Abstract
It has been reported that patients infected with nef-defective human immunodeficiency virus type 1 (HIV-1) do not progress to AIDS; however, mutations that abrogate Nef expression are not common in long-term nonprogressors (LTNPs). We postulated that Nef function might be impaired in LTNPs, irrespective of the presence or absence of detectable amino acid sequence anomalies. To challenge this hypothesis we compared in vitro function of nef alleles that were derived from three groups of Japanese patients: LTNPs, progressors, and asymptomatic carriers (ACs). The patient-derived nef alleles were subcloned into a nef-defective infectious HIV-1 molecular clone and an expression vector. We first examined Nef-dependent enhancement of infection in a single-round infectivity assay by the use of MAGNEF cells, in which Nef is required more strictly for the infection than in the parent MAGI cells. All nef alleles from LTNPs showed reduced enhancement in the infectivity of nef-defective HIV-1 mutants compared to the nef alleles of progressors or ACs. Second, we found that nef alleles from LTNPs were less efficient in CD4 downregulation than those of progressors or ACs. Third, all nef alleles from LTNPs, progressors, and ACs reduced the cell surface expression of major histocompatibility complex class I to a similar level. Last, there was no correlation between Hck-binding activity of Nef and clinical grouping. In conclusion, we detected inefficient enhancement of HIV-1 infectivity and CD4 downregulation by HIV-1 nef alleles of LTNPs. It awaits further study to conclude that these characteristics of nef alleles are the cause or the consequence of the long-term nonprogression after HIV-1 infection.
The median time to development of AIDS in most cohorts is approximately 10 years after initial exposure to the virus (36, 40). However, some human immunodeficiency virus type 1 (HIV-1)-infected persons remain clinically healthy without any drug treatment and show no decline in CD4+ T-cell counts even though they have been seropositive for more than 10 years (5, 6, 15, 21, 29, 35, 43). The mechanism that generates the long-term nonprogressors (LTNPs) is under intense investigation, because a clue to vaccine development for HIV-1 may be underlie it.
Defects in the nef gene of HIV-1 have been linked to nonprogressive infection. Following an earlier case report of an LTNP carrying only nef-defective HIV-1 (28), an Australian cohort study has also shown that six LTNPs were infected with nef-deleted HIV-1 from a single blood donor (30). Supporting this view, rhesus monkeys inoculated experimentally with a nef-defective simian immunodeficiency virus have low viral loads and normal CD4+ T-cell numbers (26). Furthermore, there are reports that document the presence of nef-defective HIV-1 genomes in LTNPs (14, 19, 32, 42). However, some reports argue against the role of nef in the establishment of long-term nonprogression because most of the nef alleles isolated from LTNPs were intact in the length of the coding region and in the tested biological function (9, 23, 24, 32, 34).
The in vitro function of nef can largely be summarized in four categories (for reviews see references 12, 13, 16, and 19): downregulation of cell surface CD4, downregulation of the class I major histocompatibility complex (MHC), stimulation of the signal transduction cascades, and enhancement of viral replication in specific cell types. More recently, an antiapoptotic effect of Nef has also been reported (46). Extensive mutagenic studies have revealed that these activities are genetically separable and mapped to different regions of Nef (reviewed in reference 19); however, it remains elusive as to which in vitro function is most critical for the in vivo pathogenicity of primate lentiviruses.
Among the many in vitro functions of Nef, the enhancement of viral infectivity appears to be associated most directly with the replication cycle of HIV-1; however, Nef is dispensable for viral replication under commonly used laboratory conditions (26). Thus, reporter cells including HeLa-CD4-LTR-βGal cells (MAGI) (27) are often preferred for analysis. In assays using reporter cells, the difference in infectivity between the nef-defective mutant and wild-type HIV is less than 10-fold in most cases (2, 3, 7, 11, 25). Considering a large statistical error in the infectivity assay, this difference seems to be subtle. We postulated that the previous failure in the detection of functional differences between the nef alleles from LTNPs and those from progressors might have been caused by this small requirement for Nef in many reporter cells. Based on the observation that the Nef-induced enhancement of HIV-1 infectivity correlates inversely with the amount of CD4 in the target cells (45), we have isolated a MAGI-derived cell line, MAGNEF, which requires nef more strictly than does the parent MAGI cell in the single-round infectivity assay (44). Here we used MAGNEF cells for the quantitative analysis of the enhancement of HIV-1 infectivity by nef genes isolated from patients with different clinical outcomes.
MATERIALS AND METHODS
Subjects.
Characterization of the patients and isolation of HIV-1 proviral nef gene from genomic DNA of peripheral blood mononuclear cells (PBMCs) by nested PCR have been described previously (47, 48). Briefly, we studied 14 Japanese hemophiliacs who were infected with HIV-1 through contaminated blood products more than 10 years before sample collection. Five were LTNPs and maintained their CD4+ cell count above 450/μl without antiretroviral therapy. Six were progressors with CD4+ cell counts below 100/μl at the time of sample collection. In the present study, we included 5 asymptomatic carriers (ACs) who were infected with HIV-1 within 3 years of sample collection and maintained their CD4+ cell count above 450/μl without antiretroviral therapy.
Isolation, cloning, and expression of the proviral nef gene.
Primers used in nested PCR were set at highly conserved regions of env and the long terminal repeat as described previously (48). PCR products were cloned into pCR-blunt Topo II (Invitrogen) and were sequenced with a BigDye sequencing kit (PE Biosystems). To facilitate subcloning, the coding sequence of nef was amplified by PCR with primer sets containing SalI and NotI sites in forward and reverse primers, respectively. A primer set used to amplify each nef gene was chosen so that the amino acid sequence of each Nef would not change after PCR amplification. After PCR amplification, the fragments were again subcloned into pCR-blunt Topo II and verified for the sequence. For Nef expression, we used pCAGGS-IRES-EGFP, which was derived from the pCAGGS eukaryotic expression vector (37) and contained the internal ribosome entry site (IRES) of encephalomyocarditis virus at the 3′ end of the cloning site, followed by the enhanced green fluorescent protein (EGFP) gene. The Nef-coding DNA fragments were cleaved out from the plasmids with SalI and NotI and were subcloned into pCAGGS-IRES-EGFP cleaved with XhoI and NotI.
