Abstract
The mouse epithelial MODE-K cell line expressing human CD46 or CD150 cellular receptors was found to be nonpermissive for measles virus (MV) replication. The virus binding and membrane fusion steps were unimpaired, but only very limited amounts of virus protein and RNA synthesized were detected after the infection. In a minigenome chloramphenicol acetyltransferase assay, MODE-K cells were as able as the permissive HeLa cells in supporting MV polymerase activity. The restriction phenotype of MODE-K cells could be alleviated by providing, in trans, either N-P-L or N-P functional protein complexes but not by P-L complexes or individual N, P, and L proteins. Several human × mouse (HeLa × MODE-K) somatic hybrid clones expressing human CD46 were isolated and found to be either nonpermissive or permissive according to their human chromosomal contents. The MV-restricted phenotype exhibited by the MODE-K cell line suggests that a cellular factor(s) can control MV transcription, possibly by stabilizing the incoming virus polymerase templates.
Members of the order Mononegavirales are among the most simple enveloped mammalian viruses which replicate within the cytosol. Their negative-strand RNA genome is used as a template for both transcription of mRNA and virus replication involving the synthesis of antigenomic and genomic RNA. For example, the measles virus (MV) genome encodes the nucleoprotein N (which associates with the genomic [and antigenomic] RNA to form the polymerase template), the phosphoprotein P, a polymerase cofactor, the large L protein (which harbors the polymerase enzymatic activities), the envelope hemagglutinin (H) and fusion (F) proteins, and the matrix (M) protein.
MV replication is regulated by the virion structure and/or the entry pathway, as shown by the much slower replication of chimeric MV having their H and F glycoprotein genes substituted with that of the vesicular stomatitis virus (VSV) G protein (51) and by the transcriptional inhibitory activity of mutated M proteins (52). Regulatory nucleic sequences also control the level of MV replication (3, 27). MV replication in a cellular host is also regulated by nonstructural viral proteins MV-C (15, 45, 56) and MV-V (55) proteins. Changes in the primary sequences of these proteins and/or L polymerase protein are associated with transcriptional impediment (53).
Much less is known on the possible involvement of cellular factors in MV replication outside the key role of the cellular receptors CD46 (12, 37) and CD150 (13, 30, 54) allowing MV entry in human cells and the potent inhibitory activity of alpha/beta interferon (IFN) (36, 39). MV gene expression and replication have been reported to be enhanced in human and simian host cells after heat shock or by the overexpression of Hsp72 (40, 57, 58). In acellular conditions, MV polymerase activity can be observed only when cytosolic fractions are present, and tubulin seems to act as a cellular cofactor of the polymerase (35). The host cell can exert late control of virus budding, as shown by the reduced amounts of virus progeny observed in murine L.CD46 cells (61), associated with a defect in MV assembly in membrane rafts (34, 60) (S. Vincent and D. Gerlier, unpublished data). Several studies have pointed out that host cells can exert control early in MV replication; MV replication is reduced in CD46-expressing chicken embryo fibroblasts or Vero cells without prior growth adaptation (14, 53) and in lymphocytes from CD46 transgenic mice (16, 28). During an initial screening of MV replication in various murine cell lines expressing human CD46 as MV receptor, we observed that CD46-expressing MODE-K epithelial cells derived from murine intestine were poorly infected. A detailed study was undertaken and revealed an early postentry block of MV replication affecting the initial transcription.
MATERIALS AND METHODS
Cells.
Human epithelial HeLa and mouse intestinal epithelial MODE-K (59) cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 6% heat-inactivated fetal calf serum (FCS), 10 mM HEPES, 2 mM glutamine, and 10 μg of gentamicin per ml at 37°C in the presence of 7% CO2. MODE-K.CD46 and MODE-K.CD150 clones were obtained by transfection using Lipofectamine reagent (Life Technologies) with pIRV.CD46 (coding for B1 CD46 isoform) and pCAGG-SLAM (coding for CD150/SLAM) (54) together with pAG475-2 (33) (coding for hygromycin resistance). Hygromycin-resistant HeLa cells were derived after transfection with pAG475-2 and selection by growth in 500 μg of hygromycin per ml. Somatic cell hybrids were derived by fusing 2 × 106 G418-resistant MODE-K cells with 2 × 106 hygromycin-resistant HeLa cells using polyethylene glycol 1500 (10, 11, 42). Briefly, the two cell types were mixed overnight in a 60-mm-diameter petri dish, washed with DMEM without FCS, and incubated with 3 ml of polyethylene glycol 1500 (Roche Molecular Biochemicals) for 1 min at room temperature. After three washes with DMEM, the cell monolayer was incubated in complete growth medium for 24 h. The cells were then detached by trypsin-EDTA treatment and seeded in six 35-mm-diameter petri dishes in the presence of 500 μg of hygromycin per ml and 2 mg of G418 per ml. Somatic cell clones were recovered after 1 month of culture and selected for dual expression of mouse major histocompatibility complex class I (MHC-I) and human CD46 using immunolabeling and flow cytometry. The hybrid cell clones were thereafter quickly tested for their permissiveness to MV and human chromosome contents.
Viruses.
Hallé (recombinant Edmonston-based tag strain of MV [43]) and chimeric MGV and MG/FV (where the reading frames of MV envelope proteins H and F were substituted by a single reading frame encoding the VSV G glycoprotein or a G/F hybrid molecule [51]) were amplified in Vero cells. The recombinant vaccinia viruses vv-N, vv-P (62), vv-L, vv-NP, vv-PL, vv-NPL (29), vv-HF (63), vv T7lacZ (1), vv-Tk, vv-T3, and vv-T7 (19) code for MV-N; MV-P; MV-L; MV-N and -P; MV-P and -L; MV-N, -P, and -L; MV-H and -F; β-galactosidase under T7 polymerase control; thymidine kinase; and T3 and T7 polymerases, respectively. After one cycle of freeze-thawing of infected cells, clarified supernatants were collected and used as virus stocks.
Infections.
Cells that had been plated and were allowed to grow overnight were infected at the indicated multiplicity of infection (MOI). After 1 h at 37°C, the cells were washed once with fresh medium and incubated at 37°C in the presence or absence of a fusion inhibitory peptide z-d-Phe-l-Phe-Gly (46) at 20 μg/ml. Virus progeny was titrated from infected cells that had been frozen and thawed once and then centrifuged at 400 × g for 5 min to discard cell debris. The supernatants were titrated by the 50% tissue culture infective dose method (25) on a Vero cell monolayer.
Antibodies.
The following antibodies and monoclonal antibodies (MAbs) were used: rabbit anti-MV-F cytoplasmic tail, produced as reported by Cathomen et al. (6); mouse anti-MV-H BH195 (18) and cl55 MAbs; anti-MV-N MAb cl25 (22); anti-MV-P, a mouse anti-MV serum; anti-MV-M MAb CLONE from Chemicon; anti-VSV-G MAb P5D4 from Sigma; anti-CD46 MAb MCI20.6 (38); anti-CD150 MAb A12 from Pharmingen-Becton-Dickinson; anti-Hsp72 MAb W27 from Santa-Cruz; and anti-mouse MHC-I H2-Kk MAb 16-3-1N (American Type Culture Collection).
