Abstract
More than 10 mature proteins processed from coronavirus gene 1-encoded polyproteins have been identified in virus-infected cells. Here, we report the identification of the most C-terminal cleavage product of the 1a polyprotein as a 16-kDa protein in infectious bronchitis virus-infected Vero cells. Indirect immunofluorescence demonstrated that the protein exhibits a distinct perinuclear punctate staining pattern, suggesting that it is associated with cellular membranes. Positive staining observed on nonpermeabilized cells indicates that the protein may get transported to the cell surface, but no secretion of the protein out of the cells was observed. Treatment of the membrane fraction prepared from cells expressing the 16-kDa protein with Triton X-100, a high pH, and a high concentration of salts showed that the protein may be tightly associated with intracellular membranes. Dual-labeling experiments demonstrated that the 16-kDa protein colocalized with the 5′-bromouridine 5′-triphosphate-labeled viral RNA, suggesting that it may be associated with the viral replication machinery. Sequence comparison of the 16-kDa protein with the equivalent products of other coronaviruses showed multiple conserved cysteine residues, and site-directed mutagenesis studies revealed that these conserved residues may contribute to dimerization of the 16-kDa protein. Furthermore, increased accumulation of the 16-kDa protein upon stimulation with epidermal growth factor was observed, providing preliminary evidence that the protein might be involved in the growth factor signaling pathway.
The 27.6-kb genome length mRNA, mRNA1, of the avian coronavirus infectious bronchitis virus (IBV) is composed of two large overlapping open reading frames (ORFs), 1a and 1b, at the 5′ unique region. It encodes a 441-kDa 1a polyprotein and a 1a/1b fusion polyprotein of 741 kDa by a frameshifting mechanism (6, 7, 8). Proteolytic processing of the 441-kDa 1a and 741-kDa 1a/1b polyproteins to smaller mature products is mediated by two proteinases, namely, a papain-like proteinase and a picornavirus 3C-like proteinase (see Fig. 1a). The papain-like proteinase is involved in cleavage of the N-terminal part of the 1a and 1a/1b polyproteins to release an 87-kDa protein and a 195-kDa protein in IBV-infected cells (23, 24, 26). Similar enzymatic activities were also identified for the first and second papain-like proteinase domains of human and murine coronaviruses (1, 2, 3, 11, 14, 17, 20, 21, 36, 45). Processing of the 1a and 1a/1b polyproteins at other positions is mediated by the 3C-like proteinase. This proteinase was shown to recognize several conserved Q-S (G and N) dipeptide bonds, resulting in the release of more than 10 mature products (4, 16, 18, 19, 25, 27, 28, 29, 31, 32, 33, 34, 36, 39, 44), and was identified as a 33-kDa protein for IBV (24, 34), a 27- or 29-kDa protein for the coronavirus MHV-A59 (30, 32, 35) and a 34-kDa protein for human coronavirus (42, 43, 44).
FIG. 1.
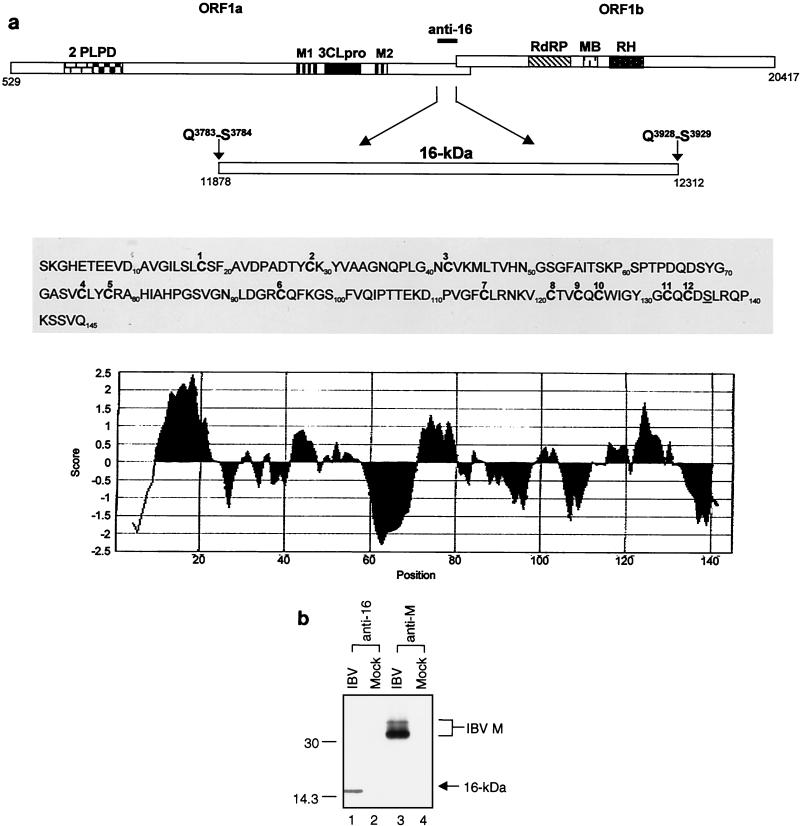
(a) Diagram of ORFs 1a and 1b illustrating the two overlapping papain-like proteinase domains (PLPDs), the 3C-like proteinase (3CLpro), the RNA-dependent RNA polymerase, the metal-binding domain, and the helicase. The Q3783-S3784 and Q3928-S3929 scissile bonds and 16-kDa viral product generated by cleavage with the 3C-like proteinase are indicated. Also shown are the IBV sequence recognized by anti-16-kDa protein serum, the stretches of hydrophobic residues (M1 and M2) encoded by ORF 1a, and the amino acid sequence and hydropathy plot (22) of the 16-kDa protein. The molecular mass of the 16-kDa protein was estimated on the basis of its migration on SDS-PAGE. The Q3783-S3784 and Q3928-S3929 residues are encoded by nucleotides 11,875 to 11,880 and 12,310 to 12,315, respectively. The 12 cysteine residues are in bold. (b) Detection of the 16-kDa protein in IBV-infected Vero cells. IBV-infected (IBV) and mock-infected (Mock) Vero cells were labeled with [35S]methionine-cysteine and harvested at 12 h postinfection. The cell debris and nuclei fractions were removed by low-speed centrifugation (1,500 × g) for 10 min, and the cytosol and membrane fractions were separated by high-speed centrifugation (150,000 × g). The cytosol and membrane fractions were immunoprecipitated with anti-16-kDa protein serum and anti-M serum. Polypeptides were separated on an SDS-15% polyacrylamide gel and detected by fluorography. The values on the left are molecular sizes in kilodaltons.
Recently, immunofluorescence and biochemical studies with region-specific antisera have suggested that 3C-like proteinase and several other cleavage products may be membrane associated and are colocalized with the viral RNA replication-transcription machinery (4, 5, 10, 34, 36, 37, 38, 41, 44). A more recent study on MHV-A59 suggested that only p28, the first cleavage product (9, 12), might play a direct role in viral RNA synthesis, together with polymerase and helicase, while other cleavage products, such as p1a-22, may segregate into different but tightly associated membrane populations that may have independent functions during viral replication (4, 5, 38). Elucidation of the exact functions of these cleavage products would be essential for understanding the replication mechanism of coronavirus. One particularly interesting protein is the most C-terminal cleavage product of the 1a polyprotein. As a cysteine-rich protein, it was predicted to be a viral growth factor-like protein (15).