Plasmid.
An infectious proviral clone of HIV-1, pNL4-3, and its nef-defective mutant, pNL-Xh, have been described previously (1). We introduced XhoI and NotI sites in pNL4-3 for insertion of the nef gene from patients as follows: DNA fragments extending from the 3′ region of env of NL4-3, nucleotides (nt) 8032 to 8785, were amplified by PCR with a forward primer, 5′-TGCTGTGCCTTGGAATGCTAG-3′, and a reverse primer, 5′-GTCGACGCGGCCGCACGCGTCTCGAGTTATAGCAAAATCCTTTCCAAGCCC-3′. The underlined sequence contains recognition sites of SalI, NotI, MluI, and XhoI from the 5′ end. The amplified DNA fragment was cloned into pCR-blunt Topo II, and the sequences were verified. From this plasmid, a DNA fragment of the carboxyl-terminal region of env was cleaved out by BamHI and SalI, and it was inserted into pNL4-3 cleaved with BamHI at nt 8465 and XhoI at nt 8892. Therefore, in the resulting plasmid, named pNL-Not, the 5′ region of nef from nt 8786 to 8892 was replaced with the recognition sites of XhoI, MluI, and NotI. The nef alleles derived from patients and NL4-3 were cleaved with SalI and NotI and inserted into pNL-Not cleaved with XhoI and NotI. As a positive control, we amplified the nef allele of pNL4-3 and inserted it into pNL-Not.
Cells and transfection.
MAGIC5 and MAGIC5D cells were CCR5-expressing derivatives of HeLa-CD4-LTR-βGal (MAGI) cells (22, 27). The cell line named MAGNEF was obtained by limiting dilution of MAGIC5 cells. Compared with the parent MAGIC5 cells, MAGNEF cells expressed a lower level of CD4 and required Nef more strictly for HIV-1 infectivity (44). By using of a phycoerythrin fluorescence quantitation kit (Becton Dickinson), the numbers of cell surface CD4 were determined as 4 × 104 in human peripheral CD4-positive T cells, 5 × 103 in MAGIC5, 5 × 105 in MAGIC5D, and 1 × 103 in MAGNEF. MAGNEF and MAGIC5D cells were cultured in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum (FCS). 293T human embryonic kidney cells were a gift from Bruce J. Mayer at the University of Connecticut and were maintained in Dulbecco's modified Eagle's medium with 10% FCS. Plasmid DNAs were transfected into 293T cells by the calcium phosphate coprecipitation method or into MAGIC5D cells with Lipofectamine 2000 (Gibco-BRL).
MAGI assay.
Infectious DNAs of HIV-1 were transfected into 293T cells by the calcium phosphate coprecipitation method. Forty-eight hours later, cultured supernatants were filtered through a 0.45-μm-pore-size filter (Whatman, Clifton, N.J.) and examined for their p24gag concentration by an HIV-1 p24 antigen enzyme-linked immunosorbent assay kit (ZeptoMatrix, Buffalo, N.Y.). We plated 104 MAGNEF cells on 96-well tissue culture plates and inoculated them with serially diluted virus stocks. Forty-eight hours after infection, cells were fixed and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactoside as described elsewhere (27). We chose wells in which the number of positive cells was about 50 for calculation of the infectivity of the virus stocks. Under this condition, the dilution of the viral stocks and the number of infected cells were in a linear range.
Flow cytometry analysis .
MAGIC5D cells were transfected with pCAGGS-IRES-EGFP-derived vectors carrying the nef alleles of patients. Thirty-six hours after transfection cells were detached from culture dishes with EDTA treatment and were suspended in phosphate-buffered saline containing 1% FCS. After being stained with saturating amounts of phycoerythrin-conjugated Leu3A monoclonal antibody specific for CD4 (Becton Dickinson) or phycoerythrin-conjugated W6/32 monoclonal antibody specific for the assembled MHC-I heavy chain β2-microglobulin complex (DAKO), cells were washed with phosphate-buffered saline and fixed with 1% formaldehyde. Fluorescence intensities of green fluorescent protein (GFP) and either CD4 or MHC-I were analyzed with a FACSCalibur flow cytometer (Becton Dickinson). After gating with GFP intensity, the geometric mean fluorescence intensity of CD4 or MHC-I was calculated with CELLQuest software (Becton Dickinson). The relative downregulation efficiency was shown by the formula (intensity of NL4-3-Nef-expressing cells)/(intensity of cells to be evaluated).
Immunoblotting.
Cells were lysed in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100), clarified by centrifugation, separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, and transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, Mass.). We used a mixture of monoclonal antibodies against p24gag and gp41env for the detection of viral proteins. Nef on the filters was detected by a pool of monoclonal antibodies provided by K. Ikuta at Osaka University (38).
GST fusion protein and in vitro binding assay.
The 5′ region of Hck coding the SH2 and SH3 domains was amplified by PCR with the following primers: forward primer, 5′-GGGCCCCCCGTCGACATGGGGTGCATGAAG-3′; reverse primer, 5′-CGCGGCCGCCCCAAGGCTTCTGGGCTTGGA-3′. The underlined sequences indicate SalI and NotI restriction sites. The PCR product was cleaved with SalI and NotI and subcloned in pGEX3X (Amersham-Pharmacia Biotech). The SH2 and SH3 domains of Hck were expressed as a fusion protein with glutathione S-transferase (GST) in transformed Escherichia coli and purified with glutathione-Sepharose beads as described previously (33). 293T cells were transfected with pCAGGS-IRES-EGFP-derived Nef expression vectors carrying the nef allele of each patient. Forty-eight hours after transfection, cells were lysed in lysis buffer, cleared by centrifugation, and incubated with equal amounts of the GST-Hck proteins bound to the glutathione beads. The binding reaction was carried out at 4°C for 12 h. After being washed six times with lysis buffer, bound proteins were separated on SDS-12.5% polyacrylamide gels and analyzed by being immunoblotted with a pool of anti-Nef monoclonal antibodies.
