Abstract
The 3′ terminus of the bamboo mosaic potexvirus (BaMV) contains a poly(A) tail, the 5′ portion of which participates in the formation of an RNA pseudoknot required for BaMV RNA replication. Recombinant RNA-dependent RNA polymerase (RdRp) of BaMV binds to the pseudoknot poly(A) tail in gel mobility shift assays (C.-Y. Huang, Y.-L. Huang, M. Meng, Y.-H. Hsu, and C.-H. Tsai, J. Virol. 75:2818-2824, 2001). Approximately 20 nucleotides of the poly(A) tail adjacent to the 3′ untranslated region (UTR) are protected from diethylpyrocarbonate modification, suggesting that this region may be used to initiate minus-strand RNA synthesis. The 5′ terminus of the minus-strand RNA synthesized by the RdRp in vitro was examined using 5′ rapid amplification of cDNA ends (RACE) and DNA sequencing. Minus-strand RNA synthesis was found to initiate from several positions within the poly(A) tail, with the highest frequency of initiation being from the 7th to the 10th adenylates counted from the 5′-most adenylate of the poly(A) tail. Sequence analyses of BaMV progeny RNAs recovered from Nicotiana benthamiana protoplasts which were inoculated with mutants containing a mutation at the 1st, 4th, 7th, or 16th position of the poly(A) tail suggested the existence of variable initiation sites, similar to those found in 5′ RACE experiments. We deduce that the initiation site for minus-strand RNA synthesis is not fixed at one position but resides opposite one of the 15 adenylates of the poly(A) tail immediately downstream of the 3′ UTR of BaMV genomic RNA.
Bamboo mosaic potexvirus (BaMV) consists of a 6,366-nucleotide-long single-stranded positive-sense RNA genome with a 5′ m7GpppG cap structure and a 3′ poly(A) tail (15, 30). Five major open reading frames (1 to 5) encode polypeptides of 155, 28, 13, 6, and 25 kDa. Besides the two major 2.0- and 1.0-kb-long subgenomic RNAs detected in infected cells (14), a satellite RNA could also be found in association with some strains of BaMV (16). The tertiary structure of the 3′ untranslated region (UTR) of BaMV RNA was determined by enzymatic and chemical structural probing and was determined to be involved in viral RNA replication (3, 27). The advantages of using BaMV as a model for studying the mechanism of viral RNA replication are as follows. (i) The full-length infectious cDNA clone has been constructed successfully (27) and can be used as a good tool for studying viral RNA replication in vivo. (ii) The Escherichia coli-overexpressed BaMV recombinant RNA-dependent RNA polymerase (RdRp) has been purified and demonstrated to be capable of transcribing the viral RNA in vitro (13). (iii) The functional RdRp extract from BaMV-infected plants was shown to specifically use short transcript templates for both minus- and plus-strand RNA syntheses in vitro (4).
In order to faithfully complete the replication cycle of positive-sense RNA viruses, it is important to initiate minus-sense RNA synthesis at the correct position to ensure the generation of full-length positive-sense progeny (12). The 3′ UTRs of positive-sense viral RNA genomes have been thought to be involved in regulating minus-sense RNA synthesis. The 3′-end tRNA-like structures of the genomic RNAs of brome mosaic virus (BMV) and turnip yellow mosaic virus (TYMV) were demonstrated to contain the signal for initiating minus-strand RNA synthesis in vitro (5, 11, 18, 22). A hairpin structure in the 3′-terminal region of turnip crinkle virus satellite RNA C was demonstrated to be involved in minus-sense RNA synthesis (23). Although some information has been obtained from studies of poliovirus showing that the poly(A) tail could be involved as a structural requirement for viral replication (17), there is only limited information available regarding the initiation of replication of those RNA viruses with a poly(A)-tailed genome.
The initiation of BMV and TYMV minus-strand RNA synthesis has been mapped to the guanylate residue opposite the penultimate residue of the 3′-terminal -CCA (2, 5, 18, 22). No empirical experiment has been done to identify the initiation site of minus-strand RNA synthesis of RNA viruses possessing poly(A)-tailed genomes. Here, we have mapped the 5′-end sequences of the minus-sense RNA genome derived from viral double-stranded RNA (dsRNA). We also mutagenized the sequence in the poly(A) tail as a means to identify the in vivo initiation site of BaMV minus-strand synthesis.
MATERIALS AND METHODS
Mutant construction.
