Abstract
Murine gammaherpesvirus 68 (γHV68 [also known as MHV-68]) establishes a latent infection in mice, providing a small-animal model with which to identify host and viral factors that regulate gammaherpesvirus latency. While γHV68 establishes a latent infection in multiple tissues, including splenocytes and peritoneal cells, the requirements for latent infection within these tissues are poorly defined. Here we report the characterization of a spontaneous 9.5-kb-deletion mutant of γHV68 that lacks the M1, M2, M3, and M4 genes and eight viral tRNA-like genes. Previously, this locus has been shown to contain the latency-associated M2, M3, and viral tRNA-like genes. Through characterization of this mutant, we found that the M1, M2, M3, M4 genes and the viral tRNA-like genes are dispensable for (i) in vitro replication and (ii) the establishment and maintenance of latency in vivo and reactivation from latency following intraperitoneal infection. In contrast, following intranasal infection with this mutant, there was a defect in splenic latency at both early and late times, a phenotype not observed in peritoneal cells. These results indicate (i) that there are different genetic requirements for the establishment of latency in different latent reservoirs and (ii) that the genetic requirements for latency depend on the route of infection. While some of these phenotypes have been observed with specific mutations in the M1 and M2 genes, other phenotypes have never been observed with the available γHV68 mutants. These studies highlight the importance of loss-of-function mutations in defining the genetic requirements for the establishment and maintenance of herpesvirus latency.
The gammaherpesviruses are lymphotropic viruses that establish a lifelong infection of the host and are associated with cellular transformation and the development of malignancies. The gammaherpesviruses include the human pathogens Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV or HHV-8) (reviewed in references 21, 24, and 27). Previous analysis of EBV and KSHV has been largely restricted to in vitro analyses given the absence of small-animal models. Murine gammaherpesvirus 68 (γHV68 or MHV-68) is a member of the gammaherpesvirus family and is closely related to EBV and KSHV (15, 16, 45). γHV68 infection of inbred mice has been established as a tractable, genetically manipulable model with which to study gammaherpesvirus pathogenesis (reviewed in references 14, 23, 25, 32, 34, 36, and 47).
A hallmark of herpesvirus infection is the ability of these viruses to establish a latent infection, during which the viral genome is present as an episome within the nucleus of the infected cell in the absence of virus replication (29). During latent infection, there is very restricted transcription of the viral genome and this latent reservoir persists throughout the lifetime of the host. The gammaherpesviruses all possess the common property of establishing a latent infection in lymphocytes (29).
Upon infection of inbred mice, γHV68 establishes a long-term latent infection that can be detected in multiple tissues, including splenocytes, peritoneal cells, and lymph nodes (9, 38, 51). γHV68 may also establish a persistent infection in the lungs (37). Within these tissues, investigators have identified γHV68 latency in multiple cell types, including B cells, macrophages, and dendritic cells, with lung epithelium a site of viral persistence (17, 37, 39, 52). The relative contribution of these different cell types to successful long-term γHV68 infection is unknown.
To understand the mechanisms by which gammaherpesviruses maintain a latent infection while evading destruction by the host immune response, research has focused on genes transcribed during latency. Multiple groups have identified candidate latency-associated transcripts expressed during γHV68 latency (7, 18, 28, 33, 46). These include (i) immunomodulatory molecules, including a high-affinity chemokine-binding protein (the M3 gene product [26, 44]) and an inhibitor of major histocompatibility complex class I-restricted antigen presentation (the K3 gene product [35]); (ii) a viral bcl-2 homolog with demonstrated antiapoptotic activity (4, 30, 49); (iii) a gene potentially involved in maintenance of the latent viral episome (gene 73 is a homolog of the KSHV latency-associated nuclear antigen, which tethers the viral episome to host chromosomes [3, 12]); (iv) a viral G protein-coupled receptor that has been demonstrated to have transforming activity in vitro (48); and (v) genes of unknown function, including the viral tRNA-like genes (7) and the M2, M8, M9, and M12/M13 genes.
