pH 8.0]) and analyzed. Double-transfected cells with pMT107 (octa-His6Ub [36]) vector were lysed in UB (500 μl/well) plus 5 mM imidazole (Im). The lysate was mixed on a rotator with 50 μl of Ni-nitrilotriacetic acid (Ni-NTA) resin for 30 min at 4°C. The slurry was applied to a GenElute column (Sigma) and centrifuged on the table spinner. The resin was successively washed four times with 500 μl of UB plus 5 mM Im, and His6-Ub conjugates were eluted with 100 μl (2× 50 μl of UB containing 500 mM Im.
The antibodies used for Western blotting were affinity-purified antiubiquitin antibody (11), anti-uH2A immunoglobulin M (IgM) monoclonal antibody (MAb) E6C5 (a gift from J. E. Celis), anti-β-tubulin IgG1 MAb TUB 2.1 (Sigma), affinity-purified anti-ISG15 antibody (32), and anti-GST IgG1 MAb B-14 (Santa Cruz Biotechnology). The rabbit anti-Avp polyclonal antiserum was raised against recombinant Avp.
Assay for cellular DUB activity.
The S-100 cell lysate was prepared from infected and noninfected HeLa cells (∼2 × 107) as follows. The cells were harvested in phosphate-buffered saline containing 1 mM MgCl2 and centrifuged at 600 × g for 10 min at 4°C. The cell pellet was suspended in 0.5 ml of ice-cold suspension buffer S (20 mM HEPES-K [pH 7.5], 10 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol [DTT], 0.5 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, 0.5% NP-40, DNase I [50 μg/ml], RNase A [DNase I], 4% glycerol) and incubated on ice for 20 min. The suspension was then subjected to three 5-s bursts of sonication, and centrifuged at 100,000 × g for 30 min in a Beckman TLA-100.4 rotor. The resulting supernatant (S-100 cell lysate) was used for the in vitro DUB assay as follows. The cell lysate was incubated with a 100 μM concentration of fluorogenic substrate zLRGG-AMC (diluted from 50 mM stock solution in dimethyl sulfoxide [DMSO]; Bachem) for 1 h at 37°C. The fluorescence of liberated AMC was measured at 345 nm and 440 nm with an Aminco Bowman S2 luminescence spectrometer.
Affinity chromatography with biot-Ubal.
The biot-Ubal was synthesized from C-terminal Ub76-diol, which has been prepared from bovine ubiquitin (Sigma) by carboxypeptidase Y-catalyzed G76 transpeptidation with 1-amino-2,3-propanediol (18; R. Cohen personal communication). The biotinylated biot-Ub-diol was prepared by acylation of ∼0.12 mM Ub-diol with 0.6 mM NHS-LC-Biotin (diluted from freshly prepared 100 mM solution in DMSO; Pierce) in 100 mM NaCl-25 mM HEPES-K, pH 7.5. After 3 h at room temperature the reaction was quenched by addition of 2-aminoethanol to 50 mM and dialyzed briefly (∼2 h) against 100 mM NaOAc, pH 5.0. The biot-Ub-diol was then purified by gel filtration on a BIO-GEL P-4 column (1 by 15 cm; Bio-Rad) equilibrated with 100 mM NaOAc, pH 5.0, and stored at −80°C. The biot-BSA was synthesized similarly. The protein biotinylation was confirmed by Western blot analysis using peroxidase-conjugated streptavidin (Sigma). The biot-Ubal was prepared by oxidation of ∼0.1 mM of biot-Ub-diol with 1 mM NaJO4 (sodium periodate, diluted from freshly prepared 100 mM solution) for 4 h at 4°C in the dark. The reaction was quenched with 10% glycerol for 30 min at 4°C, and the resulting solution of biot-Ubal was used the same day.
The S-100 cell lysate was prepared as described above except that the procedure was scaled up to ∼109 HeLa cells and 5 ml of buffer S. Prior to addition of biot-Ubal, the lysate (2 ml) was precleared by rocking with 100 μl of streptavidin-agarose (Sigma) for 30 min at 4°C. This helps to reduce the background of nonspecific proteins. The resin was then removed and 50 μl of biot-Ubal (or biot-BSA in the control) was added. After incubation at 4°C for 1 h, the lysate was mixed on a rotator with 100 μl of streptavidin-agarose for another 1 h. The slurry was then transferred to a small Poly-Prep column (Bio-Rad) and washed successively with: the following: 4 ml of buffer S− (buffer S without DNase I and RNase A); 2 ml of buffer S− with 500 mM NaCl; 0.5 ml of buffer S− containing ubiquitin (1 mg/ml); and 150 μl of buffer UB with 100 mM DTT. The agarose beads were finally extracted with SDS-loading buffer. Each fraction was then analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie blue staining. The specific proteins eluted with UB-DTT buffer were sequenced by mass spectrometry.
Mass spectrometry.
Protein sequencing using mass spectrometry (MS) was carried out as described previously (34). Briefly, the Coomassie blue-stained protein band was in-gel digested with trypsin, and the recovered peptides were analyzed with a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Voyager Elite XI; Perseptive Biosystem). The resulting peptide mass fingerprint was used for protein identification using two programs: MS-FIT (http://prospector.ucsf.edu/ucsfhtml3.2/msfit.htm) and PROFOUND (http://129.85.19.192/prowl-cgi/ProFound.exe). The protein bands that could not be identified by MALDI-MS were subjected to the nanospray MS/MS analysis with a Q-Tof instrument (Micromass, Manchester). The MS/MS spectra were transformed using MaxEnt3 (MassLynx; Micromass) and the amino acid sequences were analyzed using PepSeq (BioLynx; Micromass). Data derived from MS/MS spectra were used to search a protein database for homologous sequences (http://www.ncbi.nlm.nih.gov/blast/blast).
Synthesis of Avp inhibitors (LxggH).