Mutants derived from the full-length infectious cDNA clone pBaMV-O (27) were generated by a two-step PCR technique (20). Upstream primers (5′TAAAGACCTTTA20GGTTTCTACAGTTTTTTCCT−1AAAAAAA3′, 5′TTTTTTCCAAAG−4AAAAAAAAAA3′, 5′TTCCAAAAAAG−7AAAAAAAAAA3′, and 5′CCAAAAAAAAAAAAAAAG−16AAAAAAAAAA3′) and the universal primer (5′GTTTTCCCAGTCACGAC3′) were used to generate 149-, 122-, 118-, and 116-bp DNA fragments, respectively, which were then used as megaprimers for the second PCR with BaMV5910 (5′CCAAACCGACGTTCGCCA3′) to generate a 570-bp DNA fragment containing internal NruI and BamHI sites. (Nucleotides are numbered from the 3′-end cytosine just upstream of the poly(A) tail, and the adenylate residues of the poly(A) tail are numbered with a minus sign counting from the same C residue.) The PCR fragments were then gel purified and cloned into a T-vector plasmid (Novagen). The sequence of each mutant was verified before subcloning the segment from NruI (5964) to the 3′-end BamHI into NruI-BamHI-cut pBaMV-O. The resulting mutants were pBaMV-O/20A−1U, with a 20U−1A base pair changed to a 20A−1U base pair, and the single point mutants pBaMV-O/−4G, -O/−7G, and -O/−16G, with a guanylate residue at the 4th, 7th, and 16th position of the poly(A) tail, respectively (Fig. 1).
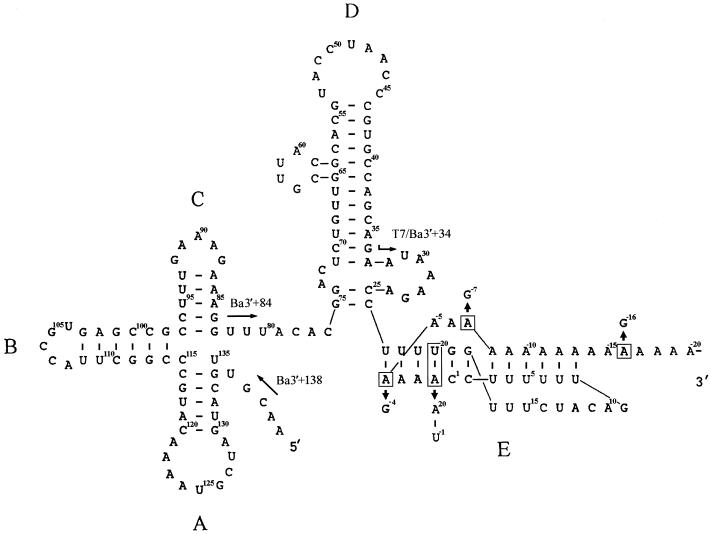
FIG. 1.
Proposed secondary structure of the 3′ UTR of BaMV RNA. The nucleotides are numbered from the 3′-end cytosine just upstream of the poly(A) tail. The arrows indicate the locations and the orientations of the primers used for PCR. The locations of the mutations in domain E are boxed and replaced with the indicated nucleotides.
DEPC footprinting analysis.
The modification at the N-7 position of adenine residues by diethylpyrocarbonate (DEPC) in footprinting analysis for structural probing was performed according to the method of Huang et al. (9). The 20-μl reaction mixture containing 1 μl of 5′-end-labeled r34/40A transcripts (10,000 cpm; 3 ng) was incubated at 30°C for 15 min, with the addition of 100 ng of RdRp and 1 μl of DEPC (Sigma; ca. 97%). The mixtures were then ethanol precipitated directly with yeast total RNAs (Boehringer Mannheim). The pellet was dissolved in 100 μl of water, phenol-chloroform extracted to remove the RdRp, and then precipitated with ethanol. The RNAs were dissolved in 20 μl of 1 M aniline (redistilled)-acetic acid (pH 4.3) solution directly and incubated at 60°C for 20 min in the dark, and then the cleaved RNA fragments were frozen at −80°C and lyophilized. The cleaved RNA fragments were resuspended in urea-containing loading buffer and resolved by electrophoretic separation on 8% denaturing (7 M urea) polyacrylamide gels.
dsRNA preparation.