In this report, we show the identification of a 16-kDa protein, representing the most C-terminal cleavage product of the IBV 1a polyprotein. Indirect immunofluorescent staining of IBV-infected Vero cells and Cos-7 cells expressing the 16-kDa protein showed a distinct perinuclear, punctate staining pattern. Confocal microscopy and lateral laser sectioning demonstrated that this protein is associated with intracellular membrane structures, and positive staining in nonpermeabilized cells suggested strongly that it gets transported to the cell surface. However, the protein cannot be secreted into the culture medium and no cleavable signal sequence was present in the N-terminal region of the protein. Treatment with detergent, an alkaline pH, and a high salt concentration demonstrated that the 16-kDa protein may be tightly associated with the intracellular membranes. In addition, dual labeling of IBV-infected cells with 5′-bromouridine 5′-triphosphate (BrUTP) and anti-16-kDa protein serum showed that the 16-kDa protein was colocalized with viral RNA, suggesting that it may be associated with the viral RNA replication machinery. Sequence comparison with other members of the Coronaviridae family showed that the presence of multiple conserved cysteine residues, and mutagenesis studies demonstrated that these cysteine residues could form interchain disulfide bonds, resulting in dimerization of the 16-kDa protein. Finally, increased accumulation of the 16-kDa protein was observed when cells expressing the protein were stimulated with epidermal growth factor (EGF), suggesting that the protein might be involved in the EGF signaling pathway.
MATERIALS AND METHODS
Virus and cells.
The egg-adapted Beaudette strain of IBV (ATCC VR-22), obtained from the American Type Culture Collection, was adapted to Vero cells and prepared as described previously (33, 34). Virus stocks were prepared after the 62nd passage by infecting monolayers of Vero cells at a multiplicity of infection of approximately 0.1 PFU/cell and harvested at 24 h postinfection. The titer of the virus preparation was determined by plaque assay on Vero cells.
Vero, Cos-7, and BHK-21 cells were grown at 37°C in 5% CO2 and maintained in Dulbecco's modified minimal essential medium (Gibco BRL, Life Technologies) supplemented with 10% newborn calf serum.
Radiolabeling of IBV-infected Vero cells and transient expression of plasmid DNAs with the vaccinia virus-T7 expression system.
Confluent monolayers of Vero cells grown on 60-mm-diameter dishes were infected with IBV at a multiplicity of infection of approximately 2 PFU/cell. The cells were labeled with [35S]methionine-cysteine at 25 μCi/ml at 4 h postinfection and harvested at 8 h postinfection.
Semiconfluent monolayers of Cos-7 cells grown on 60-mm dishes were infected with 10 PFU of vTF7-3 per cell (13) and transfected with 12 μg of plasmid DNAs with 20 μl of Lipofectin liposomal transfection reagent in accordance with the instructions of the manufacturer (Gibco, BRL). After incubation of the cells at 37°C for 7 h, the cells were incubated in methionine- and cysteine-free medium (ICN) for 30 min and labeled with 25 μCi of [35S]methionine-cysteine (35S Express Protein Labeling Mix; NEN Life Science) per ml for 13 h. The cells were scraped off the dishes in phosphate-buffered saline and recovered by centrifugation at 14,000 × g for 1 min.
Cell fractionation of [35S]methionine-cysteine-labeled cells.
Radiolabeled cells were resuspended in hypotonic buffer (1 mM Tris-HCl [pH 7.4], 0.1 mM EDTA, 15 mM NaCl) containing 2 μg of leupeptin per ml and 0.4 mM phenylmethylsulfonyl fluoride and broken by 20 strokes with a Dounce cell homogenizer. Cell debris and nuclei were removed by centrifugation at 1,500 × g for 10 min at 4°C. The membranes (postnuclear fraction) were pelleted by ultracentrifugation through a 6% sucrose cushion in a Beckman TLA 120.1 rotor (150,000 × g for 30 min at 4°C). Membrane pellets were resuspended in immunoprecipitation buffer (20 mM Tris-HCl [pH 7.4], 5 mM EDTA, 150 mM NaCl, 0.5% sodium dodecyl sulfate [SDS], 0.1% sodium deoxycholate, 0.5% Nonidet P-40), and the cytosol fraction (supernatant) was adjusted to the same volume and ion-detergent conditions with 5× immunoprecipitation buffer. Samples were precleared by centrifugation, incubated with specific antisera for 30 min at room temperature, and then incubated with protein A-Sepharose CL-4B (Sigma) for 30 min. Immunoprecipitated beads were washed three times with the immunoprecipitation buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
Radioimmunoprecipitation assay and SDS-PAGE.
SDS-PAGE was carried out with 15% polyacrylamide concentrations, and the labeled polypeptides were detected by autoradiography or fluorography of dried gels as described previously (25).
PCR.
Appropriate primers and template DNAs were used for amplification reactions with cloned Pfu DNA polymerase (Stratagene) under standard buffer conditions with 2 mM MgCl2.
Cell-free transcription and translation.
Plasmid DNAs were expressed in rabbit reticulocyte lysate with a coupled transcription-translation system (TnT; Promega) in the presence of [35S]methionine (Amersham-Pharmacia Biotech) in accordance with the instructions of the manufacturer. One microgram of plasmid DNA was incubated in a 50-μl reaction mixture with or without 5 μl of canine pancreatic microsomal membranes (Promega).
BrUTP labeling of de novo-synthesized viral RNA.
IBV-infected or mock-infected Vero cells were treated with 20 μg of actinomycin D (Sigma) at 10 h postinfection for 4 h to shut off host cell mRNA transcription. One millimolar BrUTP (Sigma) was introduced into cells by using 7.5 μl of SuperFect (Qiagen), and the cells were incubated for another 1 h before fixation and permeabilization for immunofluorescent staining as described below.
Indirect immunofluorescence microscopy and confocal microscopy.
Cells were grown on coverslips and infected with IBV or transfected with appropriate plasmid DNAs. After washing with phosphate-buffered saline containing 1 mM CaCl2 and 1 mM MgCl2 (PBSCM), the cells were fixed with 4% paraformaldehyde (in PBSCM) for 30 min at room temperature and permeabilized with 0.2% Triton X-100 (in PBSCM), followed by incubation with specific antiserum at room temperature for 2 h. Antibodies were diluted in fluorescence dilution buffer (PBSCM with 5% normal goat serum, 5% newborn calf serum, and 2% bovine serum albumin, pH 7.6). The cells were then washed with PBSCM and incubated with fluorescein isothiocyanate- or tetramethylrhodamine isothiocyanate-conjugated anti-rabbit or anti-mouse immunoglobulin G (Sigma) in fluorescence dilution buffer at 4°C for 1 h before mounting.
Confocal microscopy was performed on a Zeiss Axioplan microscope connected to a Bio-Rad MRC 1624 laser scanner equipped with an argon laser with appropriate filters. Fluorescent images were superimposed to allow fine comparison, and colocalization of green (fluorescein isothiocyanate) and red (tetramethylrhodamine isothiocyanate) signals in a single pixel produces yellow while separated signals are green or red.
Western blot analysis.
SDS-PAGE of viral polypeptides was carried out as mentioned, and viral proteins were transferred to nitrocellulose membranes (Stratagene) with a semidry transfer cell (Bio-Rad Trans-Blot SD). The membranes were blocked overnight at 4°C in blocking buffer containing 5% skim milk powder in TBST (20 mM Tris HCl [pH 7.4], 150 mM NaCl, 0.1% Tween 20) and incubated with specific antiserum diluted in blocking buffer (1:500 to 1:2,000) at room temperature for 2 h. Membranes were washed three times with TBST, further blocked in blocking buffer for 20 min at room temperature, incubated with horseradish peroxidase-conjugated anti-rabbit or anti-mouse immunoglobulin G (Dako) diluted in blocking buffer (1:2,500) at room temperature for 1 h, and then subjected to color development with detection reagents (enhanced chemiluminescence [ECL]).