The peptide-aldehydes with a general structure z-Leu-X-Gly-NH-CH2-CHO [LxggH, x= A, E, K(Me)2, where Me is methyl] were synthesized by the reduction of the corresponding peptide ethyl esters with diisobutyl aluminum hydride (DIBAL) as described previously (22). The z-Leu-Ala-Gly-Gly-OEt (where Et is ethyl) was prepared by conventional solution phase condensation of z-Leu-Ala with Gly-Gly-OEt×. Briefly, a solution of z-Leu-Ala (170 mg, 0.5 mmol), Gly-Gly-OEt × HCl (80 mg, 0.5 mmol), triethylamine (40 μl, 0.5 mmol) and the coupling agents N,N′-dicyclohexylcarbodiimide plus HOBT (0.6 mmol each) in DMSO was stirred overnight at room temperature. Then, 10 ml of CHCl3 was added and the organic layer was successively washed with 0.1 N HCl and saturated NaHCO3, dried over MgSO4, and evaporated to dryness. The residue was chromatographed on a silica gel column (2 by 30 cm; CHCl3:MeOH, 19:1) to give the pure z-Leu-Ala-Gly-Gly-OEt (200 mg, ∼87% yield). This ester (150 mg, 0.3 mmol) was dissolved in 10 ml of anhydrous CH2Cl2 at −78°C, and 2 ml of DIBAL (1 M solution in CH2Cl2) was added dropwise with a continuous stirring over a period of 15 min. The reaction mixture was stirred for 1 h at −78°C under an argon atmosphere and quenched by adding 10 ml of cold (−78°C) MeOH. The resulting emulsion was then poured into 50 ml of ice-cold 1 N HCl with vigorous stirring, and extracted with ethyl acetate. Combined organic fraction was dried over MgSO4, and evaporated to dryness. The residue was chromatographed on a semipreparative 2,2 × 25-cm RP-HPLC-C18 column (Vydac) using a linear 5 to 60% acetonitrile gradient in 1% aqueous trifluoroacetic acid. The fractions containing LaggH were identified by MALDI-MS (m/z = 435) and evaporated to give 28 mg of peptide-aldehyde (∼21% yield). The peptide-aldehydes LxggH [x = E, K(Me)2] were synthesized by a similar procedure with some modifications. Experimental details for synthesis as well as the study on the effect of these inhibitors on Ad infection will be published elsewhere. The reagents for peptide synthesis were purchased from Bachem and Aldrich.
Plasmid construction and purification of proteins.
The expression and purification of Avp by using the plasmid pT7AD23K6 (a gift from C. W. Anderson) was performed as described previously (1, 25). For the construction of pAvp and pAvp-pVIc vectors, the Avp cDNAs were amplified by PCR from pT7AD23K6 plasmid, and cloned into the BamHI/HindIII sites of mammalian expression vector pXJ41 under the control of cytomegalovirus promoter (a gift from M. Rosa). In the case of pAvp-pVIc, the downstream primer 5′-AATAAGCTTAAAAGCAACGACGACGTTTAAGAGACTGAACCCCGAGACCAACCATGTTTTTCAAGTGACA-3′ contained the sequence coding for proteinase cleavage site and peptide activator pVIc (in boldface type; the cloning sites are underlined). The C122G active-site mutants were prepared for all Avp constructs by oligonucleotide-directed mutagenesis using PCR with Pfu DNA polymerase (QuikChange; Stratagene). The primers used were 5′-CCCAACTCGGCCGCCGGTGGACTATTCTGCTGC-3′ and complementary oligonucleotide (point mutation is shown in boldface type). For the expression of His6-tagged Ub in HeLa cells we used the plasmid pMT107 coding for octa-His6Ub under control of cytomegalovirus promoter (a gift from D. Bohmann [36]). The vectors for bacterial expression of the Avp substrates His6Ub-GST and His6Ub-pVIc-GST were prepared in two steps. First, the His6Ub and His6Ub-pVIc sequences were generated from pMT107 by PCR of and were cloned into NcoI/BamHI sites of pET-11d vector (Novagen). The primers used were 5′-TTACCATGGGACACCATCACCATCACCATATGCAGATCTTCGTGAA GG-3′ (for both sequences) and 5′-CATGGATCCACCTCTGAGACGGAGTAC-3′ (for His6-Ub) or 5′-CATGGATCCTCTCTTGAGTGACTGTACACCGAGACCTACGATACCTCTGAGACGGAGTAC-3′ (for His6Ub-pVIc), which codes for the IVGL↓G cleavage site and a part of peptide pVIc sequences (in bold). Next, the GST sequence was amplified by PCR of pGEX-4T (AP Biotech) and was cloned into BamHI/EcoRI sites of pHis6Ub and pHis6Ub-pVIc vectors. The GST sequence was mutated before amplification in order to delete the natural Avp cleavage site (ML83GG→ ML83RG, QuikChange; Stratagene). The primers used were 5′-GCTGACAAGCACATGTTGCGTGGTTGTCCAAAAGAGC-3′ and the complementary oligonucleotide. For protein preparation, the pHis6Ub-GST and pHis6Ub-pVIc-GST constructs were transformed into Escherichia coli BL21lysS (Novagen). The bacteria were grown in LB-amp medium to A600∼0.4 and protein expression was induced with 0.4 mM of IPTG. After incubation at 37°C for 3 h, the bacterial lysate was prepared as described previously (25), and Im was added to 5 mM concentration. The lysate from a 1-liter culture (50 ml) was loaded on the 4 ml Ni-NTA column (Qiagen). The column was washed with 25 ml of lysis buffer (50 mM Tris, pH 8.0, 0.05% [vol/vol] Triton X-100, 15 mM NaCl, 1 mM DTT) with 5 mM Im, and the fusion proteins were eluted by stepwise addition of 5 to 10 ml of lysis buffer containing different concentration of Im (25 mM → 500 mM). The proteins (purity ∼80%) are usually eluted at 50 to 150 mM of Im. The selected fractions were pooled and subjected to the second purification on unoS cation-exchange column (Bio-Rad) resulting in the pure His6Ub-GST and pHis6Ub-pVIc-GST fusion proteins. The expression and purification of ISG15 and pro-ISG15 were performed as described previously (32) The USP8 (UBPY) construct was a gift from C. F. Draetta, the recombinant Ulp1 and Smt3-GFP proteins were a gift from C. D. Lima.
In vitro proteolysis assay.
Assays for recombinant Avp activity were carried out at 37°C in a 100 μl of assay buffer (10 mM octyl glucoside, 1 mM EDTA, 1 mM DTT, and 20 mM Tris [pH 8]) and contained protein substrate (0.1 to 1 μM), Avp (10 to 100 nM), and pVIc peptide (1 μM). The pVIc peptide (GVQSLKRRRCF) was synthesized using Fmoc chemistry. Tetra-Ub was purchased from Affiniti, Inc. To study the effect of the inhibitors, Avp was pretreated with LaggH (1 μM) or Ubal (1 μM) for 5 min at 4°C. For SDS-PAGE, 10-μl aliquots of proteolysis mixture were removed at appropriate intervals and quenched by addition of 1 μl of 0.5 M NEM in DMSO.
