Abstract
The entire nucleotide sequences of 70 hepatitis B virus (HBV) isolates of genotype B (HBV/B), including 38 newly determined and 32 retrieved from the international DNA database (DDBJ/EMBL/GenBank), were compared phylogenetically. Two subgroups of HBV/B were identified based on sequence divergence in the precore region plus the core gene, one with the recombination with genotype C and the other without it. The analysis over the entire genome of HBV/B by the SimPlot program located the recombination with genotype C in the precore region plus the core gene spanning nucleotide positions from 1740 to 1838 to 2443 to 2485. Within this genomic area, HBV/B strains with the recombination had higher nucleotide and amino acid homology to genotype C than those without the recombination (96.9 versus 91.1% in nucleotides and 97.0 versus 92.9% in amino acids). There were 29 HBV/B strains without the recombination, and they were all recovered from carriers in Japan. The remaining 41 HBV/B isolates having the recombination with genotype C were from carriers in China (12 strains), Hong Kong (3 strains), Indonesia (4 strains), Japan (3 strains), Taiwan (4 strains), Thailand (3 strains), and Vietnam (12 strains). Due to the frequency of the distribution of HBV/B without the recombination (29 of 32 isolates, or 91%) and the fact that it was exclusive to Japan, it was provisionally classified into the Bj (j standing for Japan) subgroup, and HBV/B with the recombination was classified into the Ba (a for Asia) subgroup. Virological differences between HBV/Bj and HBV/Ba may be reflected in the severity of clinical disease in the patients infected with HBV of genotype B, which seems to be under strong geographic influences in Asia.
At present, seven genotypes of hepatitis B virus (HBV) have been distinguished based on a divergence over the entire genomic sequence of >8%, and they are named with capital letters from A to G (11, 12, 18). These genotypes have distinct geographical distributions (7, 9) and may influence the biology of HBV and clinical disease in hosts. For instance, HBV of genotype C (HBV/C) induces more severe liver disease than HBV/B in Asia (6, 14), and HBV/A is associated more frequently with chronic infection than HBV/D in Europe (10).
HBV strains of the same genotype, however, may differ virologically and in the capacity to induce clinical disease. Such differences are most conspicuous in patients who are infected with HBV/B in Asia, where genotypes B and C are prevalent. HBV/B induces less severe liver disease than HBV/C in Japan and is not detected in patients with hepatocellular carcinoma (HCC) who are <30 years old, unlike in the other parts of Asia (13). Remarkably, patients infected with HBV/B in Taiwan develop HCC at a much earlier age than those with HBV/C (6).
These backgrounds point to virological differences in HBV strains of the same genotype which influence the severity and evolution of liver disease in hosts. For the purpose of sorting out such differences, the entire nucleotide sequences of 70 HBV/B isolates from Asia, including 38 identified in the present study and 32 retrieved from the international database (DDBJ/EMBL/GenBank), were compared in phylogenetic analyses. Two kinds of HBV/B emerged, one of which possessed the recombination with HBV/C over the precore (preC) region plus the core gene while the other did not. Based on HBV/B without the recombination being restricted to Japan and HBV/B with the recombination being ubiquitous in the other countries of Asia, combined with marked clinical differences among the patients infected with HBV/B in various Asian countries, the recombination with HBV/C may provide HBV/B with an increased capacity to induce liver disease.
MATERIALS AND METHODS
Determination of the complete nucleotide sequences of HBV/B isolates.
DNA was extracted from 100 μl of serum that had been stored at −20°C using a DNA extractor kit (Genome Science Laboratory, Fukushima, Japan). The complete nucleotide sequences of 38 HBV/B isolates recovered from patients with HBV-associated liver disease in various countries (7 from China, 3 from Hong Kong, 19 from Japan, 4 from Taiwan, 3 from Thailand, and 2 from Vietnam) were determined by the method reported previously (19). In brief, two overlapping fragments of 3,200 and 462 bp were amplified by PCR, and 11 overlapping HBV DNA fragments were amplified further by PCR with nested primers. Amplification was performed in a 96-well cycler (GeneAMP9600; Perkin-Elmer Cetus, Norwalk, Calif.), and the PCR products were run on electrophoresis in 3.0% (wt/vol) agarose, stained with ethidium bromide, and observed under the UV light. Standard precautions were taken for avoiding contamination during PCR. A negative control serum was also processed and included in each run to ensure specificity. Twelve overlapping HBV DNA fragments thus amplified were sequenced directly by the dideoxy method using a Dye Deoxy Terminator cycle-sequencing kit in a model 3100 fluorescent DNA sequencer (Applied Biosystems, Foster City, Calif.).
