Abstract
The prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) is an enveloped virus with a bisegmented negative-strand RNA genome whose proteomic capability is limited to four polypeptides, namely, nucleoprotein; surface glycoprotein (GP), which is proteolytically processed into GP1 and GP2; polymerase (L); and a small (11-kDa) RING finger protein (Z). Using a reverse genetic system based on the ARM strain of LCMV, we have previously shown that Z has a strong inhibitory activity on LCMV minigenome transcription and RNA replication (T. I. Cornu and J. C. de la Torre, J. Virol. 75:9415-9426, 2001). In the present study, we have identified regions and specific amino acid residues within Z which contribute to its inhibitory activity on RNA synthesis mediated by the LCMV polymerase. Z proteins from different LCMV strains had similar inhibitory activities on the expression of the LCMV ARM minigenome, whereas the Z protein of the genetically more distantly related Tacaribe virus had an approximately 10-fold lower inhibitory activity on ARM minigenome expression. Results from the use of chimera proteins between Z and Xenopus Neuralized, a nonviral RING finger protein, indicated that the structural integrity of the Z RING domain (RD) was required but not sufficient for the inhibitory activity of Z. Serial deletion mutants of the N and C termini of Z showed that the N terminus (residues 1 through 16) and C terminus (residues 79 through 90) do not contribute to the Z inhibitory activity. A highly conserved tryptophan (W) residue located at position 36 in ARM-Z, next to the second conserved cysteine (C) residue of the Z RD, also contributed to the Z inhibitory activity.
Lymphocytic choriomeningitis virus (LCMV) provides one of the most widely used model systems to study viral persistence and pathogenesis (4, 5, 22-24, 26, 31). LCMV is the prototypic member of the family Arenaviridae that includes important human pathogens such as Lassa fever virus (LV) (6, 27, 28, 37, 39).
The LCMV genome consists of two negative-sense, single-stranded RNA segments, designated L and S, with approximate sizes of 7.2 and 3.4 kb, respectively (30, 33, 36, 37). Each RNA segment has an ambisense coding strategy, encoding two proteins in opposite orientations, separated by an intergenic region (33, 37). The S RNA segment directs synthesis of the three major structural proteins: the nucleoprotein, NP (∼63 kDa) (30, 33, 37), and the two virion glycoproteins, GP-1 (40 to 46 kDa) and GP-2 (35 kDa), which are derived from the posttranslational cleavage of a precursor polypeptide, GP-C (75 kDa) (33, 37, 38, 42). Tetramers of GP-1 and GP-2 make up the spikes on the virion envelope and mediate virus interaction with the host cell surface receptor (3, 8). The L RNA segment encodes a high-molecular-mass protein, L (∼200 kDa) (36), and a small (11-kDa) RING finger protein (Z) (35). L has the characteristic motifs conserved in all the viral RNA-dependent RNA polymerases (32) and is thought to be the main viral component of the arenavirus polymerase (12, 33, 37). The NP, the most abundant viral protein in virally infected cells, associates with the viral RNA to form the nucleocapsid, which is the template for the viral RNA polymerase (12, 13). The nucleocapsid associated with the viral polymerase constitutes the viral ribonucleoprotein, which is active in virus transcription and replication (12, 33, 37). As with other negative-strand RNA viruses, this ribonucleoprotein is the minimum unit of LCMV infectivity.
A system for the rescue of an LCMV minigenome, derived from the S segment of the Armstrong (ARM) strain (ARM S MG), has been described previously. Both transcription and replication, mediated by LCMV polymerase, were reconstituted by intracellular coexpression of ARM S MG and viral proteins from transfected plasmids (20). By using this system, it was shown that the 5′ and 3′ untranslated regions, together with the intergenic region of the ARM S RNA, are cis-acting signals sufficient to allow RNA synthesis mediated by LCMV RNA polymerase (20), though it remains to be determined which of these cis-acting sequences are strictly required for viral replication and transcription. It was also demonstrated that NP and L are the minimal trans-acting viral factors required for replication and transcription of LCMV genome analogues (20). Z was not required for intracellular transcription and replication of the LCMV minigenome (20). Coexpression of Z protein exerted, in a dose-dependent manner, a potent inhibitory effect on intracellular transcription and replication of the LCMV minigenome (20).
Here, we investigate the contribution of different Z regions to the inhibition of LCMV minigenome expression. We show that the extent of the inhibition of ARM S MG expression by Z proteins from different arenaviruses is linked to the degree of their genetic relationship with ARM-Z. We present evidence that the RING finger domain (RD), although necessary, is not sufficient for the Z-mediated inhibition of ARM S MG expression. In addition, our results indicate that the N and C termini of ARM-Z, including amino acid (aa) residues 1 through 16 and 79 through 90, respectively, are not required for its inhibitory activity whereas a highly conserved tryptophan (W) residue present at position 36 in ARM-Z appears to play an important role in this inhibitory activity of Z.
MATERIALS AND METHODS
Cells and viruses.
Baby hamster kidney cells (BHK-21) were grown in Dulbecco's modified minimal essential medium supplemented with heat-inactivated (55°C for 30 min) 10% fetal calf serum, 2 mM l-glutamine, 1× tryptose-phosphate broth, 1 mM sodium pyruvate, and 0.5% glucose. Infections with LCMV were carried out using a plaque-isolated clonal virus population of strains Armstrong (ARM-5) and WE (WEc54) (25, 29), as well as Tacaribe virus (TV).
Plasmids.
Plasmids pLCMVSCAT2 (ARM S MG), pCITE-NP, pGEM-L, and pUCIRES-Z have been described previously (20).
Plasmids pUCIRES-WEZ and pCITE-TVZ express the Z proteins of LCMV strains WE and TV, respectively. The WE- and TV-Z open reading frames (ORFs) were obtained by reverse transcription (RT)-PCR using RNAs isolated from LCMV WE- and TV-infected BHK-21 cells, respectively. The predicted 270-bp (WE) and 300-bp (TV) PCR fragments were gel purified and digested with NcoI and StuI (WE) or with BamHI and XbaI (TV). The WE ORF was cloned into the NcoI-StuI sites of the plasmid pUCIRES (20), whereas the TV ORF was cloned into the BamHI-XbaI sites of plasmid pCITE2a(+) (Novagen).
Plasmid pUCIRES-MXZ contains the Z ORF of LCMV strain MX. The MX-Z ORF was excised with NcoI and SmaI from plasmid pBluescript-SKII-MXZ (15) and cloned into the NcoI-StuI sites of plasmid pUCIRES-Z (20).
Plasmids pUCIRES-Zmyc, pUCIRES-TVZmyc, and pUCIRES-LVZmyc were generated by c-myc tagging of the C termini of the LCMV Z, TV Z, and LV Z proteins, respectively, with a patch PCR approach (39). For the generation of plasmid pUCIRES-LVZmyc, the LV-Z ORF was amplified by RT-PCR with RNA isolated from LV-infected Vero cells. The 400-bp PCR fragment was gel purified, digested with NcoI and StuI, and inserted into the NcoI-StuI sites of pUCIRES. Infection of Vero cells with LV, RNA extraction, and RT-PCR to obtain LV-Z ORF were performed in biosafety level 4 facilities at the Centers for Disease Control and Prevention, Atlanta, Georgia.
Plasmid pGEM-XNeu contains a c-myc-tagged ORF of Xenopus Neuralize (XNeu) (11). The c-myc-tagged XNeu ORF was obtained from plasmid pCS2MT.XNeu (11) and cloned into the AvaI-SalI sites of plasmid pGEM-3Z (Promega).