Flow cytometry analysis.
For cell surface detection of protein, 2 × 105 cells were incubated for 30 min at 4°C in a final volume of 60 μl of DMEM-6% FCS-0.05% NaN3 containing an appropriate dilution of the antibody, in round-bottom 96-well microplates. Cells were then washed three times by centrifugation at 280 × g for 2 min and incubated for 30 min with 50 μl of phycoerythrin-labeled goat anti-mouse immunoglobulin (Ig) (Beckman-Coulter). After two washes, labeled cells were fixed in 1% paraformaldehyde diluted in ISOTON II (Beckman-Coulter) buffer. The fluorescence labeling was then measured by flow cytometry.
Virus binding assay.
The virus binding assay was described previously (37). In brief, 2 × 105 cells were incubated at 4°C for 60 min with purified MV Hallé at a final protein concentration of 50 μg/ml. Following incubation, the cells were washed, and the binding was revealed with an anti-H antibody. After incubation with a phycoerythrin-conjugated anti-Ig antibody, a flow cytometry analysis was performed.
Cell fusion assay.
A virus-based quantitative cell fusion-dependent reporter gene system detailed previously (8) was used. Briefly, the “receptor” cell partners were infected with a recombinant vaccinia virus expressing T7 RNA polymerase (MOI = 5). Simultaneously, the “fusion” cell partners were infected with vv-HF (MOI = 5) and a recombinant vaccinia virus encoding the T7 promoter linked to the lacZ gene (vv T7lacZ) (MOI = 5). After removal of nonadsorbed virus by washing, cells were resuspended in medium containing 5 μg of the fusion inhibitory peptide z-d-Phe-l-Phe-Gly per ml. This ensured the inhibition of potential fusion between neighboring fusion cells expressing functional MV receptor and the H and F proteins. After overnight incubation at 37°C and several washes at 37°C to completely remove the fusion inhibitory peptide, the cells were detached with a brief trypsin-EDTA treatment. The receptor and fusing partners were then cocultured at a ratio of 1:1 in a 96-well flat-bottom plate in the presence of cytosine arabinoside (40 μg/ml). After incubation at 37°C for 6 h, the fusion was then determined by the reporter gene activation assay for β-galactosidase using o-nitrophenyl-β-d-galactopyranoside as a colorimetric substrate.
Western blot analysis.
Infected cells, washed in phosphate-buffered saline, were lysed in radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 0.1% sodium dodecyl sulfate, 0,5% desoxycholate, 1% Triton X-100) containing a cocktail of protease inhibitors (Complete; Roche Biochemicals). After 20 min of incubation at 4°C, the lysates were centrifuged for 15 min at 12,000 × g at 4°C. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto a polyvinylidene difluoride membrane. Membranes were saturated with 5% nonfat dried milk in TBS-T (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.1% Tween 20) and incubated for 1 h with specific antibodies in TBS-T containing 1% nonfat dried milk. Immunoreactive bands were visualized by using secondary horseradish peroxidase-conjugated antibodies (Promega) and enhanced chemiluminescence (Roche Molecular Biochemicals).
RNA quantification by RT-PCR.
Total RNA was extracted from cells using SV Total RNA Isolation System from Promega. MV-N mRNA and antigenomic RNA were detected by reverse transcription-PCR (RT-PCR). cDNA was synthesized from 0.2 μg of RNA using antisense oligonucleotide 5′ GAG ATT CCT GCC ATG GCT TG 3′ (genomic positions 1601 to 1620) and C-Therm reverse transcriptase (Roche Molecular Biochemicals) (10 min at 32°C, 30 min at 70°C, and 5 min at 90°C). Amplification of the cDNA was done by adding Taq DNA polymerase (Life Technologies) and the sense primer 5′ TGC TCT GGA GCT ATG CCA TG 3′ (genomic positions 1098 to 1117) for 35 cycles (1 cycle consists of 40s at 95°C, 60s at 55°C, and 50s at 72°C). The PCR fragments were run on 2% agarose gels and visualized by ethidium bromide staining. RT-PCRs were run in parallel on each RNA sample using β-actin primers: sense primer 5′ AGG CCA ACC GCG AGA AGA TGA C 3′ and antisense primer 5′ AGC TCG TAG CTC TTC TCC AGG G 3′.
Minigenome CAT assay.
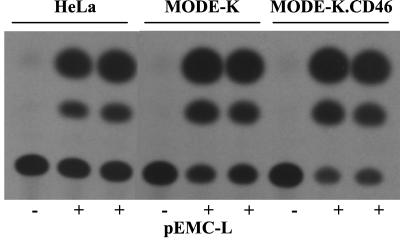
The minigenome chloramphenicol acetyltransferase (CAT) assay was described previously (43, 47). Briefly, HeLa, MODE-K, and MODE-K.CD46 cells were infected with vv-T7 at 3 to 5 MOI and transfected with 1.5 μg of pEMC-Na, 1.5 μg of pEMC-Pa, 0.5 μg of pEMC-L, and 1.5 μg of p107MV(−):CAT which is transcribed by the T7 polymerase into the MV-CAT minigenome encoding CAT. Thirty hours postinfection (p.i.), cells were lysed with reporter lysis buffer (Promega). Protein lysates were incubated with [14C]chloramphenicol and acetyl coenzyme A for 1 to 2 h. The acetylated products were resolved by thin-layer chromatography.
Human chromosome analysis.
The interspecies somatic hybrids were resuspended in RPMI and blocked in metaphase by colchicin. The cell suspension was then made hypotonic (by adding KCl) and fixed in methanol-acetic acid (3:1). After the cells were washed, they were dropped onto a cold wet slide to spread the metaphase plates. R-banding was performed after Earle pretreatment for 55 min. Classification of the human chromosomes was done according to the International Standard for Chromosome Nomenclature.
RESULTS
Postentry restriction of MV replication in mouse MODE-K.CD46 cells.