In particular, one latency-associated region of transcription contains the M2, M3, and viral tRNA-like genes. Both the M2 and M3 genes have been identified in at least two independent screens for latency-associated transcripts (18, 28, 33, 46). The M2 gene has no known homolog to any other cellular or viral gene but is known to encode a protein that is recognized by cytotoxic T lymphocytes during latent infection in BALB/c mice (18). Adoptive transfer of CD8+ T cells specific for M2, or vaccination against M2, results in a decreased latent load at early, but not late, times postinfection (42, 43). Notably, M2 is only transiently expressed in wild-type (wt) mice, during the peak of viral latency (day 14 postinfection) (42). Recent characterization of an M2-deficient γHV68 strain (γHV68-M2.Stop) has demonstrated that M2 is required for wt splenic viral titers after intraperitoneal (i.p.) infection and for wt levels of latently infected cells in the spleen following intranasal (i.n.) infection (19). The M3 gene encodes a high-affinity chemokine-binding protein (26, 44). Recently, a lacZ mutation within the M3 gene has been shown to result in a diminished frequency of latently infected cells in the spleen following i.n. infection (8). However, an M3 mutant virus containing a translational stop codon near the 5′ end of the M3 gene that ablates expression of a functional M3 protein does not have a defect in latency or reactivation (V. van Berkel, S. H. Speck, and H. W. Virgin IV, unpublished data). Based on the similarity between the phenotypes produced by a stop codon mutation in the M2 gene and the M3 lacZ mutation, the latent alterations in M3.LacZ may result from alterations in the neighboring M2 gene and not disruption of M3. This locus also contains the M1 gene. While transcription of the M1 gene has not been detected during latency, a deletion within the M1 gene results in increased reactivation from latency following i.p. infection (11).
Recently, Macrae et al. have characterized γHV76, a second gammaherpesvirus isolated in parallel with γHV68 (5, 22). This virus has a genome structure nearly identical to that of wt γHV68, except that it contains a 9.5-kb deletion that removes the M1, M2, M3, and M4 genes and eight viral tRNA-like genes (22). On the basis of their analysis of γHV76, the authors reported that this locus is important for acute pathogenesis, including acute replication and establishment of γHV68 latency in the spleen following i.n. infection. While γHV76 had a defect in splenic latency at early times, the authors only examined splenic latency after i.n. infection and did not perform a quantitative analysis of the frequency of viral genome-positive cells during latency.
Here we report the characterization of a spontaneous deletion mutant of γHV68 with a nearly identical 9.5-kb deletion at the left end of the genome that removes the M1, M2, M3, and M4 genes and all eight viral tRNA-like genes. Given the nature of this deletion, this virus lacks at least three candidate latency-associated genes. We have characterized the ability of this deletion mutant to establish a latent infection and reactivate from latency, examining splenocytes and peritoneal cells, following i.p. and i.n. infection, at early and late times. Based on these analyses, this viral mutant does not have an absolute defect in latency and reactivation but, instead, demonstrates tissue-specific and route-of-infection-dependent alterations in latency and reactivation.
MATERIALS AND METHODS
Viruses and tissue culture.
γHV68 strain WUMS (ATCC VR-1465) was used for all infections and is designated the wt γHV68 strain. γHV68 passage and tissue culture were performed as previously described (11).
Isolation of γHV68Δbp1-9473.
γHV68Δbp1-9473 was initially identified during the screen for a marker rescue virus for the M1.LacZ recombinant (see reference 11 for a description of the generation of the M1.LacZ recombinant and the M1 marker rescue virus). During this screen, candidate marker rescue isolates were identified by white plaque morphology. After two rounds of plaque purification, candidate marker rescue isolates that exhibited 100% white plaque morphology were inoculated onto monolayers of NIH 3T12 cells in six-well plates (Costar) with one-fifth to one-half of the volume picked during plaque purification. Virus was harvested 9 days postinfection and stored at −80°C. To extract DNA, samples were subjected to four freeze-thaw cycles and spun at 4°C for 30 min at 12,000 rpm in a Sorvall microcentrifuge. Samples were resuspended in Tris-EDTA (10 mM Tris, 1 mM EDTA) with 0.5% sodium dodecyl sulfate, and 0.5 mg of proteinase K per ml (TESP buffer) and incubated at 37°C for at least 8 h. Samples were subjected to at least three phenol-chloroform-isoamyl alcohol (25:24:1) extractions, followed by one chloroform-isoamyl alcohol (24:1) extraction. DNA was precipitated with 3 M sodium acetate and 2 to 3 volumes of 100% ethanol. Southern blot analysis was performed with a diagnostic EcoRV restriction digestion and a 32P-labeled M1 locus probe (bp 1,702 to 4,308 of γHV68 strain WUMS [45]). Southern blots and cloning were done in accordance with standard protocols (31). In certain cases, Southern blot assays were performed with a biotinylated probe and chemiluminescence detection (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, Md.).