Thirty-two complete nucleotide sequences of HBV/B originating in various countries (China, 5; Indonesia, 4; Japan, 13; and Vietnam, 10) were retrieved from the international DNA database (DDBJ/EMBL/GenBank).
Phylogenetic analysis.
Full-length sequences of 70 HBV/B isolates were aligned with the CLUSTAL W software program (21), and the alignment was confirmed by visual inspection. Genetic distances were estimated by the six-parameter method, and phylogenetic trees were constructed by the neighbor-joining method (17). To confirm the reliability of the phylogenetic tree, bootstrap resampling and reconstruction were carried out 1,000 times for analysis by the ODEN program of the National Institute of Genetics (Mishima, Japan) (5).
Evidence for recombination events.
Recombination was searched for by the method of Robertson et al. (16) with use of the SimPlot program and bootscanning analysis (8). Four sequences were used for detecting phylogenetically informative sites: one putative recombinant sequence (genotype Ba), one sequence representative of each of the two parental lineages considered to be involved in the recombination (genotypes Bj and C), and one sequence of a known outgroup (genotype F). Informative sites were identified where two sequences shared one nucleotide and the other two shared another. P values were calculated using Fisher's exact test.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper will appear in the DDBJ/EMBL/GenBank databases under accession no. AB073821 through AB073858.
RESULTS
HBV/B isolates that had or did not have recombination with HBV/C.
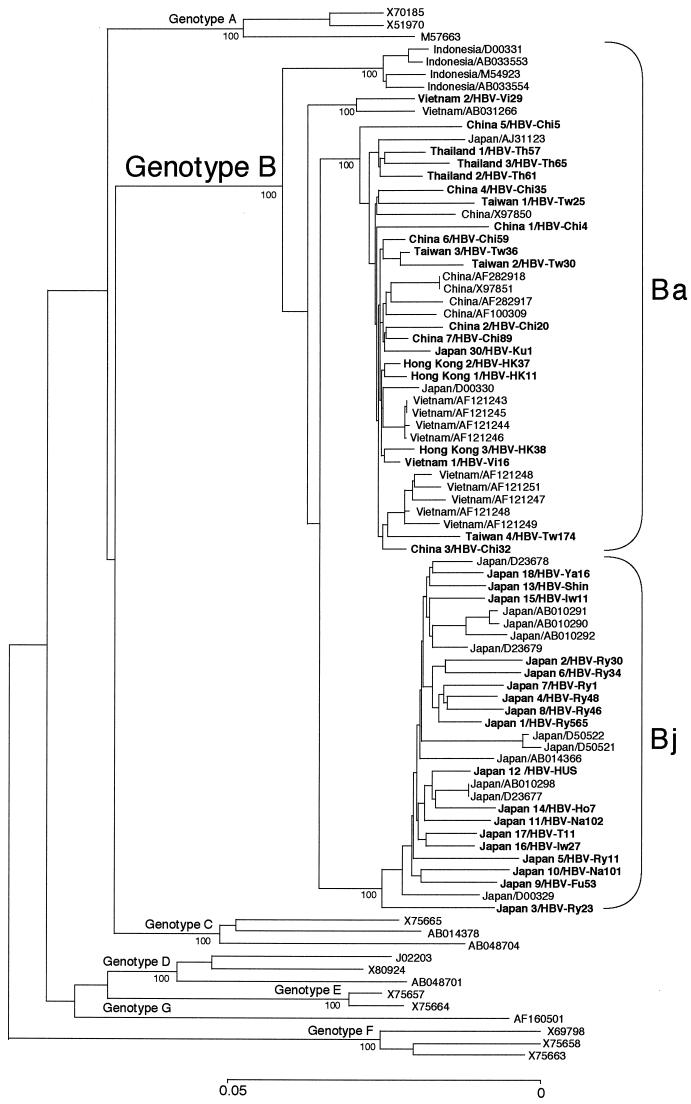
Figure 1 compares phylogenetically the entire nucleotide sequences of 70 HBV/B isolates among themselves and to 15 HBV isolates representative of the other six genotypes (A and C to G). Of the entire HBV/B sequences, 32 were retrieved from the international DDBJ/EMBL/GenBank database while the remaining 38 were determined in the present study (accession no. AB073821 to AB073858). All 70 HBV/B isolates from Asian carriers clustered on a branch of genotype B and further into four separate subbranches intrinsic to the countries where they were recovered. Notably, 29 of the 32 (91%) HBV/B isolates from Japan clustered on a single subbranch.