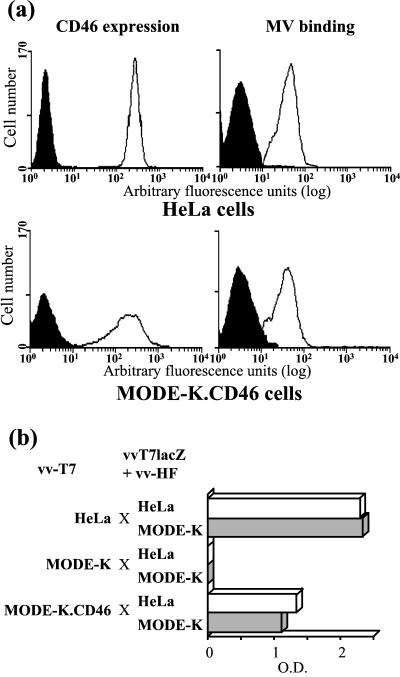
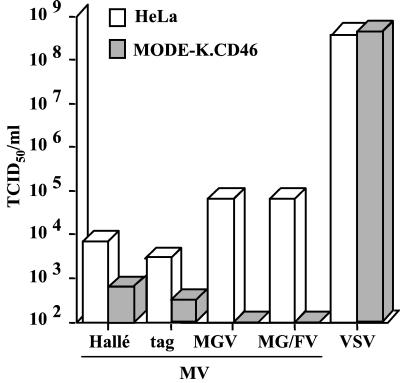
A transfected MODE-K cell clone expressing CD46 at a level similar to that of the permissive human HeLa cells was isolated and further characterized. The MV binding ability of MODE-K.CD46 cells was similar to that of HeLa cells, and MODE-K.CD46 cells were able to fuse with either HeLa or MODE-K cells expressing MV H and F glycoproteins, although apparently not as efficiently as HeLa cells did (Fig. 1). When MODE-K.CD46 cells were infected by MV Hallé strain, there were very few viral progeny, more than 1 log unit below that observed when HeLa cells were infected (Fig. 2). Similar results were obtained with the recombinant tag virus derived from the Edmonston strain. After infection with recombinant MGV and MG/FV virus, which code for the unique envelope glycoprotein G from VSV and a chimeric VSV-G/MV-F protein instead of MV H and F glycoproteins, respectively, the viral progeny from MODE-K.CD46 cells was reduced to a undetectable level. The restricted MV replication in MODE-K.CD46 cells was observed whichever cell clone was tested, as well as in MODE-K.CD150 cells (see below). This restriction was specific for MV replication machinery and likely occurs at the postentry level, since the replication of VSV, another member of the Mononegavirales order, was as efficient as that observed in HeLa cells.
FIG. 1.
Expression of CD46 by MODE-K cells allows MV binding (a) and MV-H+F-mediated cell-cell fusion (b). Human HeLa and murine MODE-K.CD46 cells were tested for expression of CD46 and MV binding ability using flow cytometry assay (a) and for the ability to fuse with HeLa (white bars) or MODE-K (grey bars) cells infected with vaccinia virus encoding MV H and F glycoproteins using a gene reporter fusion assay (b). The black histograms in panel a represent the fluorescence background of the cells in the absence of anti-H antibody. O.D., optical density.
FIG. 2.
Poor ability of MODE-K.CD46 cells to produce infectious MV. HeLa and MODE-K.CD46 cells were infected with MV Hallé, tag, MGV, MG/FV, and VSV at 1 MOI, and the virus progeny was measured 3 days (Hallé, tag, and VSV) and 9 days (MGV and MG/FV) after the infection. TCID50, 50% tissue culture infective dose.
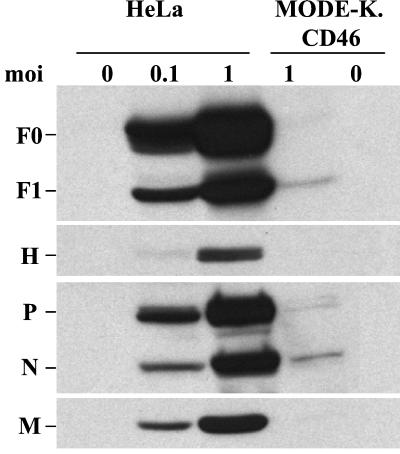
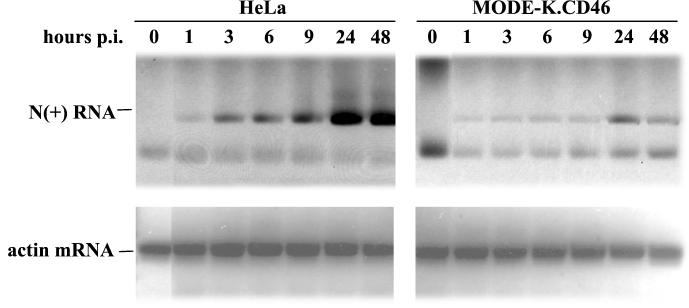
To identify the step where the virus cycle was blocked, the expression of viral proteins was examined. Small amounts of N, P, M, and F protein could be detected 2 days p.i. in MV-infected MODE-K.CD46 cells, whereas all viral proteins accumulated in HeLa cells (Fig. 3). This lack of protein expression was correlated with a very small amount of N+ RNA strand which could be detected only after 1 day p.i., whereas N+ RNA was already detected at 3 h p.i. in HeLa cells and accumulated thereafter (Fig. 4). The low signal observed at 1 h p.i. in both cells reflects the input RNA of the infecting virus.
FIG. 3.
Defect in MV protein synthesis in MODE-K.CD46 cells. HeLa and MODE-K.CD46 cells were infected by MV Hallé at 0.1 or 1 MOI, and cell extracts were analyzed 2 days p.i. for MV protein contents by Western blotting.
FIG. 4.
Defect in MV RNA synthesis in MODE-K.CD46 cells. HeLa and MODE-K.CD46 cells were infected by MV tag at 1 MOI, and MV N+ RNA was analyzed by semiquantitative RT-PCR at several time intervals. The PCR products were visualized by ethidium bromide staining. Equal amounts of RNA were used in each RT-PCR sample as shown by the levels of β-actin RNA PCR products.
Ability of MODE-K cells to support MV polymerase activity in a MV minigenome CAT assay.
MODE-K and MODE-K.CD46 cells were transfected with eucaryotic vectors encoding the minimal components required for MV polymerase activity, namely, the N, P, and L proteins and a minigenome encoding for the CAT enzymatic activity. As shown by the level of acetylated chloramphenicol (Fig. 5), both of these cell lines were as efficient as the permissive human HeLa cells in supporting MV polymerase transcription activity. It should be noted that the transcription level reflects both the transcription from the primary minigenome and the possible transcription from replicated genomes. The transfection efficiency was independently tested and found to be similar in the three cell types. Thus, there is not a general defect preventing MV polymerase activity in MODE-K cells. When tested for their ability to support the rescue of synthetic MV CAT minireplicons after infection with MV (48), the MODE-K.CD46 cells were inefficient, as expected, because they are nonpermissive (data not shown).
FIG. 5.
MV polymerase activity in HeLa and MODE-K cells measured using a MV CAT minigenome transcription assay. CAT activity from transcription of the MV CAT minigenome in HeLa, MODE-K, and MODE-K.CD46 cells in the presence (+) or absence (−) of MV-L protein. Cells were infected with vv-T7 and transfected with plasmids pEMC-N, pEMC-P, pEMC-L, and p107MV(-) encoding MV-N, -P, and -L proteins and MV CAT minigenome, respectively.
Recovery of MV protein synthesis by MV polymerase components provided in trans.