Plasmids expressing XNeu-Z chimeric proteins were as follows. (i) Plasmid pGEM-NeuΔRD/ZRD expresses a modified XNeu protein in which its RD was replaced by the LCMV-Z RD. (ii) Plasmid pUCIRES-ZΔR/NeuRD expresses a modified LCMV Z protein where its RD was replaced by the RD of XNeu. The procedures used to generate these plasmids involved the use of a combination of patch PCR and overlap extension PCR procedures.
Mutants with deletions at either the N and/or C terminus of ARM-Z were obtained by patch PCR. This was performed with Pfu polymerase (Roche), and products were gel purified, digested with NcoI and StuI, and ligated into NcoI- and StuI-digested pUCIRES.
Plasmid pUCIRES-Z-A36, containing the mutation from W to A, was obtained by overlap extension PCR in two steps. In the first step, plasmid pUCIRES-Z was used as template in two independent PCRs. PCR products 1 and 2 contained residues 1 to 39 and 33 to 90, respectively, of ARM-Z. The overlapping segment of PCR product included the mutation from W to A at the amino acid residue at position 36. In the second step, the two PCR fragments were fused by overlap extension PCR. The final 300-bp PCR product was gel purified, digested with NcoI and StuI, and cloned into the NcoI-StuI sites of pUCIRES.
Primers and detailed descriptions of the procedures used to generate the plasmids used in this study, as well as the plasmids, are available from the authors upon request.
Analysis of minigenome expression.
The analysis of LCMV minigenome expression was done essentially as described previously (9, 20). Briefly, BHK-21 cells were infected with vFT7-3 at a multiplicity of infection (MOI) of 3 and transfected with pLCMVSCAT2 (0.5 μg), pCITE-NP (1.5 μg), pGEM-L (0.1 μg), pTMI-GFP (0.1 μg), and different amounts of plasmid DNA encoding ARM-Z, different Z mutants, and other arenavirus Z proteins, as indicated in the corresponding figure legends. Plasmid pTMI-GFP was included to assess the efficiency of transfection based on green fluorescence protein expression. In all cases, the amount of DNA used in the transfections was normalized to 2.7 μg/M6 well by using the pTMI plasmid. Transfections were performed using Lipofectamine (Life Technologies). 1-β-d-Arabinofuranosylcytosine (50 μg/ml) was included to inhibit the late phase of vaccinia virus expression. Cells were resuspended in 2 ml of phosphate-buffered saline and split in two equal aliquots. After the cells were spun, one aliquot was resuspended in 50 μl of 0.25 M Tris-HCl (pH 7.5) and cell lysates were prepared and assayed for chloramphenicol acetyltransferase (CAT) activity. The second aliquot was resuspended in lysis buffer (50 mM Tris-HCl [pH 8], 62.5 mM EDTA, 1% NP-40, 0.4% deoxycholate) and used for analysis of protein expression.
Western blot assay.
Cells were harvested in lysis buffer (50 mM Tris-HCl [pH 8], 62.5 mM EDTA, 1% NP-40, 0.4% deoxycholate) and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by blotting on an Immobilon-P polyvinylidene difluoride membrane (Millipore). Expression of LCMV WEc54-Z, LCMV ARM-Z, and the tryptophan LCMV ARM-Z mutant was detected with a rabbit serum raised against LCMV-Z. The rabbit serum raised against Z was obtained by immunizing rabbits with an N-terminal peptide of ARM-Z (GQGKSREEKGTNSTNRAEI) coupled to keyhole limpet hemocyanin. LCMV MX-Z was detected with a rabbit serum raised against MX-Z (courtesy of J. Pastorek). The presence of antigen-antibody complexes was detected by incubation with an anti-rabbit peroxidase antibody (Roche).
Mutant proteins that were fused to a c-myc tag were detected with a mouse anti-c-myc antibody, followed by incubation with a peroxidase-conjugated anti-mouse antibody (Roche). Proteins were visualized by enhanced chemiluminescence (Roche).
CAT assays.
Whole-cell extracts were prepared by three freeze-thaw cycles in a dry ice-ethanol bath and a 37°C water bath. Cell extracts were clarified by centrifugation at 12,000 × g for 5 min at 4°C. Equal amounts (5 μl) of each sample were incubated for 30 min at 37°C in the presence of 0.25 M Tris (pH 7.8), 0.6 mg of acetyl coenzyme A (Roche)/ml, and 0.05 μCi of [14C]chloramphenicol (ICN). The reaction was stopped by the addition of 1 ml of ethyl acetate, and chloramphenicol was extracted by separating the phases by centrifugation. Nine hundred microliters of supernatant was dried, resuspended in 25 μl of ethyl acetate, and then analyzed by thin-layer chromatography (TLC). Samples were run for 30 min in CHCl3-methanol (95:5). The TLC plate was dried and exposed to an X-ray film or analyzed with a phosphorimager.
RESULTS
Effect of Z proteins from different LCMV strains and arenavirus species on the expression of ARM S MG.
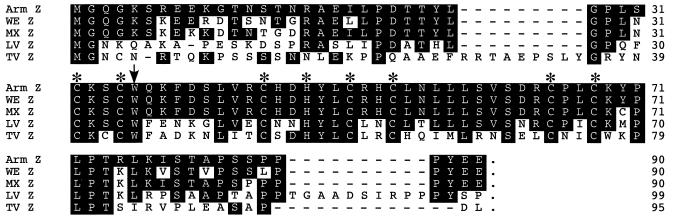
Comparison of the amino acid sequences of five different arenavirus Z genes (Fig. 1) showed a genetic relationship among these viruses consistent with that previously reported for the NP and GP genes. ARM-Z exhibited the strongest genetic relationship to the Z genes of LCMV strains MX and WE, followed by the Z genes of LV, another member of the Old World group arenavirus, and finally, the Z gene of TV, a more distantly related New World arenavirus.
FIG. 1.
Alignment of the Z protein sequences of LV, TV, and strains WE, MX, and ARM of LCMV. Sequences were aligned with respect to LCMV ARM-Z. Boxed residues are conserved with respect to ARM-Z. Asterisks indicate the Cys (C) residues involved in the formation of the RD. The conserved Trp (W) residue among arenavirus Z proteins is indicated by the arrow.
With the aim of gaining further insight into the specificity of the Z-mediated inhibition of RNA synthesis directed by the LCMV polymerase, we tested WE, MX, LV, and TV Z proteins for their potential inhibitory activity on RNA synthesis in the LCMV minigenome rescue assay.