Southern blot analysis of viral isolate 4.19 failed to demonstrate hybridization with the 32P-labeled M1 probe. Subsequent Southern blot analysis was performed with a panel of probes that revealed that this isolate did contain wt γHV68 sequences (with a probe for the gene 50 sequence from bp 67,540 to bp 68,814; data not shown). Southern blot analysis of the left end of the viral genome identified a large deletion, as shown in Fig. 1. This viral isolate was identified as γHV68Δbp1-9473, or γHV68Δ9473, as it will be referred to throughout the remainder of this report.
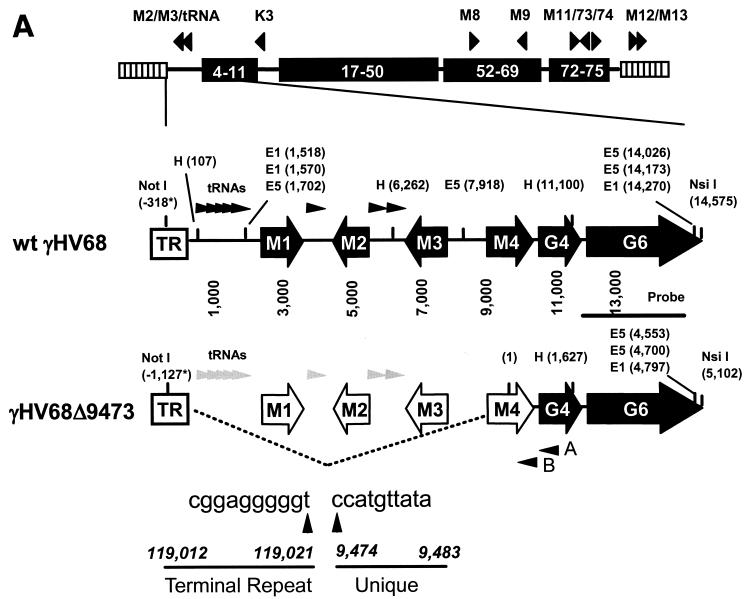
FIG.1.
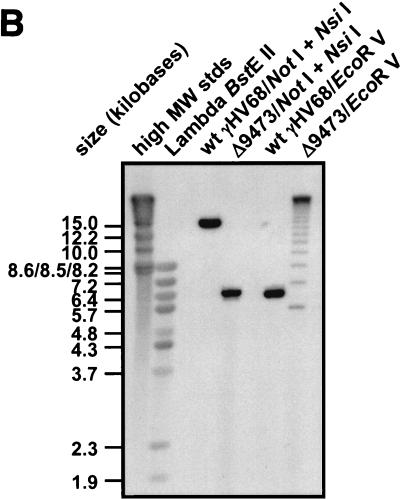
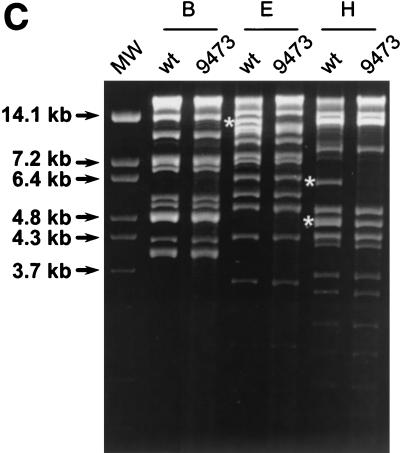
Identification of γHV68Δ9473, a spontaneous deletion mutant of γHV68. (A) Schematic of the wt γHV68 genome with candidate latency-associated transcripts indicated above the genome (see the text for details; tRNAs have been excluded for clarity) and corresponding genomic structures of wt γHV68 and γHV68Δ9473 at the left end of the viral genome. To define the structure of γHV68Δ9473, we sequenced across the junction of the γHV68Δ9473 deletion with two independent sequencing reactions and primers (primers A and B, as indicated). The sequence at this junction is indicated beneath the γHV68Δ9473 genome. Restriction sites: E5, EcoRV; E1, EcoRI; H, HindIII. TR, terminal repeat. Empty genes represent genes missing from γHV68Δ9473. The asterisks indicate that the wt γHV68 coordinate for the NotI site is based upon the available genomic sequence, with the γHV68Δ9473 coordinate for NotI based upon the sequencing of a genomic subclone. Both coordinates define the distance between the NotI site and the first base pair of the unique γHV68 sequence. (B) Southern blot analysis of wt γHV68 or γHV68Δ9473 genomic DNA restriction digested with either NotI-NsiI or EcoRV (E5), followed by hybridization with a biotinylated probe for the sequence surrounding and containing gene 6 (bp 11,100 to 14,026; indicated in Fig. 1A). Hybridization was detected with chemiluminescence detection. High-molecular-weight (MW) DNA standards (stds) (Gibco BRL) and BstEII-digested lambda DNA (New England Biolabs) were included on the blot and detected by hybridization with biotinylated lambda DNA, and fragment sizes (kilobases) are indicated to the left. (C) Comparison of restriction endonuclease digestion of 1.5 μg each of the wt γHV68 and γHV68Δ9473 genomic DNAs with BamHI (B), EcoRI (E), and HindIII (H). Digested viral DNA was electrophoresed on a 1.2% agarose gel and stained with ethidium bromide. Restriction fragments absent from γHV68Δ9473 are identified by asterisks on the agarose gel. MW, molecular weight markers.