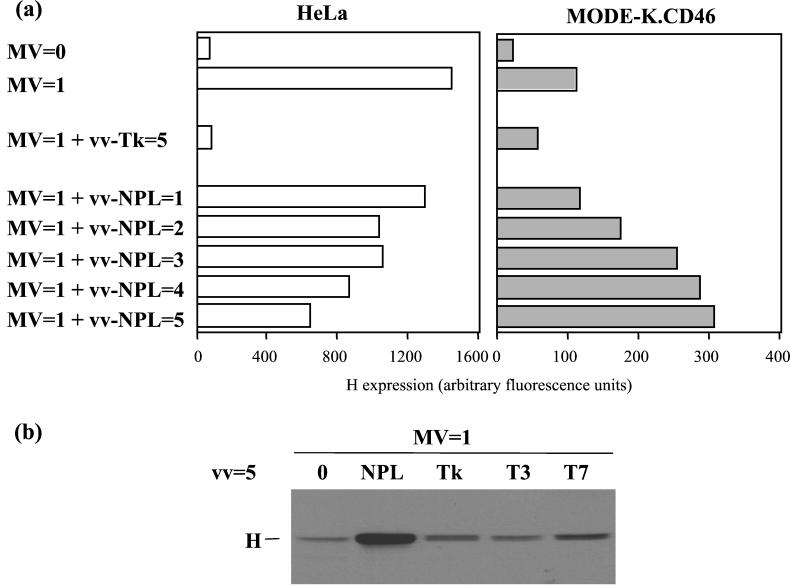
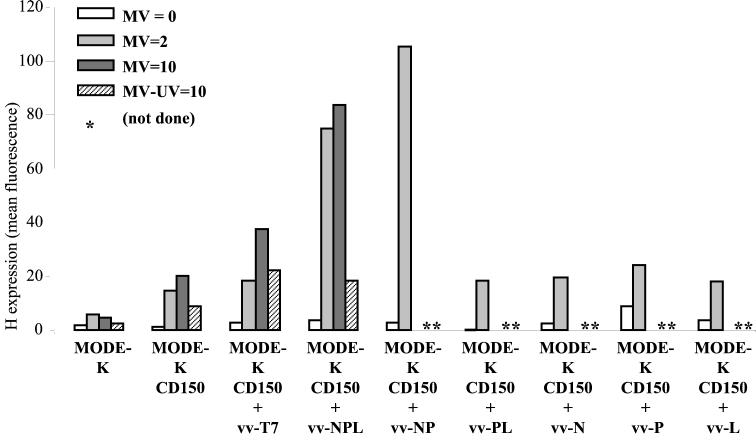
Since the MV replication block was early after infection and affected the polymerase RNA synthesis, we attempted to supplement in trans the infected cells with MV polymerase components. MODE-K.CD46 cells were infected with MV alone or together with a recombinant vaccinia virus encoding an appropriate ratio of N, P, and L proteins in complexes proficient for polymerase activity measured using a MV CAT minigenome rescuing assay (29). A dose-dependent increase of H expression was observed when the cells were coinfected with vv-NPL, whereas coinfection with vv-Tk, vv-T3, or vv-T7 did not increase the level of H expression (Fig. 6). For a control, the MV and vv-NPL coinfection of HeLa cells resulted in a dose-dependent reduction of the level of H expression. The inhibitory effect of the coinfection of HeLa cells with vv-Tk was likely due to its much higher replication level in human cells and increased cytopathic effect, an effect not observed with vv-NPL which, like the vv MVA strain, has both K1L and C7L genes deleted (29). The trans-complementation effect was further explored using MODE-K cells expressing CD150 as an alternate cellular receptor (Fig. 7). The expression of CD150 allowed a significant expression of H protein after MV infection at a MOI of 2. Increasing infectious MV input up to a MOI of 10 resulted in cell surface contents in H just above that observed after infection at a MOI of 2 (20 and 15 arbitrary units, respectively), the H excess reflecting mostly the H brought by the higher input virus particles as measured when using 10 equivalent MOI of UV-inactivated MV (9 arbitrary units). The infection of MODE-K.CD150 cells with vv-T7 resulted in some apparent increase of H expression whether the cells were coinfected or not coinfected with MV, which was inactivated or not inactivated with UV. This nonspecific effect was likely due to some vaccinia virus-induced cytopathic effect. Indeed, the infection with vv-NPL in the absence of MV or with UV-inactivated MV resulted in a similar higher level of H expression compared to that of the corresponding MV infections in the absence of any vaccinia virus coinfection. In contrast, providing N-P-L or N-P proteins in trans, using vv-NPL or vv-NP, resulted in a significant increase in H expression (Fig. 7). The isolated expression of N, P, or L protein was not able to circumvent the virus replication block with an H expression level similar to that observed after vv-T7 infection. Likewise, the expression of P-L complexes was unable to alleviate the H expression defect. As expected, UV irradiation of MV prior to MODE-K.CD150 infection resulted in the lack of significant increase in the level of H expression induced by vv-NPL (Fig. 7).
FIG. 6.
Recovery of MV protein synthesis in MODE-K.CD46 cells by MV-NPL complex provided in trans. Expression of H protein measured after cell surface immunolabeling and flow cytometry (a) (results expressed in mean fluorescence arbitrary units) or Western blotting (b) 2 days p.i. in HeLa and MODE-K.CD46 cells coinfected with MV (Hallé; MOI = 1) and recombinant vaccinia virus (MOI = 1 to 5) encoding MV-NPL proteins, thymidine kinase, or T3 or T7 polymerase.
FIG. 7.
Recovery of MV protein synthesis in MODE-K.CD150 cells by MV-NPL or MV-NP complex provided in trans. Cell surface expression of H protein (in mean fluorescence arbitrary units) on MODE-K and MODE-K.CD150 cells 2 days after coinfection with MV (Hallé, MOI = 0, 2 or 10) and vv-T7, vv-NPL, vv-NP, vv-PL, vv-N, vv-P or vv-L (MOI = 5). Sham infection using UV-inactivated MV (MV-UV) was used for a control.
Inability of heat shock to alleviate MV replication block in MODE-K.CD46 cells.
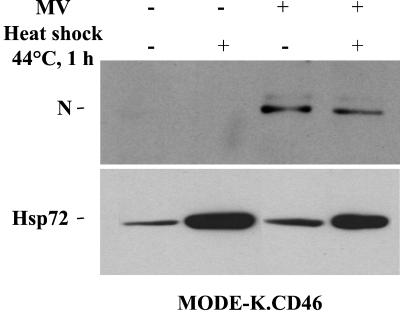
Heat shock and Hsp72 protein have been reported to enhance MV replication in permissive human and simian cells (40, 57, 58). MODE-K.CD46 cells were heated at 44°C for 1 h, resulting in an increase in the expression of endogenous Hsp72 (Fig. 8). However, after MV infection, no change in MV-N expression was observed.
FIG. 8.