BaMV-infected leaves (1 g) of Nicotiana benthamiana were ground with SET buffer (30 mM Tris-HCl [pH 8.0], 150 mM NaCl, 2 mM EDTA), 2% sodium dodecyl sulfate, and 2.5 mg of bentonite/ml (6, 10). The extract was shaken at room temperature for 10 min and centrifuged at 14,000 × g and 4°C for 2 min. The supernatant was transferred to a new tube containing 20 mg of CF-11 cellulose and 15% ethanol, and the tube was shaken for 10 min at room temperature. The cellulose mixture was centrifuged at the same speed for 2 min and washed with SET buffer containing 15% ethanol, and the procedure was repeated four times. The dsRNAs were finally eluted from the cellulose pellet with SET buffer after being shaken at room temperature for 30 min. After centrifugation at 14,000 × g and 4°C for 2 min, the supernatant containing dsRNAs was extracted with phenol-chloroform and precipitated with ethanol. The dsRNAs were dissolved in water and stored at −80°C.
Determination of terminal sequence of minus-sense RNA of BaMV by 5′ rapid amplification of cDNA ends (RACE).
The dsRNAs of BaMV were used to synthesize first-strand cDNA with primer Ba3′+138 (5′GGGCGTTGCATGATCG3′) and reverse transcriptase (Gibco-BRL) at 42°C for 30 min. The first-strand cDNA was then tailed with dCTP by terminal deoxynucleotidyl transferase (Promega) at 37°C for 30 min. After the dC-tailing reaction, the tailed cDNA was used as a template for PCR with the 5′ primer Ba3′+138 or Ba3′+84 (5′GTTTACACGGACTCTG3′) and the 3′ primer OligodG/BamHI (5′GGGGATCCGGGGGGGGGGGGGG3′). The PCR products were cloned into the pGEM T-easy vector (Promega) and sequenced.
Determination of the 5′-terminal sequence of minus-sense RNA synthesized in vitro by BaMV RdRp.
RNA synthesis in vitro with RdRp preparation was carried out in a total 50-μl reaction mixture containing 25 μl of detergent-solubilized and micrococcal-nuclease-treated RdRp preparation (4); 2 μg of RNA template; 2 mM (each) ATP, CTP, and GTP; 2 μM UTP; 0.066 μM [α-32P]UTP (3,000 Ci/mmol; Dupont-NEN); 4.8 mg of bentonite/ml; 10 mM dithiothreitol; and 10 mM MgCl2. After 1 h of incubation at 30°C, the reaction was stopped by adding 50 μl of Tris-EDTA buffer, and the products were extracted with phenol-chloroform and precipitated twice with ethanol. The 5′ RACE experiment determining the initiation sites of minus-sense transcripts was done as described above.
In vitro transcription and inoculation with protoplasts.
Plasmid DNAs were prepared from 50-ml bacterial cultures, and the mutated sequences were confirmed by sequencing. Capped genomic transcripts were generated with T7 RNA polymerase from the BamHI-linearized plasmid templates containing wild-type and mutated genomic cDNAs and analyzed as described previously (27) prior to inoculation. N. benthamiana was grown in a growth chamber under a 16-h day length at 28°C. Mesophyll protoplasts (4 × 105) of N. benthamiana were inoculated with 5 μg of transcript RNAs and incubated at 25°C for 48 h under constant illumination as described previously (25, 26, 27).
Analysis of viral products by Western and Northern blotting.
The levels of coat protein in harvested protoplasts were analyzed in Western blots, using anti-BaMV capsid protein serum as a primary antibody (14), horseradish peroxidase-labeled secondary antibody, and the chromogenic substrate 4-chloro-1-naphthol as described previously (25, 28). The results were quantified by scanning densitometry (Intelligent Quantifier; Bioimage). RNAs were extracted from the protoplasts, glyoxalated, electrophoresed through 1% agarose, and transferred to nylon membranes as described previously (25). The hybridization probe was a 32P-labeled RNA transcript complementary to 0.6 kb at the 3′ end of BaMV RNA (15).
Progeny RNA analysis.
Total RNAs were isolated from the inoculated protoplasts after 48 h of incubation and were reverse transcribed with primer Ba3′/10T (5′GCCCCGGGATCCTTTTTTTTTT3′) in a 5-μl reaction mixture containing 1 mM deoxynucleoside triphosphate, 100 mM Tris-HCl (pH 8.3), 80 mM KCl, 12 mM MgCl2, 2 mM dithiothreitol, and 10 U of SuperScript II (GIBCO/BRL). After incubation at 42°C for 30 min, 2 μl of reactant was taken to the PCR with Ba3′/10T as the downstream primer and Ba3′+138 as the upstream primer. The PCR fragments containing the sequence of the 3′ end of BaMV RNA were cloned into the T vector and sequenced.
RESULTS
Interaction between BaMV RdRp and the poly(A) sequence.