Construction of plasmids.
Plasmids pIBVM and pBP5 were described before (28, 33, 34). The IBV sequences present in these two constructs are from nucleotides 24430 to 27608 and 10752 to 12313, respectively.
Plasmid pIBV16K, which covers nucleotides 11875 to 12312 and contains a UAG termination codon immediately downstream of the viral sequence, was constructed by ligating a 437-bp PCR fragment into NcoI- and BamHI-digested pKT0. The cloning primers are LN-9 (5′-GTCTTACCATGGAGTCTAAAGGGCAT-3′) and LDX-5 (5′-GCTCCAGGATCCTATTGAACAGAAGA-3′). Plasmid pSinRep16K was cloned by ligating a 500-bp NheI- and SmaI-digested fragment from pIBV16K into XbaI- and StuI-digested pSinRep5 (Invitrogen).
Plasmid pSinRepEGFR was constructed by cloning a 3.5-kb reverse transcription-PCR fragment coding for the EGF receptor (EGFR) from total RNA extracted from HeLa cells. The XbaI- and StuI-digested 3.5-kb PCR fragment was cloned into XbaI- and StuI-digested pSinRep5. The cloning primers were EGFR-5′ (5′-GCAGCGTCTAGAATGCGACCCTCCGGG-3′) and EGFR-3′ (5′-CCGTGGAGGCCTTCATGCTCCAATAAA-3′). Plasmid pT716K was cloned by ligating a BglII- and BamHI-digested fragment from pIBV16K into BamHI-digested vector pT7φ10 (34) containing the T7 tag sequence. All subsequent mutant forms with the cysteine residues mutated to alanine were constructed by site-directed mutagenesis.
RESULTS
Identification of the 16-kDa protein in IBV-infected Vero cells.
We have previously shown that two Q-S dipeptide bonds (Q3783-S3784 and Q3928-S3929) encoded by nucleotides 11875 to 11880 and 12310 to 12315, respectively, are cleavage sites of the IBV 3C-like proteinase (27, 28). Cleavage at these two positions would result in the release of a polypeptide with a calculated molecular mass of 15.4 kDa (Fig. 1a). To identify this product, a region-specific polyclonal antiserum, anti-16-kDa protein serum, was raised in rabbits against the IBV sequence encoded between nucleotides 11878 and 12312 with protein expressed in Escherichia coli.
The initial application of this antiserum for identification of the predicated cleavage product in IBV-infected cell lysates proved difficult due to the high background level. Various ways were attempted to enrich the protein and reduce the background, and the protein was successfully identified in IBV-infected Vero cells after removing the nuclear fraction from total cell lysates by centrifugation at low speed (1,500 × g) at 4°C for 10 min. As shown in Fig. 1b, a polypeptide with an apparent molecular mass of 16 kDa was specifically identified in IBV-infected Vero cells (lane 1) but not in mock-infected cells (lane 2). The 16-kDa protein may represent the cleavage product released from the Q3783-S3784 and Q3928-S3929 dipeptide bonds. In a control experiment, the postnuclear fractions of cell lysates prepared from IBV-infected and mock-infected Vero cells were subjected to an immunoprecipitation assay with anti-M serum. The anti-M serum specifically detected the IBV membrane protein (M) in IBV-infected cells (Fig. 1b, lane 3) but not in mock-infected cells (Fig. 1b, lane 4).
Membrane association and colocalization of the 16-kDa protein with de novo-synthesized viral RNA.
Subcellular localization of the 16-kDa protein was first studied by fusing the protein with the green fluorescent protein (GFP) to gain initial clues to where the protein is localized in cells. Transfection of Cos-7 cells with pEGFP-16k showed a perinuclear, punctate staining pattern (Fig. 2a, part A), while the GFP control showed a diffuse distribution pattern (Fig. 2a, part B), suggesting that the fusion protein may be localized to intracellular membrane compartments. Indirect immunofluorescent staining of IBV-infected Vero cells fixed with 4% paraformaldehyde at 10 h postinfection with anti-16-kDa protein serum showed a staining pattern similar to that of the GFP-16-kDa fusion protein (Fig. 2a, part C). No positive staining was observed in mock-infected cells (Fig. 2a, part D).
FIG. 2.
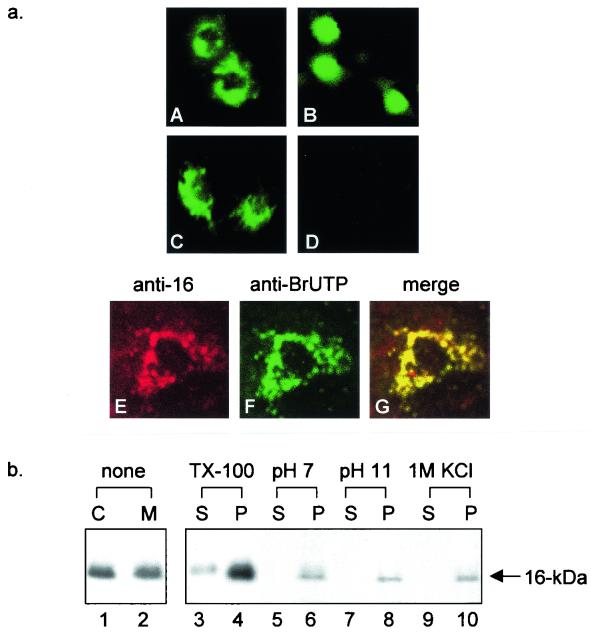
(a) Subcellular localization of the 16-kDa protein. (A) Cos-7 cells overexpressing pEGFP-16k at 16 h posttransfection showing perinuclear staining. (B) Cos-7 cells overexpressing the EGFP control vector (pEGFP) showing diffuse staining. (C) Immunofluorescent staining of IBV-infected Vero cells with anti-16-kDa protein serum (1:20) at 13 h postinfection showing distinct punctate perinuclear staining. (D) Immunofluorescent staining of mock-infected Vero cells with anti-16-kDa protein serum (1:20) at 13 h postinfection. (E) Double labeling of IBV-infected Vero cells that were transfected with BrUTP at 10 h postinfection showing the staining profile of anti-16-kDa protein serum. (F) Double labeling of IBV-infected Vero cells that were transfected with BrUTP at 10 h postinfection showing the staining pattern of anti-BrUTP serum. (G) Superimposition of images E and F. All images were taken from a Zeiss Axioplan confocal microscope. (b) Membrane association of the 16-kDa protein. Cos-7 cells expressing the 16-kDa protein were labeled with [35S]methionine-cysteine for 4 h and harvested. Cells were lysed with a Dounce homogenizer and fractionated into membrane (M) and cytosol (C) fractions at pH 7 (lanes 1 and 2) by ultracentrifugation. Polypeptides were immunoprecipitated with anti-16-kDa protein serum, separated on an SDS-15% polyacrylamide gel, and detected by fluorography. Membrane (M) pellets were resuspended in hypotonic buffer; treated with 1% Triton X-100 (TX-100), 100 mM Na2CO3, or 1 M KCl; and further fractionated into supernatant (S) and pellet (P) fractions by ultracentrifugation. Polypeptides were immunoprecipitated with anti-16-kDa protein serum, separated on an SDS-15% polyacrylamide gel, and detected by fluorography.