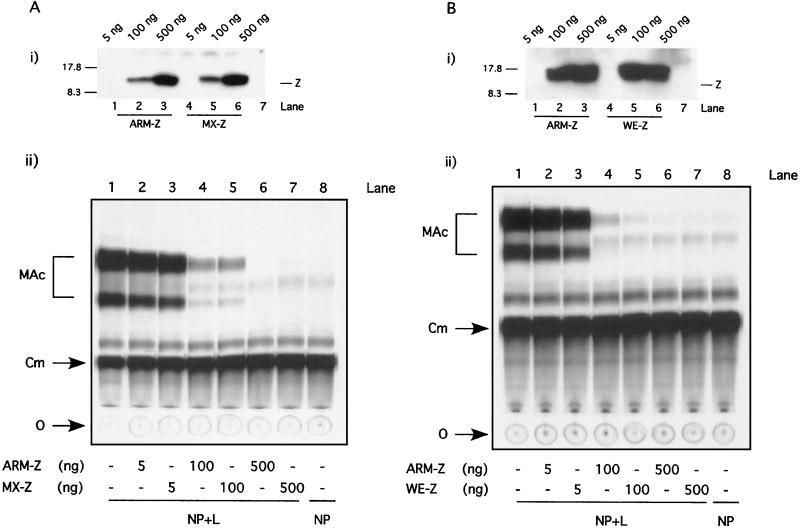
We initially examined whether Z proteins from LCMV strains MX and WE, genetically very closely related to ARM, were also able to affect the activity of the ARM S MG. We first verified under our experimental conditions that the WE and MX Z proteins could be expressed at levels similar to those for ARM-Z. For this, we transfected BHK-21 cells with increasing amounts of plasmids expressing ARM, MX, or WE Z proteins and examined Z expression levels by Western blot analysis using a rabbit serum raised against either LCMV ARM-Z or MX-Z. Both sera recognized the WE Z protein equally well. To recreate the same experimental conditions used for the minigenome rescue assay, these transfections were also done to include expression plasmids for the NP and L proteins, as well as the ARM S MG. Both MX and WE Z proteins were expressed at levels similar to those for ARM-Z (Fig. 2, panels i). Next, we tested the effect of these proteins on the expression of ARM S MG, as determined by levels of MG-mediated CAT activity. Both MX and WE Z proteins inhibited ARM S MG expression to the same extent as ARM-Z protein (Fig. 2, panels ii).
FIG. 2.
Inhibitory activity of MX- and WE-Z proteins compared to that of ARM-Z. BHK-21 cell monolayers were infected with vFT7.3 and subsequently cotransfected with ARM S MG, pCITE-NP, pGEM-L, and increasing amounts of pUCIRES-Z or pUCIRES-MXZ (A), or pUCIRES-WEZ (B). Cells were harvested at 24 h posttransfection and divided into two equivalent aliquots. One aliquot was used for analysis of Z expression by Western blot analysis (panels i). Plasmid pGEM-L was omitted in lanes 7. Size markers are indicated on the left of the gels; the position of the Z proteins is indicated to the right. The other aliquot of each cell lysate was assayed for CAT activity (panels ii) as described in Materials and Methods. O, origin; Cm, chloramphenicol; MAc, monoacetylated chloramphenicol.
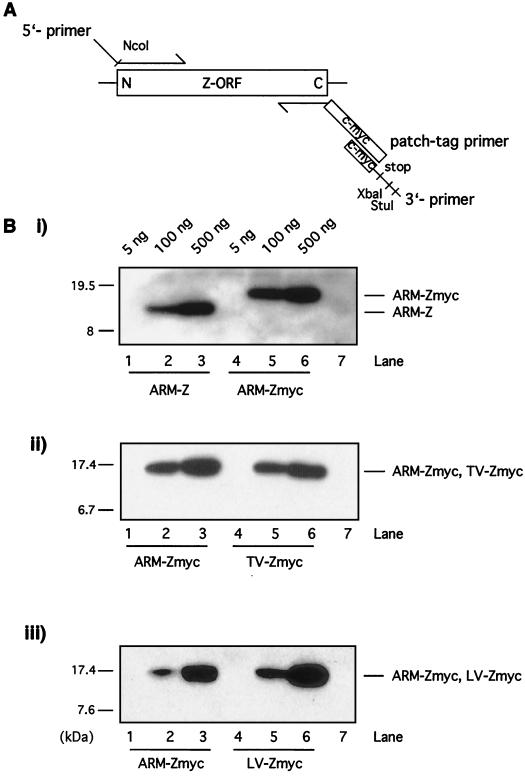
We then investigated whether the Z protein of other arenavirus species with different degrees of genetic distance from ARM could also affect the expression of ARM S MG. We tested the Z genes of LV and TV, members of New and Old World group arenaviruses, respectively. Since no antibodies were available to us for the detection of these Z proteins, both proteins were tagged at their C termini with a c-myc epitope. To ascertain that the introduction of the tag did not interfere with the function of Z being examined in our assay, we also assayed a c-myc-tagged LCMV ARM-Z as a control. The c-myc tag was added by patch PCR (Fig. 3A). Both ARM-Z and ARM-Zmyc were expressed at similar levels in transfected BHK-21 cells under the experimental conditions used for the minigenome rescue assay (Fig. 3B, panel i). This result allowed us to determine whether LV and TV Z proteins could be expressed, also under the experimental conditions of the minigenome rescue assay, at levels similar to those for ARM-Z. For this, we compared expression levels of ARM- Zmyc, LV-Zmyc, and TV-Zmyc by Western blot analysis using an antibody to the c-myc epitope used for the tagging of these Z proteins. Both LV and TV Z proteins were expressed at levels similar to those for ARM-Z (Fig. 3B, panels ii and iii).
FIG. 3.
(A) Patch PCR strategy. The c-myc tag was added by patch PCR using the following three primers: an upstream primer (5′ primer), complementary to the N terminus of the gene and containing a NcoI restriction site, designed to amplify the N terminus of the gene; a reverse primer (patch-tag primer), complementary to the C terminus of the gene, whose sequence replaced the Z stop codon by two Gly residues to form a hinge, followed by the nucleotide sequence of the 10 amino acids of the c-myc tag; and a third primer (3′ primer) with the same orientation as the patch-tag primer and containing the last 21 nucleotides of the c-myc tag, a stop codon, and the XbaI and StuI restriction sites. (B) Expression of ARM-Z, ARM-Zmyc, TV-Zmyc and LV-Zmyc proteins. BHK-21 cells were infected with vFT7.3 and cotransfected with ARM S MG, pCITE-NP, pGEM-L, and increasing amounts of the indicated plasmids: pUCIRES-Z, pUCIRES-Zmyc, pUCIRES-TVZmyc, or pUCIRES-LVZmyc. Plasmid pGEM-L was omitted in lanes 7. Cell lysates were prepared at 24 h posttransfection, resolved by SDS-16% PAGE, and assayed for expression of Z protein by Western blot analysis. (i) Detection of ARM-Z and ARM-Zmyc was performed with a rabbit anti-Z antibody. (ii) Detection of ARM-Zmyc together with TV-Zmyc was performed with a mouse anti-c-myc antibody. (iii) Detection of ARM-Zmyc together with LV-Zmyc was also performed with mouse anti-c-myc antibody. The positions of the Z proteins are indicated to the right of the gels; the molecular size markers are indicated to the left.