RESULTS
Nef-defective virus used in the assay.
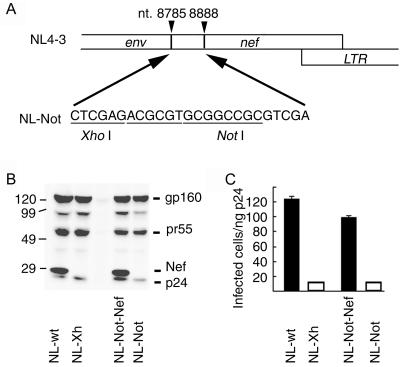
Our aim in this study was to examine whether there is any difference between the activities of Nef from LTNPs and from AIDS patients. For this goal we first constructed a pNL4-3-derived vector, named pNL-Not, which lacked the 5′ region of nef and contained restriction enzyme recognition sites for the insertion of nef alleles of patients (Fig. 1A). As a positive control we inserted the nef allele of NL4-3 into pNL-Not, generating pNL-Not-Nef. Plasmids encoding the wild-type NL4-3, a Nef-defective mutant of NL4-3 (NL-Xh), pNL-Not, and pNL-Not-Nef, were transfected to 293T cells to produce viruses. Expression of Nef and structural proteins was confirmed in cells transfected with pNL4-3 and pNL-Not-Nef but not pNL-Xh and pNL-Not (Fig. 1B). We used a pool of anti-Nef monoclonal antibodies, epitopes of which were distributed throughout Nef (38); however, we could not detect any protein in the cells transfected with pNL-Not, negating the expression of a truncated Nef protein from pNL-Not. Notably, we did not find any difference in the expression level of gp160env and pr55gag among the cells transfected with Nef-positive or Nef-negative proviral DNA.
FIG. 1.
Basic vector for the analysis of Nef enhancement of infectivity. (A) The 5′ region of nef of the HIV-1 NL4-3 strain was replaced with oligonucleotides containing XhoI and NotI restriction sites to generate NL-Not. Then nef allele of NL4-3 amplified by PCR was inserted into the XhoI/NotI site to generate NL-Not-Nef. (B) Wild-type NL4-3, nef-defective NL4-3 (NL-Xh), NL-Not, and NL-Not inserted with NL4-3 nef (NL-Not-Nef) were transfected into 293T cells for propagation of virus. 293T cells were lysed and analyzed by immunoblotting with a mixture of monoclonal antibodies against p24gag, gp41env, and Nef. Molecular standards are shown on the left. (C) Serially diluted virus stocks were inoculated into MAGNEF cells plated on 96-well dishes. Forty-eight hours later, infected cells were identified by being stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. Infectivity was normalized by the quantity of p24gag in the virus stocks. Experiments were done in triplicate and repeated three times. A representative result is shown with standard deviations.
The infectivity of these viruses was examined by use of MAGNEF indicator cells (Fig. 1C). The infectivity of NL4-3 was about 10-fold higher than that of the Nef-defective NL4-3, NL-Xh. The infectivity of NL-Not-Nef was slightly lower than that of NL4-3; however, the Nef-dependent increase in the infectivity was obvious when we compared the infectivity of NL-Not with that of NL-Not-Nef. The result validated the use of NL-Not for quantifying the Nef enhancement of infectivity.
Analysis of nef alleles from patients.
We analyzed four LTNPs (patients 1 [p1], p3, p4, and p6), six progressors (p13 to p18), and five ACs (p21, p22, p24, p25, and p26). The nef alleles were PCR amplified from PBMCs and cloned into plasmids as described previously (48). More than five independent clones were sequenced for each patient. When all of the analyzed sequences were identical (p13, p17, p18, p22, and p24), only one nef allele for each patient was further subcloned into pNL-Not and pCAGGS-IRES-EGFP. In the other cases, two representative clones were chosen for construction of expression vectors.
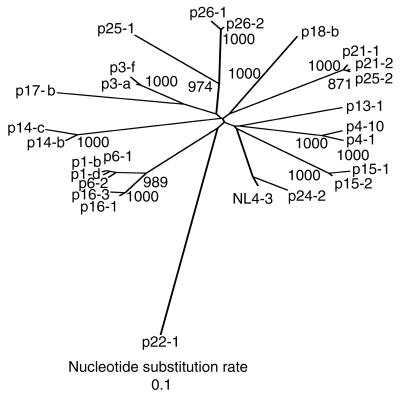
To search for any sequence motif that is characteristic for each of the three groups, we analyzed the nef sequences by creating a phylogenetic tree with the neighbor-joining method (Fig. 2). When two distinct sequences were found from any one patient, i.e., p1, p3, p4, p6, p14, p15, p16, p21, or p26, the two sequences formed a cluster, validating the same origin. The only exception was p25, whose two nef alleles clustered with either p21 or p26. This may suggest that patient 25 had suffered from repeated infection. Most importantly, nef alleles of LTNPs, progressors, and ACs did not form any clusters in the phylogenetic tree. We also created a phylogenetic tree by using the decoded amino acid sequences of the nef alleles and found that nef proteins of LTNPs, progressors, and ACs did not form any clusters. The amino acid sequences of the nef alleles and their alignment can be obtained from our web site (http://www-tv.biken.osaka-u.ac.jp/nef/).
FIG. 2.
Phylogenetic analysis of the nucleotide sequences of nef alleles from LTNPs, AIDS patients (AIDS), and asymptomatic carriers (ACs). The phylogenetic tree was constructed by the neighbor-joining method. Symbols before hyphens indicate patient codes, and numbers or letters after hyphens specify each nef clone. Values along the branches indicate the bootstrap probability. All branch lengths are drawn in accordance with their relative genetic distances.
