Abstract
A critical event in papillomavirus transformation of human cells is the inactivation of pRB by the E7 protein. E7, like many other viral oncoproteins, possesses a well-characterized LXCXE peptide motif that interacts with the pocket domain of pRB. Disruption of the LXCXE-binding cleft on pRB renders it resistant to E7 binding and inactivation. Such binding cleft mutants of pRB are capable of inducing a G1 arrest in the human papillomavirus 18-transformed HeLa cell line. We show here that the efficient inactivation of pRB in HeLa cells does not simply depend on the integrity of the LXCXE-binding cleft. Multiple site-directed mutants that alter conserved surfaces of the pRB pocket domain cause HeLa cells to accumulate in G1. We divide these mutants into two classes: those that can be bound by E7 and those that cannot. The E7 interacting mutants include changes in conserved residues that lie in a groove between the A and B halves of the pocket. Surprisingly, none of these mutants show a clear defect in any of the known mechanisms for pRB inactivation by E7. Analysis of mutants that are compromised for E7 binding reveals that this interaction depends on both the LXCXE-binding cleft and on a conserved group of lysines adjacent to the cleft. These basic amino acids on pRB define a discrete interaction point with E7. These residues most likely form ionic interactions with conserved acidic amino acids on E7 since a stable pRB/E7 interaction was restored when the lysine residues on pRB and the acidic residues on E7 were interchanged.
Inactivation of the retinoblastoma tumor suppressor protein (pRB) is a common event in human cancer (35, 42). Most frequently, pRB is inactivated by mutation of its regulators. Inactivating mutations of p16 or activating alterations of the genes encoding cyclin D1 and cyclin-dependent kinase 4 (cdk4) result in the functional inactivation of pRB. Less commonly, the gene encoding pRB (RB-1) is inactivated by direct mutation (42). Additionally, pRB is also known to be inactivated directly by virally encoded oncoproteins like E7 in high-risk papillomaviruses (14, 22, 36). High-risk papillomaviruses are detectable in 90% of cervical carcinomas, establishing the importance of human papillomaviruses (HPVs) in this tumor type (50).
Studies of the transforming properties of HPV E7 and other viral oncoproteins, such as simian virus 40 (SV40) TAg and adenovirus type 5 E1A, have revealed the requirement for a conserved motif containing the peptide sequence LXCXE (16, 41, 43). This motif is known to facilitate a tight protein-protein interaction between the viral oncoproteins and pRB (9, 13, 15, 36, 47, 48). Beyond the need for direct binding to pRB through the LXCXE motif, E7's precise mechanism of inactivation of pRB has remained elusive. Like E1A and TAg, E7 is capable of disrupting pRB-E2F protein complexes (6, 38, 49). The exact mechanism of this dissociation activity is not known, but it requires portions of the E7 protein both N and C terminal to the LXCXE motif (23, 24, 33, 39). More recently, it has also been shown that E7 possesses the ability to destabilize pRB (1, 2, 10, 27). This process involves the targeting of pRB for degradation by ubiquitination and transfer to the 26S proteasome (1, 2, 45). The extreme N terminus of E7 is required for pRB degradation since a missense mutation of H2P or addition of an N-terminal epitope tag disrupts this function (17, 23). Previous work has shown that the small pocket domain of pRB is sufficient for degradation (17). Mutations which alter the LXCXE-binding cleft prevent binding by E7 and block degradation (17).
Direct mutation of pRB and inactivation by viral proteins clearly are irreversible inactivation events. However, a reversible inactivation of pRB occurs as part of each normal cell cycle (reviewed in references 12 and 46). In late G1 two distinct cyclin-cdk complexes successively phosphorylate pRB to facilitate its inactivation. It has been shown that pRB phosphorylation begins with cyclin D-cdk4 or -cdk6 and is in turn followed by cyclin E-cdk2 (34). This regulation is not mediated by a single modification. Instead, a number of studies have shown that inactivation of pRB requires the accumulation of numerous phosphorylation events (3, 30, 32). Once phosphorylated, pRB inactivation is thought to occur by intramolecular rearrangements of flexible domains, such as the spacer and the C-pocket which are the primary locations of phosphates on inactive pRB (20, 30). The tighter binding of these phosphorylated domains to the A-B pocket domain would then displace associated factors such as E2Fs and chromatin remodeling machinery. Recent work by Harbour et al. suggests that phosphates on the C-pocket interact with a well-conserved group of lysines on the B-half of the pocket domain to exclude histone deacetylases from binding (20).
We and others have carried out a mutational analysis of the LXCXE-binding cleft on pRB (5, 7, 8, 11). We found that mutation of the LXCXE-binding cleft in pRB disrupts the ability of pRB to interact with HPV E7 proteins. Surprisingly, the binding cleft mutants retain a considerable degree of the transcriptional repression and cell cycle regulatory functions of the wild-type protein. This separation of activities is most clearly illustrated by the properties of these mutants in human papillomavirus type 18 (HPV-18)-transformed HeLa cells (11). Much work has been done to establish the importance of E6 and E7 function to maintain the proliferative state of HeLa cells (19, 26, 37). Repression of expression of these oncogenes by the E2 protein blocks growth and rapidly induces a senescent state (18). Interestingly, the expression of LXCXE cleft mutants of pRB causes HeLa cells to accumulate in the G1 phase of the cell cycle. This property is specific to the mutant forms of the protein and does not occur when wild-type pRB is expressed. Presumably, this G1 arrest reflects both the inability of the mutant pRB protein to be targeted by E7 and its ability to perform many of the normal functions of the wild-type protein.
Random mutagenesis approaches have been of limited use for the analysis of the pRB pocket, in part because the majority of mutants that are generated disrupt the overall integrity of the domain rather than selectively removing specific functions. In this respect, the ability of pRB mutants to arrest HeLa cells provides a very stringent assay, since it demands that the mutant protein be active for cell cycle control and be able to exert an effect that overrides the activity of E6 and E7. Here, we have taken advantage of this assay to screen a collection of 35 site-directed mutants of the pRB pocket for variants that induce HeLa cells to accumulate in G1. As expected, we identified several alleles that alter the LXCXE-binding cleft. In addition, we found mutants in two distinct regions of the pocket domain. One group of mutants is clustered close together in a groove between the A and B halves of the pocket. These mutant forms of pRB resemble the wild-type protein in that they interact with E2F and E7 and are targeted for degradation by the viral protein. However, these mutants appear to be poorly phosphorylated in vivo and may represent a class of “gain-of-function” alleles of RB-1. A second class of mutants identified contain changes in a well-conserved basic surface on the B half of the pocket. Our analysis suggests that these residues on pRB interact with conserved acidic amino acids on E7 and are important for a stable association between the two molecules.
MATERIALS AND METHODS
Plasmid construction.