FIG. 1.
Phylogenetic tree constructed on the complete nucleotide sequences of 70 HBV isolates of genotype B and 15 isolates representative of genotypes A, C, D, E, F, and G. Bootstrap values are shown along each main branch. The length of the horizontal bar indicates the number of nucleotide substitutions per site. Countries of origin are indicated for genotype B, and accession numbers are shown after a slash for isolates the sequences of which have been deposited in DNA databases. Isolates in boldface were sequenced in the present study.
In phylogenetic analyses of the three open reading frames designated the polymerase (P) gene, X gene, and envelope (S) gene, all 70 HBV/B isolates clustered on a branch of genotype B, as in phylogenetic analysis of the full genome sequence (Fig. 1 and data not shown).
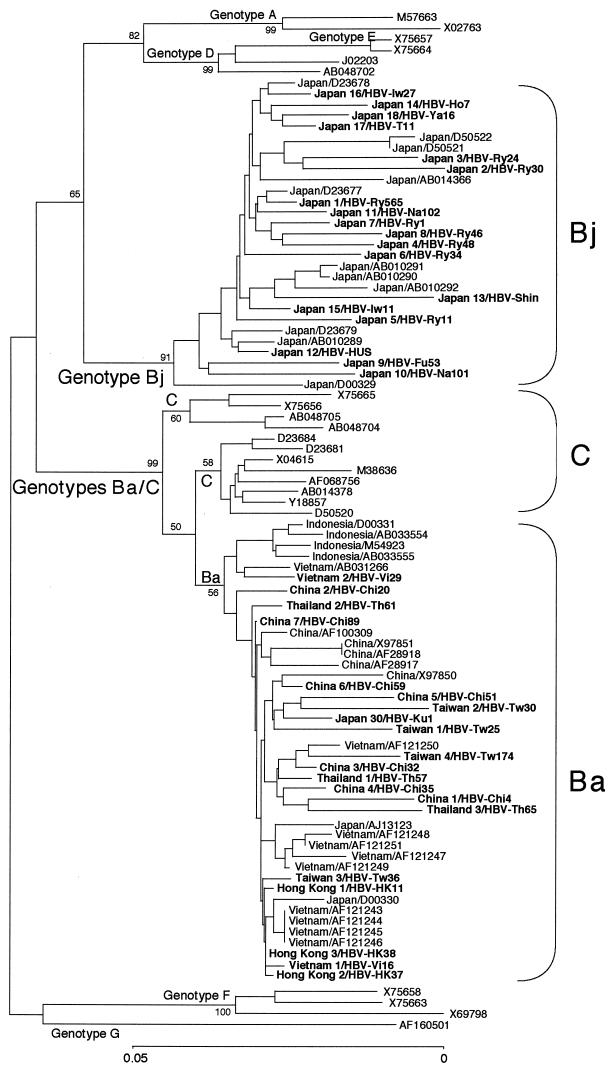
When the preC region plus the core gene sequences of 70 HBV/B isolates were analyzed phylogenetically, however, they divided into two categories based on association with HBV/C isolates (Fig. 2). The 29 HBV/B isolates from Japan were independent of HBV/C and clustered on a separate branch. By contrast, the remaining 41 HBV/B isolates from China (12 strains), Hong Kong (3 strains), Indonesia (4 strains), Taiwan (4 strains), Thailand (3 strains), and Vietnam (12 strains), as well as Japan (3 strains), clustered on another branch that joined one of the two branches bearing HBV/C isolates.