Cleavage of zLRGG-AMC by recombinant Avp was measured by liberation of fluorescent AMC at 345 nm and 440 nm. The composition of assay mixture was as described above with 20 nM Avp and 50 μM zLRGG-AMC. This fluorogenic substrate was also used for determination of Ki of the synthesized inhibitors by the Dixon method. Generally, three different substrate concentrations and six different inhibitor concentrations were used. Cleavage of the proteins by recombinant Avp was analyzed by Western blotting.
For chromatographic analysis of Avp species, 30 μg of Avp was incubated in 200 μl of assay buffer in the presence or absence of 100 μM pVIc for 15 min at 4°C. To obtain Ubal-Avp-pVIc ternary complex, 30 μg of Ubal was added and incubated for another 15 min at 4°C. The mixture was then analyzed on unoS cation-exchange column using a BioLogic FPLC system (Bio-Rad).
Structure analysis.
Analysis of 3D structures of the peptidases was performed using programs from the CCP4 suite (4) as described in the legend to Fig. 5. Insight II program was used for the construction of molecular surface of the substrate-binding cleft of Avp.
RESULTS
DUB activity in Ad-infected cells.
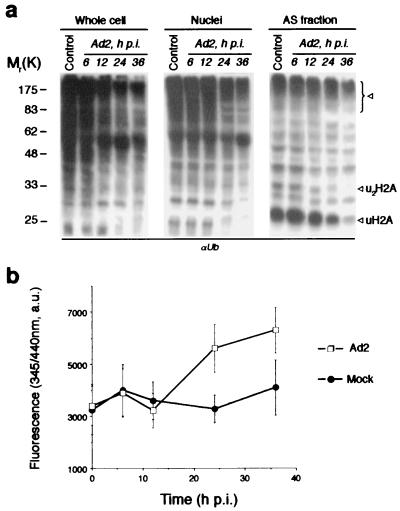
Ubiquitin-dependent protein degradation by proteasomes is an important pathway of major histocompatibility complex class I antigen presentation and antivirus defense. Several viruses have elaborate mechanisms to block this pathway (31). We asked whether Ad infection might result in a down-regulation of ubiquitin-dependent proteolysis that would be reflected by a change in the amount of the Ub conjugates in the cell. To test this, HeLa cells were infected with wild-type human Ad2, harvested at different times p.i., and fractionated into cytosolic, nuclear, and AS nuclear fractions. The same amount of cellular proteins from each fraction was resolved by SDS-PAGE and Ub conjugates were analyzed by Western blotting with αUb antibodies (Fig. 1a). A nearly complete disappearance of mono- and di-ubiquitinated histone H2A (see uH2A, ∼23K band reactive also with anti-uH2A-specific MAb, and u2H2A, ∼29K band, Fig. 1a) as well as a significant decline of high-molecular weight Ub conjugates was observed in infected cells. This effect was observed at the late phase of Ad2 infection (>20 h p.i.) and was more pronounced in nuclei and AS fractions enriched with basic nuclear proteins. We reasoned that the effect of Ad2 infection on protein ubiquitination could result from different causes such as inhibition of ubiquitin conjugation induced by viral shutoff of host protein synthesis or/and induction of DUB activity in infected cells. To discriminate between these alternatives, we measured the DUB activity using the fluorogenic substrate zLRGG-AMC, cleavable by different types of DUBs. Ad2 infection resulted in a significant (∼45%) increase in intracellular DUB activity as seen by an increased substrate cleavage and a liberation of fluorescent AMC (Fig. 1b). The DUB activity and protein deubiquitination followed a similar time course (Fig. 1), suggesting that activation of DUB may contribute to the observed decline in Ub conjugate pool during infection.
FIG. 1.
Deubiquitinating activity in Ad-infected cells. (a) Decline of the Ub conjugates pool in Ad2-infected cells. HeLa cells were infected with Ad2 (multiplicity of infection = 100), whereupon cells were harvested at the indicated time p.i. and whole cell, nuclear, and AS nuclear extracts were prepared. The same amount of the protein from each sample was resolved by SDS-PAGE and analyzed by Western blotting with anti-Ub antibodies. Lane control, extracts from mock-infected cells. Arrows indicate the Ub conjugates significantly diminished during Ad2 infection. The uH2A band was also recognized by anti-uH2A MAb E6C5 (not shown). (b) Ad2-infected cells demonstrate increased DUB activity. Whole cell lysates from Ad2- and mock-infected cells were incubated with zLRGG-AMC fluorogenic substrate (100 μM, 1 h) and the fluorescence of liberated AMC was measured at 345 nm/440 nm. Values are means ± standard deviations (error bars) from three separate experiments
Ubal-affinity chromatography.
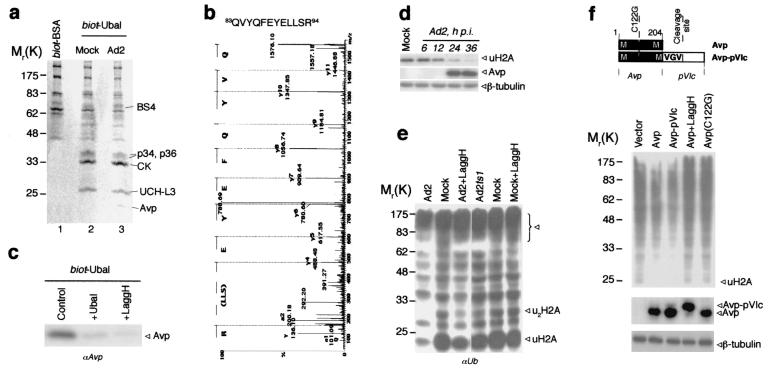
We then attempted to isolate the proteins responsible for the deubiquitinating activity from mock- and Ad2-infected cells using inhibitor affinity chromatography. Ubal has been shown to be a highly specific reversible inhibitor for several classes of DUBs (18, 20, 42). Therefore, we synthesized Ubal labeled with biotin on the ɛ-amino group of lysine residues (biot-Ubal). The cell extract (∼109 cells) was incubated with biot-Ubal (or with biot-BSA as a control) and was loaded on a streptavidin-agarose column. Bound proteins were eluted using DTT-urea, which induced the hydrolysis of thio-hemiacetal bond formed between Ubal and the cysteines of attached proteins. The proteins were then resolved by SDS-PAGE and stained with Coomassie blue (Fig. 2a). Protein pattern obtained with biot-BSA was different from ones obtained with biot-Ubal. Both mock- and Ad2-infected cells demonstrated similar patterns of Ubal-binding proteins, except for a band with a molecular weight of ∼23,000 (∼23K) in eluate from infected cells. The proteins eluted from biot-Ubal column were then sequenced by mass spectrometry.
FIG. 2.