Site-directed mutagenesis of the LXCXE cleft region by PCR has been described previously (11). Sequence changes in the B half of the pocket were constructed in an NheI-BsmI fragment, while changes in the A half were carried out in a 0.5-kb NcoI-NheI containing construct. After sequencing to ensure that only the desired substitutions had been incorporated into the mutant allele, the mutants were transferred into the full-length RB cDNA in the pCMV-neo-Bam expression vector. The LXCXE cleft mutant RB38 containing Y756A, S758L, was obtained from D. Dean (8), while the LXCXE plus the B-pocket allele RB37, containing four lysine-to-alanine substitutions, was constructed to resemble an allele that has been previously published by Harbour et al. (20). Cytomegalovirus (CMV)-RbΔcdk was made by ligating a BglII-BamHI fragment containing RbΔcdk from pECE-RbΔcdk (32) into the BamHI site of CMV-neo-Bam. CMV-E7 (type 16) and GST-E7 (type 16) have been published previously (40, 49). PCR-directed mutagenesis of a pBSK-E7 (type 16) plasmid was used to alter the sequence encoding Glu-33 to Glu-37 to five lysines. This mutant E7-EK clone was ligated into the BamHI site of pGEX-2T. Similarly, a PCR fragment containing E7 (type 18) was ligated into the BamHI site of pGEX-20T. Glutathione S-transferase (GST) fusion proteins were expressed and purified as recommended by the manufacturer (Amersham Pharmacia). CMV-HA-DP1, CMV-HA-E2F2, and CMV-βGal have been used previously (11).
Cell culture and transfections.
HeLa, C33A, and Saos-2 cells were obtained from the American Type Culture Collection and were cultured as described earlier (11). For flow cytometry experiments HeLa cells were plated at 2.5 × 106 per 10-cm dish. Each plate was transfected by calcium phosphate with 15 μg of CMV-RB or mutant expression vector and 5 μg of CMV-CD20 or pMACS Kk.II expression vector. Cells were harvested at 48 h posttransfection and processed for flow cytometry as described previously (25, 44). Where indicated, nocodazole was added at 50 ng/ml, at 24 h prior to harvesting of the cells.
Transfected HeLa cells were harvested for magnetic bead isolation on Kk microbeads essentially as described by the manufacturer (Miltenyi Biotec, Auburn, Calif.). Isolated cells were pelleted in microfuge tubes, and cells were lysed directly in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer for pRB expression analysis.
C33A cells were plated at a density of 106 per 6-cm dish. Cells were transfected with 10 μg of CMV-RB expression plasmid by calcium phosphate and lysed at 72 h posttransfection. Saos-2 cells were plated at a density of 4 × 105 cells per well of a six-well plate or 106 cells per 6-cm dish. Saos-2 cells were transfected with 4 μg of DNA by using Fugene 6 (Boehringer Mannheim) according to the manufacturer's directions.
Cell extract preparation and GST pulldown assays.
C33A cell extracts were prepared in 0.5 ml of gel shift extract buffer (25) from a 10-cm plate of cells by freeze-thaw lysis, and debris was cleared by centrifugation at 100,000 × g for 20 min. Then, 100 μl of the extract was mixed with 1 μg of GST fusion protein on ice for at least 30 min. Glutathione-Sepharose beads (10-μl bed volume) were mixed with extracts and GST fusion protein for 15 min with rocking. Beads were spun out and washed two times in gel shift extract buffer without glycerol and with only 200 mM NaCl. Washed beads were resuspended in 100 μl of SDS-PAGE sample buffer and then boiled for 5 min. Samples were resolved by SDS-8% PAGE, and pRB was detected by Western blotting with anti-pRB monoclonal antibody C36 as described previously (11).
E7 destabilization assays.
E7's ability to diminish pRB expression levels was measured similarly to the approach of Gonzalez et al. (17). Saos-2 cells were seeded in a six-well plate. Each well was transfected with 4 μg of CMV-RB (or mutant); 0.5 μg of CMV-βGal; and 0, 0.1, or 1 μg of CMV-E7 by using Fugene 6 (Boehringer Mannheim). All transfection mixes were normalized for DNA content by adding CMV-CD20. Extracts were prepared in 0.25 ml of radioimmunoprecipitation assay buffer (21) at 72 h posttransfection. Then, 50 μl of the extract was used in a liquid β-galactosidase enzyme assay to determine the relative transfection efficiency, and the remainder was mixed with SDS-PAGE sample buffer and used to determine pRB expression levels by Western blotting.
E7 competition in electrophoretic mobility shift assays.
CMV-HA-E2F2 and CMV-HA-DP1 were cotransfected into C33A cells, and extracts were prepared in gel shift extract buffer by freeze-thaw lysis. Similarly, extracts containing pRB or pRB mutants were prepared from transfected Saos-2 cells. DNA-binding reactions were prepared as described by Hurford et al. (25). Each reaction contained 0.3 μg of C33A extract, 1 to 1.5 μg of Saos-2 extract (normalized to pRB content) and, where indicated, 1 ng to 1 μg of GST protein as competitor. Binding was allowed to proceed for 10 min on ice prior to the addition of the 32P-labeled E2F probe containing a single consensus E2F binding site. Reactions were incubated a further 20 min prior to loading on a 4% acrylamide gel and then resolved at 180 V for 3 h at 4°C. Bands were visualized by autoradiography.
RESULTS
A screen for pRB mutants that are able to arrest E7-expressing cells.
Previously, we found that mutation of the LXCXE-binding cleft in pRB disrupts the ability of pRB to interact with HPV E7 proteins. Surprisingly, given the conservation of this binding site, LXCXE-binding cleft mutants retain a considerable degree of cell cycle regulatory function and are able to cause HPV-18-transformed HeLa cells to accumulate in G1 (11) (Table 1). To ensure that the accumulation of 2N DNA content cells was a true reflection of a lengthened transit time through G1 and not accelerated passage through other phases of the cell cycle, we examined the effect of the mitotic inhibitor nocodazole on the cell cycle profile. HeLa cells were transfected with CMV-CD20 and either CMV-βGal, CMV-RB (expressing wild-type, full-length human pRB), or CMV-RB9 (full-length pRB carrying three amino acid changes in the LXCXE-binding cleft). Cultures were treated with nocodazole 24 h prior to harvesting them, and cells were examined by flow cytometric analysis. As shown in Table 1, nocodazole treatment dramatically increases the percentage of cells in the G2 and M phases of the cell cycle. In agreement with our previous work, introduction of wild-type pRB into these cells causes no significant increase in G1 accumulation relative to the β-galactosidase-transfected control. Transfection of a LXCXE-binding cleft mutant, RB9, not only increases the percentage of cells in G1 but this accumulation persists in the presence of nocodazole. This indicates that many of the RB9-expressing cells are either arrested in G1 or are significantly delayed in their progression through G1. To confirm that this G1 arrest is a specific effect and not a general feature of all pRB mutants, we examined the effects of tumor-derived loss-of-function alleles such as C706F and R661W. Cell cycle phases of HeLa cells were unaffected by introduction of either mutant protein (data not shown). Thus, the ability to arrest HeLa cells is an unusual property of LXCXE cleft mutants that is not shared by either the wild-type protein or tumor-derived alleles.
TABLE 1.