Several Nidovirales ORF 1a products were recently shown to be associated with the viral RNA replication machinery, as colocalization of these products with BrUTP-labeled viral RNA at the site of viral RNA synthesis was observed (4, 5, 36, 37, 38, 40, 41). The perinuclear, punctate staining pattern of the 16-kDa protein suggested that it may be one of these proteins. To investigate this possibility, IBV-infected Vero cells were BrUTP labeled at 10 h postinfection for 4 h and double labeled with an anti-BrdU monoclonal antibody (Roche) and anti-16-kDa protein serum, showing perinuclear punctate vesicular staining patterns (Fig. 2a, parts E and F). A clear overlap between the two labeling patterns was observed (Fig. 2a, part G), suggesting that the 16-kDa protein is associated with the viral RNA replication machinery.
The subcellular localization profiles of the 16-kDa protein strongly suggest that it is associated with cellular membranes. This possibility was further explored by fractionating Cos-7 cells expressing the protein into membrane and cytosol fractions and analyzing the presence of the protein in each fraction by immunoprecipitation assay. As shown in Fig. 2b, fractionation of cell lysates into cytosol and membrane fractions after removal of the nuclei and cell debris by centrifugation at low speed showed the presence of the 16-kDa protein in both the cytosol and membrane fractions (Fig. 2b, lanes 1 and 2). Quantification by densitometry showed that approximately 51 and 49% of the protein was detected in the cytosol and membrane fractions, respectively, indicating that a certain proportion of the protein is associated with cellular membranes.
Biochemical studies were then carried out to ascertain whether the 16-kDa protein is an integral membrane protein or a peripherally associated membrane protein. The membrane fraction shown in Fig. 2b (lane 2) was treated with either 1% Triton X-100, 100 mM Na2CO3 (pH 11), or 1 M KCl (high salt); ultracentrifuged to separate the soluble contents (S) from the pellets (P); and then immunoprecipitated with anti-16-kDa protein serum. Under high-pH conditions (pH 11), membrane vesicles are converted to open membrane sheets that can be recovered by ultracentrifugation. The soluble contents of the vesicles are released into the cytosol fraction, and proteins that are peripherally associated with membranes will therefore be found in the cytosol fraction. Only proteins that are tightly associated with membranes will remain in the membrane pellets. After treatment with Na2CO3, pH 11, and 1 M KCl, the 16-kDa protein was detected solely in the pellets (Fig. 2b, lanes 6, 8, and 10, respectively). However, treatment of the same membrane pellets with 1% Triton X-100 led to the detection of a significant amount of the 16-kDa protein in the supernatant (Fig. 2b, lanes 3 and 4).
Transport of the 16-kDa protein to the cell surface.
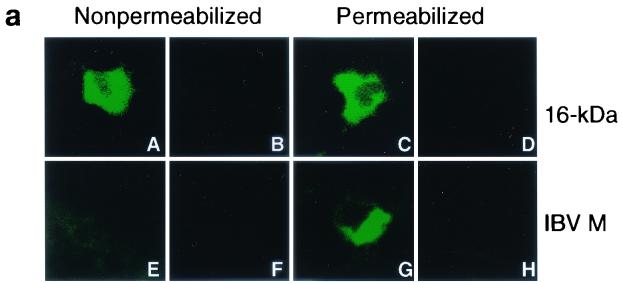
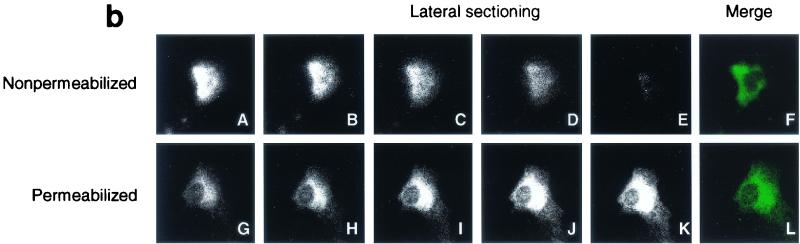
To investigate if the 16-kDa protein is transported to the cell surface, an indirect immunofluorescence assay was performed on Cos-7 cells expressing the 16-kDa protein under nonpermeabilized and permeabilized conditions. To avoid the high background level of anti-16-kDa protein serum, the protein was attached to an 11-amino-acid (MASMTGGQQMG) T7 tag at the N terminus and a highly specific anti-T7 monoclonal antibody (Novagen) was used to stain cells expressing the protein. As can be seen in Fig. 3a, positive staining was observed on nonpermeabilized Cos-7 cells (A), suggesting that the 16-kDa protein is transported to the cell surface. The IBV M protein was used as a control. Under nonpermeabilized conditions, anti-M serum did not detect any positive staining (Fig. 3a, part E) while the distinct Golgi pattern was observed on permeabilized cells (Fig. 3a, part G). Lateral laser sectioning was then performed to analyze the overall staining profile of the 16-kDa protein. A total of 35 sections were obtained from a typical positive cell, and five representative sections are shown in Fig. 3b. A strong fluorescence signal was observed in a section from the cell surface (Fig. 3b, parts A and B), and the strength of the signal was gradually reduced in sections toward the inside of the cell (Fig. 3b, parts C to E). These results suggest that the 16-kDa protein was present mainly on the cell surface when the cells were stained under nonpermeabilized conditions (Fig. 3b, part A to E). In permeabilized cells, the 16-kDa protein appeared to be localized in the perinuclear region (Fig. 3b, part L), similar to that observed in IBV-infected cells. Unlike in nonpermeabilized cells, the strength of the signal gradually increased from the section on the surface to the sections toward the inside of the cell (Fig. 3b, parts G to K).
FIG. 3.
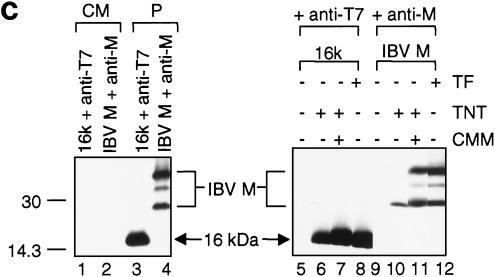
(a) Subcellular localization profiles of the 16-kDa protein. Immunofluorescent staining of Cos-7 cells expressing the T7-tagged 16-kDa protein with anti-T7 monoclonal antibody (1:200) at 6 h posttransfection showed positive staining under nonpermeabilized conditions (A) and a perinuclear staining pattern under permeabilized conditions (C). Immunofluorescent staining of Cos-7 cells expressing the IBV M protein with anti-M serum (1:30) at 6 h posttransfection showed negative staining under nonpermeabilized conditions (E) and a Golgi staining pattern under permeabilized conditions (G). Parts B, D, F, and H represent negative control cells that were transfected with an empty vector. (b) Transport of the 16-kDa protein to the cell surface. Immunofluorescent staining was performed on Cos-7 cells expressing the T7-tagged 16-kDa protein with anti-T7 monoclonal antibody (1:200) at 6 h posttransfection. Lateral sectioning on nonpermeabilized (A to F) and permeabilized (G to L) Cos-7 cells expressing the 16-kDa protein was carried out with a Zeiss Axioplan confocal microscope connected to a Bio-Rad MRC 1624 laser scanner. A total of 35 sections were taken from each cell, and 5 representative sections are presented. (c) No detection of the 16-kDa protein in the culture medium and absence of a cleavable signal sequence. Plasmids pT716k and pIBVM were transiently expressed in Cos-7 cells with the vaccinia virus-T7 system. The cells were labeled with [35S]methionine-cysteine, both the culture medium (lanes 1 and 2) and cell lysates (lanes 3 and 4) were prepared, and the labeled polypeptides were immunoprecipitated with anti-T7 and anti-M sera. Gel electrophoresis of polypeptides was performed on an SDS-15% polyacrylamide gel, and polypeptides were detected by fluorography. Plasmids pT716k and pIBVM were expressed in vitro with the TnT system in rabbit reticulocyte lysate (Promega) in the presence and absence of canine microsomal membranes (CMM) for up to 2 h at 30°C. At the same time, Cos-7 cells transfected (TF) with pT716k (lane 8) and pIBVM (lane 12) were labeled with [35S]methionine-cysteine. Immunoprecipitations were carried out with anti-T7 and anti-M sera, and the polypeptides were analyzed on an SDS-15% polyacrylamide gel and detected by fluorography. The values on the left are molecular sizes in kilodaltons.