Identification of Ad proteinase Avp as a DUB. (a) The proteins purified by Ubal-affinity chromatography. Whole cell lysates were incubated with biot-BSA or biot-Ubal (20 ng/mg of cell protein, 1 h, 4°C), and loaded on streptavidin agarose column. After extensive washing the bound proteins were eluted with DTT-urea. The eluates from Ad2 (line 1, 3) and mock-infected (line 2) cells were resolved by SDS-PAGE and stained with Coomassie blue. The proteins identified by mass spectrometry are shown. (b) The MS/MS spectrum of Avp-derived peptide obtained from 23K band (Fig. 2a, line 3). (c) Competition of Ubal and LaggH for Avp retrieval on biot-Ubal/streptavidin column. The conditions were as for Fig. 2a, but 10 times less cell lysate was used (control). Ubal (fivefold excess over biot-Ubal) or LaggH (100 μM) was added in the incubation buffer. Western blot from eluates was probed with anti-Avp antiserum. (d) Simultaneous analysis of uH2A and Avp in Ad2-infected cells. Whole cell lysates were prepared at the indicated time p.i., and analyzed by Western blotting with anti-uH2A MAb E6C5 and anti-Avp antiserum (probing with anti-β-tubulin IgG served a control). (e) Contribution of Avp activity to protein deubiquitination during Ad infection. Cells were infected with Ad2, Adts1, or Ad2 in the presence of LaggH (100 μM) for 24 h at nonpermissive temperature (39.5°C). The same amount of AS protein was analyzed by Western blotting with anti-Ub antibody. (f) Deubiquitinating activity of Avp in vivo. HeLa cells were cotransfected with His6-tagged ubiquitin and either control empty vector, or the vector with wild-type proteinase Avp, Avp-pVIc fusion, or inactive mutant Avp(C122G). In Avp-pVIc construct the pVIc activating peptide GVQSLKRRRCF is separated from proteinase sequence by MVGV↓G Avp cleavage site. After 24 h of incubation in the presence or absence of 100 μM LaggH, cells were lysed in 8 M urea, and His6Ub conjugates were isolated by using Ni-NTA resin and analyzed by Western blotting with anti-Ub antibody. The same cell lysates were also analyzed for Avp expression with anti-Avp antiserum (probing with anti-β-tubulin IgG served as a control).
A 26K band was identified as ubiquitin C-terminal hydrolase-L3 (UCH-L3). Two bands at 34K and 36K derived from the same protein that relates distantly to cysteine peptidases and contains a putative ubiquitin-associated (UBA) domain (unpublished data). A band at ∼65K appeared to be BS4, a protein with unknown function, which contains three different UBA domains. The UBA domain is a common motif in proteins of the ubiquitination pathway (15). Although the specific role of this domain is not yet determined, its recovery on the Ubal column suggests that it can interact directly with ubiquitin. A recent report that UBA domains of DNA damage-inducible proteins interact with ubiquitin, supports this conclusion (3). Interestingly, the sequence analysis of BS4 revealed also the presence of the ubiquitin homology (UbH) domain (I85-S161; 22% identity and 66% similarity to ubiquitin) with conserved residues in the positions corresponding to K48, K29, L8, I44, and V70 of ubiquitin, which are implicated in polyubiquitin chain formation and proteolytic targeting. The 30K band yielded the peptides matching cytokeratin consensus sequences. This last finding may reflect a possible interaction between ubiquitin and intracellular filaments that have been fixed by Ub-aldehyde cross-linking. The association of ubiquitin with intermediate filaments has already been reported (26).
Surprisingly, only one bona fide DUB, UCH-L3, was obtained from Ubal column. The inability to retrieve other DUBs on the Ubal column, even though some of them are known to be inhibited by Ubal, might stem from the relative instability of thio-hemiacetal bond as well as from the fact that they can be sequestered in the multimolecular complexes (20).
Deubiquitinating activity of Ad protease (Avp) in vivo.
The 23K protein that was present only in the eluate from Ad-infected cells was identified by mass spectrometry as Ad2-coded proteinase, Avp (Fig. 2b). This cysteine proteinase processes several Ad proteins and this processing is essential for the formation of mature infectious virions (39). There are about 10 copies of Avp in Ad virion (1) and in virus-infected cells Avp is recovered in very small amounts among other viral structural proteins (unpublished results). Nevertheless, its enrichment on Ubal column was sufficient to visualize the protein upon Coomassie blue staining (Fig. 2a).
The substrate specificity of Avp defined as (M,L,I)XGG↓X and (M,L,I)XGX↓G (39, 40) overlaps the DUB specificity, LRGG↓X. We therefore asked whether Avp contributes to the protein deubiquitination observed in infected cells. For this we synthesized a panel of peptide-aldehyde inhibitors of Avp having the general structure z-Leu-X-Gly-NH-CH2-CHO. These compounds inhibited recombinant Avp (Kis < 2 μM) and blocked the processing of viral proteins in infected cells (data not shown). Cell-permeable inhibitor LaggH (X = A, Ki ∼ 0.5 μM), which did not interfere with cellular DUB activity (Fig. 2e and unpublished results), was used throughout this study.
Both LaggH and nonbiotinylated Ubal were used to compete for the retrieval of proteins from Ad2-infected cells on biot-Ubal column. Pretreatment of the cell lysate with nonbiotinylated Ubal or LaggH before incubation with biot-Ubal markedly reduced the retrieval of Avp on the column (Fig. 2c). This result indicates that the proteinase binding to the affinity column requires the interaction of biot-Ubal with Avp active site. The expression of Avp, coded by the late L3 region of the Ad genome, was observed at ∼18 to 20 h p.i., which coincided with the decline of cellular ubiquitinated histone uH2A (Fig. 2d). We therefore tested whether deubiquitination of nuclear proteins results from Avp proteolytic activity or from the virus-mediated shutoff of host protein synthesis. To uncouple the two effects, Ad2ts1 virus was exploited. This virus, which codes for the ts mutant Avp(P137L) inactive at the restrictive temperature (39.5°C), can still inhibit host protein synthesis (5, 45). Cells were infected at the nonpermissive temperature and ubiquitinated proteins were assayed in AS nuclear fraction at 24 h p.i, using the same amount of total protein. Infection with Ad2ts1 did not change significantly the cellular ubiquitination pattern (Fig. 2e). Although, some decline in Ub conjugates could be observed in Ad2ts1-infected cells after 36 h p.i, the effect was much less pronounced than for the wild-type Ad2. Treatment of Ad2-infected cells with specific Avp inhibitor LaggH counteracted significantly the protein deubiquitination (Fig. 2e). LaggH did not change the ubiquitination pattern in mock-infected cells (Fig. 2e) suggesting that the inhibitor does not act on cellular DUB. It should be noted that we were unable to perform these experiments at the 36 -h time point when Ad2-induced deubiquitination activity is the strongest (see Fig. 1) since the prolonged treatment of infected cells with LaggH detached cells. Collectively, these results are consistent with our hypothesis that Avp can contribute to the increase in intracellular DUB activity during Ad2 infection.