Enhancement of viral infectivity by patient-derived nef alleles.
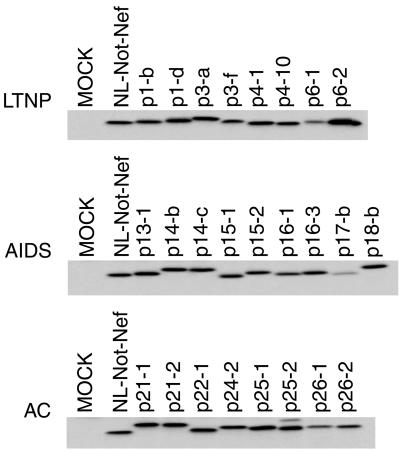
pNL-Not-derived plasmids carrying the nef allele of patients were transfected into 293T cells for the preparation of viruses. Expression of Nef was confirmed by immunoblot analysis of the transfected 293T cells. All viruses produced Nef proteins of about 30 kDa (Fig. 3). We did not find any remarkable difference in the level of Nef from LTNPs, progressors, or ACs.
FIG. 3.
Expression of Nef from recombinant HIV-1. The sequence-verified nef alleles from LTNPs, AIDS patients, and ACs were inserted into the XhoI/NotI site of pNL-Not. The infectious DNAs were transfected into 293T cells to propagate viruses. The cell lysates were analyzed by being immunoblotted with a pool of anti-Nef monoclonal antibodies.
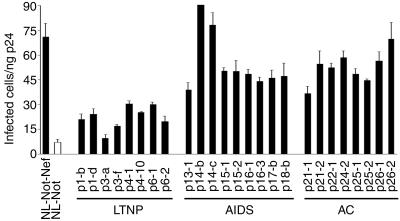
We analyzed the infectivity of these viruses by using MAGNEF indicator cells. As shown in Fig. 4, the enhancement of viral infectivity by the nef alleles of LTNPs was significantly lower than that by nef of NL4-3. The level of increase in infectivity was at most threefold by the nef alleles of LTNPs, whereas the nef alleles derived from progressors and ACs increased the infectivity at least fivefold. The difference between LTNPs and progressors or ACs was statistically significant by t test and Welch's test (P < 0.01).
FIG. 4.
Enhancement of viral infectivity by the nef alleles derived from LTNPs, AIDS patients, or ACs. The recombinant HIV-1 viruses carrying nef alleles of patients were prepared by transfection of infectious DNAs to 293T cells. Infectivity was examined as for Fig. 1. Three independent experiments were performed, and representative data with standard deviations are shown.
Downregulation of cell surface expression of CD4 by nef alleles from patients.
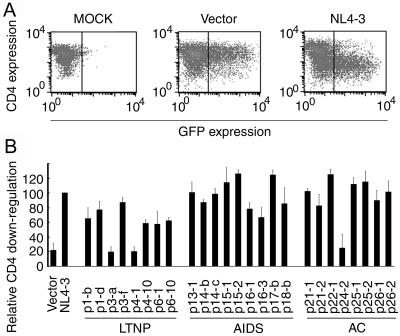
For the purpose of downregulation of cell surface expression of CD4 by nef alleles, nef alleles were inserted into the pCAGGS-IRES-EGFP vector and introduced into MAGIC5D cells. In these plasmids, a single mRNA could produce both Nef and EGFP; the coding sequence of EGFP was placed downstream to the internal ribosomal entry site. Cells were detached from dishes, and the cell surface expression of CD4 was analyzed by flow cytometry. As shown in Fig. 5A, expression of Nef of NL4-3 markedly suppressed the cell surface expression of CD4.
FIG. 5.
Downregulation of cell surface CD4 by Nef derived from LTNPs, AIDS patients, and ACs. (A) MAGIC5D indicator cells were transfected with pCAGGS (MOCK), pCAGGS-IRES-EGFP (Vector), or pCAGGS-IRES-EGFP-4-3-Nef (NL4-3). After 36 h, cells were stained with phycoerythrin-conjugated anti-CD4 monoclonal antibody and were analyzed for fluorescence intensity by FACSCaliber. The vertical bars indicate the thresholds for GFP. (B) pCAGGS-IRES-EGFP (Vector), pCAGGS-IRES-EGFP-4-3Nef (NL4-3), or expression vectors for the nef alleles from patients as indicated at the bottom were transfected and analyzed as described for panel A. After gating for the EGFP-positive cells, the expression level of CD4 was determined as geometric mean fluorescence with CELLQuest software. Relative downregulation efficiency (%) is shown by the following equation: (mean fluorescence intensity of cells expressing NL4-3 Nef)/(mean fluorescence intensity of cells to be evaluated) × 100. Averages from three independent experiments are shown with standard deviations.
Similarly, we examined the CD4 downregulation by the nef alleles from patients. In Fig. 5B, we show the downregulation efficiency as a ratio to that of NL4-3. Three of 7 nef alleles from LTNPs reduced the CD4 expression level as efficiently as the nef allele of NL4-3, 2 did so less efficiently, and the other 2 did not have any effect. Most of the nef alleles from progressors and ACs, except p24-2, reduced the CD4 level as efficiently as the nef allele of NL4-3. Thus, the nef alleles from LTNPs downregulate CD4 less efficiently than do the nef alleles of progressors and ACs. The difference was statistically significant by t test and Welch's test (P < 0.05).
Downregulation of cell surface MHC-I by nef alleles from patients.