Gel mobility shift and competition assays have been used to distinguish sites in the 3′ UTR of BaMV RNA bound by the E. coli-expressed BaMV RdRp domain, the loop region of domain D, and the poly(A) tail (9). Previous footprinting experiments using the 5′-end-labeled transcript r84/40A could not resolve protection in the poly(A) tail. To map in detail protection of the poly(A) tail by the RdRp, we used the shorter 5′-end-labeled transcript r34/40A as a probe (Fig. 1). About 20 of the 40 adenylates immediately downstream of the 3′ UTR of r34/40A were protected by 100 ng of RdRp (Fig. 2). No protection could be found on the probe with no protein added or with 400 ng of bovine serum albumin (Fig. 2). In addition, some cleavages (the nonadenylate residues G9 and C11 in the loop of the pseudoknot) were observed in the absence of RdRp but were protected in the presence of RdRp. Overall, this footprinting result suggested that the polymerase domain of BaMV (Δ893 RdRp [9]) could not only protect about 20 adenylates right after the 3′ UTR of the poly(A) tail but could also protect the loop region of the pseudoknot of BaMV RNA sequence from modification (Fig. 1).
FIG. 2.
DEPC footprint of E. coli-expressed RdRp binding site on r34/40 RNA. An autoradiograph of the aniline cleavage products on an 8% urea polyacrylamide gel is shown. Lanes 1 and 2, labeled with RNases T1 and A, were 5′-end 32P-labeled r34/40A digested with RNases T1 and A, respectively, and used as markers. Lanes 3 to 6 contain aniline cleavage products without (−) DEPC modification (lane 5) or with (+) DEPC modification in the presence of no protein, 100 ng of RdRp (lane 4), or 400 ng of bovine serum albumin (BSA; lane 6) as indicated above each lane. The positions of selected nucleotides in the r34/40A sequence are indicated on the left for reference.
A range of oligo(U) lengths at the 5′ ends of BaMV minus strands present in plants.
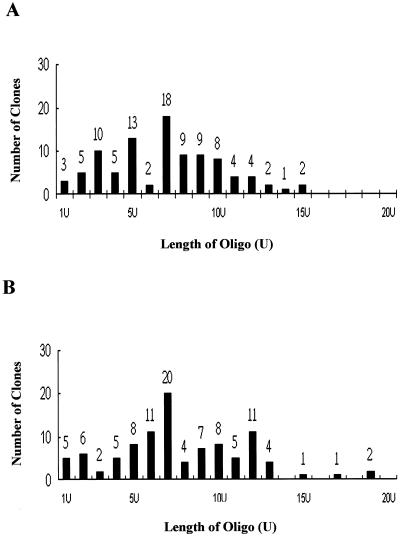
The results of the footprinting experiment suggested that the protected 20 adenylates right after the 3′ UTR could be the putative initiation sites for BaMV minus-sense RNA synthesis. In order to test this hypothesis, we used the 5′ RACE technique to identify the sequence of the 5′ end of minus-sense RNA. To obtain minus-sense RNAs from the infected plants, we extracted viral dsRNAs, which are regarded as the viral replicative intermediate containing the minus-sense RNA. After 5′ RACE amplification using primers Ba3′+138 and OligodG/BamHI, the expected fragments about 150 bp in size could not be seen clearly under the shielded UV light box. A second PCR was required to amplify them to a visible amount with the BaMV-specific internal primer Ba3′+84 (Fig. 1) and the downstream primer OligodG/BamHI. DNA fragments about 100 bp in size were eluted from the polyacrylamide gel, cloned, and sequenced. A total of 95 clones were sequenced. The results showed that about half of the clones comprised 7 to 10 uridylates at the 5′ end of minus-strand RNA (Fig. 3). Ten and 13 clones comprised three and five Us, repectively. Only two clones had the longest stretch of 15 Us. These results indicated that there was no fixed length of oligo(U) at the 5′ end of BaMV minus-strand RNA and implied that the BaMV minus-strand RNA synthesis was initiated opposite one of the adenylates of the poly(A) tail.
FIG. 3.
Summary of the sequencing results of clones derived from 5′ RACE experiments. (A) Analysis of oligo(U) at the 5′ ends of genomic minus-strand RNAs derived from viral dsRNAs synthesized in infected tissue. (B) A similar analysis of dsRNAs synthesized in vitro by RdRp assays. Each bar represents the number of clones with the same number of uridylates at the 5′ end of minus-strand RNA.
5′ ends of minus-strand RNA synthesized in vitro with RdRp preparation.