To find out whether Avp can deubiquitinate cellular proteins independently both of other viral products and of infectious cycle, we transfected the cells with plasmids that code for different variants of the proteinase, including active site cysteine (C122G) mutants (Fig. 2f). It has been shown that recombinant Avp requires as a cofactor pVIc, an 11-amino acid peptide cleaved by Avp from the C terminus of viral protein pVI (24, 41). We therefore made a construct that codes for Avp-pVIc fusion proteins encompassing MVGV↓G cleavage site necessary for proteinase autoactivation (Fig. 2f). All vectors provided high, comparable levels of protein expression in HeLa cells, with Avp-pVIc being self-cleaved (Fig. 2f). The cells were cotransfected with Avp plasmids and pHT109 plasmid encoding His6-tagged ubiquitin (36). After 24 h ubiquitinated proteins were isolated on Ni-NTA resin, resolved by SDS-PAGE, and analyzed by Western blotting with αUb antibodies (Fig. 2f). Transfection of cells with Avp or Avp-pVIc reduced the amount of recovered Ub conjugates, whereas LaggH predictably suppressed this effect. The treatment with LaggH blocks also, as expected, the processing of Avp-pVIc fusion (Fig. 2f). Transfection with inactive Avp(C122G) mutant did not affect the level of cellular ubiquitination. The Avp-pVIc construct consistently induced protein deubiquitination somewhat stronger than did the Avp construct, possibly due to increased proteinase activity of this protein. Together, these findings demonstrate that Avp can function as a DUB in vivo.
Substrate specificity of Avp in vitro.
Although for some DUBs a minimal substrate has been shown to be ubiquitin C-terminal peptide LRGG↓X (35), the majority of DUBs require the presence of the entire ubiquitin moiety for efficient cleavage. In addition, the nature of Ub-X bond and the size of X extension often restrict the specificity of DUBs (7, 42, 43). More than a dozen different viral sequences are cleaved by Avp during Ad2 maturation suggesting that Avp has lower specificity than DUBs. Accordingly, we predicted that Avp could trim other ubiquitin-like proteins, provided they contain a canonical C-terminal LRGG sequence. To examine the specificity of Avp toward Ub-related substrates we used an in vitro assay. A recombinant Avp was purified from E. coli as described previously (1, 25), and incubated with native-like or model substrates (Fig. 3a and c). The kinetics of substrate cleavage was then analyzed by Western blotting. Avp was able to hydrolyze the G76→Nɛ-K48 isopeptide bond of tetra-ubiquitin (tetra-Ub), which is the linkage found naturally in the polyUb chains formed on proteolytic substrates. Furthermore, we attempted to ascertain in vitro whether Avp can process the peptide bond of ISG15 precursor protein (pro-ISG15), the product of ISG15 gene that has an 8 amino acid carboxy-terminal extension downstream of the LRGG sequence (32). ISG15, also known as ubiquitin cross-reactive protein (UCRP), contains two ubiquitin-like domains and differs significantly from ubiquitin-derived substrates. We observed that Avp cleaves pro-ISG15 at least as efficiently as tetra-Ub (Fig. 3a). These data suggest that Avp substrate specificity is defined mainly by active site cleft and by substrate amino acids in P1-P4 positions.
FIG. 3.
Substrate specificity of Avp. (a) Cleavage of tetra-Ub and pro-ISG15 by recombinant Avp in the presence of pVIc peptide. Reactions were stopped at different times with NEM (50 mM) and analyzed by Western blotting with rabbit anti-Ub and anti-ISG15 antibodies. Where indicated, Avp was pretreated with the inhibitors LaggH (1 μM) or Ubal (1 μM) for 5 min at 4°C. In one case the activating peptide pVIc was omitted from reaction mixture. (b) Chromatographic analysis of Avp species on unoS column: Avp alone (I), Avp incubated with pVIc for 15 min (II), and Avp incubated with pVIc and Ubal for 30 min (III). (c) Cleavage of Ub-GST and Ub-pVIc-GST fusion proteins by Avp. The reaction conditions were as for Fig. 3a. The Avp cleavage sites are indicated in bold, whereas the pVIc-derived sequence is in italics. The anti-GST MAb B-14 and rabbit anti-Ub antibody were used for Western blotting.
Formation of Avp complexes with activating peptide and Ubal was demonstrated by chromatographic analysis using strong cation-exchange resin. For both Ub- and ISG15-derived substrates, the cleavage by recombinant Avp required the presence of peptide cofactor pVIc, and was inhibited by LaggH (Ki < 0.5 μM) and Ubal (Ki ≈ 1 μM). The activation of Avp (retention time [r.t.] ≈ 8 min) with positively charged pVIc peptide involves the formation of Avp-pVIc complex (r.t. ≈ 14 min) (Fig. 3b). On the other hand, the inhibition of the proteolysis by Ubal (r.t. ≈ 4 min for Ubal alone) results from the trapping of Avp-pVIc as a thio-hemiacetal compound in the ternary Ubal-Avp-pVIc complex (r.t. ≈ 13 min). The composition of Avp complexes was confirmed by Western blotting and mass spectrometry (not shown).
We next compared the deubiquitinating activity of Avp with its processing activity toward viral proteins. To retain the same structural context we used two model substrates: Ub-GST fusion protein (for DUB activity) and Ub-pVI-GST protein (for processing activity), consisting of ubiquitin (amino acids 1 to 75) and GST joined by an 11-residue linker derived from viral pVI protein (Fig. 3c). The LRGG cleavage site has been replaced in the linker by the viral IVGV↓G cleavage site, essential for the formation of pVIc cofactor peptide and for the virus maturation. This sequence is one of the most readily cleavable by Avp during Ad infection and represents therefore the optimal reference substrate for Avp processing. We noted that some cleavage of Ub-pVI-GST was observed even without pVIc peptide (Fig. 3c, right side), indicating that the part of cofactor sequence contained in the substrate can act in cis to catalyze the substrate processing by Avp. Avp cleaves both recognition sequences, viral as well as ubiquitin C-terminal, with similar efficacy. Interestingly, even though Ub-pVI-GST was a better substrate than Ub-GST, the rate of Ub-GST cleavage by Avp was similar, supporting the idea that the protein deubiquitination by Avp may be as efficient as processing of viral proteins.