After identification of this deletion, γHV68Δ9473 was plaque purified by three rounds on NIH 3T12 monolayers under methylcellulose as previously described (11). A small viral preparation was generated by infecting NIH 3T12 monolayers in a six-well plate with one-fifth of the volume of the final plaque pick. This sample was harvested at a 50% cytopathic effect (CPE) 5 days postinfection. A large viral stock was generated on NIH 3T12 monolayers infected with approximately 0.05 PFU per cell. Virus was harvested at a 50% CPE at 4 days postinfection, homogenized, clarified, and aliquoted for storage at −80°C. Stocks titers were independently determined by plaque assay at least twice. Infectious viral genomic DNA was isolated as previously described (45).
Sequence analysis.
To define the limits of the deletion contained within γHV68Δ9473, the gene 6-containing NotI-NsiI restriction fragment was subcloned from γHV68Δ9473 genomic DNA. To do this, γHV68Δ9473 genomic DNA was subjected to NotI-NsiI-XhoI triple digestion and restriction fragments between 3.7 and 7.2 kb were ligated into an EagI-PstI double-digested pBlueScriptKS+ vector lacking the NgoMIV restriction site. Miniprepped plasmids were screened by multiple restriction digestion, and the gene 6-containing plasmid was identified on the basis of the estimated size of the deletion. This plasmid was then sequenced (see below) using an alkaline lysis miniprep of plasmid DNA. Once the gene 6 sequence was confirmed, the plasmid was purified on a cesium chloride gradient and subjected to further sequencing. All sequencing reaction mixtures used the BigDye Terminator Cycle Sequencing Ready Reaction reagent (ABI PRISM Applied Biosystems, Foster City, Calif.). For each 10-μl sequencing reaction mixture, 300 to 400 ng of plasmid DNA was combined with 10 pmol of primer and 2 μl of ready reaction mix. Each sequencing reaction mixture was subjected to 30 to 50 cycles (sequencing cycle: 95°C for 10 s, 50°C for 5 s, and 60°C for 4 min) in a GeneAmp 9600 or 9700 (PE Applied Biosystems), precipitated, and submitted for sequencing. The sequence of the γHV68Δ9473 junction was obtained with two primers, i.e., primer A (5′-GGCACATGTTGTCACCATTT-3′), which hybridizes to bp 9,923 to 9,942 of wt γHV68, and primer B (sequence 5′-GAGTGTTTCCTCTTTAGACATCCAA-3′), which hybridizes to bp 9,614 to 9,638 of wt γHV68 (indicated in Fig. 1A). Sequence analyses were performed with Vector NTI version 5.0 (InforMax, Inc., Bethesda, Md.) and EditSeq 3.99 and SeqManII 3.6.0 for Windows (DNASTAR, Inc., Madison, Wis.).