In order to test whether the 5′ ends of minus-strand RNAs derived from RNA synthesis in vitro with RdRp preparation were the same as those identified with the viral dsRNAs isolated from infected plants, BaMV3′+138 containing the BaMV 3′ UTR with 40 adenylates was used as a template in the reaction. The dsRNA products were extracted after the RdRp reaction and used for 5′ RACE to determine the 5′-end sequences of the newly synthesized minus-strand RNAs. Twenty of a total of 100 clones sequenced contained a plasmid comprising the BaMV sequence with seven adenylates connected to the deoxycytidylate tail. That would suggest that about 20% of products initiated started opposite the seventh adenylate residue of the poly(A) tail during minus-strand RNA synthesis (Fig. 3). About 75% of the clones showed putative initiation sites opposite the 5th to 12th adenylate residues of the poly(A) tail. Two clones contained a string of 19 adenylates adjacent to the dC-tail. The identification of the variable length of the short oligo(U) at the 5′ end of minus-strand RNA synthesis in vitro with RdRp preparation matched that of double-stranded viral RNA isolated from infected plants. To inspect any nuclease activity that could trim the products derived from an in vitro replication assay and cause the variation of the 5′-end sequence, the 3′ ends of minus-strand RNA templates were used to synthesize the plus-strand RNAs to see any variation of the 5′ ends of the products. The sequences of all clones showed a fixed initiation site for plus-strand RNA synthesis (unpublished data). These results suggested that no activity exists in the replicase preparation that could cause the variation of the 5′-end sequence.
Multiple putative initiation sites were deduced from an in vivo replication assay.
It is possible that the variation of putative initiation sites identified by the 5′ RACE technique could be an artifact of any degradation activity introduced during the RNA isolation or the processes of the 5′ RACE. In order to confirm the results of the 5′ RACE, we replaced the first adenylate residue of the poly(A) tail of the infectious clone pBaMV-O with a uridylate residue and changed 20U to 20A in order to maintain the base pairing. We also replaced the 4th, 7th, and 16th adenylate residues of the poly(A) tail with guanylate residues. Mutations at these positions were likely to maintain the 3′ pseudoknot structure as predicted by the computer-assisted folding programs (1, 31) (Fig. 1). The maintenance of a pseudoknot structure was identified as important for the replication of BaMV RNA (27).
The resulting four mutants, BaMV-O/20A−1U, -O/−4G, -O/−7G, and -O/−16G, were transcribed and inoculated into N. benthamiana protoplasts. Total proteins and RNAs were extracted from the inoculated protoplasts after a 48-h incubation. The levels of coat protein and genomic and subgenomic RNAs accumulated in the protoplasts were analyzed by Western blotting (Fig. 4). The levels of coat protein accumulated in protoplasts were used as an indication of the replication efficiency of each mutant, as shown in Table 1. All four mutants replicated as efficiently as the wild type. These results indicated that these mutations would not interfere with the replication of BaMV RNA or revert to the sequence of the wild type.
FIG. 4.
Amplification of BaMV and its derivatives in N. benthamiana protoplasts. Protoplasts (4 × 105) were inoculated with 5 μg of transcripts from wild-type (wt) pBaMV-O and mutants labeled −4G, −7G, and −16G above each lane. The protoplasts were harvested 48 h postinoculation. (A) Total protein extracts were separated on a 14% sodium dodecyl sulfate-polyacrylamide gel, blotted, and probed with antiserum against BaMV coat protein. The blot was developed using horseradish peroxidase-linked secondary antibodies and 5-chloro-1-naphthol color reagent. (B) Sequences of the clones derived from the progeny RNAs after reverse transcription-PCR. The positions of selected nucleotides are indicated on the right for reference.
TABLE 1.
Viral accumulation level and progeny RNA analysis of BaMV and its derivatives in protoplasts of N. benthamiana
| Inoculum RNA | Replication efficiencya | Progeny RNA(s) | Ratiob |
|---|---|---|---|
| BaMV-O (wild type) | 1.00 | BaMV-O | 3/3 |
| BaMV-O/20A−1U | 0.96 ± 0.10 | BaMV-O/20A −1U | 5/5 |
| BaMV-O/−4G | 0.94 ± 0.08 | BaMV-O/−4G | 14/19 |
| BaMV-O/−4A | 5/19 | ||
| BaMV-O/−7G | 0.92 ± 0.06 | BaMV-O/−7G | 6/14 |
| BaMV-O/−7A | 8/14 | ||
| BaMV-O/−16G | 0.98 ± 0.08 | BaMV-O/−16A | 17/17 |
Coat protein accumulation in protoplasts, detected by Western blotting with anti-BaMV coat protein serum; all data are averages (± standard deviations) of at least five independent experiments.
Ratio of number of clones with indicated sequence to total number of effective clones sequenced.