LXCXE cleft mutants of pRB induce a G1 arrest in HeLa cellsa
| Transfected gene and treatment | % CD20+ cells in:
|
% CD20− cells in:
|
||||
|---|---|---|---|---|---|---|
| G1 | S | G2/M | G1 | S | G2/M | |
| β-Gal | ||||||
| None | 35.8 | 34.5 | 29.7 | |||
| Nocodazole | 8.2 | 14.2 | 77.6 | 3.9 | 12.7 | 83.4 |
| RB | ||||||
| None | 35.8 | 40.5 | 23.7 | |||
| Nocodazole | 7.6 | 26.5 | 65.9 | 4.7 | 16.3 | 79.0 |
| RB9 | ||||||
| None | 53.1 | 32.8 | 14.1 | |||
| Nocodazole | 40.6 | 14.1 | 45.3 | 10.0 | 15.4 | 74.6 |
HeLa cells were transfected with pRB mutants and CD20. Flow cytometry was used to identify transfected cells by CD20 staining, and the cell cycle phases were determined by measuring propidium iodide staining of these cells. Nocodazole (50 ng/ml) was added 24 h prior to harvesting to trap cells in M phase.
Mapping studies have shown that the pocket domain of pRB and the LXCXE motif of E7 proteins are critical for the interaction between these proteins. This mapping does not exclude the possibility that other regions of the proteins might have an important impact on the ability of E7 to inactivate pRB. Indeed, it has previously been shown that full-length E7 protein has an affinity for the pRB pocket that is ca. 100-fold greater than short LXCXE-containing peptides (31), suggesting that additional contact sites are likely to exist that are important for the interaction. We reasoned that any mutation that prevents the inactivation of pRB by E7, without compromising pRB's cell cycle arrest functions, might also cause a G1 accumulation in the HeLa cell assay. We therefore used this assay to screen a collection of 35 mutants in the pocket domain of pRB. The mutants were created in the context of full-length human pRB by site-specific mutation of residues that are conserved in pRB homologs between species and are predicted to be on the surface of the pocket. On average, these mutants contain two or three substitutions per allele. Mutants were transfected into HeLa cells and analyzed by fluorescence-activated cell sorting for their ability to induce an increase in G1 cells (summarized in Table 2).
TABLE 2.
Arrest of HeLa cells by pRB mutantsa
| Allele | Mutation(s) |
|---|---|
| Wild type | None |
| RB4 | Y709A |
| RB5 | Y709F and K713A |
| RB6 | Y756F and N757A |
| RB7 | K722E, K729E, and K740E |
| RB8 | Y709A, K713A, I753A, N757A, and M761A |
| RB9 | I753A, N757A, and M761A |
| RB10 | Y709A and K713A |
| RB11 | L660A, T664A, and R668A |
| RB12 | K652A and R656A |
| RB13 | R544A and K548A |
| RB17 | Y709A, K713A, I753A, N757A, M761A, K722E, K729E, and K740E |
| RB19 | Q575A and S576A |
| RB28a | K713A, K720A, K722A, K729A, K740A, and K745A |
| RB28b | K713A, K715A, K722A, K729A, K740A, K745A, and K765A |
| RB37 | K713A, K720A, K722A, and K729A |
| RB38 | Y756A and S758L |
| RbΔcdk | T246A, T350A, S601A, S605A, S781A, S787A, S788A, S800A, S804A, T814A, and T819A |
RB mutants were transfected into HeLa cells as in Table 1. Cell cycle analysis was performed as described previously. Mutants that caused a 5% or greater increase in cells with 2N DNA content are listed.
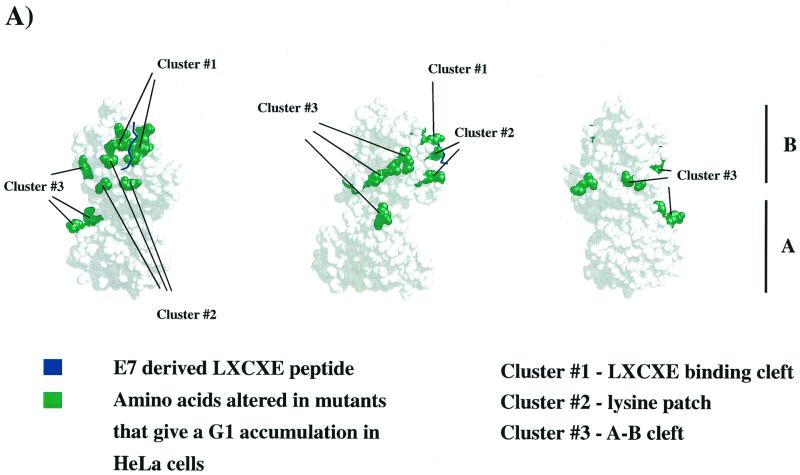
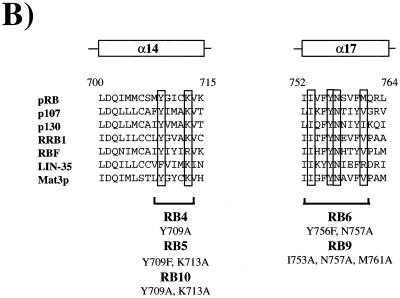
From this screen we found multiple RB-1 alleles that gave a consistent increase in the proportion of G1 cells. The position of these mutations on the crystal structure suggested that these mutants might represent three distinct surfaces of the pocket domain (see Fig. 1A). First, as expected, multiple mutations affecting well-conserved residues in the LXCXE-binding groove gave a robust increase in G1 cells (see Table 2 and Fig. 1B). The changes in RB4, RB5, RB6, RB8, RB9, RB10, and RB38 were all designed to eliminate residues that are predicted to contact the LXCXE peptide. As described previously, each of these mutants is impaired for its ability to interact with E7 (reference 11 and data not shown).
FIG. 1.
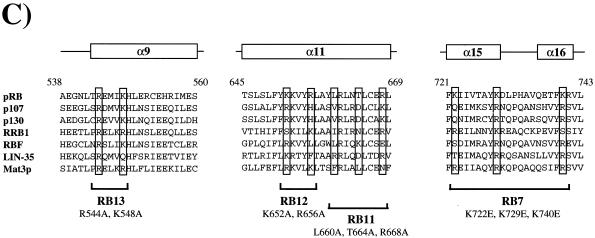
Mutations that cause an increase in G1 HeLa cells target three well-conserved surfaces of the pRB pocket domain. (A) Structure of the pRB small pocket domain. Amino acids whose alteration results in a G1 increase in HeLa cells are colored green. Each view of the pocket domain is rotated 90° relative to its nearest neighbor. The A and B halves of the pRB pocket are indicated to the right of panel A. (B and C) Sequence alignments of structural segments of the pRB protein whose alterations induce an accumulation of 2N DNA cells in HeLa. The sequences are derived from human pRB, p107, and p130, as well as Zea maize RRB1, Drosophila melanogaster RBF, Caenorhabditis elegans LIN-35, and Clamydomonas rheinhardtii Mat3p. The amino acid position numbers refer to human pRB. The secondary structure of these segments of pRB is designated above each pileup and is derived from the crystal structure. Previous mutations that have been reported to disrupt binding to the LXCXE motif of E7 are boxed in panel B. Mutations that antagonize E7 binding or function in this study are boxed in panel C; locations of the various arresting mutants are also indicated.