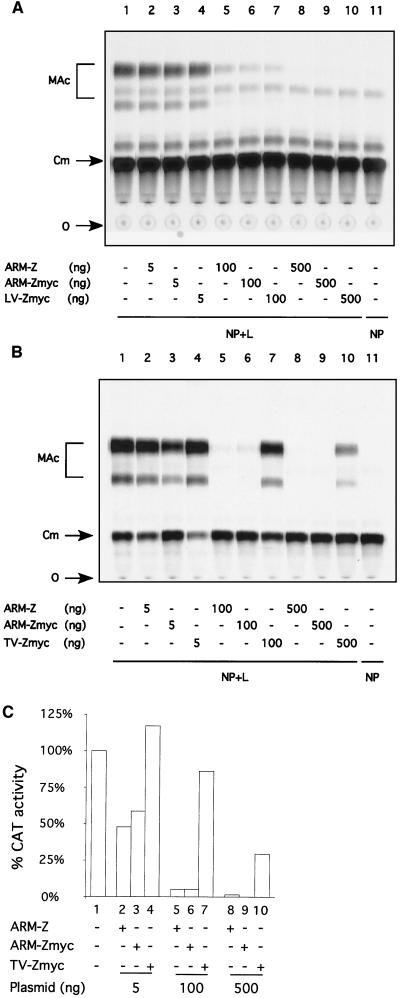
We next assessed the effect, compared to that of ARM-Z, of increasing amounts of the ARM-Zmyc, LV-Zmyc, and TV-Zmyc proteins on the expression of the LCMV minigenome. Both ARM-Z and ARM-Zmyc had the same ability to inhibit ARM S MG expression (Fig. 4 A through C, compare lanes 2, 5, and 8 with lanes 3, 6, and 9). Expression of similar levels of ARM-Zmyc or LV-Zmyc proteins resulted in the same degree of inhibition as for ARM S MG expression (Fig. 4A, compare lanes 3, 6, and 9 with 4, 7, and 10). In contrast, TV-Zmyc had a significantly lower inhibitory effect on the expression of ARM S MG than ARM-Zmyc (Fig. 4B, compare lanes 3, 6, and 9 with 4, 7, and 10). Quantitation of TLC results with a phosphorimager indicated that the inhibitory effect caused by transfecting 100 ng of TV-Zmyc plasmid was about 16-fold lower than that caused by the same amount of ARM-Zmyc plasmid (Fig. 4C). Levels of CAT activity in cells transfected with 500 ng of plasmid ARM-Zmyc were within background values (Fig. 4B and C, compare lanes 9 and 11), whereas lysates of cells transfected with 500 ng of plasmid TV-Zmyc exhibited about 30% of the CAT activity detected in the absence of Z (Fig. 4B and C, compare lanes 1, 9, and 10).
FIG. 4.
Comparison of LV-Z and TV-Z inhibitory activities with respect to ARM-Z. BHK-21 cell monolayers were infected with vFT7.3 and cotransfected with ARM S MG, pCITE-NP, pGEM-L, and increasing amounts of pUCIRES-Z, pUCIRES-Zmyc, pUCIRES-LVZmyc (A), or pUCIRES-TVZmyc (B), as indicated on the corresponding charts. Plasmid pGEM-L was omitted in lanes 11. Cell lysates were assayed for CAT activity as described in Materials and Methods. O, origin; Cm, chloramphenicol; MAc, monoacetylated chloramphenicol. (C) Levels of CAT activity were quantitated with a phosphorimager. The percent conversion obtained in the absence of L was considered to be nonspecific background; for the quantitation of CAT, we subtracted this background conversion value from the conversion values obtained from the other samples.
Contribution of the RD to Z-mediated inhibition of ARM S MG expression.
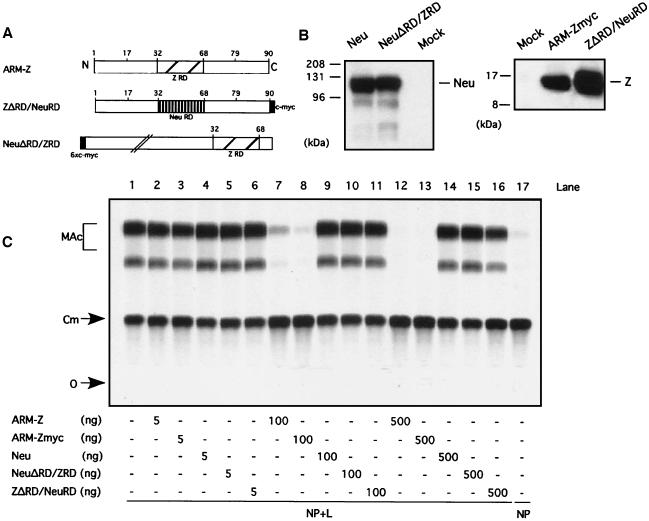
We have shown that mutations that disrupt the structure of the RD of ARM-Z result in elimination of the ARM-Z-mediated inhibition of ARM S MG expression. Likewise, RING finger proteins unrelated to LCMV-Z did not have any inhibitory effect on ARM S MG expression. However, the sole contribution of the RD to the Z inhibitory activity was not examined. The RDs of the LCMV ARM, MX, and WE Z proteins have identical amino acid sequences (Fig. 1). In contrast, the RDs of TV Z and LV Z proteins have 21 of 37 (56%) and 13 of 37 (35%) aa substitutions, respectively, with respect to ARM (Fig. 1). This led us to consider the possibility that the Z RD could specify mainly the inhibitory activity of LCMV-Z. To address this question, we sought to test the inhibitory activity of the ARM-Z RD per se in isolation from the rest of the ARM-Z protein. With this aim, we constructed chimeric proteins between ARM-Z and XNeu, a nonviral RING finger protein (11). In these chimeric proteins, the corresponding RDs were swapped (Fig. 5A). NeuΔRD/ZRD and ZΔRD/NeuRD correspond to proteins where the RDs of Neu and ARM-Z were replaced by the RDs of ARM-Z and Neu, respectively. This approach also allowed us to address the question of whether an RD from a protein unrelated to Z could completely, or partially, substitute for the Z RD with respect to the Z-mediated inhibitory activity of LCMV minigenome expression.
FIG. 5.
(A) Schematic of the different chimeric protein constructs. A c-myc tag (black box) was added by patch PCR to the C terminus of ZΔRD/NeuRD. The ARM-Z RD is indicated by a hatched box, whereas the Neu RD is indicated by a box with vertical stripes. Numbers refer to the amino acid position in ARM-Z. Neu and Z ORFs are about 600 and 90 aa residues long, respectively, although the RDs of both Z and Neu are similar in size. The schematic for NeuΔRD/ZRD is shown at a smaller scale than that for ZΔRD/NeuRD. (B) Neu and NeuΔRD/ZRD, as well as the ARM-Z chimera ZΔRD/NeuRD, are stably expressed. BHK-21 cell monolayers were infected with vFT7.3 and cotransfected with ARM S MG, pCITE-NP, pGEM-L, and 500 ng of pGEM-XNeu, pGEM-XNeuΔRD/ZRD, pUCIRES-Zmyc, or pUCIRES-ZΔRD/NeuRD. Lysates were harvested 24 h posttransfection and subjected to SDS-10% PAGE (for Neu proteins) and SDS-16% PAGE. Proteins were visualized by Western blot analysis with a mouse anti-c-myc antibody. Molecular size markers are indicated to the left of the gels; the positions of Neu and Z proteins are indicated to the right. (C) Expression of ARM S MG is not inhibited by either Neu or the chimeric proteins NeuΔRD/ZRD and ZΔRD/NeuRD. BHK-21 cell monolayers were infected with vFT7.3 and cotransfected with ARM S MG, pCITE-NP, pGEM-L, and increasing amounts of pUCIRES-Z, pUCIRES-Zmyc, pGEM-Neu, pGEM-NeuΔRD/ZRD, or pUCIRES-ZRD/NeuRD, as indicated below the gels. Plasmid pGEM-L was omitted in lane 17. Cell lysates were assayed for CAT activity as described in Materials and Methods. O, origin; Cm, chloramphenicol; MAc, monoacetylated chloramphenicol.