After confirming that the 16-kDa protein can transported to the cell surface, we tested if it is a secretory protein. Cos-7 cells were transfected with pT716K and labeled with [35S]methionine. The labeled culture medium was collected, the concentration of ion and detergents was adjusted with 2× radioimmunoprecipitation assay buffer, and immunoprecipitation was done with anti-T7 serum. As shown in Fig. 3c, the 16-kDa protein was not detected in the culture medium (lane 1). However, the protein was detected in lysates prepared from the same transfected cells (Fig. 3c, lane 3), indicating that the protein was efficiently expressed. In a control experiment, IBV M protein was detected only in the cell lysates (Fig. 3c, lane 4) but not in the culture medium (Fig. 3c, lane 2).
The Kyte-and-Doolittle (22) hydropathy plot of the 16-kDa protein (Fig. 1a) showed that the protein is rather hydrophobic, especially in its N-terminal region. To determine if this region contains a classical cleavable signal sequence, the size of the 16-kDa protein expressed in vitro in reticulocyte lysates in the presence or absence of the canine microsomal membranes was compared with the size of the protein expressed in intact cells. To facilitate detection of the potential cleavage event, the construct containing the 11-amino-acid T7 tag was used in these experiments. As shown in Fig. 3c, the efficient detection and comigration of the 16-kDa protein synthesized in vitro (lanes 6 and 7) and in intact cells (lane 8) indicated that no cleavable signal sequence was present in the 16-kDa protein. Once again, IBV M was used as a control, showing the glycosylation of the protein in the presence of canine microsomal membranes (Fig. 3c, lane 11).
Presence of multiple conserved cysteine residues in the 16-kDa protein and the counterparts of other coronaviruses and dimerization of the 16-kDa protein.
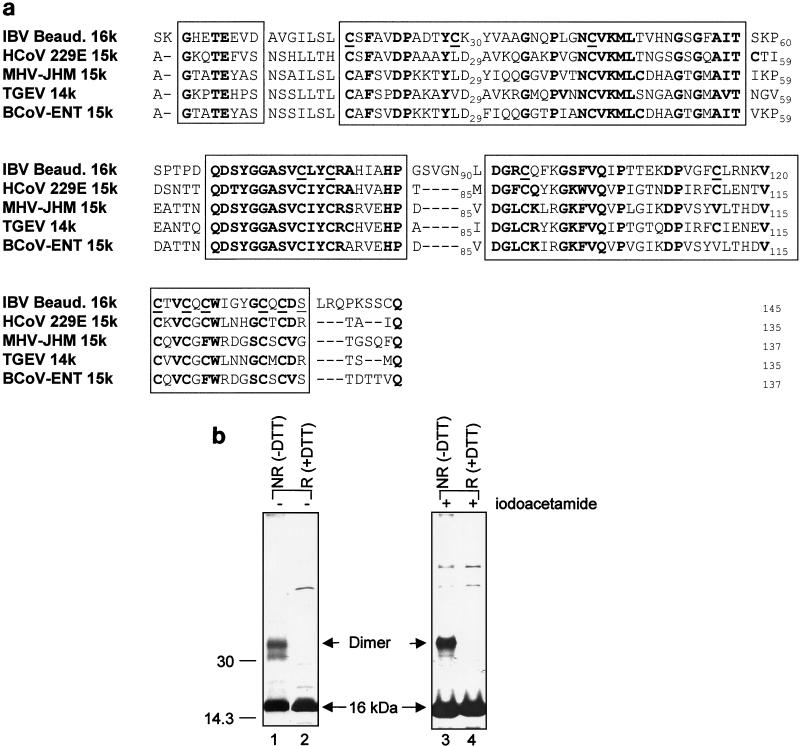
The C-terminal region of the coronavirus 1a polyprotein was known as the cysteine-rich region due to the presence of multiple cysteine residues. In IBV, 12 cysteine are found in the 16-kDa protein (Fig. 1a). Among them, 11 are conserved among the counterparts of four other coronaviruses (Fig. 4a). These conserved cysteine residues may form inter- or intrachain disulfide bonds, resulting in homodimer or oligomer formation. This possibility was investigated by analysis of the protein on a nonreducing SDS-polyacrylamide gel. As shown in Fig. 4b, a band migrating at approximately 33 kDa and representing a dimer of the 16-kDa protein was observed under nonreducing conditions (lane 1) but not under reducing conditions (lane 2). Interestingly, a band slightly below the 33-kDa dimer was also observed (Fig. 4b, lane 1). The identity of this band is uncertain, but it may represent a different conformation of the dimer. No other oligomers were observed (Fig. 4b, lane 1). To rule out possible cross-linking of the 12 cysteine residues, a parallel experiment was done in duplicate. Lysates were prepared in buffer containing 10 mM iodoacetamide (Sigma). As shown in Fig. 4b, the 33-kDa dimer of the 16-kDa protein was once again detected specifically under nonreducing conditions (lane 3) and not under reducing conditions (lane 4).
FIG. 4.
(a) Comparison of the amino acid sequence of the 16-kDa protein with those of the equivalent proteins of other coronaviruses. The products included in this comparison are the HcoV-229E 15-kDa protein, the MHV-JHM 15-kDa protein, the porcine transmissible gastroenteritis virus (TGEV) 14-kDa protein, and the bovine coronavirus (BCoV-ENT) 15-kDa protein. The conserved regions are in bold and outlined, and the cysteine residues are in bold and underlined. The residue (Ser-136) putatively involved in phosphorylation is outlined. (b) Dimerization of the 16-kDa protein. Plasmid pT716K was transiently expressed in Cos-7 cells with the vaccinia virus-T7 system. Cells were labeled with [35S]methionine-cysteine, lysates were prepared in TGP buffer (1% Triton X-100, 10% glycerol, 50 mM HEPES [pH 7.4], 1 mM sodium vanadate, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml) with (lanes 1 and 2) or without (lanes 3 and 4) 10 mM iodoacetamide (Sigma), and proteins were immunoprecipitated with anti-T7 antibody in duplicate. Gel electrophoresis of the polypeptides was performed on an SDS-15% polyacrylamide gel in the presence [R (+DTT)] or absence [NR (−DTT)] of the reducing agent dithiothreitol. Polypeptides were detected by fluorography. The values on the left are molecular sizes in kilodaltons.