FIG. 2.
Phylogenetic tree constructed on the sequences of the preC region plus the core gene of 70 HBV isolates of genotype B and representatives of the other six genotypes. Bootstrap values are shown along each main branch. The length of the horizontal bar indicates the number of nucleotide substitutions per site. Countries of origin and accession numbers are shown for the isolates of genotype B. Isolates in boldface were sequenced in the present study.
According to distinct geographical distributions of B/C recombinants and nonrecombinants, the HBV/B isolates having the recombination with genotype C were assigned the genotype Ba (a standing for Asia) and those without it were assigned the genotype Bj (j for Japan) provisionally.
Extent of recombination with genotype C in HBV/Ba isolates.
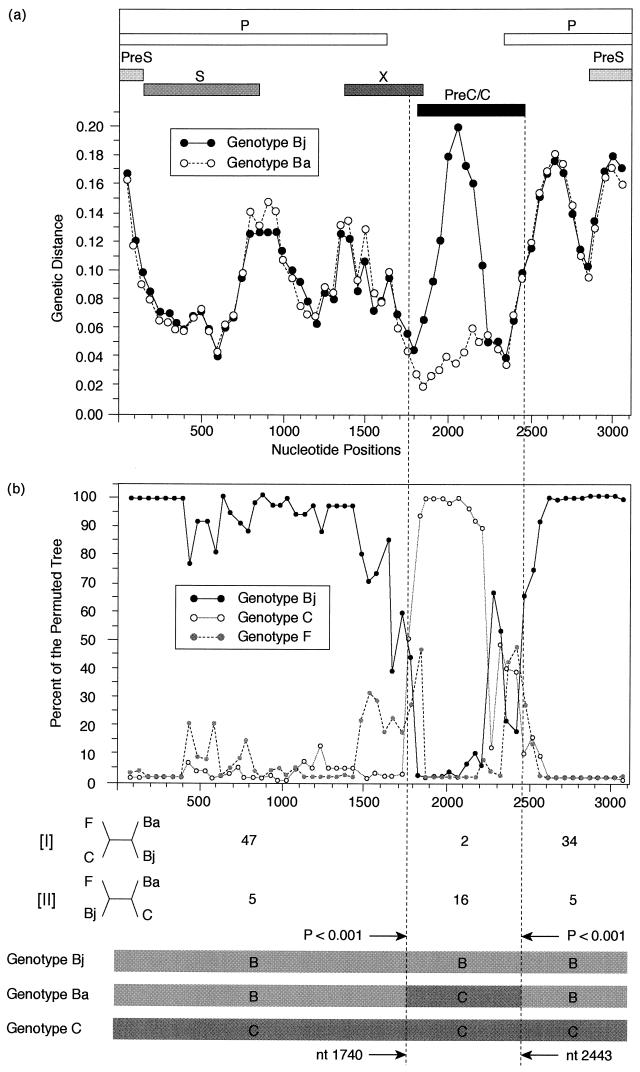
Figure 3a compares the genetic distances, measured against the standard HBV/C isolate (AB014378), among 10 isolates of genotype Bj and 10 isolates of genotype Ba over the entire genome. The profiles of genotypes Bj and Ba are comparable, except in the preC region plus the core gene. Within the preC-core, therefore, HBV isolates of genotype Ba were closer to HBV/C than those of genotype Bj.
FIG.3.