Induction of Hsp72 by heat shock does not alleviate the defect of MV protein synthesis in MODE-K.CD46 cells. MV-N and Hsp72 protein expression in MODE-K.CD46 cells treated (+) or not treated (−) with heat shock for 1 h at 44°C and infected for 2 days by MV (+) (Hallé; MOI = 1) is shown.
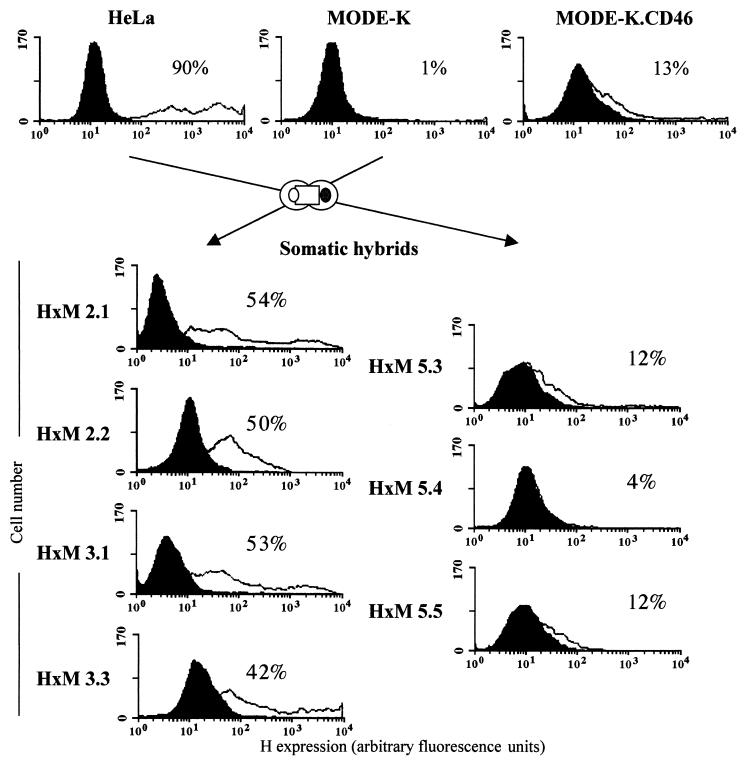
HeLa × MODE-K somatic hybrids display either blocked or permissive MV replication phenotype.
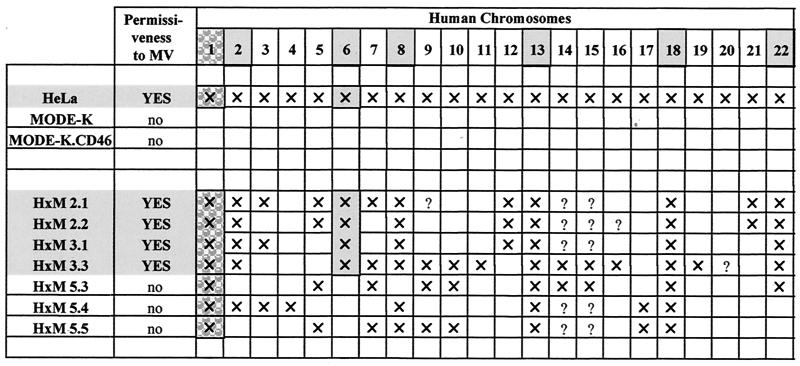
HeLa × MODE-K (H×M) somatic hybrid cell clones, isolated on the basis of dual expression of human CD46 and murine MHC-I, were tested for their ability to support MV infection. As shown on Fig. 9, four of seven clones exhibited a permissive phenotype, with more than 50% of cells expressing H protein 2 days p.i. The three other clones exhibited a nonpermissive phenotype similar to that of MODE-K.CD46 cells. Since human × mouse somatic cell hybrids are characterized by variable contents of human chromosomes, the permissiveness of some H×M hybrids indicate that one (or a combination of several) human cellular factor(s) can alleviate the MV replication block of the MODE-K cells. Indeed, as expected from the chromosome location of the CD46 gene, they all contained chromosome 1 (Table 1). The permissive phenotype was associated with the presence of chromosomes 2, 6, 8, 13, 18 and 22, pointing to these chromosomes as encoding the putative cellular factor(s) enabling MV replication in the MODE-K cellular context. The fact that chromosome 6 was present only in the permissive clone makes it the best candidate, but the possibility that the human factor(s) results from a complex interaction of proteins encoded by distinct chromosomes cannot be excluded.
FIG. 9.
Somatic hybrid clones of MODE-K and human cells exhibit either blocked or permissive MV protein synthesis. HeLa and MODE-K cells were fused, and somatic cell hybrids were selected for dual expression of human CD46 and mouse MHC-I proteins. Isolated hybrid clones were infected with MV (Hallé; MOI = 1) and expression of cell surface H protein was measured after immunolabeling and flow cytometry. H expression level is shown by the shift of the white curve to the right and by percent positive cells. The black curves correspond to the autofluorescence level of each cell clone in the absence of anti-H antibody.
TABLE 1.
Human chromosome contents of HeLa × MODE-K (HxM) somatic hybridsa
CD46-encoding chromosome 1 (stippling with a pattern) and chromosomes associated with permissiveness (light grey stippling) are indicated. Symbols: ×, chromosome detected; ?, not determined.
DISCUSSION
Here, we reported a detailed host restriction phenotype for a member of the Mononegavirales order characterized by an early block of MV transcription which can be trans-complemented by either MV N-P and MV N-P-L complexes or by a human cellular factor(s).
There are several pieces of data that support the hypothesis that the restriction of MV replication in the mouse MODE-K cell line is caused by a defect in MV RNA synthesis. MV replication block was observed irrespective of the cell entry pathway, namely, CD46 (12, 37) and CD150 (13, 30, 54) cellular receptor-mediated pathway or VSV-G pH-dependent endocytic pathway (4), as shown by the lack of replication of recombinant MGV and MG/FV viruses. Indeed, both CD46- and CD150-mediated MV binding and fusion activity could be detected when these receptors were expressed in MODE-K cells. Furthermore, the ability of N-P or N-P-L protein complexes to alleviate this block in MODE-K.CD46 and MODE-K.CD150 cells implies that the MV nucleocapsids from incoming infectious virus have entered the cytosol of MODE-K cells.