Involvement of multiple cysteine residues in dimerization of the 16-kDa protein.
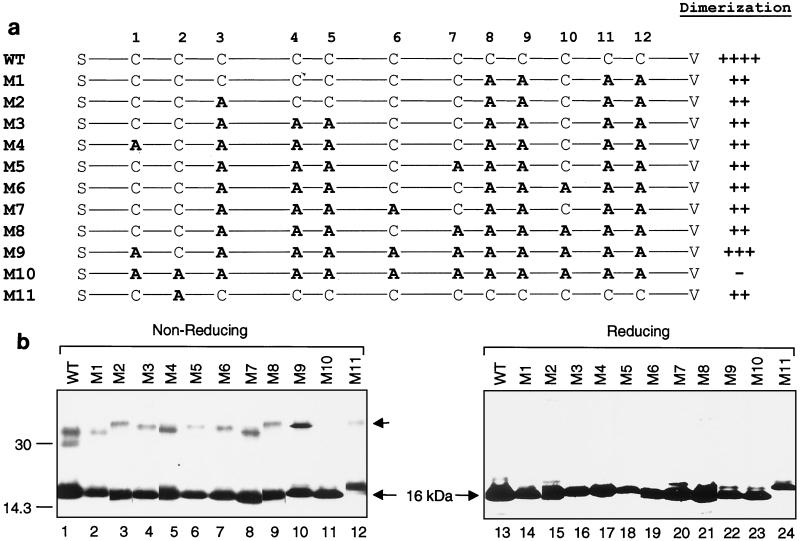
Mutagenesis of the 12 cysteine residues in the 16-kDa protein was then carried out to determine which residue(s) is responsible for the dimerization of the protein. Eleven mutant forms containing mutations of different cysteine resides to alanine were made and expressed in cells (Fig. 5a). The effects of these mutations on the dimerization of the protein are shown in Fig. 5b and summarized in Fig. 5a. As can be seen, various degrees of dimerization were observed with all of the mutant forms except M10, which contains mutations of all of the 12 cysteine residues (Fig. 5a and b). A significant reduction in dimerization was observed with M1 (Fig. 5b, lane 2) and M11, which contain mutations of the four cysteine residues (Cys-8, -9, -11, and -12) at the C-terminal end of the protein and the second cysteine only, respectively. These results indicate that these residues may play a role in the dimerization of the protein. Interestingly, the levels of dimerization were not drastically affected in other mutant forms, including M9, which contains mutations of 11 cysteine residues (Fig. 5b).
FIG. 5.
(a) Summary of the effects of mutations of the conserved cysteine residues on dimerization of the 16-kDa protein. The cysteine residues mutated to alanines are in bold, and the degree of dimerization is indicated by the number of plus signs for each construct. (b) Dimerization of the 16-kDa protein and its mutant forms. Plasmids pT716K (WT) and mutant constructs M1 to M11 were transiently expressed in Cos-7 cells with the vaccinia virus-T7 system. The cells were labeled with [35S]methionine-cysteine, lysates were prepared, and the labeled polypeptides were immunoprecipitated with anti-T7 antibody in duplicate. Gel electrophoresis of the polypeptides was performed on an SDS-15% polyacrylamide gel under either nonreducing (lanes 1 to 12) or reducing (lanes 13 to 24) conditions. Polypeptides were detected by fluorography. The position of the monomeric 16-kDa protein is indicated by the long arrow, and that of the dimers is indicated by the short arrow. The values on the left are molecular sizes in kilodaltons.
Upregulation of the 16-kDa protein in EGF-stimulated cells.
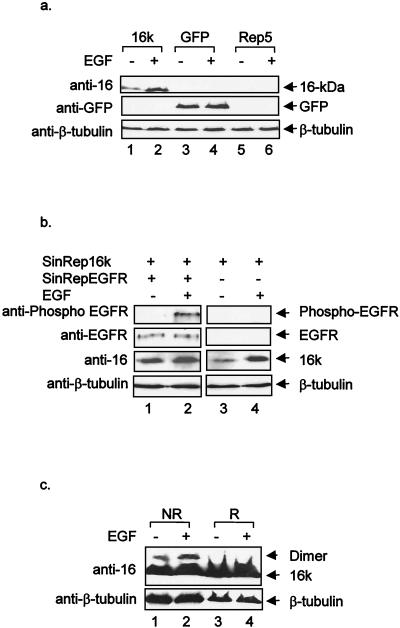
The 16-kDa protein was initially predicted to be a viral growth factor-like protein due to its cysteine-rich property and the arrangement of the cysteine residues in the molecule (15). The observations that it could be transported to the cell surface and form homodimers and potential intrachain disulfide bonds prompted us to examine the response of the protein to the EGF. It was reasoned that if the protein could function as a viral growth factor, it might compete with EGF for the EGFR. To achieve high transfection efficiency, a transient, noncytotoxic Sindbis virus expression system was used (Invitrogen). For this purpose, the 16-kDa protein was cloned into the Sindbis virus expression vector and expressed in BHK-21 cells with helper RNA, which encodes the Sindbis virus structural proteins for the assembly of pseudovirions. Meanwhile, GFP was used to monitor transfection efficiency and as a negative control. At 36 h posttransfection, when approximately 85 to 95% of the cells transfected with the GFP construct produced the protein, cells were starved in serum-free medium for 12 h before being stimulated with 50 ng of EGF per ml for 30 min and then harvested for Western blot analysis. As can be seen in Fig. 6a, efficient expression of the 16-kDa protein was observed. Upon EGF stimulation, a significant increase in the amounts of the 16-kDa protein was detected (Fig. 6a, lanes 1 and 2). Blotting of the same membrane with a commercially available anti-GFP antibody (Clontech) showed no obvious increase in GFP expression upon EGF stimulation (Fig. 6a, lane 3 and 4). To rule out a discrepancy between the amounts of the samples loaded, the membrane was then probed with anti-β-tubulin antibody, showing that similar amounts of β-tubulin were detected (Fig. 6a, lanes 1 to 6).
FIG. 6.
(a) Effect of EGF on expression of the 16-kDa protein. Cells were transfected by electroporation with RNAs transcribed from plasmids (as indicated above the lanes) and stimulated with 50 ng of EGF per ml for 30 min at 48 h postinfection. Cells were harvested, and gel electrophoresis of total proteins was performed on an SDS-15% polyacrylamide gel. The proteins were transferred to nitrocellulose membranes; blotted with anti-16-kDa protein, anti-GFP, and anti-β-tubulin sera; and detected with an ECL + Plus Western blotting detection kit. (b) Cotransfection of the 16-kDa protein and human EGFR. Cells were cotransfected by electroporation with RNAs transcribed from plasmids pSinRep16k and pSinRepEGFR and stimulated with 50 ng of EGF per ml for 30 min at 48 h posttransfection (lanes 1 and 2). Cells transfected with RNA transcribed from plasmid pSinRepGFP were included as a control. Cells were harvested, and gel electrophoresis of the total proteins was performed on an SDS-15% polyacrylamide gel. The proteins were transferred to nitrocellulose membranes; blotted with anti-phosphorylated EGFR, anti-EGFR, anti-16-kDa protein, and anti-β-tubulin sera; and detected with an ECL + Plus Western blotting detection kit. (c) Effect of EGF stimulation on dimerization of the 16-kDa protein. Cells were transfected by electroporation with RNA transcribed from plasmid pSinRep16k and stimulated with 50 ng of EGF per ml at 48 h posttransfection. Lysates were prepared in TGP buffer, and gel electrophoresis of total proteins was performed on an SDS-15% polyacrylamide gel under either nonreducing (NR) or reducing (R) conditions. The proteins were transferred to nitrocellulose membranes, blotted with anti-16-kDa protein serum and anti-β-tubulin serum, and detected with an ECL + Plus Western blotting detection kit.
