Abstract
The G glycoprotein of human respiratory syncytial virus (RSV) was identified previously as the viral attachment protein. Although we and others recently showed that G is not essential for replication in vitro, it does affect the efficiency of replication in a cell type-dependent fashion and is required for efficient replication in vivo. The ectodomain of G is composed of two heavily glycosylated domains with mucin-like characteristics that are separated by a short central region that is relatively devoid of glycosylation sites. This central region contains a 13-amino acid segment that is conserved in the same form among RSV isolates and is overlapped by a second segment containing four cysteine residues whose spacings are conserved in the same form and which create a cystine noose. The conserved nature of the cystine noose and flanking 13-amino acid segment suggested that this region likely was important for attachment activity. To test this hypothesis, we constructed recombinant RSVs from which the region containing the cysteine residues was deleted together with part or all of the conserved 13-amino acid segment. Surprisingly, each deletion had little or no effect on the intracellular synthesis and processing of the G protein, the kinetics or efficiency of virus replication in vitro, or sensitivity to neutralization by soluble heparin in vitro. In addition, neither deletion had any discernible effect on the ability of RSV to infect the upper respiratory tract of mice and both resulted in a 3- to 10-fold reduction in the lower respiratory tract. Thus, although the G protein is necessary for efficient virus replication in vivo, this activity does not require the central conserved cystine noose region.
Human respiratory syncytial virus (RSV), the prototype of the Pneumovirus genus in the paramyxovirus family, is the most important viral etiologic agent of pediatric respiratory disease worldwide (6). Its genome consists of a single, negative-sense RNA of 15,190 to 15,225 nucleotides (for the three strains sequenced to date) encoding 10 major subgenomic mRNAs and 11 viral proteins. Three of these proteins, G, F, and SH, are transmembrane virion glycoproteins. The G glycoprotein has been identified as the viral attachment protein (25) although, as described below, under certain conditions it is completely dispensable for efficient replication in vitro. The F protein mediates membrane fusion and also appears to be able to serve as an auxiliary attachment protein (17, 21, 22, 34, 36). Expression of the SH protein can be eliminated with little effect on virus replication in vitro and in vivo and its function remains unknown (3, 41), although its counterpart in simian virus 5 was recently shown to function as an antagonist of apoptosis (16).
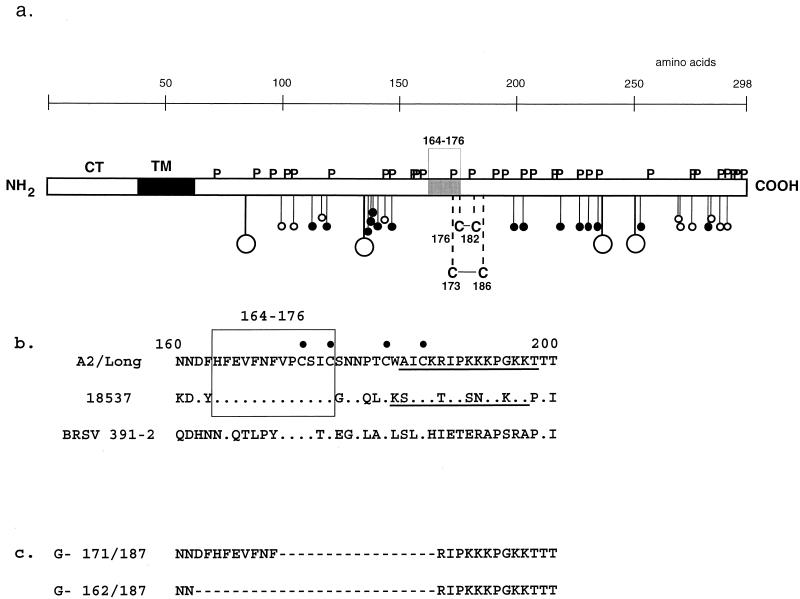
RSV G is a highly glycosylated type II transmembrane protein with a single hydrophobic region near the N terminus which serves as both signal sequence and membrane anchor. The protein backbone is 292 to 299 amino acids long, depending on the virus strain (19, 33), and has an Mr of approximately 33 kDa. In comparison, the mature form of G migrates during gel electrophoresis in the presence of sodium dodecyl sulfate (SDS) as a wide band between 84 to 90 kDa (6). This difference in gel mobility is due to extensive glycosylation of the mature G protein, which affects both the molecular mass of the glycoprotein and its interaction with SDS. Most of the carbohydrate moieties on the G protein are O-linked sugars; there are more than 70 serine and threonine potential acceptor sites, and analysis of an engineered F-G chimeric protein expressed by recombinant baculovirus in insect cells provided evidence of 23 to 24 O-linked side chains (39). Figure 1a is a diagram of the G protein of strain A2 showing the positions of the 25 serine and threonine residues that are most likely to contain O-linked sugar, based on sequence context and predicted surface accessibility (15). The number of potential acceptor sites for N-linked carbohydrate chains can differ between different strains of RSV, from four likely sites in strain A2 to eight in the Long strain (6). The N- and O-linked glycosylation sites are clustered in two regions of the protein, with amino acid content reminiscent of mucins. These mucin-like domains constitute most of the ectodomain and are highly divergent in amino acid sequence between the RSV antigenic subgroups and individual isolates. These two domains are separated by a short central domain that is relatively devoid of potential carbohydrate acceptor sites and contains a stretch of 13 amino acids (positions 164 to 176 in the amino acid sequence of the G protein of strain A2) that is conserved in the same form for different strains and subgroups of RSV. This region overlaps four closely spaced cysteine residues (positions 173, 176, 182, and 186 in strain A2) that form disulfide bonds in the pattern 1-4 and 2-3 and create a cystine noose (Fig. 1a) (12, 19, 24). Monoclonal antibody-resistant mutants have been isolated in which point mutations within the 13-amino acid conserved region occurred (38) or residues 176, 182, and 186 were substituted singly or 182 and 186 were substituted together (27, 31, 38). Nonetheless, the high degree of conservation of the conserved segment and overlapping cystine noose made these structures obvious candidates for involvment in receptor binding.