Structural basis for deubiquitinating activity of Avp.
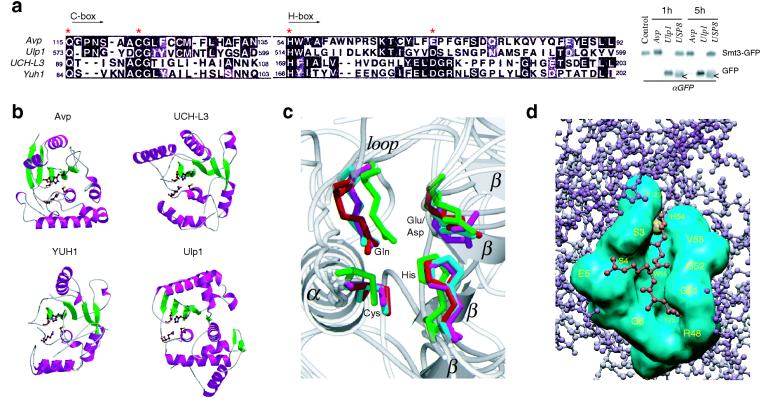
Because our results demonstrate that Avp functions as a DUB, we examined whether there is structural similarity between Avp and one of the known DUBs (Fig. 4a). Although the comparison of the catalytic core residues reveals some similarity of Avp to UCH family of DUBs (UCH-L3 and YUH1), the only significant structural match observed was, as previously reported (21, 28), to the Ulp1 catalytic domain. This ubiquitin-like protein protease belonging to the Avp family is involved in Smt3/SUMO deconjugation (21). However, unlike Ulp1, Avp did not cleave Smt3-GFP fusion in our in vitro assays (blot in Fig. 4a, some negligible cleavage of Smt3-GFP could be observed only after 5 h of incubation with Avp).
FIG. 4.
Structural comparison of Avp with UCH and Ulp families of enzymes. (a) Sequence alignment of the catalytic core domains (Cys and His boxes) of Avp, human UCH-L3, yeast YUH1, and yeast Ulp1 (MEGALIGN [6]). Asterisks indicate the residues of the catalytic triad and Gln residue of the oxyanion hole. Western blot shows the comparative cleavage of Smt3-GFP fusion by Avp, Ulp1, and deubiqutinating enzyme USP8. White arrows show the protein present in crude USP8 preparation cross-reacting with anti-GFP antibody. (b) Ribbon diagrams of the peptidases (PDB identifiers 1AVP, 1UCH, 1CMX, 1EUV). The side chains of the catalytic residues and Gln residue of the oxyanion hole are shown. Figure 4b and c were prepared with MOLSCRIPT (19) and RASTER3D (27). (c) Comparison of the peptidase active sites. Superposition of the active site residues, obtained with the program LSQMAN (CCP4 suite [4]). Avp is shown in magenta, UCH-L3 s shown in green, YUH1 s shown in cyan, and Ulp1 s shown in red. Secondary structure elements of Avp are labeled. (d) Model of the binding of ubiquitin C-terminal peptide LRG-Glz (red) to the active-site cleft of Avp (cyan). The active site tetrads of Avp and YUH1 (in the complex with Ubal) were aligned, and the residues involved in Ubal binding by Avp were identified by determination of the contacts between LRG-Glz peptide and Avp (CCONTACT, CCP4 suite [4]).
It has been already noted that four proteases: Avp, UCH-L3, YUH1 and Ulp1, show common structural features of the papain-like fold (8, 17, 18, 28) (Fig. 4b). Topologically, however, Avp is similar to Ulp1 (Topp, CCP4 suite [4]); both enzymes are H/C proteases (by ordering of C and H boxes), whereas UCH-L3 and YUH1 are C/H proteases. Superposition of the active sites of the proteases shows significant overlap of the catalytic residues, a characteristic feature of many cysteine proteases (Fig. 4c). The alignment of the active sites of Avp and YUH1 in the complex with Ubal places the C-terminal Ubal peptide LRG-Glz precisely in the substrate-binding cleft of Avp, allowing the determination of all amino acids constituting the S1-S4 substrate binding sites of Avp (Fig. 4d). These residues (labeled in yellow) were used for the construction of the molecular surface of the substrate-binding cleft of Avp (cyan). The model predicts a covalent bond between Ubal C-terminal carbon atom and the sulfur atom of Avp Cys122 (shown as yellow sphere), at least seven hydrogen bonds (including one between oxygen atom of Glz and Gln115 of the oxyanion hole), and numerous van der Waals contacts. Similar to YUH1 (18), the active-site cleft of Avp is bounded on one side by the N-terminal residues of enzyme and on the other side by the residues close to the active-site histidine. The S1 pocket of Avp is formed mainly by Ala120 (conserved), Val53 (consensus), and Trp55 (conserved) with participation of the active-site residues. The main specificity determinant, the S2 pocket, is formed by Trp55 (found at the same place as Tyr167 in S2 pocket of YUH1), Ser4 (consensus), Ser3 (consensus), and Val53. The S3 pocket comprises Gly52 (conserved), Gly53 (conserved), and Trp55 interacting with the backbone atoms of Arg74 (Ubal) at the P3 position, and Glu5 (conserved), Ser3, and Ser4 showing some interaction with Ubal sidechain of Arg74. Another important specificity determinant, the S4 pocket, is formed by Gln6 (consensus), Gly51, and Glu5 interacting with the main chain atoms of the P4 residue, and by Arg48 (conserved, positioned similarly to Asn225 in the S4 pocket of YUH1), Gly51, and Gly52, which have extensive contacts with Ubal sidechain of Leu73. The observed similarities in the organization of the active site cleft of Avp and YUH1 may explain the similar cleavage sequence specificities of these proteases.
DISCUSSION
The invasion strategy of many viruses involves the synthesis of viral gene products that mimic the functions of the cellular proteins and thus interfere with the key cellular processes. It is plausible to think that following the appearance of the ubiquitin system in Eukaria several viruses, which infect eukaryotic cells, have acquired ubiquitin-associated functions. The first hint that viruses might have something to do with ubiquitination machinery came from the observation that some viruses code for the homologues of ubiquitin or Ub-conjugating enzymes (10, 14) or include ubiquitin and ubiquitinated proteins in the composition of virus particles (37, 38). Accumulating data on the virus interference with ubiquitin system reveal at least three distinct strategies used by viruses: (i) utilization of the protein ubiquitination for cellular membrane remodeling that is required for enveloped virus budding/internalization (12, 37), (ii) recruitment of Ub-conjugating enzymes for targeted ubiquitination and degradation of cellular proteins, first shown for papillomavirus E6 protein (33), and (iii) recruitment of DUBs. To date only one example of this third strategy has been reported: the interaction of herpes simplex virus (HSV-1) protein Vmw110 with cellular deubiquitinating protease HAUSP, which is required for efficient activation of gene expression by Vmw110 and stimulation of virus replication (9). Although the precise mechanism of HAUSP function is unknown, the fact that it is closely related to D-Ubp-64E, a negative regulator of gene silencing (9, 43), suggests that it can participate in reorganization of chromatin structure. The association of deubiquitinating activity with retrovirus particles has also been suggested (37).