Second, we found that mutations in a group of well-conserved lysine residues located close to the LXCXE-binding cleft (see Fig. 1C) were also able to arrest HeLa cells in G1 (31). These gave an increase in the G1 population that was comparable to that seen with the LXCXE cleft mutants. This “lysine patch” has been noted previously and has been proposed to interact with the C terminus of pRB subsequent to phosphorylation of pRB by cyclin-dependent kinases (20). Some of the more extensive lysine patch mutants (e.g., RB28a, RB28b, and RB37) include mutation of K713, a residue that is predicted to contact the LXCXE peptide (31), although mutation of it alone has no effect on E7 binding (data not shown). However, G1 accumulation is also seen with other mutants, such as RB7, in which it only affects lysine residues not previously implicated in contact with the E7 peptide (K722, K729, and K740).
Third, a lower but reproducible level of G1 accumulation was observed with mutations that are distant from the LXCXE-binding cleft (RB11, RB12, RB13, and RB19). These changes are located close to a groove that separates the A and B halves of the pocket domain. The residues changed in RB11, RB12, and RB13 are closely clustered on the crystal structure and contain substitutions of basic amino acids that have been highly conserved throughout the evolution of the RB gene (Fig. 1C). Indeed, several of these residues are as well conserved between pRB homologs of different species as the residues that we have shown to be required for stable association with the LXCXE peptide (Fig. 1), suggesting that this cluster of residues provides an important aspect of pRB function or regulation.
The RB-1 alleles selected in the HeLa cell assay fall into two classes based on their E7-binding properties.
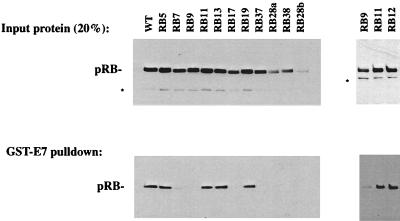
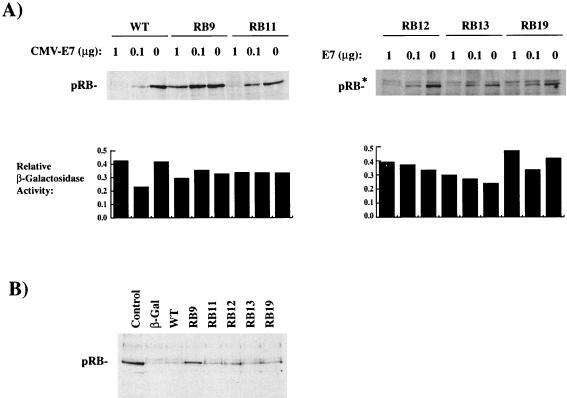
As a first step in the characterization of these mutants, we tested whether the encoded proteins interact with E7. Interactions between pRB and E7 were tested in GST-E7 “pulldown” experiments. To do this, CMV-driven expression constructs for these mutants were transfected into C33A cells. Extracts were then prepared and analyzed by SDS-PAGE and Western blotted to quantify expression levels (Fig. 2, upper panels). Extracts were also mixed with GST-E7 (type 16) protein, and mutant pRB bound to E7 was detected by Western blotting (Fig. 2, lower panels). Mutants encoded by RB28a and RB28b routinely displayed reduced expression relative to wild-type pRB. Mutant RB12 was analyzed on a separate gel (rightmost panels) than the others and is shown accompanied by reiterations of RB9 and RB11 expression and binding to provide negative and positive controls, respectively.
FIG. 2.
There are two classes of HeLa arresting mutants based on their ability to bind to E7. C33A cells were transfected with plasmids encoding the indicated mutants, and extracts were prepared. (Upper panels) Expression levels of the various mutants were analyzed by Western blotting. (Lower panels) The ability of the different mutant forms of pRB to bind E7 was quantified by mixing extracts with GST-E7 and Western blotting to detect the amount of pRB precipitated with GST-E7. Mutant RB12 was analyzed separately and displayed with positive (RB11) and negative (RB9) controls in the rightmost panels. The asterisk marks a proteolytic breakdown product of pRB.
Since mutations in the LXCXE-binding groove that prevent binding of E7 were known to arrest HeLa cells in G1, we anticipated that some of the new alleles selected in the screen might show a similar defect. Indeed, RB7, RB17, RB28a, RB28b, and RB37 (each containing changes in the lysine patch adjacent to the LXCXE-binding cleft) were defective in binding to E7. Although RB28a and RB28b are expressed at reduced levels, compared to other mutants, prolonged exposure of Western blots failed to reveal even trace amounts bound to GST-E7 (type 16).
Surprisingly, not all of the mutants that were selected by their ability to increase the G1 population of HeLa cells were defective in binding to GST-E7. RB11, RB12, RB13, and RB19, which contain substitutions in the A-B interface region, showed a robust interaction with GST-E7 (type 16) that was similar to that of wild-type pRB. Since HeLa cells express type 18 E7, we also tested binding to a GST-E7 (type 18) and obtained the same results (data not shown). Thus, the RB-1 mutants that altered cell cycle progression in HeLa cells seem to separate cleanly into two classes: (i) mutations in the LXCXE-binding cleft and/or the nearby lysines that failed to bind in a stable manner to E7 and (ii) mutations in the A-B interface region that retained E7 binding.
RB11, RB12, RB13, and RB19 interact with E2F, and these complexes are disrupted efficiently by E7.
We first examined the properties of RB11, RB12, RB13, and RB19, the subset of RB mutants that are able to increase the G1 population of HeLa cells but retain the ability to bind to E7. E7 has been proposed to inactivate pRB in two distinct ways. It has been found to associate with pRB and to disrupt pRB-E2F complexes (6, 38, 49). In addition, E7 is able to target pRB for ubiquitin-mediated proteolysis (17, 23). This process can be easily monitored by using cotransfection assays, since the coexpression of E7 with pRB causes a rapid reduction in pRB levels (1, 2, 10, 27). This assay was used to test whether the RB mutants might be resistant to E7-induced degradation.
Increasing amounts of CMV-E7 expression vector proportionately reduced the expression of wild-type pRB, but not to mutant RB9, an LXCXE cleft mutant that fails to bind to E7 and serves as a positive control for resistance to E7 (Fig. 3A). As shown in Fig. 3A, the expression levels of RB11, RB12, RB13, and RB19 are all strongly reduced by cotransfection with E7, and none resembles the resistant phenotype of RB9.
FIG. 3.
E7 destabilizes pRB mutants in the A-B cleft. (A) Saos-2 cells were cotransfected with RB, E7, and β-galactosidase expression vectors. Extracts were Western blotted for pRB expression levels and assayed for β-galactosidase activity to indicate the transfection efficiency. The amounts of cotransfected E7 expression plasmid are indicated above each lane. The β-galactosidase activity levels are within a twofold variation. A nonspecific, cross-reacting band is indicated by an asterisk. (B) Expression levels of wild-type or mutant pRB in transfected and sorted HeLa cells. RB transfected Saos-2 cell extracts were included in the leftmost lane as a positive control.
It is possible that this transfection-based degradation assay may not entirely recapitulate the degradation of exogenously introduced pRB in HeLa cells. For this reason, we coexpressed pRB with a cell surface marker in HeLa cells, isolated the transfected cells by magnetic bead sorting, and assessed the levels of pRB by Western blotting (Fig. 3B). Expression of endogenous pRB is nearly undetectable in the negative control β-galactosidase-transfected cells. The wild-type-transfected cells show a slight increase over the negative control, while the LXCXE cleft mutant RB9 shows abundant expression of pRB. As before, the levels of RB11, RB12, RB13, and RB19 are greatly diminished relative to RB9, and these clearly represent a different class of mutants from the LXCXE cleft mutants. However, the levels of the RB11, RB12, RB13, and RB19 are slightly elevated over that of wild-type pRB. It is feasible that this slight difference may be sufficient to give a modest accumulation of G1 phase cells, but the significance of this small difference is difficult to assess.