FIG. 1.
Structure of the RSV strain A2 G protein (a), amino acid sequence of the central conserved cystine noose (b), and amino acids deleted from rRSV (c). (a) The 298-amino acid G protein is shown as an open rectangle. The segment of amino acid sequence that is conserved in the same form between the two RSV antigenic subgroups (amino acids 164 to 176) is shaded. Potential acceptor sites for N-linked carbohydrate (Asn-X-Ser/Thr, where X is not Pro) are shown as stalks with large circles. The most likely predicted serine and threonine acceptor sites, based on the presence of serine or, preferably, threonine in a sequence context predicted to have exposed and extended secondary structure with a general lack of hydrophilic and bulky residues (15), are shown as stalks with small open and filled circles, respectively. The closest predicted O-linked sugars to either side of the central conserved cystine noose would involve Thr-147 and Thr-199. Direct biochemical analysis confirmed an absence of sugars involving amino acids 152 to 187 (12). Proline and conserved cysteine residues are indicated by P and C, respectively, and the disulfide bonding pattern of the cysteine residues is indicated (12). TM, hydrophobic signal anchor; CT, cytoplasmic tail. (b) Sequence spanning the central region of the RSV-A2 G protein (amino acids 160 to 200 are shown) and alignment with the corresponding region of strain 18537 of subgroup B (18537) and bovine RSV (BRSV) strain 391-2. Periods denote amino acid identity, bullets above the sequence indicate the four conserved cysteine residues, and the 13-amino acid sequence that is conserved among the human strains is boxed. A previously described (11) putative heparin-binding domain discussed in the text is underlined. (c) Amino acid sequences of the corresponding regions of mutant viruses G-171/187 and G-162/187. Deleted residues are indicated by dashes.
Although the cellular receptor for RSV has not been identified, there is evidence that cell surface glycosaminoglycans (GAGs) are important for efficient infection, at least in immortalized cell lines in vitro. Viral infectivity can be inhibited by preincubation of virus with soluble GAGs such as heparin, which presumably saturate binding sites on the virion or by prior depletion of cell surface GAGs by enzymatic treatment or genetic mutation (2, 11, 14, 21, 23). The major mediator of this interaction is the G glycoprotein (23), although F also appears to be able to bind GAGs (10, 21). Immediately downstream of the cystine noose is a cluster of positively charged amino acids that shares similarity with heparin-binding domains from other viral and mammalian proteins. Analysis of synthetic peptides spanning the G ectodomain provided evidence that amino acids 184 to 198 contain the major GAG-binding site (11). However, this site could be deleted from recombinant virus without affecting infectivity or sensitivity to neutralization by soluble GAGs (36); thus, the sites involved in GAG binding and in G protein function in general remain to be identified.
A biologically derived RSV vaccine candidate was found to have sustained a spontaneous deletion of the SH and G genes, indicating that neither is essential for replication in vitro under appropriate conditions (22). This was confirmed with recombinant virus in which the G gene was deleted alone or in combination with SH (34, 36). However, the deletion of G affected the efficiency of replication in vitro in a cell type-dependent fashion (36). The G-deletion virus was less efficient for infection of HEp-2 cells, and this restriction appeared to be mainly at the level of attachment and entry, with a minor, secondary effect on the level of packaging. In contrast, in Vero cells, the G-deletion virus replicated with an efficiency similar to that of its wild-type parent, indicating that all aspects of replication necessary for infectivity and the production of infectious particles can take place efficiently in the absence of G. Importantly, however, the G protein is necessary for efficient infection in mice and humans (22, 36). In the study reported here, we sought to determine whether the contribution of G to efficient virus replication in HEp-2 cells and in mice depends on the conserved cystine noose structure.
MATERIALS AND METHODS
Plasmid construction.
Two mutants of the G protein were constructed, G-171/187 and G-162/187, designated according to the amino acids whose codons were deleted from the G open reading frame (ORF). To mutate the G ORF, a pUC19 plasmid (pUC19-GFM2) containing the G, F, and M2 genes, present as a fragment bounded by PacI and BamHI sites that occur naturally in the RSV strain A2 genome, was subjected to PCR with Vent DNA polymerase (New England Biolabs) by the method of Byrappa et al. (4). Oligonucleotides were designed such that they would prime the GFM2 plasmid in opposite directions to produce a linear plasmid incorporating the desired mutations. The 5′-phosphorylated oligonucleotides used were as follows: G-171/187 and G-162/187 forward, 5′-AGAATACCAAACAAAAAACCAGGAAAGAAAACCACTACC-3′ (nucleotides [nt] 5250 to 5288); G-171/187 reverse, 5′-AAAGTTGAACACTTCAAAGTGAAAATCATTATTGGG-3′ (nt 5198 to 5163); G-162/187 reverse, 5′-ATTATTGGGTTTGCTTGGTGGTTTGTTTTGGCG-3′ (nt 5171 to 5139).