In vitro replication.
NIH 3T12 cells were plated at 1.2 × 105 cells per well in six-well plates (Costar) 2 days prior to infection. At the time of infection, NIH 3T12 cells (6 × 105 to 1 × 106) were infected with 0.05 PFU per cell (multiplicity of infection = 0.05) of either wt γHV68 or γHV68Δ9473 in a 200-μl volume with plates rocked every 15 min for 1 h at 37°C. After this hour, complete Dulbecco’s modified Eagle medium (DMEM) was added to the monolayer. This time was defined as time zero postinfection. At the time of harvesting, medium was harvested from cultures for the supernatant fraction. The cell-associated fraction was collected by adding 1 ml of fresh, complete DMEM to the well and harvesting the monolayer with a cell lifter. Samples were stored at −80°C. Prior to plaque assay, cell-associated fractions were subjected to three freeze-thaw cycles; titers of supernatant samples were determined without further freeze-thaw cycles.
Mice, infections, and organ harvesting.
Female C57BL/6 mice were purchased from Jackson Laboratories or the National Cancer Institute and used at 8 to 15 weeks of age. Mice were maintained at Washington University, St. Louis, Mo., in accordance with all University and Federal guidelines. Mice were placed under metofane anesthesia prior to infection, as well as before sacrifice by cervical dislocation. Alternatively, mice were sacrificed by asphyxiation with carbon dioxide. All mice were infected with 106 PFU by i.p. injection in 0.5 ml of complete DMEM or with 4 × 105 PFU by i.n. injection in 40 μl of complete DMEM. Upon sacrifice, organs were placed in 1 ml of complete DMEM on ice and frozen at −80°C. Resident peritoneal cells were harvested by peritoneal lavage with 10 ml of complete DMEM containing 1% fetal calf serum. Sentinel mice were assayed every 3 months and were negative for adventitious mouse pathogens by serology.
Plaque assay.
Plaque assays were performed with NIH 3T12 monolayers under a Noble agar overlay as described previously (11). Organs in which titers were to be determined were thawed and homogenized with either a Ten Broeck tissue grinder or 1.0-mm silica beads and four 1-min rounds of mechanical disruption at maximum speed in a Mini-Beadbeater-8 (Biospec Products, Bartlesville, Okla.) (19). The limit of detection for this assay is 50 PFU/ml per organ (log10 1.7).
Limiting-dilution ex vivo reactivation assay.
Detection of γHV68 reactivation from latency was performed as previously described (11, 50, 51). Briefly, serial dilutions of latently infected cells were plated onto mouse embryonic fibroblast monolayers and scored for CPE after at least 21 days of coculture. To detect preformed infectious virus, parallel samples were mechanically disrupted as previously described (50). This disruption procedure kills >99% of the cells with, at most, a twofold effect on the titer of preformed infectious virus (50, 51). For all of the assays reported in this paper, disrupted samples were uniformly negative.
Limiting-dilution nested PCR detection of γHV68 genome-positive cells.