We then tested the effect of EGFR overexpression on the accumulation of the 16-kDa protein upon stimulation by EGF. The 3.5-kb human EGFR was cloned into the Sindbis virus expression vector, coexpressed with the 16-kDa protein in BHK cells, and stimulated with EGF at 48 h posttransfection. As observed in Fig. 6b, similar levels of the unphosphorylated EGFR were detected in both stimulated and nonstimulated cells (lanes 1 and 2) and phosphorylated EGFR was detected by anti-phosphorylated EGFR antibody (Transduction Laboratories) in EGF-stimulated cells only (lane 2). However, coexpression of the EGFR with the 16-kDa protein reduced the upregulatory effects of EGF on the accumulation of the 16-kDa protein. As can be seen, only a moderate increase in the 16-kDa protein was observed in cells coexpressing the EGFR and the 16-kDa protein upon stimulation with EGF (Fig. 6b, lanes 1 and 2). Once again, the blot was probed with anti-β-tubulin antibody and equal amounts of β-tubulin were detected (Fig. 6b, lanes 1 to 4). These results support the notion that the increased accumulation of the 16-kDa protein in EGF-stimulated cells is a specific reaction.
To verify that dimerization of the 16-kDa protein is affected upon EGF stimulation, cells were stimulated with EGF and harvested for Western blot analysis. Upon EGF stimulation, both monomeric and dimeric forms of the 16-kDa protein were increased (Fig. 6c, lanes 1 and 2). The blot was probed with anti-β-tubulin, showing that equal amounts of β-tubulin were detected (Fig. 6c, lanes 1 to 4). These results suggested that EGF stimulation did not affect the dimerization of the 16-kDa protein.
DISCUSSION
Coronavirus encodes a cysteine-rich polypeptide as a part of the 1a and 1a/1b polyproteins. Proteolytic processing of these polyproteins by the virus-encoded 3C-like proteinase results in the release of this polypeptide from its precursor as a mature viral nonstructural protein. In this communication, it was identified as a 16-kDa protein in IBV-infected Vero cells. Biochemical and functional characterization of the 16-kDa protein demonstrated that the protein may be associated with cellular membranes, may colocalize with de novo-synthesized viral RNA, and can form homodimers via multiple conserved cysteine residues.
Previous studies have shown that proteolytic events mediated by the 3C-like proteinase might take place in the cytosol (34), and individual cleavage products would be targeted to their respective membrane compartments after being released from the precursor. In this study, the subcellular localization pattern of the 16-kDa protein and its colocalization with BrUTP-labeled viral RNA indicated that the protein may be associated with the viral RNA replication complex in the perinuclear region. The perinuclear punctate staining pattern observed is similar to those reported for MHV-A59 (4, 5, 41). Studies with arterivirus have provided convincing evidence that the viral RNA replication complex in which the replicase subunits are associated is the site of viral RNA synthesis (40). As the uncleaved precursor protein of the coronavirus polymerase cannot support RNA synthesis (37), the association of the 16-kDa protein with the viral RNA replication complex indicates direct involvement of the protein in viral RNA replication.
Sequence analysis and comparison revealed that the 16-kDa protein and the equivalent products of other coronaviruses contain multiple conserved cysteine residues. Systematic mutagenesis and biochemical studies done in this investigation demonstrated that some of these cysteine residues are involved in the formation of interchain disulfide bonds, resulting in the dimerization of the protein. However, we were unable to ascertain which residue(s) is absolutely required for the formation of these interchain disulfide bonds and how many disulfide bonds are formed during dimerization of the protein, as none of the mutant forms (except M10, in which all 12 cysteine residues are deleted) abolished the dimerization of the protein. Based on the differential effects of individual residues on the dimerization of the protein, it is likely that residues 8, 9, 11, and 12 play a more dominant role than other residues. Another unresolved issue is whether these conserved cysteine residues can form intrachain disulfide bonds and, if so, which residue(s) is involved. Analysis of the wild-type 16-kDa protein by nonreducing SDS-PAGE detected a band migrating more rapidly than the dimer. The same band was not detected in any mutant forms. It is likely that this band may represent the 16-kDa protein with different conformation due to the formation of intrachain disulfide bonds.
The 16-kDa protein was previously postulated to be a growth factor-like protein due to its cysteine-rich properties, as well as the arrangement of the cysteine residues (15). Positive staining was observed on nonpermeabilized cells expressing the 16-kDa protein, confirming that the protein could be transported to the cell surface (Fig. 3). Failure to detect the protein in the culture medium indicates that the protein could not be secreted out of the cells. A recent report showed that the equivalent cleavage product of the MHV-A59 1a polyprotein was occasionally found close to the plasma membrane. Electron microscopy confirmed the endocytic nature of the protein (41), suggesting that the product is associated with membranes of endocytic origin. As the 16-kDa protein is obviously a nonstructural protein, it is tempting to speculate that the detection of the endosome-associated 16-kDa protein could be due to receptor-mediated endocytosis of the 16-kDa protein located on the cell surface. It would therefore be intriguing to determine if the protein can interact with the growth factor receptor, as such an interaction would be essential for regulation of the death and survival of virus-infected cells in order to enable the completion of viral replication and the spread of viral infection. The observations that the accumulation of the 16-kDa protein could be significantly increased by EGF stimulation and that the protein tends to form homodimers suggest that it is involved in the EGF signaling pathway. This argument would be further supported by coexpression of the 16-kDa protein with the EGFR, showing that EGF-stimulated accumulation of the 16-kDa protein was limited by expression of the EGFR. However, neither physical interaction of the 16-kDa protein with the EGFR nor obvious stimulation-inhibition of cell growth upon overexpression of the 16-kDa protein was observed (data not shown).
How could EGF up-regulate accumulation of the 16-kDa protein? Three possibilities were considered. First, stimulation with EGF could result in dimerization of the 16-kDa protein, which, in turn, may stabilize the protein. As shown in Fig. 6c, analysis of the dimerization rates of the protein with and without stimulation of the cells with EGF showed that no difference in the dimerization of the protein occurred, ruling out this possibility. Similarly, EGF stimulation could lead to phosphorylation of the 16-kDa protein, altering the conformation of the protein. Computer prediction and preliminary mutagenesis data revealed that Ser-136 could be phosphorylated (data not shown). Once again, this phosphorylation was not affected by EGF stimulation. No Tyr residue in the 16-kDa protein could be phosphorylated, as shown by these analyses. The final possibility considered was that EGF stimulation could alter the subcellular distribution patterns of the 16-kDa protein. One potential consequence, for example, is to increase transport of the protein to the cell surface. Systematic analysis of this possibility was hindered by the fact that the viral expression systems used could dramatically affect cell morphology at late stages of expression. Further studies are required to explain the underlying mechanism, as well as its functional implications for coronavirus replication and pathogenesis.
Acknowledgments
We thank H. Y. Xu for excellent technical assistance and Z. L. Wen, C. E. Ang, and M. Huang for helpful suggestions and discussions.
This work was supported by a grant from the National Science and Technology Board of Singapore.