The linear amplified plasmids were isolated by agarose gel electrophoresis, self-ligated, and transformed into DH10B competent cells (Life Technologies). The presence of the mutations was confirmed by restriction digest and nucleotide sequence analysis. The mutated G ORFs were excised from the GFM2 plasmids by using PacI and StuI, which cut on either side of the G gene, and inserted into fresh GFM2 to ensure that no secondary mutations from the PCR were present in the F and M2 genes. The PacI-BamHI fragment of these mutated GFM2 plasmids was inserted into the PacI-BamHI window of D51, which contains the upstream end of the RSV antigenomic cDNA from the T7 promoter and leader region to the end of the SH gene. This reconstructed the D50 plasmid, which contains the antigenomic cDNA from the T7 promoter and leader region to the end of the M2 gene (35). Full-length antigenomic cDNAs were then assembled by inserting the BamHI-MluI fragment of D39 (7, 20) into the mutant D50 plasmids.
Recovery of recombinant RSV (rRSV).
Transfections were performed essentially as described previously (7). Briefly, monolayers of HEp-2 cells in 6-well dishes were simultaneously infected with 3 focus-forming units per cell of a recombinant vaccinia virus (MVA strain) expressing T7 RNA polymerase (MVA-T7) (42) and transfected with a mixture of plasmids encoding the RSV N, P, L, M2-1, and antigenome (wild type or mutant), each under the control of the T7 promoter (0.4, 0.3, 0.2, 0.1, and 1.0 μg, respectively) using LipofectACE (Life Technologies). The transfection-infection mixture was removed after 18 h of incubation at 32°C and replaced with fresh medium (OptiMEM; Gibco) supplemented with 4% fetal bovine serum. After 48 h, the clarified supernatants were passaged onto fresh HEp-2 cells and incubated at 37°C.
Viral titers were determined by plaque assay in Vero cells for 6 days under 0.8% methylcellulose followed by fixation with 80% methanol. Plaques were visualized by incubation with a cocktail of three murine anti-RSV F monoclonal antibodies, followed by incubation with goat anti-mouse IgG coupled to horseradish peroxidase, followed by color development with 4CN substrate (Kirkegaard & Perry Laboratories) as described previously (28).
Western blot analysis.
Cell pellets from infected cells were disrupted by addition of 2× sample buffer (100 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 0.2% bromophenol blue, 200 mM dithiothreitol) and centrifugation through Qiashredders (Qiagen). Approximately 1.5 × 105 cell equivalents of each infected cell extract were subjected to electrophoresis on SDS-4 to 20% polyacrylamide gels (Invitrogen) and transferred to polyvinylidene difluoride membrane (Invitrogen). Blots were incubated with rabbit antiserum raised against either purified RSV or a peptide spanning amino acids 186 to 201 of the G protein. Viral proteins were visualized by secondary incubation with horseradish peroxidase-coupled goat anti-rabbit immunoglobulin G antibodies followed by chemiluminescence (Amersham).
Plaque reduction assay.
Virus dilutions were preincubated in OptiMEM containing the indicated concentrations (see Fig. 4) of heparin (Sigma) for 15 min at room temperature, and titers were determined on HEp-2 and Vero cells as described previously (36).
FIG. 4.
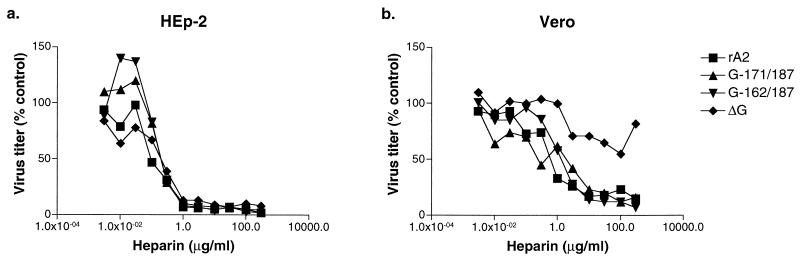
Heparin sensitivity of mutant rRSVs. Wild-type rRSV, central domain mutant G-171/187 or G-162/187, or the previously described ΔG mutant lacking the G gene (36) was incubated with serial dilutions of heparin, and the titer was determined by plaque assay. Shown are the averages of duplicate wells.
RESULTS
Construction of rRSV-containing mutations in the central domain of the G glycoprotein.