We determined the frequency of cells containing the γHV68 genome with a previously described nested PCR assay with modifications as described below (51, 52). This assay detects and amplifies gene 50 of γHV68 with ca. single-copy sensitivity in a background of 104 uninfected NIH 3T12 cells. Peritoneal cells and splenocytes were harvested from latently infected mice and frozen in 10% dimethyl sulfoxide at −80°C. At the time of assay, cells were thawed, counted, resuspended in an isotonic solution, and diluted in a background of 104 uninfected NIH 3T12 cells. A 5-μl volume of cell suspension was plated into 5 μl of lysis buffer in a 96-well PCR plate (MWG Biotech, High Point, N.C.), and plates were sealed with adhesive PCR foil (Eppendorf Scientific, Westbury, N.Y.) (51, 52). After 6 h of cell lysis at 56°C, proteinase K was inactivated at 95°C for 20 min. A 10-μl volume of round one PCR cocktail (25 mM KCl, 10 mM Tris-HCl [pH 9.0], 1.5 mM MgCl2, 0.4 mM deoxynucleoside triphosphates, 2 ng of the round 1 primers per μl, 0.1 U of Taq per μl) was added directly to each well and subjected to the initial round of PCR (51, 52). Subsequently, 10 μl of round two PCR cocktail (150 mM KCl, 30 mM Tris-HCl [pH 9.0], 1.5% Triton X-100, 4.5 mM MgCl2, 0.6 mM deoxynucleoside triphosphates, 4 ng of the round 2 primers per μl, 0.1 U of Taq per μl) was added directly to each well and subjected to the second round of nested PCR. All cell lysis procedures and PCRs were performed under previously described conditions (51, 52) in a PrimusHT thermal cycler (MWG Biotech). During all reactions, 150 N of pressure was applied to PCR plates to reduce evaporation from the wells. Products were analyzed by ethidium bromide staining of a 1.5 or 2.0% agarose gel, and all PCR products had the size anticipated for this PCR. For all manipulations of latently infected cells, aerosol-resistant barrier tips were used. Twelve PCRs were performed for each cell dilution with six serial threefold dilutions per sample per experiment. With each set of samples, 6 water controls and 18 controls for PCR sensitivity were included (10 copies, 1 copy, and 0.1 copy of pBamHIN plasmid diluted into a background of 104 uninfected cells [51, 52]). Sensitivity controls for this assay detected and amplified gene 50 at 93.9% (10 copies), 45.5% (1 copy), 6.7% (0.1 copy), and 0% (H2O controls). Every assay had at least four of six reactions positive at 10 copies.
Statistical analyses.
All data were analyzed with GraphPad Prism version 3.02 for Windows (GraphPad Software, San Diego, Calif.; www.graphpad.com). Titer data were statistically analyzed with the nonparametric Mann-Whitney test. For all limiting-dilution analysis of latency and reactivation, data were subjected to nonlinear regression (with a sigmoidal dose-response algorithm to fit the data). In this analysis, (i) top and bottom values were set at constant values of 100 and 0, respectively; (ii) there was no weighting of the data; and (iii) data were fitted with a total of 300 segments. Frequencies of viral genome-positive cells and ex vivo reactivation were obtained with the nonlinear regression fit of the data to determine the cell number at which 63.2% of the wells were positive. Based on the Poisson distribution, this is the frequency at which there is at least one event present in a population. Statistical significance was determined by performing a paired t test on frequencies of wt γHV68 and γHV68Δ9473. Reactivation efficiency, defined as the fraction of γHV68 genome-positive cells that reactivated from latency, was calculated by taking the quotient of the frequency of ex vivo reactivation divided by the frequency of viral genome-positive cells with statistical significance analyzed by paired t test.
RESULTS
Isolation and characterization of γHV68Δ9473.
During a screen for candidate marker rescue viruses of the γHV68-M1.LacZ mutant (11), we identified a virus that lacked the LacZ expression cassette but also failed to hybridize to a 32P-labeled probe specific for the M1 locus by Southern blot analysis (data not shown). This isolate was named γHV68Δ9473. Additional Southern blot analyses demonstrated that γHV68Δ9473 genomic DNA did not hybridize to probes spanning the M1, M2, and M3 genes of γHV68 (data not shown; the probe coordinates were bp 107 to 1,702, 1,702 to 4,308, and 4,632 to 7,913 of the γHV68 WUMS sequence [45]). While γHV68Δ9473 genomic DNA failed to hybridize to these sequences, it did hybridize to probes for gene 6 (bp 11,100 to 14,026) and gene 50 (bp 67,540 to 68,814) (data not shown), suggesting that this viral isolate was a γHV68 mutant with a deletion at the left end of the viral genome.