Sequence analysis of these progeny RNAs would provide information about the putative initiation site of minus-strand RNA synthesis. If the minus-strand RNA initiates opposite one of the nucleotides upstream of the mutation site, then the mutation will not be preserved in the progeny. On the other hand, if the initiation site is located opposite one of the nucleotides downstream of the mutation site, then the progeny RNAs will keep the mutation. Total RNAs harvested after protoplast inoculation and a 48-h incubation were taken for reverse transcription-PCR and cloned into the T vector. The number of adenylates in the poly(A) tail of each clone was critical in counting the effective clones, since the guanylate residue at the poly(A) tail could be primed with oligo(dT). Therefore, the effective clones of BaMV-O/−4G should be reverse transcribed with primer primed downstream of the mutation site and comprise at least 14 residues of the poly(A) tail (the primer with 10 deoxythymidylate residues plus the sequence from the mutation site to the last nucleotide of the 3′ UTR). Thus, there must be at least 17 and 26 adenylates, respectively, for BaMV-O/−7G and BaMV-O/−16G to be counted as effective clones. Under this criterion, all five clones derived from BaMV-O/20A−1U showed the preservation of the mutation sites, indicating that maintaining the pseudoknot structure is important and that the initiation site should be opposite one of the nucleotides downstream of the −1U of this mutant. A total of 17 effective clones were derived from BaMV-O/−16G, and all showed a revertant genotype with adenylate at the 16th position of the poly(A) tail. This result indicated that the minus-strand RNA synthesis should initiate upstream of the 16th adenylate residue of the poly(A) tail. These results matched the 5′ RACE mapping data showing that there were <15 uridylate residues found in the most-5′ ends of the minus-strand RNAs.
Since the identification of multiple putative initiation sites resulted from the 5′-end mapping data of the minus-strand RNA, the progeny derived from BaMV-O/−4G and BaMV-O/−7G would be expected to be a mixture of mutant and revertant. A total of 19 effective clones derived from BaMV-O/−4G comprised 14 mutants and 5 revertants. These results indicated that only a few progeny used the first three adenylates to initiate the minus-strand RNA and that most of the progeny would initiate opposite one of the nucleotides after the fourth adenylate residue of the poly(A) tail. Fourteen effective clones derived from BaMV-O/−7G comprising six and eight clones of mutants and revertants, respectively, indicated that similar numbers of progeny would initiate the minus-strand RNA at the 1st to 7th and 8th to 15th adenylate residues of the poly(A) tail. Taken together, these results matched quite well with the 5′ RACE mapping data showing that all the putative initiation sites are within 15 adenylates and with no fixed position for minus-strand RNA synthesis.
DISCUSSION
It is important for a positive-sense RNA virus to initiate its minus-strand RNA synthesis at the correct position to faithfully pass its genetic material to the next generation (12). BMV and TYMV, which have a tRNA-like structure with 3′-terminal -CCA, initiate their minus-strand RNA opposite the penultimate cytidylate residue with GTP (2, 5, 18, 22). Thereafter, the progeny are synthesized using the minus-strand as a template and ending with -CC at the very 3′ end. The last nucleotide of the genome, the adenylate residue, will be added by tRNA nucleotidyltransferase in the infected cells (19). These processes ensure that the entire genetic information can pass faithfully to the progeny.
The poly(A)-tailed virus white clover mosaic potexvirus has been reported to have 100 to 300 adenylates at the 3′ end of its genomic RNA (7). We have determined that BaMV RNA contains about 90 to 170 adenylates at the 3′ end by resolving it on a sequencing gel after the complete digestion of the 32pCp-labeled virion RNAs with RNase A (data not shown). Therefore, the initiation of minus-strand RNA synthesis of the poly(A)-tailed virus, with a variable number of adenylates (ranging from 90 to 170 in BaMV RNA) at the 3′ end, will be different from those of BMV and TYMV, which have a fixed end. Since the poly(A)-tailed RNA virus comprises hundreds of adenylates, it might be difficult for RdRp binding to the 3′ UTR, as in BaMV (9), to initiate the minus-strand synthesis at the very end of the poly(A) tail hundreds of nucleotides downstream. The footprint data of BaMV RdRp complexed with the RNA containing part of the 3′ UTR sequence and 40 adenylates showed that only about 20 adenylates immediately downstream of the 3′ UTR were protected (Fig. 2). It is more reasonable that a viral RdRp binds on the 3′ UTR and initiates the negative-strand RNA synthesis opposite the nearby nucleotide instead of looking for the very 3′-end nucleotide. In this case, the 5′ end of the minus strand of BaMV would contain a short stretch of uridylates instead of runs of hundreds of Us. Our 5′ RACE experiment clearly demonstrates that there are <15 Us at the 5′ end of the negative strand. Similar experiments have been performed with clover yellow vein potyvirus (24) and bovine coronavirus (8), showing a short stretch of 8 to 20 Us at the 5′ end of the negative-strand RNAs. However, a long stretch of poly(U) ranging from 60 to 80 residues found in replicative forms or replicative intermediates of Semliki Forest viruses was proposed to be a template for poly(A) synthesis of progeny RNAs (21).