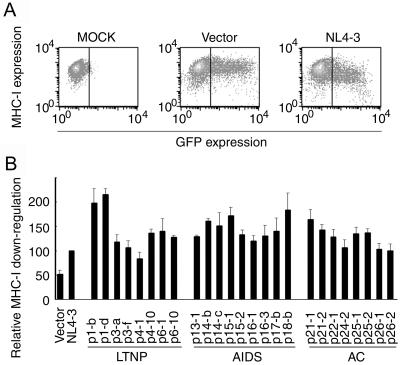
We also examined the downregulation of MHC-I (Fig. 6). All nef alleles reduced the surface expression level of MHC-I, at least as efficiently as did NL4-3-derived nef. Most importantly, we did not observe any difference in the level of MHC-I downregulation among LTNPs, progressors, and ACs.
FIG. 6.
Downregulation of cell surface MHC-I by Nef derived from LTNPs, AIDS patients, and ACs. (A) MAGIC5D indicator cells were transfected with pCAGGS (MOCK), pCAGGS-IRES-EGFP (Vector), or pCAGGS-IRES-EGFP-4-3Nef (NL4-3). After 36 h, cells were stained with phycoerythrin-conjugated anti-MHC-I monoclonal antibody and analyzed for fluorescence intensity by FACSCalibur. The vertical bars indicate the thresholds for EGFP. (B) pCAGGS-IRES-EGFP (Vector), pCAGGS-IRES-EGFP-4-3Nef (NL4-3), or expression vectors for the nef alleles from patients as indicated at the bottom were transfected and analyzed as described for panel A. After gating for EGFP-positive cells, the expression level of MHC-I was determined as geometric mean fluorescence with CELLQuest software. Relative downregulation efficiency (%) is shown by the following equation: (mean fluorescence intensity of cells expressing NL4-3 Nef)/(mean fluorescence intensity of cells to be evaluated) × 100. Averages from three independent experiments are shown with standard deviations.
Binding of Nef to Hck.
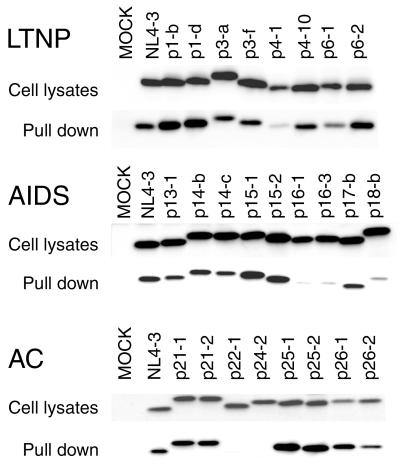
Lastly, we examined the binding of patient-derived Nef to Hck in vitro (Fig. 7). Nef proteins derived from patients were expressed in 293T cells and assayed for the binding to GST-tagged Hck, purified from E. coli. Nef proteins that showed reduced binding to Hck were p4-1, p16-1, p16-3, p18-b, p22-1, and p24-2. Thus, the ability of Nef to bind Hck was not correlated with patient clinical status.
FIG. 7.
Binding of Hck to the Nef proteins derived from LTNPs, AIDS patients, and ACs. 293T cells were transfected with expression plasmids encoding the nef alleles derived from patients. Two days after transfection, cells were lysed in lysis buffer, cleared by centrifugation, and incubated with GST-Hck SH2-SH3. Proteins bound to the GST fusion proteins were separated by SDS-polyacrylamide gel electrophoresis and were analyzed by being immunoblotted with a pool of anti-Nef monoclonal antibodies.
DISCUSSION
Despite several reports of nef deletion in LTNPs (28, 30, 32, 42), deterioration of nef function is not regarded as a common feature among LTNPs, because the deletion of nef alleles is found only in a minor fraction of LTNPs (9, 23, 34). Previously, we also found deletion of a nef allele only in one patient among seven Japanese LTNPs (48). In this study, we asked whether the other nef alleles without gross deletion differed from those of progressors or ACs in biological or biochemical activities.
The results of our analyses are summarized in Table 1. The nef alleles of LTNPs were inefficient in the enhancement of infectivity and in CD4 downregulation. However, these two activities were not always correlated in the nef alleles of progressors or ACs. For example, nef p24-2, which was derived from an AC, enhanced infectivity efficiently but downregulated CD4 only at a marginal level. Similarly, we could not find any correlation when we expanded the comparison to the four parameters. These findings are in agreement with previous reports proposing that the four nef functions are genetically separable and mapped to different regions of nef (reviewed in reference 19).
TABLE 1.
Summary of the analyses
| Group | Clonea | Enhancement of infectivityb | CD4 down- regulationc | MHC-I down- regulationd | Binding to Hcke |
|---|---|---|---|---|---|
| LTNPs | p1-b | + | + | ++ | ++ |
| p1-d | + | + | ++ | ++ | |
| p3-a | − | − | ++ | + | |
| p3-f | + | ++ | + | + | |
| p4-1 | + | − | +/− | +/− | |
| p4-10 | + | + | ++ | + | |
| p6-1 | + | +/− | ++ | + | |
| p6-2 | + | +/− | ++ | ++ | |
| AIDS | p13-1 | ++ | ++ | ++ | + |
| p14-b | +++ | ++ | ++ | + | |
| p14-c | +++ | ++ | ++ | + | |
| p15-1 | ++ | ++ | ++ | ++ | |
| p15-2 | ++ | +++ | ++ | + | |
| p16-1 | ++ | + | + | +/− | |
| p16-3 | ++ | + | + | +/− | |
| p17-b | ++ | +++ | ++ | + | |
| p18-b | ++ | ++ | ++ | +/− | |
| ACs | p21-1 | ++ | ++ | ++ | + |
| p21-2 | ++ | + | ++ | + | |
| p22-1 | ++ | +++ | + | − | |
| p24-2 | ++ | +/− | + | − | |
| p25-1 | ++ | ++ | ++ | ++ | |
| p25-2 | ++ | +++ | ++ | ++ | |
| p26-1 | ++ | + | + | + | |
| p26-2 | +++ | ++ | + | + |
Numbers before hyphens indicate patient codes, and numbers or labels after hyphens specify the nef clone.
Enhancement of infectivity is scored by the relative infectivity to NL-Not-Nef (Fig. 4): −, <30%; +, 30 to 60%; ++, 60 to 90%; +++, >90%.