To examine the contribution of the central region of G (Fig. 1a and b) to RSV infectivity and replication, we constructed two antigenomic cDNAs from which nucleotides in this region were deleted (Fig. 1c). From one antigenomic cDNA, designated G-171/187, we deleted 51 nt (positions 5199 to 5249) encoding a 17-amino acid segment (residues 171 to 187) that includes the four cysteines and part of the 13-amino acid conserved domain. From the second, G-162/187, we deleted 78 nt (5172 to 5249) encoding a 26-amino acid segment (residues 162 to 187) encompassing the cysteine residues as well as the complete conserved 13-amino acid sequence. rRSV containing either of the two central region deletions was recovered, and the two mutant viruses displayed cytopathic effects and plaque morphologies similar to those of wild-type rRSV (data not shown). The G gene and flanking region in each mutant virus were amplified from purified viral RNA by reverse transcription-PCR and sequenced, confirming that each virus contained the engineered mutation and was free of adventitious mutations.
Growth of central domain mutant viruses in culture.
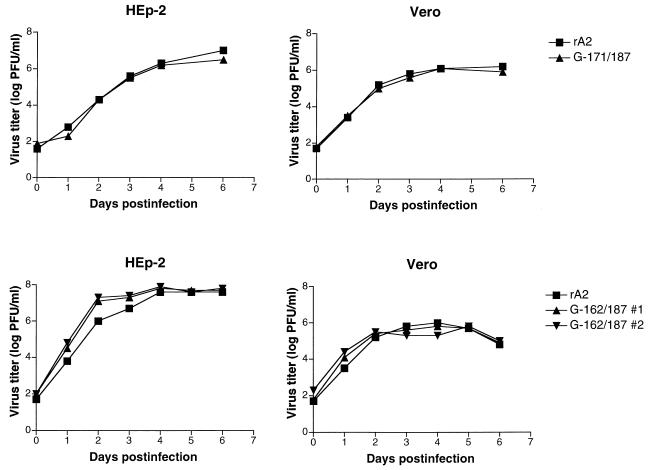
We previously showed that mutant rRSV that lacked G (ΔG) showed a markedly decreased ability to replicate in HEp-2 cells compared to that of wild-type rRSV (rA2), although there was no difference between the two viruses in Vero cells (36). This provided an in vitro assay for the functional status of the G protein. To examine whether the two deletions within the central domain affected the efficiency of virus replication in vitro, HEp-2 and Vero cell monolayers were infected at a multiplicity of infection (MOI) of 0.1 PFU per cell of either mutant or recombinant wild-type rA2, and samples were harvested daily for 6 days postinfection and assayed for virus titer by plaque assay. The G-171/187 and G-162/187 mutants replicated as efficiently as rA2 virus in both cell lines tested (Fig. 2).
FIG. 2.
Growth analysis of the central domain mutants G-171/187 and G-162/187 in cell culture. Monolayers of HEp-2 or Vero cells were infected with an MOI of 0.1 PFU per cell of wild-type rA2, G-171/187, or two independent preparations (#1 and #2) of G-162/187. Samples were harvested daily and titers were determined by plaque assay. Each point represents the average of duplicate cultures.
Viral protein expression by the central domain mutant viruses.
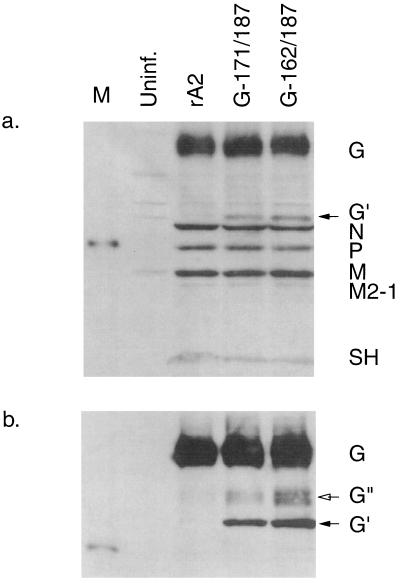
We next examined the expression of viral proteins by the mutant viruses. HEp-2 cells were infected with the mutant viruses at an MOI of 3 PFU per cell. Cell extracts were prepared 48 h postinfection and subjected to Western blot analysis using a rabbit polyclonal serum that was prepared against gradient-purified RSV. As shown in Fig. 3a, this antiserum detected the major RSV structural proteins N, P, M, M2-1, G, and SH and showed that the levels of expression of these proteins were very similar between the two mutant viruses and the wild-type rA2 parent. This particular antiserum did not react detectably with F protein in the Western blot: this probably reflects an inability to recognize denatured F protein, since the same antiserum reacted efficiently with F in a radio-immunoprecipitation assay (not shown).
FIG. 3.
Western blot analysis of RSV protein expression in cells infected by the central domain mutants G-171/187 and G-162/187. HEp-2 cells were infected with rA2, G-171/187, or G-162/187 at an MOI of 3 PFU per cell. Cell extracts were harvested 48 h postinfection and analyzed by Western blotting using an antiserum against whole virus (a) or against a peptide spanning amino acids 184 to 201 of the G protein (b). Viral proteins are indicated on the right. The two arrows indicate putative incompletely glycosylated forms of G, specifically, one containing only N-linked sugars (G′, filled arrow) and one containing an incomplete complement of O-linked sugars (G", open arrow).