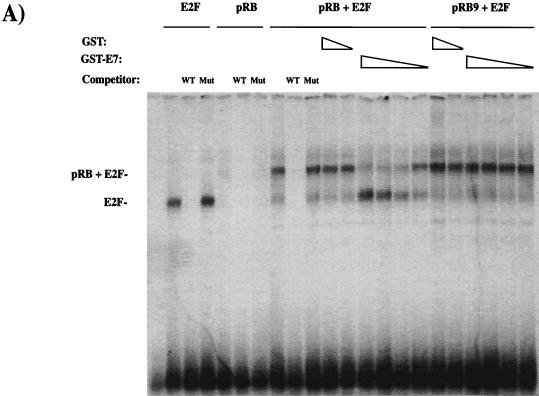
Since there was no obvious defect in E7-induced destabilization of pRB, we examined whether E7 was able to disrupt pRB-E2F complexes formed by the mutant pRB proteins. To test this, we devised a gel shift assay in which pRB-E2F complexes are assembled in vitro. CMV-E2F2 and CMV-DP1 expression constructs were transfected into C33A cells (that lack pRB), and extracts were prepared that contained an abundance of E2F site binding activity that was detectable by electrophoretic mobility shift assays (see Fig. 4A). Saos-2 cells, which also lack pRB and are arrested by ectopic overexpression of pRB, were transfected with RB expression constructs, and the lysates of these cells provided an abundant source of active pRB. For these experiments the small quantity of the pRB-containing Saos-2 extract that was used gave no detectable free E2F and negligible amounts of E2F-pocket protein complexes (Fig. 4A). However, when this extract was mixed with the E2F-containing C33A cell extracts prior to electrophoresis, the free E2F activity from the C33A extract associated with the pRB present in the Saos-2 extracts, generating an easily detectable pRB-E2F complex. The pRB-E2F complex could be supershifted by addition of the appropriate antibodies and was competed for by the addition of unlabeled E2F oligonucleotides, but not by corresponding mutant oligonucleotides (Fig. 4A and data not shown).
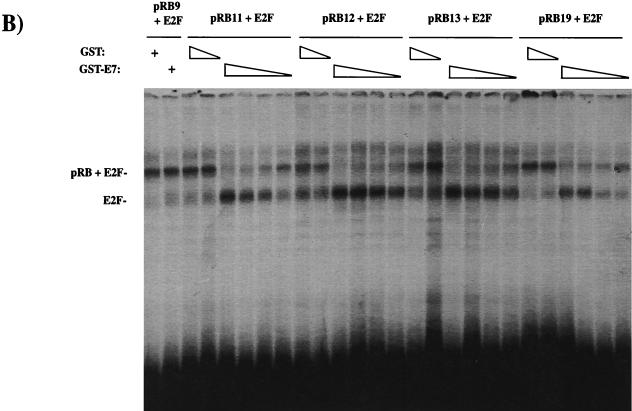
FIG. 4.
E7 competes for E2F binding to mutants in the A-B cleft. C33A extracts from cells transfected with E2F2 and DP1 expression vectors were mixed with Saos-2 extracts from RB transfectants. Interactions between E2F and pRB proteins were monitored by creating slower-migrating complexes (indicated at the left of each panel). E7 competition was detected by the ability of GST-E7 to block the formation of pRB-E2F complexes. The amounts of GST used as a competitor were 1 μg and 1 ng. The amounts of GST-E7 used were 1 μg, 100 ng, 10 ng, and 1 ng. The specificity of the complexes detected here was confirmed by the ability of a wild-type E2F site-containing cold competitor to disrupt these complexes, whereas a mutant cold competitor leaves the complexes unaffected. The sensitivity of wild-type pRB to GST-E7 competition and the resistance of pRB9 to GST-E7 are shown in panel A. (B) The sensitivity of the mutants in the A-B cleft region, along with the reiterated control of pRB9 with 1 μg of GST or GST-E7. (C) pRB expression levels in the extracts used in the experiments shown in panel B.
Purified GST or GST-E7 proteins were added to this binding reaction, and the effects of E7 on pRB-E2F complex formation were examined. The interaction between wild-type pRB and E2F was resistant to GST over a range of 3 orders of magnitude. The addition of GST-E7 blocked the appearance of pRB-E2F complexes in a quantitative manner, with 1 μg or 100 ng of GST-E7 giving almost complete competition, 10 ng of GST-E7 giving a partial effect, and 1 ng of recombinant GST-E7 having little effect. As a control, Saos-2 extracts containing pRB9 (a mutant in the LXCXE-binding cleft that fails to interact with E7) forms pRB-E2F complexes that are unaffected by the addition of GST-E7.
When mutants RB11, RB12, RB13, and RB19 were tested in this assay, each of these proteins formed E2F complexes that were disrupted by GST-E7 (Fig. 4B). The disruption of pRB-E2F complexes formed by RB11, RB12, and RB13 was indistinguishable from the disruption of wild-type pRB-containing complexes. We noted that the pRB-E2F complexes formed by RB19 were marginally more resistant to GST-E7, but this may be due to the slightly higher levels of pRB protein seen with this mutant (Fig. 4C).
We conclude that E7 is able to bind to the pRB variants encoded by RB11, RB12, RB13, and RB19 and that it can disrupt the complexes that these mutants form with E2F. Moreover, E7 appears to target these pRB variants for degradation. Thus, the G1 accumulation that these mutant proteins cause in HeLa cells does not seem to be caused by a wholescale resistance to inactivation by E7. Possible explanations of the properties of these mutants are outlined in the Discussion.
Lysine residues on the B pocket interact with acidic amino acids on E7.
We next examined the mutant alleles that failed to bind in a stable manner to GST-E7. Several of these alleles affect residues in the LXCXE-binding cleft and are known to disrupt E7 binding (11). However, this group of mutants includes RB37, RB28a, and RB28b, which contain a combination of substitutions that have previously been proposed to affect the lysine patch on the pRB pocket (5, 20). Since this allele includes a mutation of lysine 713, a residue that is proposed to contribute LXCXE binding (31), it was unclear whether the G1 arrest seen with this allele was due to the mutation of the lysine patch or to a synergistic combination of the LXCXE-binding cleft and the lysine patch. However, we observed that RB7, an allele that mutated just three lysine residues (K722E, K729E, and K740E), behaved in a similar way (RB7 increased the G1 population of HeLa cells but was compromised for E7 binding), raising the possibility that these lysine residues might be important for E7 binding.
This result is intriguing given that HPV E7, SV40 TAg, and adenovirus E1A (Fig. 5A) contain a cluster of acidic amino acids immediately C terminal to the LXCXE motif. We have previously suggested that the orientation of the E7-derived LXCXE peptide in the cocrystal with the small pocket of pRB indicates that these acidic amino acids would be close to the conserved basic patch on pRB (11). Mutation of lysines 722, 729, and 740 also disrupted the interaction between pRB and either TAg or E1A (data not shown).