We first assessed whether Neu wild type (wt), as well as the chimeric proteins, could be expressed at levels similar to those of ARM-Z. For this, we used c-myc-tagged versions of Neu and ARM-Z, as well as the chimeric proteins. This approach facilitated the comparison of the expression levels among these proteins by Western blot analysis using the same antibody to the tag. The use of the same amount of each plasmid resulted in similar levels of expression of each of the four proteins, namely, Neu, ARM-Z, NeuΔRD/ZRD, and ZΔRD/NeuRD (Fig. 5B). We then tested the ability of the chimeric proteins to inhibit the expression of ARM S MG. As previously shown, ARM-Z and ARM-Zmyc proteins exhibited the same ability to inhibit ARM S MG expression (Fig. 5C, compare lanes 2, 7, and 12 with 3, 8, and 13). Neu failed to exhibit any inhibitory activity at the highest concentration of plasmid used (Fig. 5C, compare lanes 1 and 14). Likewise, NeuΔRD/ZRD and ZΔRD/NeuRD did not inhibit LCMV minigenome expression at the highest concentration of plasmid used (Fig. 5C, compare lane 1 with lanes 15 and 16).
Inhibitory activity of ARM Z proteins containing N- and C-terminal deletions.
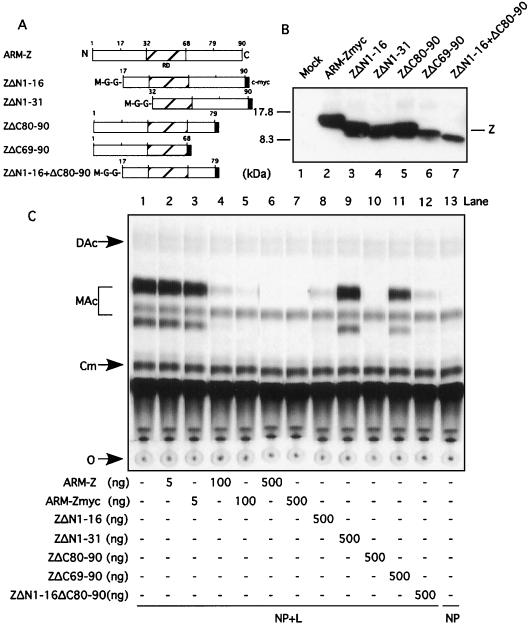
With the aim of further defining the regions of the Z protein involved in its inhibitory activity, we constructed a series of Z deletion mutants and tested them in the minigenome rescue assay. These mutants (Fig. 6A) included two N-terminal deletions containing aa residues 1 through 16 (ZΔN1-16) and aa 1 through 31 (ZΔN1-31); two C-terminal deletions including aa 80 through 90 (ZΔC80-90) and aa 69 through 90 (ZΔC69-90); and a mutant with both an N-terminal (aa 1 through 16) and a C-terminal (aa 80 through 90) deletion, designated ZΔN1-16+ΔC80-90. These truncations in the Z protein could affect their recognition by the rabbit anti-Z antibody. To overcome this problem and be able to have a reliable comparison of the expression levels of the different mutants, we generated these mutants containing a c-myc tag at their C termini.
FIG. 6.
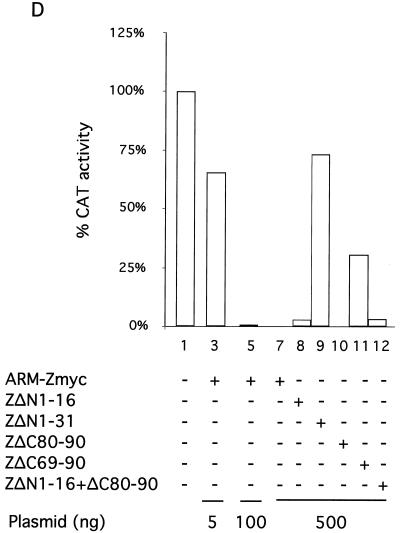
(A) A schematic of the different ARM-Z deletion mutants is shown. In all mutants, a c-myc tag (black box) was introduced by patch PCR at the corresponding C terminus. We also introduced a Met, followed by a Gly hinge, at the N terminus of each deletion mutant. (B) ARM-Z deletion mutants are stably expressed. BHK-21 cell monolayers were infected with vFT7.3 and cotransfected with 0.5 μg of pUCIRES-Z or the corresponding deletion mutants, as indicated at the bottom. Lysates were harvested 24 h posttransfection and subjected to SDS-PAGE. Proteins were visualized by Western blot analysis with a mouse anti-c-myc antibody. Molecular size markers are indicated to the left of the gels; the position of the Z protein is indicated to the right. (C) Proteins with large deletions at either the N or C terminus of ARM-Z lose their ability to inhibit pLCMVSCAT2 minigenome expression. BHK-21 cell monolayers were infected with vFT7.3 and cotransfected with ARM S MG, pCITE-NP, pGEM-L, and increasing amounts of pUCIRES-Z, pUCIRES-Zmyc, or 500 ng of each deletion mutant as indicated on the bottom of the panel. Plasmid pGEM-L was omitted in lane 13. Cell lysates were assayed for CAT activity as described in Materials and Methods. O, origin; Cm, chloramphenicol; MAc, monoacetylated chloramphenicol; DAc, diacetylated chloramphenicol. (D) Quantification of CAT activity measured by a phosphorimager is shown. The percent conversion obtained in the absence of L was considered to be nonspecific background; for the quantitation of CAT, we subtracted this background conversion value from the conversion values obtained from the other samples.
Expression levels of ARM-ZΔN1-16, ARM-ZΔN1-31, and ARM-ZΔC80-90 were similar to those of ARM-Zmyc (Fig. 6B, compare lanes 2 through 5). Expression levels of mutants ARM-ZΔC69-90 and ARM-ZΔN1-16+ΔC80-90 were slightly lower than those seen with ARM-Zmyc (Fig. 6B, compare lane 2 with lanes 6 and 7). Nevertheless, these expression levels were well within the range previously found to be required for inhibition of ARM S MG expression by ARM-Z. Deletion of the first 31 N-terminal aa or last 21 C-terminal aa resulted in Z proteins with about 30 and 70%, respectively, of the inhibitory activity exhibited by Z wt (Fig. 6C and D, compare lanes 5, 9, and 11). In contrast, Z proteins lacking the first 16 N-terminal aa or the last 11 C-terminal aa had an inhibitory activity on the expression of ARM S MG similar to that of the Z wt (Fig. 6C and D, compare lanes 7, 8, and 10). Deletion of the first 16 N-terminal aa together with the last 11 C-terminal aa resulted in a Z protein with an inhibitory activity similar to that for the Z lacking only the first 16 N-terminal aa (Fig. 6C and D, compare lanes 8 and 12).
Role of a highly conserved W residue in arenavirus Z protein in Z-mediated inhibition of RNA synthesis mediated by LCMV polymerase.