(a) Differences in genetic distances compared for the standard HBV isolate of genotype C among isolates of genotypes Bj and Ba. The average genetic distance from the reference genome of genotype C (accession no. D23684) of 10 HBV isolates of genotype Bj without the B-C recombination (Japan 1 to 10 [Fig. 1]) and that of 10 isolates of genotype Ba (Thailand 1, Thailand 2, China 1, China 2, Taiwan 1, Taiwan 2, Hong Kong 1, Hong Kong 2, Vietnam 1, and Vietnam 2 [Fig. 1]) are plotted over the entire genome. The average genetic distances were calculated using the SimPlot program (8) with a window size of 200 bp and a step size of 50 bp. PreC/C, preC region plus the core gene. (b) Resolution of recombinant events in HBV genomes of distinct genotypes was determined by the method of Robertson et al. (16) using the SimPlot program and bootscanning analysis (8). Three representative HBV isolates of genotypes Bj (Japan 9), C (partner of recombination; accession no. D23684), and F (outgroup; accession no. X75658) were compared against a representative of genotype Ba (Taiwan 1) over the entire genome with a window size of 200 bp and a step size of 50 bp (gap strip off; 100 bootstrap replicates and neighbor-joining tree analysis). The dotted vertical lines indicate breakpoints determined by chi-square maximization (the nucleotide positions are shown with solid arrows) with the numbers of phylogenetically informative sites. The numbers of informative sites shared by the topology ([I] and [II]) are shown at the bottom.
To locate the breakpoint of recombination events more precisely, the SimPlot program and bootscanning analysis (8) were carried out by the method described in Material and Methods. Figure 3b depicts the resolution of the breakpoint of B-C recombination events based on the four entire HBV genomes of genotypes Ba, Bj, C, and F. The B-C recombination breakpoints were estimated at nucleotides (nt) 1740 and 2443, respectively, by mapping the informative site and chi-square maximization (16).
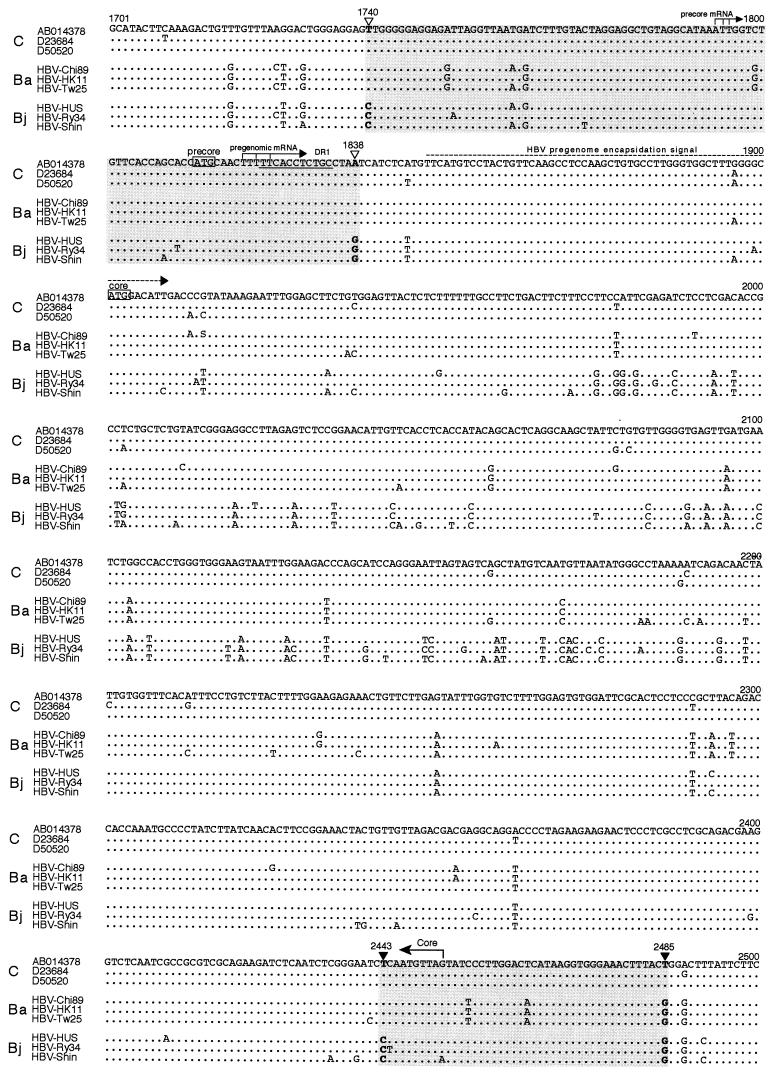
When all 41 isolates of genotype Ba were examined for the breakpoints of recombination events, the 5′ terminus was located within nt 1740 to 1838 and the 3′ terminus was located within nt 2443 to 2485 (Fig. 4). The recombination with genotype C in all genotype Ba strains covered the major part of the preC region plus the core gene and spanned 606 to 746 bp.