In this study we demonstrate the deubiquitinating activity of Ad proteinase Avp. Analysis of the Avp function in vitro and in vivo showed that the cleavage of Ub conjugates by this enzyme is determined mainly by ubiquitin C-terminal LRGG sequence, which corresponds to the Avp consensus cleavage site (M,L,I)XGX↓X. This conclusion is further supported by the similar Avp inhibition efficiency found with Ubal and with LxggH tetrapeptide inhibitors. Compared with the classical DUBs, which have a complex mechanism of substrate recognition leading to high specificity (7, 42, and 43), Avp seems to function as a low-specificity and low-selectivity DUB. Attempting to identify cellular targets which are deubiquitinated upon overexpression of Avp in HeLa cells, we observed that, although Avp seems to contribute to the processing of histone uH2A, some histone deubiquitination could also be observed in Ad2ts1-infected cells at nonpermissive temperature, when the mutant Avp is inactive (5, 39, 45). This indicates that deubiquitination of uH2A does not depend solely on Avp but some additional mechanisms might be involved, such as for example, inhibition of ubiquitin conjugation induced by viral shutoff of host protein synthesis. It has been reported that the encephalomyocarditis virus also induces the loss of uH2A in infected cells, although a general increase rather than decrease in Ub conjugation was observed (11).
The low specificity of Avp suggests that the enzyme may deubiquitinate various cellular as well as Ad ubiquitinated proteins. In this work we did not attempt identification of viral targets but it can be envisaged that Avp activity could protect viral proteins against Ub-dependent proteasome degradation. Consequently, Avp deubiquitinating activity in Ad-infected cells could lead to inhibition of antigen presentation by major histocompatibility complex class I molecules. Alternatively, Ad may take advantage of the deubiquitination of some cellular targets by Avp. In this context, it is intriguing that Avp shows significant structural similarity to Yersinia pestis virulence effector YopJ (30). YopJ is translocated from the bacteria into the host cell, where it binds to mitogen-activated protein kinase (MAPK) kinases (MKKs) and blocks their activation via phosphorylation. This results in inhibition of the MAPK/NF-κB pathways and attenuation of the host immune response. Based on the similarity of YopJ to the Ulp1 the authors suggested that YopJ functions as SUMO-1-specific protease in yet-to-be-identified SUMO-1-dependent MAPK/NF-κB pathway (30). On the other hand, the similarity of YopJ and Avp suggests that YopJ may also be a DUB that antagonizes the ubiquitination of the proteins of MAPK signaling cascade, like it probably does the DUB faf (16). Whether YopJ functions similarly to Avp may be elucidated by identification of targets of both enzymes but we know already that contrary to YopJ, Avp is not a desumoylating enzyme.
Some insight into the possible substrates of Avp was provided by our observation that recombinant Avp is capable of processing ISG15 precursor (proISG15) in vitro. This ubiquitin-like protein, which is induced by alpha/beta interferons, attaches covalently to target proteins in the cascade of reactions analogous to ubiquitin ligation (11, 23, 29). The induction of ISG15 and the formation of ISG15-conjugates seem to be important determinants for antivirus activity of interferons. This is supported by the recent observation that NS1 protein of influenza B virus inhibits conjugation of ISG15 to cellular proteins (44). Since the targets of ISG15 have not been identified, it is not clear how ISG15 conjugation affects virus infection. Interestingly, the attachment of ISG15 directs proteins to intermediate filaments (23). Our preliminary data show that Avp, like ISG15 conjugates, binds to the cytokeratin network. It is thus possible that Avp could cleave these conjugates as part of the viral strategy of interferon evasion. Future experiments should resolve this issue.
In this work, we have identified a novel function of Ad proteinase, which is deubiquitination. This unusual property of virus processing enzyme might be not limited to Ad proteinase. It should be noted that other viruses such as African swine fever virus (ASFV) and vaccinia virus have been shown to code for the proteinases with catalytic core domains similar to that of Avp and which have similar cleavage specificity (2, 21). Moreover, the ASFV proteinase has been considered as belonging to Ulp family of proteases (2). If these viral enzymes could act as DUBs, this would reflect the acquisition of an advantageous property by the viruses and may indicate the importance of ubiquitin pathways in viral infection.
Acknowledgments
We thank C. W. Anderson, D. Bohmann, C. F. Draetta, M. Hochstrasser, C. D. Lima, M. Rosa, and J. E. Celis for the gifts of expression vectors, proteins, and antibodies; J.-F. Hernandez, A. Gonzalez de Peredo, D. Lascoux, E. Thouvenin, L. Costenaro, and D. A. Skoufias for assistance in some experiments; and R. Cohen for helpful discussions. M.Y.B. thanks F. Pellegrini for help in preparation of the Avp image.
M.Y.B. was a recipient of a Poste Rouge of the CNRS. This work was supported in part by a grant from the Fondation pour la Recherche Médicale and by grant GM47426 to A.L.H.