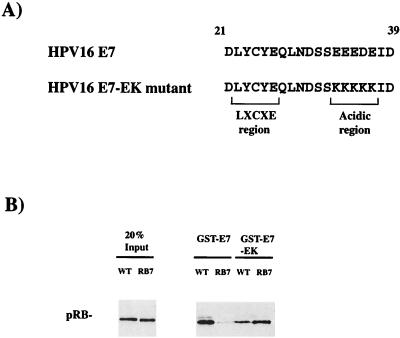
FIG. 5.
Basic residues on the surface of the pRB pocket domain make important contacts with acidic residues on E7. (A) A well-conserved region of acidic amino acids C-terminal to the LXCXE motif were altered to be lysines in the E7-EK mutant. (B) Binding of wild-type- and RB7-encoded forms of pRB to GST-E7 or the GST-E7-EK mutant was determined by Western blotting pRB that coprecipitated with the indicated GST fusion proteins.
While the notion that these conserved acidic residues might interact with the conserved lysine patch on pRB is attractive, the proximity of K722, K729, and K740 to the LXCXE-binding cleft raises a concern. It is possible that mutation of K722, K729, and K740 might, in some unintended way, compromise the LXCXE-binding cleft. Thus, the lack of E7 binding by the RB7 mutant might reflect a structural change in the LXCXE-binding site rather than a positive role for the lysine patch in E7 binding.
To distinguish between these possibilities, we tested whether the charged residues provide an ionic interaction between pRB and E7 by interchanging the charged amino acids on the respective molecules. A mutant E7 was constructed in which the acidic glutamate residues at positions 33, 34, 35, and 37 and the aspartate residue at position 36 were changed to lysine residues (E7-EK, Fig. 5A). If mutation of the lysine residues disrupts the LXCXE cleft of pRB, then the changes in E7 should have no effect on the interaction. If, on the other hand, there is an ionic interaction between the charged amino acids, then the complementary changes might restore the interaction.
As shown in Fig. 2 and 5B, RB7 with lysine-to-glutamate changes at amino acids 722, 729, and 740 is defective in binding to GST-E7 relative to wild-type pRB. In addition, GST-E7-EK shows reduced binding to wild-type pRB, relative to GST-E7. However, a strong interaction was observed when pRB7 and GST-E7-EK were incubated together. This interaction indicates that the intrinsic ability of pRB to bind to E7 was not destroyed by mutation of K722, K729, and K740, and it reveals the discrete nature of the contact between these charged residues. A derivative of the RB7 charge swap allele containing an additional mutation in the LXCXE-binding cleft (RB17) was unable to bind to GST-E7-EK (data not shown), confirming that the interaction still depended on the integrity of the LXCXE-binding cleft. We conclude that the lysine patch provides a second binding site that is distinct from the LXCXE-binding cleft and contributes to the affinity of the pRB-E7 interaction.
DISCUSSION
Disruption of the LXCXE cleft region of the pRB pocket renders these mutants resistant to E7 inactivation (11). LXCXE cleft mutants retain sufficient function, at least in the context of protein overexpression, to be able to arrest HeLa cells in G1. In this report we have extended this analysis to include a more extensive set of RB-1 mutants. Here we show that the ability of E7 to override pRB does not depend simply on the LXCXE-binding cleft; indeed, there are at least two other classes of RB-1 mutants that give a similar phenotype.
In addition to the LXCXE-binding site, the lysine patch appears to be important for E7 to associate with pRB. We infer that the ability of lysine patch mutants to block cell cycle progression in HeLa cells stems from the fact that these mutants are not efficiently targeted by E7. Charge-swap experiments suggest that the lysine patch interacts with a stretch of acidic amino acids that are just C-terminal to the LXCXE motif and are well conserved in viral oncoproteins.
In some respects, the discovery of this binding site is surprising. Single amino acid substitutions of E26 in the LXCXE motif have been shown to disrupt E7 binding (36). In addition, mutational analysis of the LXCXE-binding cleft shows that this eliminates the vast majority of the pRB-viral oncoprotein interaction (5, 7, 8, 11). A recent study that examined the interaction of the pRB pocket with an N-terminal fragment of SV40 TAg revealed that this region of TAg is in an extended conformation (29). Kim et al. (29) suggest that the lysine residues on the B portion of the pRB pocket may be too distant to make a significant contact. However, since the fragment of TAg in these crystals is truncated just C-terminal to the acidic sequence, it is not clear that this region is correctly represented.
It should be noted that none of the studies cited above preclude the possibility of an additional contact site that is necessary but not sufficient for the pRB-E7 interaction. Indeed, several other lines of evidence strongly suggest that additional contacts are likely to exist. Competition experiments have shown that CR2-derived peptides have only a fraction of the pRB-binding activity of the full-length E7 protein (28). Dissociation constants that were determined by using the small pocket domain of pRB confirm that E7 has a 100-fold-higher affinity for the pRB pocket than a 9mer peptide containing only the LXCXE (31). At least one additional contact has been mapped to the C-terminal zinc finger domain of E7 and is believed to interact with the C-pocket of pRB (39). In functional studies of E1A mutants, the deletion of amino acids 133 to 142, which removes the acidic sequence, strongly reduced the transformation properties of E1A (47). A previous effort to investigate this region of E7 has shown that deletion of amino acids 35 to 37 reduces the ability of E7 to be phosphorylated by casein kinase II (36). The ΔEDE mutant used in this study may retain the ability to interact with pRB through the two-glutamate residues that remain at positions 33 and 34.
In addition to the LXCXE cleft and the nearby lysine patch, we find that mutations in a groove near to the interface between the A and B halves of the pocket also generate forms of pRB that affect HeLa cell cycle progression. Three such alleles—RB11, RB12, and RB13—contain changes in a small group of charged amino acids that are clustered together on the crystal structure and have been highly conserved during evolution. These mutant proteins possess many of the functions of the wild-type pRB; they efficiently repress E2F-dependent transcription and arrest Saos-2 cells in G1 when overexpressed (data not shown). Surprisingly, analysis of these RB-1 mutants failed to reveal any pronounced abnormality in their interaction with E7. Not only was E7 able to bind to these mutants, but it was able to efficiently compete their interaction with E2F and to target them for degradation. Nonetheless, these mutants clearly represent a different class of alleles than the LXCXE-binding cleft and lysine patch mutants (see Table 3).
TABLE 3.
Resistance of pRB mutants to E7a
| Region | Allele | Mutation(s) | % G1 increase in HeLa cells | Score
|
||
|---|---|---|---|---|---|---|
| E7 binding | E2F-E7 competition | E7-induced degradation | ||||
| None | Wild type | None | 0 | + | + | + |
| LXCXE cleft | RB9 | I753A, N757A, and M761A | 24.0 | − | − | − |
| A-pocket | RB13 | R544A and K548A | 8.9 | + | + | + |
| A-pocket | RB19 | Q575A and S576A | 13.8 | + | + | + |
| B-pocket | RB7 | K722E, K729E, and K740E | 21.9 | +/− | +/− | +/− |
| B-pocket | RB11 | L660A, T664A, and R668A | 11.1 | + | + | + |
| B-pocket | RB12 | K652A and R656A | 19.4 | + | + | + |
| Multiple | RbΔcdk | T246A, T350A, S601A, S605A, S781A, S787A, S788A, S800A, S804A, T814A, and T819A | 6.7 | + | ND | ND |
RB mutants were transfected into HeLa cells as in Table 1. Cell cycle analysis was performed as described previously. The percent increase in cells with 2N DNA content over that of the control is shown. Binding to HPV-16 E7, competition for binding between E7 and E2F, and E7-induced degradation are scored as “+” for the wild-type level interaction, “+/−” for an intermediate effect, and “−” for resistance. Amino acid numbers for the RbΔcdk allele refer to the mouse sequence. ND, not determined.