REFERENCES
- 1.Baker, S. C., C.-K. Shieh, L. H. Soe, M.-F. Chang, D. M. Vannier, and M. M. C. Lai. 1989. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J. Virol. 63:3693-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, S. C., K. Yokomori, S. Dong, R. Carlise, A. E. Gorbalenya, E. V. Koonin, and M. M. C. Lai. 1993. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J. Virol. 67:6056-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonilla, P. J., S. A. Hughes, and S. R. Weiss. 1997. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus mouse hepatitis virus strain A59. J. Virol. 71:900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bost, A. G., R. H. Carnahan, X. T. Lu, and M. R. Denison. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 74:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bost, A. G., E. Prentice, and M. R. Dension. 2001. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 285:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boursnell, M. E. G., T. D. K. Brown, I. J. Foulds, P. F. Green, F. M. Tomely, and M. M. Binns. 1987. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 68:57-77. [DOI] [PubMed] [Google Scholar]
- 7.Brierley, I., M. E. G. Boursnell, M. M. Binns, B. Bilimoria, V. C. Blok, T. D. K. Brown, and S. C. Inglis. 1987. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 6:3779-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brierley, I., P. Digard, and S. C. Inglis. 1989. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell 57:537-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denison, M. R., and S. Pearlman. 1986. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J. Virol. 60:12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denison, M. R., and S. Pearlman. 1987. Translation and processing of MHV-A59 virion RNA in reticulocyte lysates and infected cells. Adv. Exp. Med. Biol. 218:155-156. [DOI] [PubMed] [Google Scholar]
- 11.Denison, M. R., W. J. M. Spaan, Y. van der Meer, C. A. Gibson, A. C. Sims, E. Prentice, and X. T. Lu. 1999. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J. Virol. 73:6862-6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong, S. H., and S. C. Baker. 1994. Determination of the p28 cleavage site recognized by the first papain-like cysteine proteinase of murine coronavirus. Virology 204:541-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, H. Q., J. J. Schiller, and S. C. Baker. 1996. Identification of the polymerase polyprotein products p72 and p65 of the murine coronavirus MHV-JHM. Virus Res. 45:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17:4847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grotzinger, C., G. Heusipp, J. Ziebuhr, U. Harms, J. Suss, and S. G. Siddell. 1996. Characterization of a 105-kDa polypeptide encoded in gene 1 of the human coronavirus HCV 229E. Virology 222:227-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herold, J., A. E. Gorbalenya, V. Thiel, B. Schelle, and S. G. Siddell. 1998. Proteolytic processing at the amino terminus of human coronavirus 229E gene 1-encoded polyproteins: identification of a papain-like proteinase and its substrate. J. Virol. 72:910-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heusipp, G., C. Grotzinger, J. Herold, S. G. Siddell, and J. Ziebuhr. 1997. Identification and subcellular localization of a 41 kDa, polyprotein 1ab processing product in human coronavirus 229E-infected cells. J. Gen. Virol. 78:2789-2794. [DOI] [PubMed] [Google Scholar]
- 19.Heusipp, G., U. Harms, S. G. Siddell, and J. Ziebuhr. 1997. Identification of an ATPase activity associated with a 71-kilodalton polypeptide encoded in gene 1 of the human coronavirus 229E. J. Virol. 71:5631-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes, S. A., P. J. Bonilla, and S. R. Weiss. 1995. Identification of the murine coronavirus p28 cleavage site. J. Virol. 69:809-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanajanahaluethai, A., and S. C. Baker. 2000. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 74:7911-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 23.Lim, K. P., and D. X. Liu. 1998. Characterization of the two overlapping papain-like proteinase domains encoded in gene 1 of the coronavirus infectious bronchitis virus and determination of the C-terminal cleavage site of an 87-kDa protein. Virology 245:303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim, K. P., L. F. P. Ng, and D. X. Liu. 2000. Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus Avian infectious bronchitis virus and characterization of the cleavage products. J. Virol. 74:1674-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, D. X., I. Brierley, K. W. Tibbles, and T. D. K. Brown. 1994. A 100-kilodalton polypeptide encoded by open reading frame (ORF) 1b of the coronavirus infectious bronchitis virus is processed by ORF 1a products. J. Virol. 68:5772-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, D. X., K. W. Tibbles, D. Cavanagh, T. D. K. Brown, and I. Brierley. 1995. Identification, expression, and processing of an 87 kDa polypeptide encoded by ORF 1a of the coronavirus infectious bronchitis virus. Virology 208:48-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, D. X., and T. D. K. Brown. 1995. Characterization and mutational analysis of an ORF-1a-encoding proteinase domain responsible for proteolytic processing of the infectious bronchitis virus 1a/1b polyprotein. Virology 209:420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, D. X., H. Y. Xu, and T. D. K. Brown. 1997. Proteolytic processing of the coronavirus infectious bronchitis virus 1a polyprotein: identification of a 10-kilodalton polypeptide and determination of its cleavage sites. J. Virol. 71:1814-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, D. X., S. Shen, H. Y. Xu, and S. F. Wang. 1998. Proteolytic mapping of the coronavirus infectious bronchitis virus 1b polyprotein: evidence for the presence of four cleavage sites of the 3C-like proteinase and identification of two novel cleavage products. Virology 246:288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, X. T., Y. Lu, and M. R. Denison. 1996. Intracellular and in vitro translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59. Virology 222:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, X. T., A. C. Sims, and M. R. Denison. 1998. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J. Virol. 72:2265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, Y., X. T. Lu, and M. R. Denison. 1995. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J. Virol. 69:3554-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng, L. F. P., and D. X. Liu. 1998. Identification of a 24-kDa polypeptide processed from the coronavirus infectious bronchitis virus 1a polyprotein by the 3C-like proteinase and determination of its cleavage sites. Virology 242:388-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng, L. F. P., and D. X. Liu. 2000. Further characterization of the coronavirus infectious bronchitis virus 3C-like proteinase and determination of a new cleavage site. Virology 272:27-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinon, J. D., R. R. Mayreddy, J. D. Turner, F. S. Khan, P. J. Bonilla, and S. R. Weiss. 1997. Efficient autoproteolytic processing of the MHV-A59 3C-like proteinase from the flanking hydrophobic domains requires membranes. Virology 230:309-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiller, J. J., A. Kanjanahaluethai, and S. C. Baker. 1998. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology 242:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi, S. T., J. J. Schiller, A. Kanjanahaluethai, S. C. Baker, J.-W. Oh, and M. M. C. Lai. 1999. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J. Virol. 73:5957-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sims, A. C., J. Ostermann, and M. R. Denison. 2000. Mouse hepatitis virus replicase proteins associate with two distinct populations of intracellular membranes. J. Virol. 74:5647-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tibbles, K. W., D. Cavanagh, and T. D. K. Brown. 1999. Activity of a purified His-tagged 3C-like proteinase from the coronavirus infectious bronchitis virus. Virus Res. 60:137-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Meer, Y., H. van Tol, J. K. Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. M. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziebuhr, J., J. Herold, and S. G. Siddell. 1995. Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J. Virol. 69:4331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziebuhr, J., G. Heusipp, and S. G. Siddell. 1997. Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J. Virol. 71:3992-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziebuhr, J., and S. G. Siddell. 1999. Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab. J. Virol. 73:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziebuhr, J., V. Thiel, and A. E. Gorbalenya. 2001. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involved two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 276:33220-33232. [DOI] [PMC free article] [PubMed] [Google Scholar]