In addition, Western blot analysis was performed using an antiserum raised against a synthetic peptide spanning amino acids 184 to 201 of the G protein (Fig. 3b). This confirmed that the levels of expression of the G protein were similar between the two mutants and rA2, although each mutant directed the accumulation of two smaller forms of G (Fig. 3b). These smaller forms represent processing intermediates: the first, more abundant form of greater electrophoretic mobility corresponds to G protein containing N-linked sugars but not O-linked sugars, and the second, heterodisperse smear corresponds to G protein that contains N-linked sugars and only a partial complement of O-linked sugars (8, 40). Most of the G protein produced by each mutant virus was of approximately the same electrophoretic mobility as that of wild-type rA2. The observation that only small fractions of the G protein expressed by the two central domain mutants were present as processing intermediates indicates that each of the deletions had little effect on posttranslational processing and transport of the G protein.
Heparin sensitivity of the conserved region mutant viruses.
We previously showed that ΔG was insensitive to neutralization by heparin in a plaque reduction assay using Vero cells, whereas its wild-type parent was very sensitive (36). To examine whether the two deletions in the conserved region of G altered the heparin sensitivity of RSV, we evaluated the mutant viruses in an assay in which virus was incubated with soluble heparin and infectivity was enumerated by plaque assay. The G-171/187 and G-162/187 viruses were inhibited by heparin in both HEp-2 and Vero cells (Fig. 4) at concentrations and to extents similar to those in wild-type rA2. In comparison, plaque formation of ΔG in Vero cells was only partially inhibited even at the highest concentrations of heparin used (Fig. 4b).
Growth of the conserved region mutant viruses in mice.
In previous work, deletion of the G gene greatly reduced RSV replication in the upper and lower respiratory tract of mice, such that only a few PFU were recovered, and it was unclear whether this represented a very low level of replication or carryover from the inoculation (36). Therefore, this model provides a stringent test of the functional status of G protein in vivo. BALB/c mice were inoculated intranasally with 106 PFU of G-171/187, G-162/187, ΔG, or the wild-type rA2 parent. Four days postinfection, the nasal turbinates and lungs of infected mice were harvested and viral titers were determined by plaque assay (Table 1). Each of the two central domain mutant viruses grew in the upper respiratory tract with an efficiency that was indistinguishable from that of wild-type rA2, while ΔG was not detectable. In the lower respiratory tract, the titers of the G-171/187 and G-162/187 viruses were approximately 10-fold and 3-fold lower, respectively, than that of rA2, compared with the >250-fold lower titer of ΔG. Since the G-162/187 virus contained the more extensive deletion and was not markedly different from wild-type rA2, the combined deletion of the conserved 13-amino acid segment and the overlapping cysteine domain did not greatly affect virus replication in BALB/c mice.
TABLE 1.
Growth of G mutant rRSV in the upper and lower respiratory tracts of BALB/c mice
| Virusa | Nasal turbinates
|
Lungs
|
||
|---|---|---|---|---|
| % Shedding | Mean titer ± SEMb | % Shedding | Mean titer ± SEMb | |
| rA2 | 100 | 3.4 ± 0.10 (A) | 100 | 5.1 ± 0.05 (A) |
| ΔG | 0 | <2.0 (B) | 67 | 2.7 ± 0.33 (B) |
| G-171/187 | 100 | 3.5 ± 0.09 (A) | 100 | 4.1 ± 0.06 (C) |
| G-162/187 | 100 | 3.4 ± 0.08 (A) | 100 | 4.6 ± 0.04 (D) |
BALB/c mice in groups of six were administered 106 PFU of the indicated virus intranasally under light anesthesia on day 0 and were sacrificed on day 4. Nasal turbinates and lungs were harvested, and virus titers were determined by plaque assay. The levels of detection in the upper and lower respiratory tracts were 2.0 and 1.7 log10 PFU/g, respectively.
Mean peak titers (log10 PFU per gram of tissue) were assigned to statistically similar groups by Duncan's multiple range test (α = 0.05). Means in each column which show different Duncan grouping letters are significantly different. SEM, standard error of the mean.
DISCUSSION
Most of the ectodomain of the G protein of RSV is composed of two large mucin-like domains that are separated by a short central domain. This region contains a 13-amino acid sequence that is conserved in the same form among all human RSV isolates and that overlaps a segment containing four cysteine residues whose positioning is conserved in the same form among human, bovine, and ovine RSV strains. This domain was an obvious candidate to be involved in the attachment activity of G or, as described below, in other accessory functions that have been ascribed to G. In the present study, we constructed two rRSVs from which 17 or 26 amino acids were deleted, thus removing the cystine noose together with the adjacent conserved segment.
Each deletion had little effect on the intracellular synthesis and processing of the G protein, based on its electrophoretic profile; there was a very small subpopulation of molecules that appeared to be incompletely O glycosylated, but most of the protein was indistinguishable by SDS-polyacrylamide gel electrophoresis from that expressed by the wild-type parent. Thus, the intracellular transport and processing of G was little affected by deletions within the central domain and the loss of the cystine noose. This is consistent with the idea that the G protein has a comparatively extended structure in which the various domains are relatively independent of each other with regard to requirements for folding and processing and the generation of infectious virus.