While further work is needed to determine the defect that is responsible for the properties of RB11, RB12, and RB13, some clues do exist. During the course of these experiments we noted that the expression of pRB mutants that lack cdk phosphorylation sites gives a modest G1 accumulation in HeLa cells that is similar to the effect seen with RB11, RB12, RB13, and RB19 (Table 3). pRB is normally inactivated by cdk phosphorylation, and these mutants have previously been proposed to represent a constitutively active form of pRB. This phosphorylation site mutant is not defective in binding viral oncoproteins (data not shown). The implication of this result is that mutations that extend or raise the activity of pRB can affect the HeLa cell cycle and increase the percentage of G1 cells, even though the mutant proteins are targeted by E7. Interestingly, Western blots of RB11, RB12, and RB13 expressed in subconfluent cultures of C33A cells showed an altered mobility on low-percentage SDS-PAGE, with a larger proportion of hypophosphorylated pRB than is seen with wild-type or the LXCXE cleft mutant RB9 (data not shown). This suggests that, although these mutations do not affect phosphorylation sites, the changes in RB11, RB12, and RB13 may affect the kinetics of pRB phosphorylation, perhaps generating “gain-of-function” RB-1 alleles. It is intriguing that Chan et al. have recently demonstrated that pRB is acetylated by p300 and that the lysine-to-glutamine mutations at residues 873 and 874 of pRB alter the phosphorylation of pRB in cycling cells (4). A number of mutations that we have studied in the A and B halves of the pocket alter conserved lysines and, if there is a general interrelationship between the acetylation of lysine residues and phosphorylation of pRB, it is quite plausible that these might affect the kinetics of pRB inactivation.
A second observation that may be relevant here is that the levels of RB11, RB12, and RB13 measured in sorted populations of transfected HeLa cells appear to be slightly higher than the wild-type pRB (Fig. 3B). Although this change is quite minor compared with the increase in the levels of the LXCXE cleft mutants, it is unclear what degree of pRB stabilization might be necessary to alter the cell cycle phasing of HeLa cells. Potentially, quite small changes in the levels of pRB might be significant if compounded by a change in the rate of inactivation. We suggest that the G1 accumulation caused by RB11, RB12, RB13, and RB19 may be due to a combination of small changes that slightly raise and extend the activity of pRB in HeLa cells.
There are several types of mutants that might be anticipated to score in the HeLa cell cycle arrest assay that we have not yet found. For example, mutants that bind to E7 but are not efficiently degraded or RB-1 mutants that form complexes with E2F that are not readily disrupted by E7 would be predicted to allow pRB to block the HeLa cell cycle. One of the limitations of the HeLa assay is that only RB-1 mutants that retain the ability to arrest the cell cycle are revealed, and this limits the spectrum of mutants that can be found. If the mutations that block the interaction with E7 or the degradation of pRB are generated by changing residues that are required for pRB function, then these mutants would not be active in HeLa cells. Nevertheless, one of the conclusions that can be drawn from this study is that the ability of E7 to inactivate pRB does not depend on a single surface. Instead, changes in several distinct surfaces on the protein can affect this process. This may have some therapeutic implications, since molecules that target these surfaces may be able to prevent E7 from efficiently inactivating pRB and may be able to restore some level of pRB function to virus-transformed cells. In this initial survey, we have examined mutations in just a small number of residues that are the most highly conserved on the surface of pRB. These represent only a small faction of the surface of the protein, and it is likely that further mutagenesis of this domain will reveal additional interesting classes of E7-resistant RB-1 mutants.
ADDENDUM
While this manuscript was being revised, Brown and Gallie (V. D. Brown and B. L. Gallie, Mol. Cell. Biol. 22:1390-1401, 2002) demonstrated that the conserved lysine patch surrounding the LXCXE cleft is necessary for a stable interaction between pRB and TAg.
Acknowledgments
We thank Bill Kaelin and Doug Dean for pRB expression constructs used in this study. We are also greatly indebted to Karl Munger for discussions and for generously providing E7 encoding plasmids. Advice from past and present members of the Molecular Oncology Lab at the MGH Cancer Center is also greatly appreciated.
F.A.D. is a fellow of the Leukemia and Lymphoma Society. This work was supported by funding from the NIH to N.J.D.
REFERENCES
- 1.Berezutskaya, R. A., B. Yu, P. Morozov, P. Raychaudhuri, and S. Bagchi. 1997. Differential regulation of the pocket domains of the retinoblastoma family proteins by the HPV16 E7 oncoprotein. Cell Growth Differ. 8:1277-1286. [PubMed] [Google Scholar]
- 2.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. [PubMed] [Google Scholar]
- 3.Brown, V. D., R. A. Phillips, and B. L. Gallie. 1999. Cumulative effect of phosphorylation of pRB on regulation of E2F activity. Mol. Cell. Biol. 19:3246-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, H. M., M. Krstic-Demonacos, L. Smith, C. Demonacos, and N. B. La Thangue. 2001. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat. Cell Biol. 3:667-674. [DOI] [PubMed] [Google Scholar]
- 5.Chan, H. M., L. Smith, and N. B. La Thangue. 2001. Role of LXCXE motif-dependent interactions in the activity of the retinoblastoma protein. Oncogene 20:6152-6163. [DOI] [PubMed] [Google Scholar]
- 6.Chellappan, S., K. B. Kraus, B. Kroger, K. Munger, P. M. Howley, W. C. Phelps, and J. R. Nevins. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and the human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 89:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, T.-T., and J. Y. J. Wang. 2000. Establishment of irreversible growth arrest in myogenic differentiation requires the RB LXCXE-binding function. Mol. Cell. Biol. 20:5571-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahiya, A., M. R. Gavin, R. X. Luo, and D. C. Dean. 2000. Role of the LXCXE binding site in Rb function. Mol. Cell. Biol. 20:6799-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeCaprio, J. A., J. W. Ludlow, J. Figge, J. Y. Shew, C. M. Huang, W. H. Lee, E. Marsilio, E. Paucha, and D. M. Livingston. 1988. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell 54:275-283. [DOI] [PubMed] [Google Scholar]
- 10.Demers, G. W., E. Espling, J. B. Harry, B. G. Etscheid, and D. A. Galloway. 1996. Abrogation of growth arrest signals by human papillomavirus type 16 E7 is mediated by sequences required for transformation. J. Virol. 70:6862-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dick, F. A., E. Sailhamer, and N. J. Dyson. 2000. Mutagenesis of the pRB pocket domain reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol. Cell. Biol. 20:3715-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 13.Dyson, N., P. Guida, K. Munger, and E. Harlow. 1992. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J. Virol. 66:6893-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 15.Ewen, M. E., J. W. Ludlow, E. Marsilio, J. A. DeCaprio, R. C. Millikan, S. H. Cheng, E. Paucha, and D. M. Livingston. 1989. An N-terminal transformation-governing sequence of SV40 large T antigen contributes to the binding of both p110Rb and a second cellular protein, p120. Cell 58:257-267. [DOI] [PubMed] [Google Scholar]