REFERENCES
- 1.Anderson, C. W. 1990. The proteinase polypeptide of adenovirus serotype 2 virions. Virology 177:259-272. [DOI] [PubMed] [Google Scholar]
- 2.Andres, G., A. Alejo, C. Simon-Mateo, and M. L. Salas. 2001. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276:780-787. [DOI] [PubMed] [Google Scholar]
- 3.Bertolaet, B. L., D. J. Clarke, M. Wolff, M. H. Watson, M. Henze, G. Divita, and S. I. Reed. 2001. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 8:417-422. [DOI] [PubMed] [Google Scholar]
- 4.CCP4. 1994. The CCP4 suite: programs for protein crystallography. Acta Cryst. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 5.Chen, P. H., D. A. Ornelles, and T. Shenk. 1993. The adenovirus L3 23-kilodalton proteinase cleaves the amino-terminal head domain from cytokeratin 18 and disrupts the cytokeratin network of HeLa cells. J. Virol. 67:3507-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clewley, J. P., and C. Arnold. 1997. MEGALIGN. The multiple alignment module of LASERGENE. Methods Mol. Biol. 70:119-129. [PubMed] [Google Scholar]
- 7.D'Andrea, A., and D. Pellman. 1998. Deubiquitinating enzymes: a new class of biological regulators. Crit. Rev. Biochem. Mol. Biol. 33:337-352. [DOI] [PubMed] [Google Scholar]
- 8.Ding, J., W. J. McGrath, R. M. Sweet, and W. F. Mangel. 1996. Crystal structure of the human adenovirus proteinase with its 11 amino acid cofactor. EMBO J. 15:1778-1783. [PMC free article] [PubMed] [Google Scholar]
- 9.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guarino, L. A. 1990. Identification of a viral gene encoding a ubiquitin-like protein. Proc. Natl. Acad. Sci. USA 87:409-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas, A. L., P. Ahrens, P. M. Bright, and H. Ankel. 1987. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem. 262:11315-11323. [PubMed] [Google Scholar]
- 12.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 14.Hingamp, P. M., J. E. Arnold, R. J. Mayer, and L. K. Dixon. 1992. A ubiquitin conjugating enzyme encoded by African swine fever virus. EMBO J. 11:361-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann, K., and P. Bucher. 1996. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci. 21:172-173. [PubMed] [Google Scholar]
- 16.Isaksson, A., F. A. Peverali, L. Kockel, M. Mlodzik, and D. Bohmann. 1997. The deubiquitination enzyme fat facets negatively regulates RTK/Ras/MAPK signalling during eye development. Mech. Dev. 68:59-67. [DOI] [PubMed] [Google Scholar]
- 17.Johnston, S. C., C. N. Larsen, W. J. Cook, K. D. Wilkinson, and C. P. Hill. 1997. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 Å resolution. EMBO J. 16:3787-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston, S. C., S. M. Riddle, R. E. Cohen, and C. P. Hill. 1999. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 18:3877-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structure. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 20.Lam, Y. A., W. Xu, G. N. DeMartino, R. E. Cohen. 1997. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385:737-740. [DOI] [PubMed] [Google Scholar]
- 21.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246-251. [DOI] [PubMed] [Google Scholar]
- 22.Lin, C. H., S. Chen, D. S. Kwon, J. K. Coward, and C. T. Walsh. 1997. Aldehyde and phosphinate analogs of glutathione and glutathionylspermidine: potent, selective binding inhibitors of the E. coli bifunctional glutathionylspermidine synthetase/amidase. Chem. Biol. 4:859-866. [DOI] [PubMed] [Google Scholar]
- 23.Loeb, K. R., and A. L. Haas. 1994. Conjugates of ubiquitin cross-reactive protein distribute in a cytoskeletal pattern. Mol. Cell. Biol. 14:8408-8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangel, W. F., W. J. McGrath, D. L. Toledo, and C. W. Anderson. 1993. Viral DNA and a viral peptide can act as cofactors of adenovirus virion proteinase activity. Nature 361:274-275. [DOI] [PubMed] [Google Scholar]
- 25.Mangel, W. F., D. L. Toledo, M. T. Brown, J. H. Martin, and W. J. McGrath. 1996. Characterization of three components of human adenovirus proteinase activity in vitro. J. Biol. Chem. 271:536-543. [DOI] [PubMed] [Google Scholar]
- 26.Mayer, R. J., J. Lowe, G. Lennox, M. Landon, K. MacLennan, and F. J. Doherty. 1989. Intermediate filament-ubiquitin diseases: implications for cell sanitization. Biochem. Soc. Symp. 55:193-201. [PubMed] [Google Scholar]
- 27.Merritt, E. A., and D. G. Bacon. 1997. RASTER3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 28.Mossessova, E., and C. D. Lima. 2000. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5:865-876. [DOI] [PubMed] [Google Scholar]
- 29.Narasimhan, J., J. L. Potter, and A. L. Haas. 1996. Conjugation of the 15-kDa interferon-induced ubiquitin homolog is distinct from that of ubiquitin. J. Biol. Chem. 271:324-330. [DOI] [PubMed] [Google Scholar]
- 30.Orth, K., Z. Xu, M. B. Mudgett, Z. Q. Bao, L. E. Palmer, J. B. Bliska, W. F. Mangel, B. Staskawicz, and J. E. Dixon. 2000. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290:1594-1597. [DOI] [PubMed] [Google Scholar]
- 31.Ploegh, H. L. 1998. Viral strategies of immune evasion. Science 280:248-253. [DOI] [PubMed] [Google Scholar]
- 32.Potter, J. L., J. Narasimhan, L. Mende-Mueller, and A. L. Haas. 1999. Precursor processing of pro-ISG15/UCRP, an interferon-beta-induced ubiquitin-like protein. J. Biol. Chem. 274:25061-25068. [DOI] [PubMed] [Google Scholar]
- 33.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 34.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 35.Stein, R. L., Z. Chen, and F. Melandri. 1995. Kinetic studies of isopeptidase T: modulation of peptidase activity by ubiquitin. Biochemistry 34:12616-12623. [DOI] [PubMed] [Google Scholar]
- 36.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 37.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 97:12945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webb, J. H., R. J. Mayer, and L. K. Dixon. 1999. A lipid modified ubiquitin is packaged into particles of several enveloped viruses. FEBS Lett. 444:136-139. [DOI] [PubMed] [Google Scholar]
- 39.Weber, J. M. 1995. Adenovirus endopeptidase and its role in virus infection. Curr. Top. Microbiol. Immunol. 199:227-235. [DOI] [PubMed] [Google Scholar]
- 40.Webster, A., S. Russell, P. Talbot, W. C. Russell, and G. D. Kemp. 1989. Characterization of the adenovirus proteinase: substrate specificity. J. Gen. Virol. 70:3225-3234. [DOI] [PubMed] [Google Scholar]
- 41.Webster, A., R. T. Hay, and G. Kemp. 1993. The adenovirus protease is activated by a virus-coded disulphide-linked peptide. Cell 72:97-104. [DOI] [PubMed] [Google Scholar]
- 42.Wilkinson, K. D., and M. Hochstrasser. 1998. The deubiquitinating enzymes, p. 99-125. In J.-M. Peters, J. R. Harris, and D. Finley (ed.), Ubiquitin and the biology of the cell. Plenum Press, New York, N.Y.
- 43.Wilkinson, K. D. 1997. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 11:1245-1256. [DOI] [PubMed] [Google Scholar]
- 44.Yuan, W., and R. M. Krug. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, Y., and R. J. Schneider. 1994. Adenovirus inhibition of cell translation facilitates release of virus particles and enhances degradation of the cytokeratin network. J. Virol. 68:2544-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]