As described previously, the G protein is completely dispensable for efficient infection of Vero cells in vitro (34, 36). However, it greatly increases the efficiency of infection of HEp-2 cells in vitro and is necessary for efficient infection in BALB/c mice. This fact provided the basis for a stringent assay for the function of the G protein in cell culture and in vivo. Each central domain mutant virus replicated in vitro as efficiently as wild-type rRSV. In vivo, the G-162/187 virus, from which the greater number of amino acids was deleted, replicated as efficiently as wild-type RSV in the upper respiratory tract and was only reduced threefold in the lower respiratory tract. Thus, under conditions in which the G protein is essential for efficient growth, such as the growth of mice, the conserved domain and cystine noose are not required for efficient growth.
We also compared the sensitivities of these viruses to neutralization by a brief incubation with soluble heparin, a model GAG. GAG-mediated neutralization of infectivity is not strictly specific to the G protein, since the F protein also can bind to GAGs such as heparin (10), albeit with significantly less affinity than G (21). However, we previously noted that in Vero cells, wild-type RSV was sensitive to neutralization with heparin whereas the ΔG mutant was more resistant (36). This indicated that, under these conditions, a G-specific GAG effect could be monitored. By this assay, the G-171/187 and G-162/187 mutant viruses displayed the same degree of sensitivity to GAG as wild-type RSV.
The G protein was recently shown to be a mimic of fractalkine (37), a proinflammatory CX3C chemokine that exists in soluble and membrane-bound forms that mediate leukocyte migration and adhesion. Cysteines 182 and 186 in the cystine noose of G represent the CX3C chemokine motif (37). The G protein appears to be able to bind to the fractalkine receptor CX3CR1 and thereby initiate productive infection in vitro (37). Thus, binding to CX3CR1 might represent one of several means by which RSV can bind to cells, with other reported examples including the GAGs mentioned above as well as intercellular adhesion molecule-1 (1). CX3CR1 is found on neural and lymphoid cells as well as on Vero cells, and mRNA for murine CX3CR1 also was very abundant in mouse lung, although the cell types involved have not been identified (9). However, the results of the present study indicated that removal of the CX3C motif from the G protein had little or no effect on infectivity and virus replication in vitro or in vivo. Thus, at a gross level, the availability of the CX3C motif to bind to CX3CR1 did not appear to make a significant independent contribution to RSV replication.
Fractalkine mimicry by the G protein might play a role in RSV infection that is not evident from gross virologic analysis alone. For example, the G protein might alter or interfere with fractalkine-mediated immune responses such as pulmonary cell trafficking. In addition, CX3CR1 has been identified on mouse monocytes, natural killer cells, and neutrophils (P. M. Murphy, personal communication), the presence of which might render these cells susceptible to RSV infection and functional perturbation. Possible effects on the immune response and lung pathology will be investigated in future work. It also is possible that fractalkine mimicry by the G protein operates inefficiently in the mouse, a nonnatural host, and hence that the replication of these deletion viruses should be examined in primates.
The cysteine noose region also has been suggested to have structural similarity with the fourth subdomain of the type I tumor necrosis factor (TNF) receptor (24). This particular domain of the receptor may play a role in TNF binding, although binding was not affected by its deletion (5, 18, 26). TNF appears to play a protective role in RSV infection in culture and in mice (29), and it is tempting to speculate that soluble G protein could function as a TNF antagonist. However, the present study indicates that this function does not have a significant effect on RSV replication in mice.
It is noteworthy that the deletions described here, together with the previously described heparin-binding domain deletions (36), span amino acids 162 to 200, accounting for most of the central domain that separates the two mucin-like domains (Fig. 1). By the process of elimination, this suggests that one or both of the two large mucin-like domains, previously thought to be relatively unimportant spacers, might suffice for G-mediated attachment in vitro and in vivo. Furthermore, monoclonal antibody-resistant mutants have been isolated that have truncations or frameshifts that eliminate the C-terminal region of G, with the largest mutant lacking the G amino acids downstream of position 193 (27, 30, 32). Since position 193 overlaps positions 162 to 200, which were deleted or substituted in this study or previously (36), it may be that G function, at least in vitro, can be provided by the upstream, membrane-proximal mucin-like domain.
Recently, Gorman et al. showed that a series of peptides containing residues 154 to 170 of RSV G could efficiently inhibit RSV cytopathic effect in an in vitro assay in which cells were infected with RSV in the presence of peptide and cytopathic effect was scored 5 days later (13). Alanine replacements and truncations indicated that residues 166 to 170 were critical for the inhibitory effect. The implication was that this peptide mimicked a sequence important in initiating infection, presumably involving the attachment step. This is in contrast to the present study, in which deletion of residues 162 to 187 did not impair virus replication in vitro and in vivo and had no effect on cytopathology during multistep growth in HEp-2 and Vero cells (data not shown). One possibility is that the inhibitory effect of the peptides described previously by Gorman et al. did not occur at the level of virus attachment to the host cell but rather involved some unanticipated mechanism. For example, the peptides might bind to the F protein, perhaps by mimicking an authentic G-F interaction or through chemical affinity and in so doing inhibit F function. The reverse genetics demonstration in the present report that this protein domain can be deleted without affecting virus growth in vitro or in vivo is compelling evidence that it is nonessential.