- 16.Figge, J., T. Webster, T. F. Smith, and E. Paucha. 1988. Prediction of similar transforming regions in simian virus 40 large T, adenovirus E1A, and myc oncoproteins. J. Virol. 62:1814-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez, S. L., M. Stremlau, X. He, J. R. Basile, and K. Munger. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 75:7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodwin, E. C., and D. DiMaio. 2000. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc. Natl. Acad. Sci. USA 97:12513-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodwin, E. C., L. K. Naeger, D. E. Breiding, E. J. Androphy, and D. DiMaio. 1998. Transactivation-competent papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J. Virol. 72:3925-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harbour, J., R. Luo, A. Dei Santi, A. Postigo, and D. Dean. 1999. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 98:859-869. [DOI] [PubMed] [Google Scholar]
- 21.Harlow, E., P. Whyte, B. J. Franza, and C. Schley. 1986. Association of adenovirus early region 1A proteins with cellular polypeptides. Mol. Cell. Biol. 6:1579-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heck, D. V., C. L. Yee, P. M. Howley, and K. Munger. 1992. Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc. Natl. Acad. Sci. USA 89:4442-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helt, A.-M., and D. A. Galloway. 2001. Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J. Virol. 75:6737-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, P. S., D. R. Patrick, G. Edwards, P. J. Goodhart, H. E. Huber, L. Miles, V. M. Garsky, A. Oliff, and D. C. Heimbrook. 1993. Protein domains governing interactions between E2F, the retinoblastoma gene product, and human papillomavirus type 16 E7 protein. Mol. Cell. Biol. 13:953-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurford, R., D. Cobrinik, M.-H. Lee, and N. Dyson. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11:1447-1463. [DOI] [PubMed] [Google Scholar]
- 26.Hwang, E. S., D. J. D. Riese, J. Settleman, L. A. Nilson, J. Honig, S. Flynn, and D. DiMaio. 1993. Inhibition of cervical carcinoma cell line proliferation by the introduction of a bovine papillomavirus regulatory gene. J. Virol. 67:3720-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, D., D. Thompson, and K. Munger. 1997. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology 239:97-107. [DOI] [PubMed] [Google Scholar]
- 28.Jones, R. E., R. J. Wegrzyn, D. R. Patrick, N. L. Balishin, G. A. Vuocolo, M. W. Riemen, J. D. Defeo, V. M. Garsky, D. C. Heimbrook, and A. Oliff. 1990. Identification of HPV-16 E7 peptides that are potent antagonists of E7 binding to the retinoblastoma suppressor protein. J. Biol. Chem. 265:12782-12785. [PubMed] [Google Scholar]
- 29.Kim, H.-Y., B.-Y. Ahn, and Y. Cho. 2001. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 20:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knudsen, E. S., and J. Y. J. Wang. 1997. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol. Cell. Biol. 17:5771-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, J.-O., A. A. Russo, and N. P. Pavletich. 1998. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature 391:859-865. [DOI] [PubMed] [Google Scholar]
- 32.Lukas, J., T. Herzinger, K. Hansen, M. C. Moroni, D. Resnitzky, K. Helin, S. I. Reed, and J. Bartek. 1997. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 11:1479-1492. [DOI] [PubMed] [Google Scholar]
- 33.Mavromatis, K. O., D. L. Jones, R. Mukherjee, C. Yee, M. Grace, and K. Munger. 1997. The carboxyl-terminal zinc-binding domain of the human papillomavirus E7 protein can be functionally replaced by the homologous sequences of the E6 protein. Virus Res. 52:109-118. [DOI] [PubMed] [Google Scholar]
- 34.Mittnacht, S. 1998. Control of pRB phosphorylation. Curr. Opin. Genet. Dev. 8:21-27. [DOI] [PubMed] [Google Scholar]
- 35.Mulligan, G., and J. Jacks. 1998. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 14:223-229. [DOI] [PubMed] [Google Scholar]
- 36.Munger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naeger, L. K., E. C. Goodwin, E. S. Hwang, R. A. DeFilippis, H. Zhang, and D. DiMaio. 1999. Bovine papillomavirus E2 protein activates a complex growth-inhibitory program in p53 negative HT-3 cervical carcinoma cells that includes repression of cyclin A and cdc25A phosphatase genes and accumulation of hypophosphorylated retinoblastoma protein. Cell Growth Differ. 10:413-422. [PubMed] [Google Scholar]
- 38.Pagano, M., M. Durst, S. Joswig, G. Draetta, and P. Jansen-Durr. 1992. Binding of the human E2F transcription factor to the retinoblastoma protein but not to cyclin A is abolished in HPV-16-immortalized cells. Oncogene 7:1681-1686. [PubMed] [Google Scholar]
- 39.Patrick, D. R., A. Oliff, and D. C. Heimbrook. 1994. Identification of a novel retinoblastoma gene product binding site on human papillomavirus type 16 E7 protein. J. Biol. Chem. 269:6842-6850. [PubMed] [Google Scholar]
- 40.Phelps, W. C., K. Munger, C. L. Yee, J. A. Barnes, and P. M. Howley. 1992. Structure-function analysis of the human papillomavirus type 16 E7 oncoprotein. J. Virol. 66:2418-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phelps, W. C., C. L. Yee, K. Munger, and P. M. Howley. 1988. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell 53:539-547. [DOI] [PubMed] [Google Scholar]
- 42.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 43.Stabel, S., P. Argos, and K. Philipson. 1985. The release of growth arrest by microinjection of adenovirus E1A DNA. EMBO J. 4:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Heuvel, S., and E. Harlow. 1993. Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262:2050-2054. [DOI] [PubMed] [Google Scholar]
- 45.Wang, J., A. Sampath, P. Raychaudhuri, and S. Bagchi. 2001. Both Rb and E7 are regulated by the ubiquitin proteasome pathway in HPV-containing cervical tumor cells. Oncogene 20:4740-4749. [DOI] [PubMed] [Google Scholar]
- 46.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 47.Whyte, P., H. E. Ruley, and E. Harlow. 1988. Two regions of the adenovirus early region 1A proteins are required for transformation. J. Virol. 62:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whyte, P., N. M. Williamson, and E. Harlow. 1989. Cellular targets for transformation by the adenovirus E1A proteins. Cell 56:67-75. [DOI] [PubMed] [Google Scholar]
- 49.Wu, E. W., K. E. Clemens, D. V. Heck, and K. Munger. 1993. The human papillomavirus E7 oncoprotein and the cellular transcription factor E2F bind to separate sites on the retinoblastoma tumor suppresoor protein. J. Virol. 67:2402-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.zur Hausen, H. 1999. Immortalization of human cells and their malignant conversion by high risk papillomavirus genotypes. Semin. Cancer Biol. 9:405-411. [DOI] [PubMed] [Google Scholar]