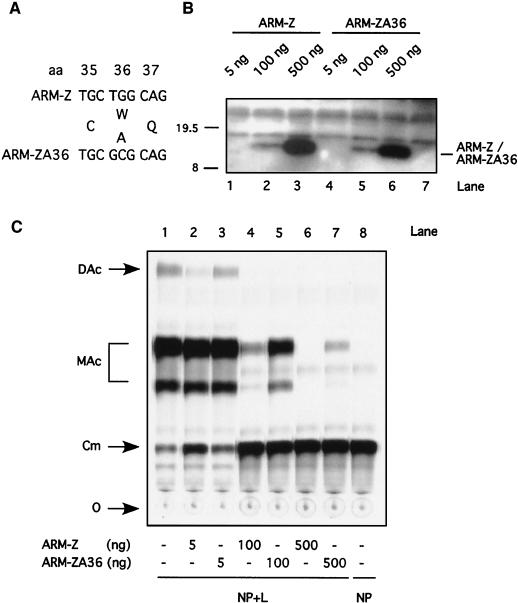
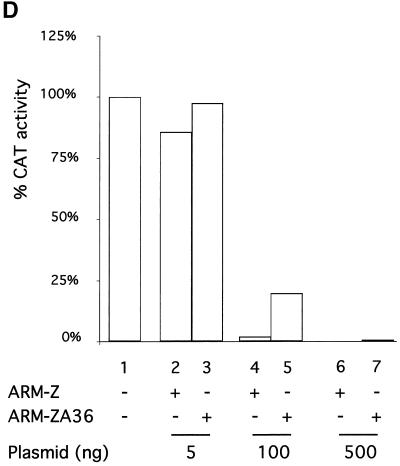
Only the few residues thought to maintain the RING finger structural scaffold are conserved among different unrelated RING proteins. A subset of RING proteins implicated in protein degradation has been shown to have a conserved W residue at a position where other hydrophobic amino acids are otherwise found. Inspection of the arenavirus Z sequences currently available also revealed the presence of a conserved W next to the second conserved cysteine (C) residue of the RD (Fig. 1). We therefore explored the requirement of this conserved W for the Z-mediated inhibition of ARM S MG expression. For this purpose, we generated an LCMV ARM-Z mutant (ARM-ZA36) in which the conserved W residue was replaced by alanine (A) (Fig. 7A). We first verified that both ARM-Z and ARM-ZA36 could be expressed at similar levels (Fig. 7B). We then tested the inhibitory activity of ARM-ZA36 in the minigenome rescue assay. The single substitution of the A for the conserved W resulted in a Z protein with a significantly reduced ability to inhibit the expression of the ARM S MG (Fig. 7C and D, compare lanes 4 and 5).
FIG. 7.
(A) Nucleotide sequence of the Z-A36 mutant is shown. Z-A36 contains two point mutations at codon position 36 (T to G and G to C), resulting in the amino acid change from Trp (W) to Ala (A). (B) Expression levels of Z-A36 are similar to those of Z wt. BHK-21 cells were infected with vFT7.3 and cotransfected with ARM S MG, pCITE-NP, pGEM-L, and different amounts of pUCIRES-Z or pUCIRES-Z-A36 as indicated above the gel. Plasmid pGEM-L was omitted in lane 7. Cell lysates were analyzed by Western blot analysis with a rabbit anti-Z antibody as described in Materials and Methods. The position of the Z proteins is indicated to the right of the gels, and molecular size markers are indicated to the left. (C) Inhibitory activity of Z-A36 is shown. Aliquots of the same cell lysates described for panel B were assayed for CAT activity as described in Materials and Methods. O, origin; Cm, chloramphenicol; MAc, monoacetylated chloramphenicol; DAc, diacetylated chloramphenicol. (D) Quantification by phosphorimager of the results shown in panel C. The percent conversion obtained in the absence of L was considered to be nonspecific background; for the quantitation of CAT, we subtracted this background conversion value from the conversion values obtained from the other samples.
DISCUSSION
The L segments of all arenaviruses genetically characterized so far direct the synthesis of a small RING finger protein called Z. The degree of genetic conservation among arenavirus Z proteins shows overall a similar trend to that previously reported based on the sequence of the NP and GP viral genes. However, the role of the Z protein in the arenavirus life cycle is at present poorly understood. There is not any obvious counterpart of Z present among other known negative-strand RNA viruses, which precludes inference of its potential functions based on findings for other viruses. Biochemical studies have shown that Z is a structural protein of the virion and also suggested that Z might be the arenavirus counterpart of the matrix protein found in other negative-strand RNA viruses (33, 34). Moreover, results from in vitro transcription and immunodepletion studies implicated the TV-Z in both mRNA synthesis and genome replication (14). In contrast, Z was not required for RNA replication and transcription of ARM S MG mediated by the LCMV polymerase. Moreover, Z exhibited a dose-dependent inhibitory effect on both transcription and replication of ARM S MG (9). Similar findings have now been documented for TV-Z as well (21). The reasons for the discrepancy between results from in vitro transcription assays using cell extracts from TV-infected patients and those obtained with LCMV and TV RNA analogs remain to be clarified. The contribution to viral RNA synthesis of Z-interacting cellular factors may have influenced the results obtained following Z immunodepletion and reconstitution of infected cell extracts. It is worth noting that during the natural course of LCMV infection, we could not detect Z protein prior to 24 h postinfection, which postdated the onset of synthesis of L RNA (9). Thus, the expression of Z during the progression from the early to late phases of the LCMV life cycle may be highly regulated. Z may play different roles during the life cycle of LCMV. Its expression at low levels early during the life cycle of LCMV could have a negative regulatory effect on RNA synthesis that might contribute to the restricted replication and noncytopathic properties of many arenaviruses. Increased levels of Z protein at later times in the LCMV life cycle could be required for viral maturation and budding. Expression of high levels of Z protein earlier than needed during the viral life cycle could have a very strong negative impact on virus multiplication. This possibility was likely recreated in our minigenome rescue assay, where Z was expressed from the onset of viral RNA synthesis.
Z has been also shown to interact with the promyelocytic leukemia protein, leading to the relocation of promyelocytic leukemia nuclear bodies to the cytoplasm, which has been proposed to be responsible for the noncytolytic nature of LCMV (1, 2). Z also interacts with the eukaryotic initiation factor 4E (eIF4E), which has been implicated in the repression of protein synthesis in an eIF4E-dependent manner (7). Together, these observations raise intriguing questions about a spectrum of potential functions played by the Z protein in the biology of arenavirus.
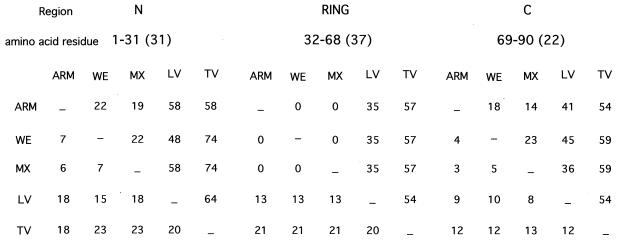
We have shown that the potent inhibitory effect of Z on intracellular transcription and replication of the LCMV minigenome required the structural integrity of the RD (9). The linear amino acid structure of LCMV-Z could be seen as composed of three modules: the N-terminal part comprising aa residues 1 through 31, the RD containing aa residues 32 through 68, and the C-terminal part including aa residues 69 through 90. Despite some minor differences in protein size, the same overall structure is also seen in other arenavirus Z proteins. The LCMV ARM, MX, and WE strains have complete amino acid identity within the Z RD, whereas the N- and C-terminal domains exhibit a lower level (about 80%) of amino acid identity (Fig. 1 and 8). The Z proteins of these three LCMV strains exhibited a similar inhibitory activity (Fig. 2), which may reflect a dominant role of the RD in the inhibitory activity of Z. Consistent with this view, the reduced inhibitory activity of TV-Z on ARM S MG expression (Fig. 4) may reflect the lower amino acid identity (43%) between the RDs of ARM and TV Z proteins. However, results obtained with the chimera proteins NeuΔRD/ZRD and ZΔRD/NeuRD (Fig. 5) indicate that the RD, although necessary, is not sufficient for Z-mediated inhibition of RNA synthesis mediated by the LCMV polymerase. In addition, results obtained with N- and C-terminal serial deletion mutants (Fig. 6) suggest that residues located within amino acid positions 16 through 31 and 69 through 79 of Z also contribute to its inhibitory activity on ARM S MG expression. It is worth noting that ARM and TV Z proteins have low amino acid identity within these two regions (Fig. 1 and 8). ARM and TV Z proteins differed in 12 of 16 and 7 of 11 residues comprising regions 16 through 31 and 69 through 79, respectively. In addition, within region 16 through 31, TV-Z has a 9-aa insertion with respect to ARM-Z.