Downregulation of CD4 is scored by the relative efficiency to that by NL4-3 Nef (Fig. 5): −, <30%; +/−, 30 to 60%; +, 60 to 90%; ++, 90 to 120%; +++, >120%.
Downregulation of MHC-I is scored by the relative efficiency to that by NL4-3 Nef (Fig. 6): −, <50%; +/−, 50 to 80%; +, 80 to 120%; ++, >120%.
Binding affinity of Nef to Hck is scored from the data derived from Fig. 7.
To the best of our knowledge, three groups have performed experiments similar to ours (8, 18, 23). Huang et al. replaced the nef allele of HIV-1 HXB2 with those of LTNPs (23). They did not find any difference in the enhancement of infectivity between the 10 recombinant viruses carrying LTNPs nef and the parent HXB2 virus by using MAGI cells. In our hands, the enhancement of HIV-1 infectivity by Nef is usually less than 10-fold in MAGI cells, which hampers the detection of subtle differences in Nef activity. Therefore, the use of MAGNEF cells, which express a lesser amount of CD4 than do MAGI cells (44), might have enabled us to detect the difference in Nef activity between LTNPs and progressors. Carl et al. constructed NL4-3-based recombinant viruses carrying nef alleles of a single LTNP (8). These viruses showed reduced infectivity both in a single-round infectivity assay with MAGI cells and in PBMCs. Geffin et al. also constructed NL4-3-based recombinant viruses carrying nef alleles of two pediatric LTNPs and examined their infectivity in herpesvirus saimiri-transformed primary human T cells (18). Again, the authors found that nef alleles of two LTNPs enhance HIV-1 infectivity less efficiently than the nef alleles of progressors. Although the number of the examined LTNPs is still small, these reports are in agreement with our finding that nef alleles of LTNPs enhanced HIV-1 infectivity less efficiently than did those of progressors and ACs in a single-round infectivity assay.
It has been reported that some nef alleles derived from LTNPs are defective in CD4 downregulation (7, 32). Mariani and coworkers reported that one patient among four LTNPs carried nef alleles that did not downregulate CD4 expression (32). Carl and coworkers reported a similar observation with nef alleles from one LTNP. Although we also found that Nef-induced CD4 downregulation was abrogated in two nef alleles from LTNPs (Fig. 5), three downregulated CD4 to a level comparable to that of the wild-type Nef. To reach a conclusion, we need to expand the number of LTNPs and to set up more quantitative assays.
The ability to downregulate cell surface MHC-I expression was retained in all nef alleles (Fig. 6). Carl and coworkers recently reported that nef alleles from progressors downregulate MHC-I less efficiently than those from ACs or LTNPs (7). The discrepancy between their results and ours may arise from the difference in cell types: we used MAGI-derived MAGIC5D cells, whereas Carl et al. used Jurkat T cells. Nevertheless, our data agree with respect to the lack of association of defective MHC-I downregulation with the establishment of LTNPs.
Binding to the SH3 domain of Hck is another function of Nef that has been associated with the Nef-dependent increase in HIV-1 infectivity (41); however, subsequent studies have shown that Nef binding to the SH3 domain-containing proteins is required for efficient downregulation of MHC-I (20, 31). Furthermore, it has been shown that dominant-negative mutants of Hck inhibit downregulation of MHC-I (10). In agreement with these previous reports, we found that the efficiency of Hck binding correlated with the level of MHC-I downregulation induced by each Nef (Table 1).
Our results of the sequence analysis of nef alleles from Japanese LTNPs were in complete agreement with those of previous studies denying the presence of a common sequence anomaly in LTNPs (9, 24, 34). We paid particular attention to the 9 sequence motifs in Nef that have been associated with biochemical or biological features (19); however, no mutation was commonly found in any patient group (www-tv.biken.osaka-u.ac.jp/nef/). Moreover, sequences from each group are found to be interdigitated in the phylogenetic tree (Fig. 2), confirming previous reports (9, 24, 34). Future development of a program that predicts the structure-function relationship of Nef may decipher this discrepancy; however, at present our infectivity assay that uses the patient nef gene will help to predict the prognosis of HIV-infected patients.
Nef is known to have many functions, and the number of functions is still increasing. For example, it has been recently shown that Nef binds to Pak to protect the infected cells from apoptosis (4, 17, 39, 46). Therefore, further efforts should be directed to seek a function of nef that correlates better with the clinical course. Finally, a positive correlation in vitro does not give evidence that the deteriorated Nef function causes the long-term nonprogression. Further study is required to examine whether these characteristics of nef alleles of LTNPs are the cause or the consequence of the long-term nonprogression after HIV-1 infection.
Acknowledgments
We thank A. Adachi, K. Ikuta, A. Kojima, T. Kurata, J. Miyazaki, and C. Aiken for materials and helpful discussions.
The research was sponsored in part by a Grant for International Health Cooperation Research (12C-1) from the Ministry of Health and Welfare, Japan, and Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists (M.T.).