Taken together, these results suggest that a positive-sense RNA virus with a poly(A)-tailed genome would use the poly(A) tail as a template to initiate minus-strand RNA synthesis. However, only a short stretch of poly(A) sequence connected to the 3′ UTR is necessary for initiation. Our working model for the initiation of BaMV minus-strand RNA synthesis calls for RdRp interaction with the 3′ UTR, including the potexviral conserved hexamer motif (ACNUAA) and part of the stem sequence (9, 29) and about 20 nucleotides of the poly(A) sequence immediately downstream of the 3′ UTR, according to the footprinting data. This model was also supported by the results derived from an in vivo experiment which showed that about 13 to 25 adenylates following the BaMV 3′ UTR were involved in maintaining the integrity of the pseudoknot structure and were required for efficient viral RNA replication in protoplasts (27). Once the RdRp has interacted with the 3′ UTR of BaMV RNA, the RdRp would initiate the minus strand opposite one of the adenylates within the pseudoknot, perhaps selectively in the loop region of the pseudoknot, which corresponds to the most favored position for initiation (Fig. 1). The occurrence of multiple putative initiation sites of minus-strand RNA synthesis could be due to the pseudoknot structure with the variable size of the loop comprising different numbers of adenylates. It is also possible that the pseudoknot structure was partially opened when the RdRp interacted with the poly(A) sequence and initiated opposite one of the adenylates. Therefore, any one of the adenylates in the opened region could be used to initiate the minus-strand RNA.
Finally, when the minus strands were used as templates to synthesize the progeny RNAs, a different-length short stretch of As would be at the very 3′ end of the genomic RNA due to the variation of initiation sites of the minus-strand RNA synthesis. These progeny RNAs must be good templates for subsequent polyadenylation up to about 90 to 170 adenylates, as found in the BaMV virion RNAs. Therefore, the enzyme functioning in polyadenylation must recognize these short stretches of poly(A)-tailed genomes and maintain the structural integrity of the virion RNA. The newly synthesized progeny RNAs with short stretches of As could be very useful in the future as substrates in searching for the enzymes that are involved in polyadenylation.
Acknowledgments
We thank Muthukumar Nadar at National Chung-Hsing University for editorial help.
This work was supported by grants from National Science Council Projects NSC 90-2311-B-005-014 and 90-2311-B-005-015.
REFERENCES
- 1.Abrahams, J. P., M. van den Berg, F. H. D. Batenburg, and C. W. A. Pleij. 1990. Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucleic Acids Res. 18:3035-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman, M. R., and C. C. Kao. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286:709-720. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, C.-P., and C.-H. Tsai. 1999. Structural and functional analysis of the 3′ untranslated region of bamboo mosaic potexvirus genomic RNA. J. Mol. Biol. 288:555-565. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, J.-H., M.-P. Ding, Y.-H. Hsu, and C.-H. Tsai. 2001. The partial purified RNA-dependent RNA polymerases from bamboo mosaic potexvirus and potato virus X infected plants containing the template-dependent activities. Virus Res. 80:41-52. [DOI] [PubMed] [Google Scholar]
- 5.Deiman, B. A. L. M., A. K. Koenen, P. W. G. Verlaan, and C. W. A. Pleij. 1998. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J. Virol. 72:3965-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dulieu, P., and M. Bar-Joseph. 1989. Rapid isolation of double stranded RNA segments from disulphide crosslinked polyacrylamide gels. J. Virol. Methods 24:77-84. [DOI] [PubMed] [Google Scholar]
- 7.Guilford, P. J., D. L. Beck, and R. L. S. Forster. 1991. Influence of the poly(A) tail and putative polyadenylation signal on the infectivity of white clover mosaic potexvirus. Virology 182:61-67. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann, M. A., and D. A. Brian. 1991. The 5′ end of coronavirus minus-strand RNAs contains a short poly(U) tract. J. Virol. 65:6331-6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, C.-Y., Y.-L. Huang, M. Meng, Y.-H. Hsu, and C.-H. Tsai. 2001. Sequences at the 3′ untranslated region of the bamboo mosaic potexvirus RNA interact with the viral RNA-dependent RNA polymerase. J. Virol. 75:2818-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jelkman, W., R. R. Martin, and E. Maiss. 1989. Cloning of four plant viruses from small quantities of double-stranded RNA. Phytopathology 79:1250-1253. [Google Scholar]