FIG.4.
Nucleotide alignments over a sequence spanning nt 1701 to 2500. They cover the putative breakpoints of recombination events in three isolates (each) representative of genotypes C (accession no. AB014378, D23684, and D50520), Ba (HBV-Chi89, HBV-HK11, and HBV-Tw25), and Bj (HBV-HUS, HBV-Ry34, and HBV-Shin). The 5′-terminal breakpoints of recombination events (within nt 1740 to 1838) are flanked by open triangles, and the 3′-terminal breakpoints (within nt 2443 to 2485) are flanked by solid triangles. The positions of various genes and elements, as well as the start of pregenomic mRNA, are indicated above the sequence.
Nucleotide and amino acid homology within the entire preC region plus the core gene between the HBV isolates of genotypes Bj and Ba.
Table 1 shows the mean pairwise nucleotide and amino acid homology as percentages, within the entire preC region plus the core gene, among the 18 isolates of genotype Bj and the 20 isolates of genotype Ba; their sequences were determined in this study. Within the genomic area in comparison, the HBV isolates of genotype Bj had no homology in either nucleotide or amino acid sequences to those of genotypes from A to G, except those of genotype Bj. The 18 HBV isolates of genotype Bj possessed a homology in nucleotide sequences of 96.5% and a homology in amino acid sequences of 96.6%. By remarkable contrast, the 20 HBV isolates of genotype Ba had homologies in both nucleotide and amino acid sequences not only among themselves (98.8 and 98.9%, respectively) but also to the isolate of genotype C (96.9 and 97.0%, respectively); they had homologies to the 18 HBV isolates of genotype Bj of only 91.1% in nucleotide sequences and 92.9% in amino acid sequences, however.
TABLE 1.
Comparison of percent homology in nucleotide and amino acid sequences within the preC region plus core gene between the 18 HBV isolates of genotype Bj and the 20 of genotype Baa
| Genotype | Accession no. | % Homology with HBV isolates of genotype Bb
|
|
|---|---|---|---|
| Bj (n = 18) | Ba (n = 20) | ||
| A | X70185 | 90.8 (92.6) | 89.6 (92.4) |
| Bj (n = 18) | 96.5 (96.6) | 91.1 (92.9) | |
| Ba (n = 20) | 91.1 (92.9) | 98.8 (98.9) | |
| C | D23684 | 90.9 (91.1) | 96.9 (97.0) |
| D | J02203 | 91.6 (92.5) | 91.1 (93.4) |
| E | X75657 | 90.9 (91.5) | 90.4 (93.0) |
| F | X75663 | 88.9 (91.5) | 89.8 (91.6) |
| G | AF160501 | 89.1 (91.9) | 89.6 (91.9) |
Each of the 18 isolates of genotype Bj and the 20 of genotype Ba was compared against the representative HBV isolates of genotypes A, C, D, E, F, and G, as well as among themselves. The mean percent homology of nucleotide sequence is shown, with the mean percent homology of amino acid sequence in parentheses.
Sequences of the 18 HBV isolates of genotype Bj and the 20 of genotype Ba were determined in the present study (Fig. 1). High homologies are in boldface type.
DISCUSSION
The most salient result of the present study is that HBV isolates of genotype B common in Asia are classified into two categories, one of which has the recombination with genotype C while the other does not. Comparison of 41 HBV/B isolates having the recombination to those without it located the 5′- and 3′-terminal breakpoints within nt 1740 to 1838 and 2443 to 2485, respectively. The recombination was restricted to the preC region plus the core gene.