REFERENCES
- 1.Adachi, A., N. Ono, H. Sakai, K. Ogawa, R. Shibata, T. Kiyomasu, H. Masuike, and S. Ueda. 1991. Generation and characterization of the human immunodeficiency virus type 1 mutants. Arch. Virol. 117:45-58. [DOI] [PubMed] [Google Scholar]
- 2.Aiken, C. 1997. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 71:5871-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, L., Z. Du, M. Rosenzweig, J. U. Jung, and R. C. Desrosiers. 1997. A role for natural simian immunodeficiency virus and human immunodeficiency virus type 1 nef alleles in lymphocyte activation. J. Virol. 71:6094-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arora, V. K., R. P. Molina, J. L. Foster, J. L. Blakemore, J. Chernoff, B. L. Fredericksen, and J. V. Garcia. 2000. Lentivirus Nef specifically activates Pak2. J. Virol. 74:11081-11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchbinder, S., and E. Vittinghoff. 1999. HIV-infected long-term nonprogressors: epidemiology, mechanisms of delayed progression, and clinical and research implications. Microbes Infect. 1:1113-1120. [DOI] [PubMed] [Google Scholar]
- 6.Cao, Y., L. Qin, L. Zhang, J. Safrit, and D. D. Ho. 1995. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 332:201-208. [DOI] [PubMed] [Google Scholar]
- 7.Carl, S., R. Daniels, A. J. Iafrate, P. Easterbrook, T. C. Greenough, J. Skowronski, and F. Kirchhoff. 2000. Partial “repair” of defective NEF genes in a long-term nonprogressor with human immunodeficiency virus type 1 infection. J. Infect. Dis. 181:132-140. [DOI] [PubMed] [Google Scholar]
- 8.Carl, S., T. C. Greenough, M. Krumbiegel, M. Greenberg, J. Skowronski, J. L. Sullivan, and F. Kirchhoff. 2001. Modulation of different human immunodeficiency virus type-1 Nef functions during progression to AIDS. J. Virol. 75:3657-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catucci, M., G. Venturi, L. Romano, P. E. Valensin, and M. Zazzi. 2000. Analysis of the HIV-1 nef gene in five intravenous drug users with long-term nonprogressive HIV-1 infection in Italy. J. Med. Virol. 60:294-299. [PubMed] [Google Scholar]
- 10.Chang, A. H., M. V. O'Shaughnessy, and F. R. Jirik. 2001. Hck SH3 domain-dependent abrogation of Nef-induced class 1 MHC down-regulation. Eur. J. Immunol. 31:2382-2387. [DOI] [PubMed] [Google Scholar]
- 11.Chowers, M. Y., C. A. Spina, T. J. Kwoh, N. J. Fitch, D. D. Richman, and J. C. Guatelli. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J. Virol. 68:2906-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, K. L., and D. Baltimore. 1999. HIV's evasion of the cellular immune response. Immunol. Rev. 168:65-74. [DOI] [PubMed] [Google Scholar]
- 13.Cullen, B. R. 1998. HIV-1 auxiliary proteins: making connections in a dying cell. Cell 93:685-692. [DOI] [PubMed] [Google Scholar]
- 14.Deacon, N. J., A. Tsykin, A. Solomon, K. Smith, M. Ludford-Menting, D. J. Hooker, D. A. McPhee, A. L. Greenway, A. Ellett, and C. Chatfield. 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270:988-991. [DOI] [PubMed] [Google Scholar]
- 15.Easterbrook, P. J., and L. K. Schrager. 1998. Long-term nonprogression in HIV infection: methodological issues and scientific priorities. Report of an international European Community-National Institutes of Health Workshop, The Royal Society, London, England, November 27-29, 1995. Scientific Coordinating Committee. AIDS Res. Hum. Retrovir. 14:1211-1228. [DOI] [PubMed] [Google Scholar]
- 16.Emerman, M., and M. H. Malim. 1998. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science 280:1880-1884. [DOI] [PubMed] [Google Scholar]
- 17.Fackler, O. T., X. Lu, J. A. Frost, M. Geyer, B. Jiang, W. Luo, A. Abo, A. S. Alberts, and B. M. Peterlin. 2000. p21-activated kinase 1 plays a critical role in cellular activation by Nef. Mol. Cell. Biol. 20:2619-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geffin, R., D. Wolf, R. Muller, M. D. Hill, E. Stellwag, M. Freitag, G. Sass, G. B. Scott, and A. S. Baur. 2000. Functional and structural defects in HIV type 1 nef genes derived from pediatric long-term survivors. AIDS Res. Hum. Retrovir. 16:1855-1868. [DOI] [PubMed] [Google Scholar]
- 19.Geyer, M., O. T. Fackler, and B. M. Peterlin. 2001. Structure-function relationships in HIV-1 Nef. EMBO Rep. 2:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg, M. E., A. J. Iafrate, and J. Skowronski. 1998. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 17:2777-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenough, T. C., M. Somasundaran, D. B. Brettler, R. M. Hesselton, A. Alimenti, F. Kirchhoff, D. Panicali, and J. L. Sullivan. 1994. Normal immune function and inability to isolate virus in culture in an individual with long-term human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retrovir. 10:395-403. [DOI] [PubMed] [Google Scholar]
- 22.Hachiya, A., S. Aizawa, M. Tanaka, Y. Takahashi, S. Ida, H. Gatanaga, Y. Hirabayashi, A. Kojima, M. Tatsumi, and S. Oka. 2001. Rapid and simple phenotypic assay for drug susceptibility of human immunodeficiency virus type 1 by using CCR5-expressed HeLa/CD4+ cell clone 1-10 (MAGIC-5). Antimicrob. Agents Chemother. 45:495-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, Y., L. Zhang, and D. D. Ho. 1995. Biological characterization of nef in long-term survivors of human immunodeficiency virus type 1 infection. J. Virol. 69:8142-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, Y., L. Zhang, and D. D. Ho. 1995. Characterization of nef sequences in long-term survivors of human immunodeficiency virus type 1 infection. J. Virol. 69:93-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iafrate, A. J., S. Carl, S. Bronson, C. Stahl-Hennig, T. Swigut, J. Skowronski, and F. Kirchhoff. 2000. Disrupting surfaces of nef required for downregulation of CD4 and for enhancement of virion infectivity attenuates simian immunodeficiency virus replication in vivo. J. Virol. 74:9836-9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kestler, H. W., III, D. J. Ringler, K. Mori, D. L. Panicali, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65:651-662. [DOI] [PubMed] [Google Scholar]