To further characterize the deletion present in γHV68Δ9473, we performed a Southern blot analysis on wt γHV68 and γHV68Δ9473 genomic DNAs with either NotI-NsiI or EcoRV digestion, followed by hybridization with a biotinylated gene 6 probe. Since gene 6 is predicted to encode the single-stranded DNA binding protein of γHV68, it is very likely an essential gene and thus would be present in any replication-competent γHV68 mutant. NotI cuts within the terminal repeat of gene 6, and NsiI cuts at the 3′ terminus of gene 6, allowing analysis of the structure of the left end of the viral genome (Fig. 1A). Whereas the gene 6 probe hybridized to the expected ca. 14.9-kb NotI-NsiI fragment with DNA prepared from wt γHV68, it hybridized to a ca. 6.4-kb fragment with γHV68Δ9473 genomic DNA (Fig. 1B). The latter result is consistent with a large deletion at the left end of the γHV68Δ9473 viral genome. Southern blot analysis of EcoRV-digested DNA demonstrated the presence of the expected 6.1-kb fragment in wt γHV68 DNA (i.e., cutting at the EcoRV sites located upstream of gene 6 [bp 7,918] and near the 3′ end of the gene 6 open reading frame [bp 14,026]). In contrast, the gene 6 probe hybridized to multiple fragments present in the EcoRV-digested DNA prepared from γHV68Δ9473 (Fig. 1B). This banding pattern is consistent with the loss of the EcoRV sites at bp 1,702 and 7,918 (Fig. 1A), which would result in EcoRV fragments containing gene 6 sequences and a variable number of terminal repeats. The presence of a variable number of terminal repeats is due to the absence of any EcoRV sites within the terminal repeat (note that linear viral DNA purified from virions was used in this analysis).
Genomic restriction fragment analysis with BamHI, EcoRI, and HindIII showed that the only detectable alterations in the γHV68Δ9473 genomic structure were those associated with the deletion present at the left end of the viral genome (Fig. 1C, white asterisks) (16, 45). These alterations included (i) the absence of the EcoRI B fragment (a 12.7-kb fragment in wt γHV68 that is not detected in γHV68Δ9473 since the deletion results in a fusion with the terminal repeats and digestion generates multiple submolar species that are not detectable in the ethidium bromide-stained gel shown in Fig. 1C), (ii) the absence of the HindIII J fragment (a 4.8-kb fragment in wt γHV68 that is not detected in γHV68Δ9473 since deletion results in fusion with the terminal repeats and thus the presence of multiple submolar species that are not detected in the ethidium bromide-stained gel), and (iii) the absence of the HindIII E fragment (a 6.2-kb fragment in wt γHV68 that is completely absent in γHV68Δ9473). It should be noted that in this analysis there were no apparent differences between the BamHI restriction fragments of wt γHV68 and γHV68Δ9473. This result was expected, given that the BamHI A1 fragment containing M1 through M4 is a large restriction fragment directly fused to the terminal repeat sequence, a fragment that should be represented by submolar heterogeneous fragments due to the presence of a variable number of terminal repeats. This fragment is not identifiable in either wt γHV68 or γHV68Δ9473 DNA. Additional restriction fragments that are fusions with the terminal repeat in wt γHV68 and, as a result, did not appear on this agarose gel include the HindIII U2 and EcoRI N1 fragments. The EcoRI U fragment is a 53-bp fragment that would also not be detected by the analysis presented in Fig. 1C.