HBV strains of certain genotypes in which a part of the open reading frame is replaced by the corresponding part of those of the other genotypes have been reported (2, 3). With a small number of recombinant strains available, it is not clear how frequently they occur and influence the evolution and pathogenicity of HBV. It may come as a surprise that HBV isolates of the “genuine” genotype B, without the recombination with genotype C, were found to be restricted to Japan and accounted for 29 of the 32 (91%) carriers of HBV/B there. By remarkable contrast, all 38 of the HBV/B isolates from the other countries in Asia, including China, Hong Kong, Indonesia, Taiwan, Thailand, and Vietnam, possessed the recombination with genotype C. Taken together, the evidence indicates that the authentic genotype B is intrinsic to Japan while the genotype B recombined with genotype C is ubiquitous in the other countries in Asia. Based on such exclusive geographical distributions, the genotype Bj is proposed for HBV/B isolates without the recombination with HBV/C and the genotype Ba is proposed for those with it.
How the recombination between genotypes B and C occurs and why it is not evenly distributed over Asian countries are not known. The prevalence of persistent infection, much more frequent in the other Asian countries than in Japan in the past, would have accelerated coinfection with HBV strains of genotypes B and C there and resulted in the recombination between them. Whether HBV/B with the recombination would have an advantage for replication and persistence in hosts over either HBV/B or HBV/C is not known. It is beyond doubt, however, that HBV/B with the recombination with HBV/C survived HBV of the genuine genotype B and has overtaken it in the individuals who were doubly infected originally.
Recombination between different genotypes occurs frequently in retroviruses, represented by human immunodeficiency virus type 1 (8). For RNA viruses, homologous recombination and template switching have been proposed as the mechanisms for intragenic recombination (8). The recombination between HBV isolates of distinct genotypes might not be irrelevant to the replication of HBV, which involves the reverse transcription of an RNA intermediate (20). The packaging of a single pregenome into the nucleocapsid of HBV, however, does not leave room for the recombination to occur between two pregenomes. Given many lines of solid evidence for recombination between distinct HBV genotypes (2, 3), the theory of HBV replication would need to be modified to explain it.
Another point of note that has surfaced in this study is the region of recombination in all 41 B-C hybrid strains, which is invariably in the preC region plus the core gene spanning 606 to 746 bp. It should be pointed out that this recombination region encompasses the DR1 region and encapsidation signal of the HBV pregenome. The DR1 region covers a hot spot (nt 1885 to 1915) for genomic recombination and is frequently integrated into the host genome (15). Of possible relevance to this, a cellular protein with the capacity to bind with this hot spot in HBV DNA has been reported and is proposed as a putative recombinogenic protein (1). Combined, this genomic region covering DR1 would be a candidate site for intragenic recombination between HBV isolates of distinct genotypes, as well as for intergenic recombination between HBV and the host genome.
In Asia, where genotypes B and C account for essentially all HBV infections (13), these two genotypes have been compared for any clinical differences. There are unequivocal lines of evidence to indicate that infection with HBV/C can induce more severe liver disease than HBV/B (6, 14). Seroconversion from hepatitis B e antigen to the corresponding antibody occurs earlier in HBV/B than HBV/C carriers, which is implicated in different severities of liver disease developing in them (13, 14). Putative cytotoxic-T-cell epitopes are presumed on the product of the HBV core gene (4). Insofar as the recombination with genotype C in HBV isolates of genotype Ba was restricted to the preC region plus the core gene, cytotoxic-T-cell epitopes of HBV/C would be more efficiently expressed in them than in HBV of the genuine genotype B (Bj) for an enhanced disease-inducing capacity. The development of HCC in young carriers in Taiwan (6) may well be attributed to the recombination of HBV/B with HBV/C.
Acknowledgments
This study was supported in part by grants from the Japanese Ministry of Education, Science, Sports and Culture (12670506 and 11691222).