- 27.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirchhoff, F., T. C. Greenough, D. B. Brettler, J. L. Sullivan, and R. C. Desrosiers. 1995. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 332:228-232. [DOI] [PubMed] [Google Scholar]
- 29.Learmont, J., B. Tindall, L. Evans, A. Cunningham, P. Cunningham, J. Wells, R. Penny, J. Kaldor, and D. A. Cooper. 1992. Long-term symptomless HIV-1 infection in recipients of blood products from a single donor. Lancet 340:863-867. [DOI] [PubMed] [Google Scholar]
- 30.Learmont, J. C., A. F. Geczy, J. Mills, L. J. Ashton, C. H. Raynes-Greenow, R. J. Garsia, W. B. Dyer, L. McIntyre, R. B. Oelrichs, D. I. Rhodes, N. J. Deacon, and J. S. Sullivan. 1999. Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. A report from the Sydney Blood Bank Cohort. N. Engl. J. Med. 340:1715-1722. [DOI] [PubMed] [Google Scholar]
- 31.Mangasarian, A., V. Piguet, J. K. Wang, Y. L. Chen, and D. Trono. 1999. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of nef selectively regulate MHC-I trafficking. J. Virol. 73:1964-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mariani, R., F. Kirchhoff, T. C. Greenough, J. L. Sullivan, R. C. Desrosiers, and J. Skowronski. 1996. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J. Virol. 70:7752-7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuda, M., S. Tanaka, S. Nagata, A. Kojima, T. Kurata, and M. Shibuya. 1992. Two species of human CRK cDNA encode proteins with distinct biological activities. Mol. Cell. Biol. 12:3482-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michael, N. L., G. Chang, L. A. d'Arcy, C. J. Tseng, D. L. Birx, and H. W. Sheppard. 1995. Functional characterization of human immunodeficiency virus type 1 nef genes in patients with divergent rates of disease progression. J. Virol. 69:6758-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montefiori, D. C., G. Pantaleo, L. M. Fink, J. T. Zhou, J. Y. Zhou, M. Bilska, G. D. Miralles, and A. S. Fauci. 1996. Neutralizing and infection-enhancing antibody responses to human immunodeficiency virus type 1 in long-term nonprogressors. J. Infect. Dis. 173:60-67. [DOI] [PubMed] [Google Scholar]
- 36.Munoz, A., M. C. Wang, S. Bass, J. M. Taylor, L. A. Kingsley, J. S. Chmiel, and B. F. Polk. 1989. Acquired immunodeficiency syndrome (AIDS)-free time after human immunodeficiency virus type 1 (HIV-1) seroconversion in homosexual men. Multicenter AIDS Cohort Study Group. Am. J. Epidemiol. 130:530-539. [DOI] [PubMed] [Google Scholar]
- 37.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-200. [DOI] [PubMed] [Google Scholar]
- 38.Otake, K., Y. Fujii, T. Nakaya, Y. Nishino, Q. Zhong, K. Fujinaga, M. Kameoka, K. Ohki, and K. Ikuta. 1994. The carboxyl-terminal region of HIV-1 Nef protein is a cell surface domain that can interact with CD4+ T cells. J. Immunol. 153:5826-5837. [PubMed] [Google Scholar]
- 39.Renkema, G. H., A. Manninen, and K. Saksela. 2001. Human immunodeficiency virus type 1 Nef selectively associates with a catalytically active subpopulation of p21-activated kinase 2 (PAK2) independently of PAK2 binding to Nck or beta-PIX. J. Virol. 75:2154-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rutherford, G. W., A. R. Lifson, N. A. Hessol, W. W. Darrow, P. M. O'Malley, S. P. Buchbinder, J. L. Barnhart, T. W. Bodecker, L. Cannon, L. S. Doll, S. D. Holmberg, J. S. Harrison, M. F. Rogers, D. Werdegar, and H. W. Jaffe. 1990. Course of HIV-I infection in a cohort of homosexual and bisexual men: an 11 year follow up study. BMJ 301:1183-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saksela, K., G. Cheng, and D. Baltimore. 1995. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 14:484-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salvi, R., A. R. Garbuglia, A. Di Caro, S. Pulciani, F. Montella, and A. Benedetto. 1998. Grossly defective nef gene sequences in a human immunodeficiency virus type 1-seropositive long-term nonprogressor. J. Virol. 72:3646-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheppard, H. W., W. Lang, M. S. Ascher, E. Vittinghoff, and W. Winkelstein. 1993. The characterization of nonprogressors: long-term HIV-1 infection with stable CD4+ T-cell levels. AIDS 7:1159-1166. [PubMed] [Google Scholar]
- 44.Tobiume, M., M. Takahoko, M. Tatsumi, and M. Matsuda. 2001. Establishment of a MAGI-derived indicator cell line that detects the Nef enhancement of HIV-1 infectivity with high sensitivity. J. Virol. Methods 97:151-158. [DOI] [PubMed] [Google Scholar]
- 45.Tobiume, M., K. Tokunaga, E. Kiyokawa, M. Takahoko, N. Mochizuki, M. Tatsumi, and M. Matsuda. 2001. Requirement of Nef for HIV-1 infectivity is biased by the expression levels of Env in the virus-producing cells and CD4 in the target cells. Arch. Virol. 146:1739-1751. [DOI] [PubMed] [Google Scholar]
- 46.Wolf, D., V. Witte, B. Laffert, K. Blume, E. Stromer, S. Trapp, P. D'Aloja, A. Schurmann, and A. S. Baur. 2001. HIV-1 Nef associated PAK and PI3 kinases stimulate Akt-independent Bad-phosphorylation to induce anti-apoptotic signals. Nat. Med. 7:1217-1224. [DOI] [PubMed] [Google Scholar]
- 47.Yamada, T., and A. Iwamoto. 1999. Expression of a novel Nef epitope on the surface of HIV type 1-infected cells. AIDS Res. Hum. Retrovir. 15:1001-1009. [DOI] [PubMed] [Google Scholar]
- 48.Yamada, T., and A. Iwamoto. 2000. Comparison of proviral accessory genes between long-term nonprogressors and progressors of human immunodeficiency virus type 1 infection. Arch. Virol. 145:1021-1027. [DOI] [PubMed] [Google Scholar]