Thus, our data show that residues 162 to 187 are not required for efficient virus replication in vitro and in mice. However, the high degree of conservation of this domain among human RSV isolates, as well as that of the cystine noose among both human and bovine RSV isolates, suggests that this region plays some other important role in its natural host. Fractalkine mimicry might account for the CX3C domain, but the remainder of the sequence of the central domain bears little resemblance to fractalkine. Thus, the basis for the high level of conservation of amino acids 164 to 176 and Cys-173 and Cys-176 remains unexplained and may be related to some accessory function that remains to be identified.
Acknowledgments
The authors thank Stephen Whitehead, Cai-Yen Firestone, Myron Hill, and Kim Tran for assistance with the animal studies and Brian Murphy for reviewing the manuscript.
This study was part of a continuing program of research and development with Wyeth-Lederle Vaccines and Pediatrics through CRADA contract AI-000087.
REFERENCES
- 1.Behera, A. K., H. Matsuse, M. Kumar, X. Kong, R. F. Lockey, and S. S. Mohapatra. 2001. Blocking intercellular adhesion molecule-1 on human epithelial cells decreases respiratory syncytial virus infection. Biochem. Biophys. Res. Commun. 280:188-195. [DOI] [PubMed] [Google Scholar]
- 2.Bourgeois, C., J. B. Bour, K. Lidholt, C. Gauthray, and P. Pothier. 1998. Heparin-like structures on respiratory syncytial virus are involved in its infectivity in vitro. J. Virol. 72:7221-7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukreyev, A., S. S. Whitehead, B. R. Murphy, and P. L. Collins. 1997. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J. Virol. 71:8973-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrappa, S., D. K. Gavin, and K. C. Gupta. 1995. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 5:404-407. [DOI] [PubMed] [Google Scholar]
- 5.Chen, P. C., G. C. DuBois, and M. J. Chen. 1995. Mapping the domain(s) critical for the binding of human tumor necrosis factor-alpha to its two receptors. J. Biol. Chem. 270:2874-2878. [DOI] [PubMed] [Google Scholar]
- 6.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1443-1485. In D. M. Knipe, and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 7.Collins, P. L., M. G. Hill, E. Camargo, H. Grosfeld, R. M. Chanock, and B. R. Murphy. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc. Natl. Acad. Sci. USA 92:11563-11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, P. L., and G. Mottet. 1992. Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus: altered O-glycosylation in the presence of brefeldin A. J. Gen. Virol. 73:849-863. [DOI] [PubMed] [Google Scholar]
- 9.Combadiere, C., J. Gao, H. L. Tiffany, and P. M. Murphy. 1998. Gene cloning, RNA distribution, and functional expression of mCX3CR1, a mouse chemotactic receptor for the CX3C chemokine fractalkine. Biochem. Biophys. Res. Commun. 253:728-732. [DOI] [PubMed] [Google Scholar]
- 10.Feldman, S. A., S. Audet, and J. A. Beeler. 2000. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J. Virol. 74:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldman, S. A., R. M. Hendry, and J. A. Beeler. 1999. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J. Virol. 73:6610-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorman, J. J., B. L. Ferguson, D. Speelman, and J. Mills. 1997. Determination of the disulfide bond arrangement of human respiratory syncytial virus attachment (G) protein by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Protein Sci. 6:1308-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorman, J. J., J. L. McKimm-Breschkin, R. S. Norton, and K. J. Barnham. 2001. Antiviral activity and structural characteristics of the nonglycosylated central subdomain of human respiratory syncytial virus attachment (g) glycoprotein. J. Biol. Chem. 276:38988-38994. [DOI] [PubMed] [Google Scholar]
- 14.Hallak, L. K., P. L. Collins, W. Knudson, and M. E. Peeples. 2000. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 271:264-275. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, J. E., O. Lund, N. Tolstrup, A. A. Gooley, K. L. Williams, and S. Brunak. 1998. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj. J. 15:115-130. [DOI] [PubMed] [Google Scholar]
- 16.He, B., G. Y. Lin, J. E. Durbin, R. K. Durbin, and R. A. Lamb. 2001. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J. Virol. 75:4068-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heminway, B. R., Y. Yu, Y. Tanaka, K. G. Perrine, E. Gustafson, J. M. Bernstein, and M. S. Galinski. 1994. Analysis of respiratory syncytial virus F, G, and SH proteins in cell fusion. Virology 200:801-805. [DOI] [PubMed] [Google Scholar]
- 18.Hwang, C. D., M. Gatanaga, E. K. Innins, R. S. Yamamoto, G. A. Granger, and T. Gatanaga. 1991. A 20 amino acid synthetic peptide of a region from the 55 kDa human TNF receptor inhibits cytolytic and binding activities of recombinant human tumour necrosis factor in vitro. Proc. R. Soc. Lond. B Biol. Sci. 245:115-119. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, P. R., M. K. Spriggs, R. A. Olmsted, and P. L. Collins. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. USA 84:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juhasz, K., S. S. Whitehead, P. T. Bui, J. M. Biggs, C. A. Boulanger, P. L. Collins, and B. R. Murphy. 1997. The temperature-sensitive (ts) phenotype of a cold-passaged (cp) live attenuated respiratory syncytial virus vaccine candidate, designated cpts530, results from a single amino acid substitution in the L protein. J. Virol. 71:5814-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karger, A., U. Schmidt, and U. J. Buchholz. 2001. Recombinant bovine respiratory syncytial virus with deletions of the G or SH genes: G and F proteins bind heparin. J. Gen. Virol. 82:631-640. [DOI] [PubMed] [Google Scholar]