FIG. 8.
Triangular matrix summarizing the total number (lower left) and percentage (upper right) of amino acid substitutions among different arenavirus Z proteins. The Z ORF was divided into three regions covering the amino acid residues indicated. Numbers within parentheses indicate the total number of amino acids included in each region.
Evidence indicates that many RING finger proteins can play a key role in the control of intracellular protein degradation by acting as ubiquitin (Ub) protein ligases (or E3s) (17, 18). E3s provide specificity to Ub conjugation by facilitating the transfer of Ub from a Ub-conjugating enzyme (Ubc or E2) to ubiquitination targets. A subset of RING proteins implicated in protein degradation has been shown to have a conserved W residue where other hydrophobic amino acids are otherwise found. Interestingly, we noticed the presence of a highly conserved W residue among the arenavirus Z proteins which is located next to the second conserved C residue of the Z RD (Fig. 1). Substitution of this W residue by A resulted in a LCMV-Z protein with reduced inhibitory activity (Fig. 7). One could then speculate that the Z inhibitory effect might be related to its potential E3 activity. This E3 activity would provide Z with the capability to specifically target for degradation proteins required at the early stages of RNA synthesis mediated by LCMV polymerase. Nevertheless, others and ourselves have failed to demonstrate an E3 activity associated with Z (9, 19). However, we cannot rule out the possibility that Z has an E3 activity that requires interaction with a different E2 not used in these assays or that Z itself is not a target for ubiquitination.
Recent findings have shown that regions around the first zinc-binding site of the Z RD interact with the dorsal surface of eIF4E (19). This interaction of Z with eIF4E significantly reduces the affinity of eIF4E for its substrate, the 5′,7-methyl guanosine cap of mRNA, and leads to inhibition of translation (19). Acting as a translational repressor, Z could affect expression of viral, or cellular, proteins required for viral RNA synthesis. This attractive model is difficult to reconcile with the observation that overexpression of Z did not affect cap-dependent expression of LCMV NP and L proteins (9). This model would predict a similar translational repressor activity for ARM and TV Z proteins, which contradicts the observed lower inhibitory activity of TV-Z on ARM S MG expression (Fig. 4).
It seems reasonable to think that the reduced inhibitory activity of TV-Z on ARM S MG expression reflects the influence of the amino acid differences between TV and LCMV-Z proteins on the interactions underlying Z-mediated inhibition of RNA synthesis by the LCMV polymerase. However, we tested TV-Z with a heterologous (LCMV) minigenome system. We therefore cannot rule out the possibility that the Z protein of TV also failed to efficiently inhibit RNA synthesis mediated by the TV polymerase, reflecting intrinsic differences between the properties of LCMV and TV Z proteins.
Homotypic viral interference can be readily demonstrated with several arenaviruses, including LCMV (41) and TV (16). This phenomenon is not strictly strain specific, as illustrated by the existence of interference among different pairs of LCMV strains. Heterotypic interference between arenaviruses has occasionally been reported, and the degree of this interference appears to be correlated with the genetic relationship between the viruses (10, 41). It is therefore tempting to speculate about the possibility that increased expression of Z protein during the virus life cycle might contribute to block replication of an additional infection by a genetically closely related arenavirus. Superinfection exclusion could also influence arenavirus evolution and help explain the observed population partitioning in the field, resulting in the maintenance of an independent evolutionary lineage of the same strain within a small geographic range.
When we compared the inhibitory activity associated with the different arenavirus Z proteins and their corresponding mutants, we exerted great care to minimize the confounding factor related to differences in levels of expression of the Z proteins being compared. This was facilitated by the use of tagged Z proteins after we verified that the c-myc tag used did not influence the activity of the Z under investigation. The elucidation of the precise mechanisms whereby Z exerts its inhibitory activity on viral RNA synthesis will contribute to a better understanding of the molecular biology of arenaviruses. It may also provide new insights aimed at the development of efficient antiviral strategies for highly pathogenic viruses such as LV and other arenaviruses known to cause hemorrhagic fevers. Identifying the domains and amino acid residues contributing to the inhibitory activity of Z on viral RNA synthesis will facilitate this task. The results presented here represent a first and necessary step toward such a goal.
Acknowledgments
We are indebted to J. Pastorek for the MX-Z cDNA and the rabbit serum to MX-Z, G. Deblandre for the XNeu DNA, and members of the Special Pathogens Branch at the CDC for conducting the RT-PCR experiment under biosafety level 4 conditions to obtain the full-length ORF of the LV-Z.
This work was supported by NIH grants AI47140 (J.C.T.) and AI09484 (T.I.C.).
Footnotes
This is publication 14641-NP from The Scripps Research Institute.
REFERENCES
- 1.Borden, K. L., E. J. Campbell Dwyer, and M. S. Salvato. 1998. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J. Virol. 72:758-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borden, K. L., E. J. Campbell Dwyer, and M. S. Salvato. 1997. The promyelocytic leukemia protein PML has a pro-apoptotic activity mediated through its RING domain. FEBS Lett. 418:30-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borrow, P., and M. B. Oldstone. 1994. Mechanism of lymphocytic choriomeningitis virus entry into cells. Virology 198:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Borrow, P., and M. B. A. Oldstone. 1997. Lymphocitic choriomeningitis virus, p. 593-627. In N. Nathanson (ed.), Viral pathogenesis, 1st ed., vol. 1. Lippincott-Raven, Philadelphia, Pa.
- 5.Buchmeier, M. J., and A. J. Zajac. 1999. Lymphocitic choriomeningitis virus, p. 575-605. In R. Ahmed and J. Chen (ed.), Persistent viral infections, 1st ed., vol. 1. John Wiley & Sons, New York, N.Y.