The order of genes in the MV genome and the reduced reinitiation at every intergenic region result in the optimal balance between the three proteins involved in the polymerase activity. The first gene encodes the N protein, which is required in large amounts to encapsidate the 15,894-nucleotide MV genome and antigenome. The P protein is synthesized in smaller amounts because of the downstream position of the gene, the transcription into the two mRNAs (P/C and V), and the translation of the P/C mRNA into P and C proteins (2, 7). The most downstream gene encodes the L protein, which is therefore synthesized in limited amounts. Overall, a single ribonucleoparticle contains an estimated 2,600 N, 300 P, and 40 L proteins for each genome. The importance of this N/P/L balance for transcription and replication of genomic RNA has been demonstrated in MV reverse genetic studies (43, 47, 48). Although encapsidation and concomitant replication of MV genomes require a constant supply of N, P, and L proteins, transcription is favored when they are in limited supply (24, 32). This likely explains why increasing amounts of N, P, and L proteins, provided in trans, reduced virus protein synthesis in HeLa cells, which are permissive for MV. The trans-complementation of restricted MV RNA synthesis in the MODE-K cell lines by N-P-L or N-P complexes suggests that N-P-L or N-P complexes may act by maintaining (stabilizing?) the incoming N-RNA genome template proficient for polymerase activity. The lack of increase in MV protein expression when the MOI was increased from 2 to 10 argues for this hypothesis, with every incoming infectious ribonucleoparticle being similarly destabilized. The lack of effect of providing N protein alone can be explained by the chaperone role of P to prevent the aggregation and nuclear accumulation of N protein and to favor viral RNA encapsidation (27, 31, 50). The L protein tends to be unstable and is unable to bind to the viral RNA or to the N-RNA template. It has to form complexes with the P protein in order to recognize the N-RNA template, as shown for the related VSV (20). Likewise, complexes of L-P and N-P proteins from Sendai virus are required for in vitro polymerase activity (26). The inability of P-L complexes to alleviate the RNA synthesis block in MODE-K cells indicates that P-L complexes from the incoming virions are not the primary target of the cellular defect.
What could be the cellular factor(s) responsible for the MV restriction phenotype of MODE-K cells? It cannot be attributed to the synthesis of alpha/beta IFN because (i) VSV, which is highly sensitive to inhibition (44), replicates normally in MODE-K cells, (ii) cells should be pretreated with IFN in order to be efficient in blocking incoming virus replication, and (iii) if mouse IFN synthesis were involved, one would expect that all human × mouse somatic cell hybrids exhibit the restrictive phenotype because only human chromosomes are lost at random. The Hsp72 protein has been associated with increased MV replication (40, 57, 58). This highly conserved protein (a single amino acid difference between mouse and human species) was detected in MODE-K.CD46 cells. Submitting these cells to a heat shock resulted in a large increase in Hsp72 expression without enhancement of MV replication. This indicates that the phenotype of MODE-K cells is unlikely to be related to Hsp72. The permissive or nonpermissive phenotype of human × mouse somatic cell hybrid according to their contents in human chromosome suggest that a human factor(s) can act in trans to allow efficient natural MV transcription. A limited number of mouse cell lines expressing either human CD46 or CD150 has been tested so far for their MV permissiveness, and they exhibit various phenotypes, from permissiveness to restriction in MV replication at various stages of the virus infection cycle (16, 28, 37, 61). The phenotype of MODE-K cells may reflect the property of a subset of mouse tissues, since MV growth in some tissues of CD46 transgenic mice is restricted at an early stage (28).
The observation that MODE-K cells efficiently support MV polymerase-driven RNA synthesis in a CAT minigenome assay indicates that transcription does work in these cells. What the minigenome experiment and the trans-complementation have in common is (i) vaccinia virus infection, which we can exclude as a complementing factor from the controls and (ii) a high level of N-P(-L) protein synthesis. Therefore, the native N-P synthesized in the cytoplasm seems to be the crucial point. Therefore, we can conclude that in the natural MV infection, RNA synthesis is restricted in MODE-K cells expressing one of the human MV receptors but can be overcome either by producing native N-P complexes or by human cellular factors (hybrid cells). Why N-P proteins have to be supplied in the cytoplasm is then a matter of speculation. One possibility is that they become inactivated when the nucleocapsid enters into the mouse cell cytoplasm, either because of additional posttranslational modifications (e.g., phosphorylation or dephosphorylation, which have been shown to be important for VSV [21, 49]) and/or because of enhanced dissociation of the polymerase-template complexes. Alternatively, the lack of association with a putative cellular cofactor acting, e.g., as a N(-P?)-RNA stabilizer, cannot be excluded. Significant amounts of additional native N-P proteins, likely outweighing the incoming N-P proteins from the few infectious particles, would either provide template building blocks in active state or increase the N(-P?) to template dissociation time according to the mass action law. A cell site-specific inactivation of natural MV RNA synthesis occurring within the vicinity of the plasma membrane is also possible. Indeed, a change in the virus entry site from plasma membrane (for MV) to acidic endosomal compartment (for recombinant MGV and MG/FV) is associated with a much reduced replication kinetics (51; D. Gerlier, unpublished data). The Sendai virus polymerase exhibits a much poorer transcription processivity in vitro than in vivo, indicating that the living cell environment is more critical for transcription than for replication of the virus genome (23). In the case of the respiratory syncytial virus, the processivity of transcription is ensured by the virus-encoded M2-1 protein (17). It is possible that MV replication fails in MODE-K cells because of the lack of efficient processivity during transcription. Besides the inhibitory activity of M protein (5, 9, 41, 52), nothing is known about the molecular events leading to the stop and start of Mononegavirales polymerase activities during budding from the host cell and fusion within the target cell, respectively. Identifying the cellular factor(s) responsible for the restriction phenotype of the MODE-K cell line will help in unraveling the regulation of MV RNA synthesis.
Acknowledgments
We thank R. Drillien, M. Billeter, H. Y. Naim, R. Cattaneo, D. Kaiserlian, C. Muller, and F. Wild for providing useful reagents; G. Varior-Krishnan for her contribution; C. Rabourdin-Combe for her support in the initiation of this work; and H. Vidal and E. Dusserre for useful advice. vCB21R-lacZ was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, from C. C. Broder, P. E. Kennedy, and E. A. Berger. The flow cytometry studies were done using the facilities of the Centre Commun d'Imagerie de Laennec.
This work was performed with financial support from the Commission of European Communities (RTD programme “Quality of Life and Management of Living Resources” [QLK2-CT2001-01225]) and from the Ministère de l'Education Nationale de la Recherche et de la Technologie (PRFMMIP).
The content of this publication does not necessarily reflect the views of the Commission of European Communities and in no way anticipates the Commission's future policy in this area.
REFERENCES
- 1.Alkhatib, G., C. C. Broder, and E. A. Berger. 1996. Cell type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J. Virol. 70:5487-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellini, W. J., G. Englund, S. Rozenblatt, H. Arnheiter, and C. D. Richardson. 1985. Measles virus P gene codes for two proteins. J. Virol. 53:908-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumberg, B. M., J. Chan, and S. A. Udem. 1991. Function of paramyxovirus 3′ and 5′ end sequences: in theory and practice, p. 235-248. In D. W. Kingsbury (ed.), The paramyxoviruses. Plenum Press, New York, N.Y.