- 11.Kao, C. C., and J.-H. Sun. 1996. Initiation of minus-strand RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: use of oligoribonucleotide primers. J. Virol. 70:6826-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 13.Li, Y.-I., Y.-M. Cheng, Y. L. Huang, C.-H. Tsai, Y.-H. Hsu, and M. Meng. 1998. Identification and characterization of the Escherichia coli-expressed RNA-dependent RNA polymerase of bamboo mosaic virus. J. Virol. 72:10093-10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, N.-S., F. Z. Lin, T. Y. Huang, and Y.-H. Hsu. 1992. Genome properties of bamboo mosaic virus. Phytopathology 82:731-734. [Google Scholar]
- 15.Lin, N.-S., B.-Y. Lin, N.-W. Lo, C.-C. Hu, T.-Y. Chow, and Y.-H. Hsu. 1994. Nucleotide sequence of the genomic RNA of bamboo mosaic potexvirus. J. Gen. Virol. 75:2513-2518. [DOI] [PubMed] [Google Scholar]
- 16.Lin, N.-S., and Y.-H. Hsu. 1994. A satellite RNA associated with bamboo mosaic potexvirus. Virology 202:707-714. [DOI] [PubMed] [Google Scholar]
- 17.Melchers, W. J. G., J. G. J. Hoenderop, H. J. Bruins Slot, C. W. A. Pleij, E. V. Pilipenko, V. I. Agol, and J. M. D. Galama. 1997. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J. Virol. 71:686-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, W. A., J. J. Bujarski, T. W. Dreher, and T. C. Hall. 1986. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J. Mol. Biol. 187:537-546. [DOI] [PubMed] [Google Scholar]
- 19.Rao, A. L., T. W. Dreher, L. E. Marsh, and T. C. Hall. 1989. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc. Natl. Acad. Sci. USA 86:5335-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 21.Sawicki, D. L., and P. J. Gomatos. 1976. Replication of Semliki Forest virus: polyadenylate in plus-strand RNA and polyuridylate in minus-strand RNA. J. Virol. 20:446-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh, R. N., and T. W. Dreher. 1997. Turnip yellow mosaic virus RNA-dependent RNA polymerase: initiation of minus strand synthesis in vitro. Virology 233:430-439. [DOI] [PubMed] [Google Scholar]
- 23.Song, C., and A. E. Simon. 1995. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 254:6-14. [DOI] [PubMed] [Google Scholar]
- 24.Tacahashi, Y., and I. Uyeda. 1999. Restoration of the 3′end of potyvirus RNA derived from poly(A)-deficient infectious cDNA clones. Virology 265:147-152. [DOI] [PubMed] [Google Scholar]
- 25.Tsai, C.-H., and T. W. Dreher. 1991. Turnip yellow mosaic virus RNAs with anticodon loop substitutions that result in decreased valylation fail to replicate efficiently. J. Virol. 65:3060-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai, C.-H., and T. W. Dreher. 1992. Second-site suppressor mutations assist in studying the function of the 3′ noncoding region of turnip yellow mosaic virus RNA. J. Virol. 66:5190-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai, C.-H., C.-P. Cheng, C.-W. Peng, B.-Y. Lin, N.-S. Lin, and Y.-H. Hsu. 1999. Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of bamboo mosaic potexvirus RNA. J. Virol. 73:2703-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiland, J. J., and T. W. Dreher. 1989. Infectious TYMV RNA from cloned cDNA: effects in vitro and in vivo of point substitutions in the initiation codons of two extensively overlapping ORFs. Nucleic Acids Res. 17:4675-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White, K. A., J. B. Bancroft, and G. A. Mackie. 1992. Mutagenesis of a hexanucleotide sequence conserved in potexvirus RNAs. Virology 189:817-820. [DOI] [PubMed] [Google Scholar]
- 30.Yang, C. C., J. S. Liu, C. P. Lin, and N.-S. Lin. 1997. Nucleotide sequence and phylogenetic analysis of a bamboo mosaic potexvirus isolated from common bamboo (Bambusa vulgaris McClure). Bot. Bull. Acad. Sin. 38:77-84. [Google Scholar]
- 31.Zuker, M. 1989. Computer prediction of RNA structure. Methods Enzymol. 180:262-288. [DOI] [PubMed] [Google Scholar]