- 6.Buckley, S. M., J. Casals, and W. G. Downs. 1970. Isolation and antigenic characterization of Lassa virus. Nature 227:174.. [DOI] [PubMed] [Google Scholar]
- 7.Campbell Dwyer, E. J., H. Lai, R. C. MacDonald, M. S. Salvato, and K. L. Borden. 2000. The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J. Virol. 74:3293-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, W., M. D. Henry, P. Borrow, H. Yamada, J. H. Elder, E. V. Ravkov, S. T. Nichol, R. W. Compans, K. P. Campbell, and M. B. Oldstone. 1998. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282:2079-2081. [DOI] [PubMed] [Google Scholar]
- 9.Cornu, T. I., and J. C. de la Torre. 2001. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J. Virol. 75:9415-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damonte, E. B., S. E. Mersich, and C. E. Coto. 1983. Response of cells persistently infected with arenaviruses to superinfection with homotypic and heterotypic viruses. Virology 129:474-478. [DOI] [PubMed] [Google Scholar]
- 11.Deblandre, G. A., E. C. Lai, and C. Kintner. 2001. Xenopus neuralized is a ubiquitin ligase that interacts with Xdelta1 and regulates notch signaling. Dev. Cell 1:795-806. [DOI] [PubMed] [Google Scholar]
- 12.Fuller-Pace, F. V., and P. J. Southern. 1989. Detection of virus-specific RNA-dependent RNA polymerase activity in extracts from cells infected with lymphocytic choriomeningitis virus: in vitro synthesis of full-length viral RNA species. J. Virol. 63:1938-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuller-Pace, F. V., and P. J. Southern. 1988. Temporal analysis of transcription and replication during acute infection with lymphocytic choriomeningitis virus. Virology 162:260-263. [DOI] [PubMed] [Google Scholar]
- 14.Garcin, D., S. Rochat, and D. Kolakofsky. 1993. The Tacaribe arenavirus small zinc finger protein is required for both mRNA synthesis and genome replication. J. Virol. 67:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibadulinova, A., V. Zelnik, L. Reiserova, E. Zavodska, M. Zatovicova, F. Ciampor, S. Pastorekova, and J. Pastorek. 1998. Sequence and characterisation of the Z gene encoding ring finger protein of the lymphocytic choriomeningitis virus MX strain. Acta Virol. 42:369-374. [PubMed] [Google Scholar]
- 16.Gimenez, H. B., and R. W. Compans. 1980. Defective interfering Tacaribe virus and persistently infected cells. Virology 107:229-239. [DOI] [PubMed] [Google Scholar]
- 17.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 18.Joazeiro, C. A., S. S. Wing, H. Huang, J. D. Leverson, T. Hunter, and Y. C. Liu. 1999. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286:309-312. [DOI] [PubMed] [Google Scholar]
- 19.Kentsis, A., E. C. Dwyer, J. M. Perez, M. Sharma, A. Chen, Z. Q. Pan, and K. L. Borden. 2001. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol. 312:609-623. [DOI] [PubMed] [Google Scholar]
- 20.Lee, K. J., I. S. Novella, M. N. Teng, M. B. A. Oldstone, and J. C. de la Torre. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 74:3470-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López, N., R. Jácamo, and M. T. Franze-Fernández. 2001. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J. Virol. 75:12241-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oldstone, M. B. 1991. Molecular anatomy of viral persistence. J. Virol. 65:6381-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oldstone, M. B. 1989. Viral persistence. Cell 56:517-520. [DOI] [PubMed] [Google Scholar]
- 24.Oldstone, M. B. 1998. Viral persistence: mechanisms and consequences. Curr. Opin. Microbiol. 1:436-441. [DOI] [PubMed] [Google Scholar]
- 25.Oldstone, M. B., R. Ahmed, M. J. Buchmeier, P. Blount, and A. Tishon. 1985. Perturbation of differentiated functions during viral infection in vivo. I. Relationship of lymphocytic choriomeningitis virus and host strains to growth hormone deficiency. Virology 142:158-174. [DOI] [PubMed] [Google Scholar]
- 26.Oldstone, M. B., R. Ahmed, J. Byrne, M. J. Buchmeier, Y. Riviere, and P. Southern. 1985. Virus and immune responses: lymphocytic choriomeningitis virus as a prototype model of viral pathogenesis. Br. Med. Bull. 41:70-74. [DOI] [PubMed] [Google Scholar]
- 27.Parodi, A. S., C. E. Coto, M. Boxaca, S. Lajmanovich, and S. Gonzalez. 1966. Characteristics of Junin virus, etiological agent of Argentine hemorrhagic fever. Arch. Gesamte Virusforsch. 19:393-402. [DOI] [PubMed] [Google Scholar]
- 28.Peters, C. J., M. B. Buchmeier, P. E. Rollin, and T. G. Ksiazek. 1996. Arenaviruses, p. 1521-1551. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 29.Riviere, Y., R. Ahmed, P. Southern, and M. B. Oldstone. 1985. Perturbation of differentiated functions during viral infection in vivo. II. Viral reassortants map growth hormone defect to the S RNA of the lymphocytic choriomeningitis virus genome. Virology 142:175-182. [DOI] [PubMed] [Google Scholar]
- 30.Riviere, Y., R. Ahmed, P. J. Southern, M. J. Buchmeier, F. J. Dutko, and M. B. Oldstone. 1985. The S RNA segment of lymphocytic choriomeningitis virus codes for the nucleoprotein and glycoproteins 1 and 2. J. Virol. 53:966-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salvato, M., P. Borrow, E. Shimomaye, and M. B. Oldstone. 1991. Molecular basis of viral persistence: a single amino acid change in the glycoprotein of lymphocytic choriomeningitis virus is associated with suppression of the antiviral cytotoxic T-lymphocyte response and establishment of persistence. J. Virol. 65:1863-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salvato, M., E. Shimomaye, and M. B. Oldstone. 1989. The primary structure of the lymphocytic choriomeningitis virus L gene encodes a putative RNA polymerase. Virology 169:377-384. [DOI] [PubMed] [Google Scholar]
- 33.Salvato, M. S. 1993. The Arenaviridae, 1st ed., vol. 1. Plenum Press, New York, N.Y.
- 34.Salvato, M. S., K. J. Schweighofer, J. Burns, and E. M. Shimomaye. 1992. Biochemical and immunological evidence that the 11 kDa zinc-binding protein of lymphocytic choriomeningitis virus is a structural component of the virus. Virus Res. 22:185-198. [DOI] [PubMed] [Google Scholar]
- 35.Salvato, M. S., and E. M. Shimomaye. 1989. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology 173:1-10. [DOI] [PubMed] [Google Scholar]
- 36.Singh, M. K., F. V. Fuller-Pace, M. J. Buchmeier, and P. J. Southern. 1987. Analysis of the genomic L RNA segment from lymphocytic choriomeningitis virus. Virology 161:448-456. [DOI] [PubMed] [Google Scholar]
- 37.Southern, P. J. 1996. Arenaviridae: the viruses and their replication, p. 1505-1551. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 38.Southern, P. J., M. K. Singh, Y. Riviere, D. R. Jacoby, M. J. Buchmeier, and M. B. Oldstone. 1987. Molecular characterization of the genomic S RNA segment from lymphocytic choriomeningitis virus. Virology 157:145-155. [DOI] [PubMed] [Google Scholar]
- 39.Speir, R. W., O. Wood, H. Liebhaber, and S. M. Buckley. 1970. Lassa fever, a new virus disease of man from West Africa. IV. Electron microscopy of Vero cell cultures infected with Lassa virus. Am. J. Trop. Med. Hyg. 19:692-694. [DOI] [PubMed] [Google Scholar]
- 40.Squinto, S. P., T. H. Aldrich, R. M. Lindsay, D. M. Morrissey, N. Panayotatos, S. M. Bianco, M. E. Furth, and G. D. Yancopoulos. 1990. Identification of functional receptors for ciliary neurotrophic factor on neuronal cell lines and primary neurons. Neuron 5:757-766. [DOI] [PubMed] [Google Scholar]
- 41.Welsh, R. M., and C. J. Pfau. 1972. Determinants of lymphocytic choriomeningitis interference. J. Gen. Virol. 14:177-187. [DOI] [PubMed] [Google Scholar]
- 42.Wright, K. E., R. C. Spiro, J. W. Burns, and M. J. Buchmeier. 1990. Post-translational processing of the glycoproteins of lymphocytic choriomeningitis virus. Virology 177:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]