- 4.Brown, J. C., W. W. Newcomb, and S. Lawrenz-Smith. 1988. pH-dependent accumulation of the vesicular stomatitis virus glycoprotein at the ends of intact virions. Virology 167:625-629. [PubMed] [Google Scholar]
- 5.Carroll, A. R., and R. R. Wagner. 1979. Role of the membrane (M) protein in endogenous inhibition of in vitro transcription by vesicular stomatitis virus. J. Virol. 29:134-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cattaneo, R., K. Kaelin, K. Baczko, and M. A. Billeter. 1989. Measles virus editing provides an additional cysteine-rich protein. Cell 56:759-764. [DOI] [PubMed] [Google Scholar]
- 8.Christiansen, D., P. Devaux, B. Reveil, A. Evlashev, B. Horvat, J. Lamy, C. Rabourdin-Combe, J. H. M. Cohen, and D. Gerlier. 2000. Octamerization enables soluble CD46 receptor to neutralize measles virus in vitro and in vivo. J. Virol. 74:4672-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinton, G. M., S. P. Little, F. S. Hagen, and A. S. Huang. 1978. The matrix (M) protein of vesicular stomatitis virus regulates transcription. Cell 15:1455-1462. [DOI] [PubMed] [Google Scholar]
- 10.Davidson, R. L., and P. S. Gerald. 1976. Improved techniques for the induction of mammalian cell hybridization by polyethylene glycol. Somatic Cell Genet. 2:165-176. [DOI] [PubMed] [Google Scholar]
- 11.Davidson, R. L., K. A. O'Malley, and T. B. Wheeler. 1976. Polyethylene glycol-induced mammalian cell hybridization: effect of polyethylene glycol molecular weight and concentration. Somatic Cell Genet. 2:271-280. [DOI] [PubMed] [Google Scholar]
- 12.Dörig, R. E., A. Marcil, A. Chopra, and C. D. Richardson. 1993. The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 75:295-305. [DOI] [PubMed] [Google Scholar]
- 13.Erlenhoefer, C., W. J. Wurzer, S. Loffler, S. Schneider-Schaulies, V. ter Meulen, and J. Schneider-Schaulies. 2001. CD150 (SLAM) is a receptor for measles virus but is not involved in viral contact-mediated proliferation inhibition. J. Virol. 75:4499-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escoffier, C., and D. Gerlier. 1999. Infection of chicken embryonic fibroblasts by measles virus: adaptation at the virus entry level. J. Virol. 73:5220-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escoffier, C., S. Manie, S. Vincent, C. P. Muller, M. Billeter, and D. Gerlier. 1999. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J. Virol. 73:1695-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evlashev, A., H. Valentin, P. Rivailler, O. Azocar, C. Rabourdin-Combe, and B. Horvat. 2001. Differential permissivity to measles virus infection of human and CD46-transgenic murine lymphocytes. J. Gen. Virol. 82:2125-2129. [DOI] [PubMed] [Google Scholar]
- 17.Fearns, R., and P. L. Collins. 1999. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 73:5852-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fournier, P., N. H. Brons, G. A. Berbers, K. H. Wiesmuller, B. T. Fleckenstein, F. Schneider, G. Jung, and C. P. Muller. 1997. Antibodies to a new linear site at the topographical or functional interface between the hemagglutinin and fusion proteins protect against measles encephalitis. J. Gen. Virol. 78:1295-1302. [DOI] [PubMed] [Google Scholar]
- 19.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao, Y., and J. Lenard. 1995. Cooperative binding of multimeric phosphoprotein (P) of vesicular stomatitis virus to polymerase (L) and template: pathways of assembly. J. Virol. 69:7718-7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, Y., and J. Lenard. 1995. Multimerization and transcriptional activation of the phosphoprotein P of the vesicular stomatitis virus by casein kinase-II. EMBO J. 14:1240-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giraudon, P., C. Gerald, and T. F. Wild. 1984. A study of measles virus antigens in acutely and persistently infected cells using monoclonal antibodies: differences in the accumulation of certain viral proteins. Intervirology 21:110-120. [DOI] [PubMed] [Google Scholar]
- 23.Gubbay, O., J. Curran, and D. Kolakofsky. 2001. Sendai virus genome synthesis and assembly are coupled: a possible mechanism to promote viral RNA polymerase processivity. J. Gen. Virol. 82:2895-2903. [DOI] [PubMed] [Google Scholar]
- 24.Hall, W. W., D. Genius, and V. ter Meulen. 1977. The effect of cycloheximide on the replication of measles virus. J. Gen. Virol. 35:579-582. [DOI] [PubMed] [Google Scholar]
- 25.Hierholzer, J. C., and R. A. Killington. 1996. Quantitation of virus, p. 35-46. In B. W. J. Mahy and H. O. Kangro (ed.), Virology methods manual. Academic Press, London, United Kingdom.
- 26.Horikami, S. M., J. Curran, D. Kolakofsky, and S. A. Moyer. 1991. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horikami, S. M., and S. A. Moyer. 1995. Structure, transcription and replication of measles virus. Curr. Top. Microbiol. Immunol. 191:35-50. [DOI] [PubMed] [Google Scholar]
- 28.Horvat, B., P. Rivailler, G. Varior-Krishnan, A. Cardoso, D. Gerlier, and C. Rabourdin-Combe. 1996. Transgenic mice expressing human measles virus (MV) receptor CD46 provide cells exhibiting different permissivities to MV infection. J. Virol. 70:6673-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howley, P. M., B. Lafont, D. Spehner, K. Kaelin, M. A. Billeter, and R. Drillien. 1999. A functional measles virus replication and transcription machinery encoded by the vaccinia virus genome. J. Virol. Methods 79:65-74. [DOI] [PubMed] [Google Scholar]
- 30.Hsu, E. C., C. Iorio, F. Sarangi, A. A. Khine, and C. D. Richardson. 2001. CDw150 (SLAM) is a receptor for lymphotropic strain of measles virus and may account for the immunosuppressive properties of this virus. Virology 279:9-21. [DOI] [PubMed] [Google Scholar]
- 31.Huber, M., R. Cattaneo, P. Spielhofer, C. Orvell, E. Norrby, M. Messerli, J. C. Perriard, and M. A. Billeter. 1991. Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology 185:299-308. [DOI] [PubMed] [Google Scholar]
- 32.Lamb, R. A., and D. Kolakofsky. 1996. Paramyxoviridae: the viruses and their replication, p. 1177-1204. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Field's virology, 3rd ed. Lippincott-Raven, New York, N.Y.