REFERENCES
- 1.Aoki, H., K. Kajino, Y. Arakawa, and O. Hino. 1996. Molecular cloning of a rat chromosome putative recombinogenic sequence homologous to the hepatitis B virus encapsidation signal. Proc. Natl. Acad. Sci. USA 93:7300-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bollyky, P. L., A. Rambaut, P. H. Harvey, and E. C. Holmes. 1996. Recombination between sequences of hepatitis B virus from different genotypes. J. Mol. Evol. 42:97-102. [DOI] [PubMed] [Google Scholar]
- 3.Bowyer, S. M., and J. G. Sim. 2000. Relationships within and between genotypes of hepatitis B virus at points across the genome: footprints of recombination in certain isolates. J. Gen. Virol. 81:379-392. [DOI] [PubMed] [Google Scholar]
- 4.Chisari, F. V., and C. Ferrari. 1995. Hepatitis B virus immunopathogenesis. Annu. Rev. Immunol. 13:29-60. [DOI] [PubMed] [Google Scholar]
- 5.Ina, Y. 1994. ODEN: a program package for molecular evolutionary analysis and database search of DNA and amino acid sequences. Comput. Appl. Biosci. 10:11-12. [DOI] [PubMed] [Google Scholar]
- 6.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 2000. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 118:554-559. [DOI] [PubMed] [Google Scholar]
- 7.Lindh, M., A. S. Andersson, and A. Gusdal. 1997. Genotypes, nt 1858 variants, and geographic origin of hepatitis B virus—large-scale analysis using a new genotyping method. J. Infect. Dis. 175:1285-1293. [DOI] [PubMed] [Google Scholar]
- 8.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magnius, L. O., and H. Norder. 1995. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology 38:24-34. [DOI] [PubMed] [Google Scholar]
- 10.Mayerat, C., A. Mantegani, and C. Frei. 1999. Does hepatitis B virus (HBV) genotype influence the clinical outcome of HBV infection? J. Viral Hepatitis 6:299-304. [DOI] [PubMed] [Google Scholar]
- 11.Norder, H., A. M. Courouce, and L. O. Magnius. 1994. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 198:489-503. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto, H., F. Tsuda, H. Sakugawa, R. I. Sastrosoewignjo, M. Imai, Y. Miyakawa, and M. Mayumi. 1988. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J. Gen. Virol. 69:2575-2583. [DOI] [PubMed] [Google Scholar]
- 13.Orito, E., T. Ichida, H. Sakugawa, M. Sata, N. Horiike, K. Hino, K. Okita, T. Okanoue, S. Iino, E. Tanaka, K. Suzuki, H. Watanabe, S. Hige, and M. Mizokami. 2001. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology 34:590-594. [DOI] [PubMed] [Google Scholar]
- 14.Orito, E., M. Mizokami, H. Sakugawa, K. Michitaka, K. Ishikawa, T. Ichida, T. Okanoue, H. Yotsuyanagi, and S. Iino, et al. 2001. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Hepatology 33:218-223. [DOI] [PubMed] [Google Scholar]
- 15.Pineau, P., A. Marchio, M. G. Mattei, W. H. Kim, J. K. Youn, P. Tiollais, and A. Dejean. 1998. Extensive analysis of duplicated-inverted hepatitis B virus integrations in human hepatocellular carcinoma. J. Gen. Virol. 79:591-600. [DOI] [PubMed] [Google Scholar]
- 16.Robertson, D. L., B. H. Hahn, and P. M. Sharp. 1995. Recombination in AIDS viruses. J. Mol. Evol. 40:249-259. [DOI] [PubMed] [Google Scholar]
- 17.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 18.Stuyver, L., S. De Gendt, C. Van Geyt, F. Zoulim, M. Fried, R. F. Schinazi, and R. Rossau. 2000. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J. Gen. Virol. 81:67-74. [DOI] [PubMed] [Google Scholar]
- 19.Sugauchi, F., M. Mizokami, E. Orito, T. Ohno, H. Kato, S. Suzuki, Y. Kimura, R. Ueda, L. A. Butterworth, and W. G. Cooksley. 2001. A novel variant genotype C of hepatitis B virus identified in isolates from Australian Aborigines: complete genome sequence and phylogenetic relatedness. J. Gen. Virol. 82:883-892. [DOI] [PubMed] [Google Scholar]
- 20.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 21.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]