- 22.Karron, R. A., D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. L. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, B. R. Murphy, and M. S. Sidhu. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 94:13961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krusat, T., and H. J. Streckert. 1997. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch. Virol. 142:1247-1254. [DOI] [PubMed] [Google Scholar]
- 24.Langedijk, J. P., B. L. de Groot, H. J. Berendsen, and J. T. van Oirschot. 1998. Structural homology of the central conserved region of the attachment protein G of respiratory syncytial virus with the fourth subdomain of 55-kDa tumor necrosis factor receptor. Virology 243:293-302. [DOI] [PubMed] [Google Scholar]
- 25.Levine, S., R. Klaiber-Franco, and P. R. Paradiso. 1987. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J. Gen. Virol. 68:2521-2524. [DOI] [PubMed] [Google Scholar]
- 26.Lie, B. L., D. Tunemoto, H. Hemmi, Y. Mizukami, H. Fukuda, H. Kikuchi, S. Kato, and N. Numao. 1992. Identification of the binding site of 55kDa tumor necrosis factor receptor by synthetic peptides. Biochem. Biophys. Res. Commun. 188:503-509. [DOI] [PubMed] [Google Scholar]
- 27.Martinez, I., J. Dopazo, and J. A. Melero. 1997. Antigenic structure of the human respiratory syncytial virus G glycoprotein and relevance of hypermutation events for the generation of antigenic variants. J. Gen. Virol. 78:2419-2429. [DOI] [PubMed] [Google Scholar]
- 28.Murphy, B. R., A. V. Sotnikov, L. A. Lawrence, S. M. Banks, and G. A. Prince. 1990. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3-6 months after immunization. Vaccine 8:497-502. [DOI] [PubMed] [Google Scholar]
- 29.Neuzil, K. M., Y. W. Tang, and B. S. Graham. 1996. Protective role of TNF-alpha in respiratory syncytial virus infection in vitro and in vivo. Am. J. Med. Sci. 311:201-204. [DOI] [PubMed] [Google Scholar]
- 30.Rueda, P., T. Delgado, A. Portela, J. A. Melero, and B. Garcia-Barreno. 1991. Premature stop codons in the G glycoprotein of human respiratory syncytial viruses resistant to neutralization by monoclonal antibodies. J. Virol. 65:3374-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rueda, P., B. Garcia-Barreno, and J. A. Melero. 1994. Loss of conserved cysteine residues in the attachment (G) glycoprotein of two human respiratory syncytial virus escape mutants that contain multiple A-G substitutions (hypermutations). Virology 198:653-662. [DOI] [PubMed] [Google Scholar]
- 32.Rueda, P., C. Palomo, B. Garcia-Barreno, and J. A. Melero. 1995. The three C-terminal residues of human respiratory syncytial virus G glycoprotein (Long strain) are essential for integrity of multiple epitopes distinguishable by antiidiotypic antibodies. Viral Immunol. 8:37-46. [DOI] [PubMed] [Google Scholar]
- 33.Sullender, W. M., M. A. Mufson, L. J. Anderson, and G. W. Wertz. 1991. Genetic diversity of the attachment protein of subgroup B respiratory syncytial viruses. J. Virol. 65:5425-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Techaarpornkul, S., N. Barretto, and M. E. Peeples. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 75:6825-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teng, M. N., and P. L. Collins. 1999. Altered growth characteristics of recombinant respiratory syncytial viruses which do not produce NS2 protein. J. Virol. 73:466-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teng, M. N., S. S. Whitehead, and P. L. Collins. 2001. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 289:283-296. [DOI] [PubMed] [Google Scholar]
- 37.Tripp, R. A., L. P. Jones, L. M. Haynes, H. Zheng, P. M. Murphy, and L. J. Anderson. 2001. CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nat. Immunol. 2:732-738. [DOI] [PubMed] [Google Scholar]
- 38.Walsh, E. E., A. R. Falsey, and W. M. Sullender. 1998. Monoclonal antibody neutralization escape mutants of respiratory syncytial virus with unique alterations in the attachment (G) protein. J. Gen. Virol. 79:479-487. [DOI] [PubMed] [Google Scholar]
- 39.Wathen, M. W., P. A. Aeed, and A. P. Elhammer. 1991. Characterization of oligosaccharide structures on a chimeric respiratory syncytial virus protein expressed in insect cell line Sf9. Biochemistry 30:2863-2868. [DOI] [PubMed] [Google Scholar]
- 40.Wertz, G. W., M. Krieger, and L. A. Ball. 1989. Structure and cell surface maturation of the attachment glycoprotein of human respiratory syncytial virus in a cell line deficient in O glycosylation. J. Virol. 63:4767-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitehead, S. S., A. Bukreyev, M. N. Teng, C. Y. Firestone, M. St. Claire, W. R. Elkins, P. L. Collins, and B. R. Murphy. 1999. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 73:3438-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wyatt, L. S., B. Moss, and S. Rozenblatt. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202-205. [DOI] [PubMed] [Google Scholar]