- 33.Long, E. O., S. Rosen-Bronson, D. R. Karp, M. Malnati, R. P. Sekaly, and D. Jaraquemada. 1991. Efficient cDNA expression vectors for stable and transient expression of HLA-DR in transfected fibroblast and lymphoid cells. Hum. Immunol. 31:229-235. [DOI] [PubMed] [Google Scholar]
- 34.Manié, S. N., S. Debreyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moyer, S. A., S. C. Baker, and S. M. Horikami. 1990. Host cell proteins required for measles virus reproduction. J. Gen. Virol. 71:775-783. [DOI] [PubMed] [Google Scholar]
- 36.Mrkic, B., J. Pavlovic, T. Rulicke, P. Volpe, C. J. Buchholz, D. Hourcade, J. P. Atkinson, A. Aguzzi, and R. Cattaneo. 1998. Measles virus spread and pathogenesis in genetically modified mice. J. Virol. 72:7420-7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naniche, D., G. Varior-Krishnan, F. Cervoni, T. F. Wild, B. Rossi, C. Rabourdin-Combe, and D. Gerlier. 1993. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 67:6025-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naniche, D., T. F. Wild, C. Rabourdin-Combe, and D. Gerlier. 1992. A monoclonal antibody recognizes a human cell surface glycoprotein involved in measles virus binding. J. Gen. Virol. 73:2617-2624. [DOI] [PubMed] [Google Scholar]
- 39.Naniche, D., A. Yeh, D. Eto, M. Manchester, R. M. Friedman, and M. B. Oldstone. 2000. Evasion of host defenses by measles virus: wild-type measles virus infection interferes with induction of alpha/beta interferon production. J. Virol. 74:7478-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parks, C. L., R. A. Lerch, P. Walpita, M. S. Sidhu, and S. A. Udem. 1999. Enhanced measles virus cDNA rescue and gene expression after heat shock. J. Virol. 73:3560-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinney, D. F., and S. U. Emerson. 1982. In vitro synthesis of triphosphate-initiated N-gene mRNA oligonucleotides is regulated by the matrix protein of vesicular stomatitis virus. J. Virol. 42:897-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pontecorvo, G. 1975. Production of mammalian somatic cell hybrids by means of polyethylene glycol treatment. Somatic Cell Genet. 1:397-400. [DOI] [PubMed] [Google Scholar]
- 43.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasmussen, L., and L. B. Farley. 1975. Inhibition of Herpesvirus hominis replication by human interferon. Infect. Immun. 12:104-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reutter, G. L., C. Cortese-Grogan, J. Wilson, and S. A. Moyer. 2001. Mutations in the measles virus C protein that up regulate viral RNA synthesis. Virology 285:100-109. [DOI] [PubMed] [Google Scholar]
- 46.Richardson, C. D., A. Scheid, and P. W. Choppin. 1980. Specific inhibition of paramyxovirus and myxovirus replication by oligopeptides with amino acid sequences similar to those at the N-termini of the F1 or HA2 viral polypeptides. Virology 105:205-222. [DOI] [PubMed] [Google Scholar]
- 47.Schneider, H., P. Spielhofer, K. Kaelin, C. Dotsch, F. Radecke, G. Sutter, and M. A. Billeter. 1997. Rescue of measles virus using a replication-deficient vaccinia-T7 vector. J. Virol. Methods 64:57-64. [DOI] [PubMed] [Google Scholar]
- 48.Sidhu, M. S., J. Chan, K. Kaelin, P. Spielhofer, F. Radecke, F. Schneider, M. Masurekar, P. C. Dowling, M. A. Billeter, and S. A. Udem. 1995. Rescue of synthetic measles virus minireplicons: measles genomic termini direct efficient expression and propagation of a reporter gene. Virology 208:800-807. [DOI] [PubMed] [Google Scholar]
- 49.Spadafora, D., D. M. Canter, R. L. Jackson, and J. Perrault. 1996. Constitutive phosphorylation of the vesicular stomatitis virus P protein modulates polymerase complex formation but is not essential for transcription or replication. J. Virol. 70:4538-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spehner, D., R. Drillien, and P. M. Howley. 1997. The assembly of the measles virus nucleoprotein into nucleocapsid-like particles is modulated by the phosphoprotein. Virology 232:260-268. [DOI] [PubMed] [Google Scholar]
- 51.Spielhofer, P., T. Bachi, T. Fehr, G. Christiansen, R. Cattaneo, K. Kaelin, M. A. Billeter, and H. Y. Naim. 1998. Chimeric measles viruses with a foreign envelope. J. Virol. 72:2150-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suryanarayana, K., K. Baczko, V. ter Meulen, and R. R. Wagner. 1994. Transcription inhibition and other properties of matrix proteins expressed by M genes cloned from measles viruses and diseased human brain tissue. J. Virol. 68:1532-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeda, M., A. Kato, F. Kobune, H. Sakata, Y. Li, T. Shioda, Y. Sakai, M. Asakawa, and Y. Nagai. 1998. Measles virus attenuation associated with transcriptional impediment and a few amino acid changes in the polymerase and accessory proteins. J. Virol. 72:8690-8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatsuo, H., N. Ono, K. Tanaka, and Y. Yanagi. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature 406:893-897. [DOI] [PubMed] [Google Scholar]
- 55.Tober, C., M. Seufert, H. Schneider, M. A. Billeter, I. C. Johnston, S. Niewiesk, V. ter Meulen, and S. Schneider-Schaulies. 1998. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J. Virol. 72:8124-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valsamakis, A., H. Schneider, P. G. Auwaerter, H. Kaneshima, M. A. Billeter, and D. E. Griffin. 1998. Recombinant measles viruses with mutations in the C, V, or F gene have altered growth phenotypes in vivo. J. Virol. 72:7754-7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasconcelos, D., E. Norrby, and M. Oglesbee. 1998. The cellular stress response increases measles virus-induced cytopathic effect. J. Gen. Virol. 79:1769-1773. [DOI] [PubMed] [Google Scholar]
- 58.Vasconcelos, D. Y., X. H. Cai, and M. J. Oglesbee. 1998. Constitutive overexpression of the major inducible 70 kDa heat shock protein mediates large plaque formation by measles virus. J. Gen. Virol. 79:2239-2247. [DOI] [PubMed] [Google Scholar]
- 59.Vidal, K., I. Grosjean, J. P. Revillard, C. Gespach, and D. Kaiserlian. 1993. Immortalization of mouse intestinal epithelial cells by the SV40-large T gene. Phenotypic and immune characterization of the MODE-K cell line. J. Immunol. Methods 166:63-73. [DOI] [PubMed] [Google Scholar]
- 60.Vincent, S., D. Gerlier, and S. N. Manié. 2000. Measles virus assembly within membrane rafts. J. Virol. 74:9911-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vincent, S., D. Spehner, S. Manie, R. Delorme, R. Drillien, and D. Gerlier. 1999. Inefficient measles virus budding in murine L.CD46 fibroblasts. Virology 265:185-195. [DOI] [PubMed] [Google Scholar]
- 62.Wild, T. F., A. Bernard, D. Spehner, and R. Drillien. 1992. Construction of vaccinia virus recombinants expressing several measles virus proteins and analysis of their efficacy in vaccination of mice. J. Gen. Virol. 73:359-367. [DOI] [PubMed] [Google Scholar]
- 63.Wild, T. F., E. Malvoisin, and R. Buckland. 1991. Measles virus: both the haemagglutinin and fusion glycoproteins are required for fusion. J. Gen. Virol. 72:439-442. [DOI] [PubMed] [Google Scholar]