To define the boundaries of this deletion, we subcloned the NotI-NsiI fragment containing gene 6 from γHV68Δ9473 and sequenced the viral genome surrounding the predicted boundary. This γHV68Δ9473 genomic subclone contained approximately 1,100 bp of the terminal repeat sequence, from the NotI site at bp 119,106 through bp 119,424, followed by more of the terminal repeat sequence, from bp 118,212 to bp 119,021 (data not shown; the coordinates are based on the γHV68 WUMS sequence [45]). Sequence analysis also identified a fusion between bp 9,474 of the unique sequence and bp 119,021 of the terminal repeat (Fig. 1A). This deletion removes bp 1 through 9,473 of the unique sequence of wt γHV68, deleting the M1, M2, and M3 genes and all eight viral tRNA-like genes, as well as the 5′ 1.1 kb of the M4 gene (the M4 gene coordinates are bp 8,409 to 9,785) (Fig. 1A). On the basis of these analyses, γHV68Δ9473 is a γHV68 isolate that lacks the M1, M2, and M3 genes and most of the M4 gene, as well as all eight viral tRNA-like genes.
Lytic replication of γHV68Δ9473.
Based upon the ability of γHV68Δ9473 to grow on NIH 3T12 fibroblasts, the M1, M2, M3, M4 and viral tRNA-like genes are not essential for lytic virus replication. To determine whether γHV68Δ9473 exhibited any defect in lytic replication, we analyzed the ability of γHV68Δ9473 to undergo multiple rounds of replication in NIH 3T12 fibroblasts. In this analysis, γHV68Δ9473 replicated in vitro comparably to wt γHV68, as measured by viral titers in both the cell-associated and supernatant fractions of infected cells (Fig. 2A and B). In addition, virus recovered from γHV68Δ9473-infected cultures at 126 h postinfection was genotypically identical to parental γHV68Δ9473 on the basis of NotI-NsiI Southern blot analysis (data not shown). These data demonstrate that the M1, M2, M3, M4, and viral tRNA-like genes are dispensable for efficient lytic replication in NIH 3T12 fibroblasts.
FIG. 2.
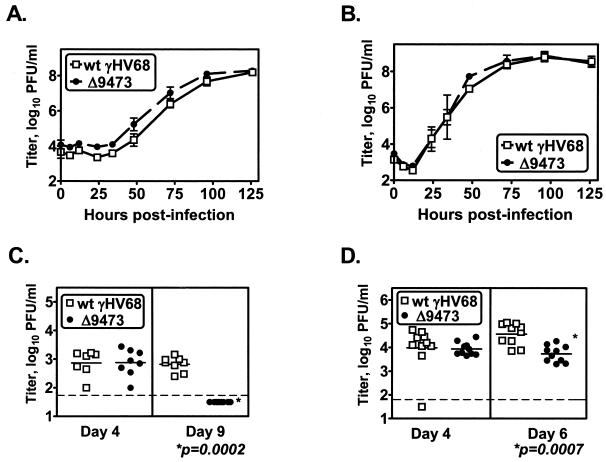
γHV68Δ9473 replicates comparably to wt γHV68 in vitro but has decreased acute-phase titers in vivo. NIH 3T12 monolayers were infected with 0.05 PFU of wt γHV68 or γHV68Δ9473 per cell, and supernatant (A) or cells (B) were harvested at the indicated times and titers were determined by plaque assay. The data shown are from two independent experiments and are plotted as mean titers (log10) ± SEM. Acute-phase titers were quantitated by plaque assay following infection of C57BL/6 mice with 4 × 105 to 106 PFU of either wt γHV68 (open squares) or γHV68Δ9473 (closed circles). Animals were harvested at the times indicated, and the graphs show spleen titers following i.p. infection (C) or lung titers following i.n. infection (D). Statistically significant differences in γHV68Δ9473 infection were detected at day 9 p.i. after i.p. infection and day 6 p.i. after i.n. infection, as indicated. The data shown were compiled from two or three independent experiments for each time point, with three to five mice per group per experiment. Each symbol represents the viral titer of an individual mouse. The horizontal dashed line indicates the limit of detection by this assay (50 PFU/ml, log10 1.7).
We extended this analysis to examine acute-phase virus replication in vivo with wt γHV68 and γHV68Δ9473. Animals infected with γHV68Δ9473 had wt splenic titers following i.p. infection at day 4 postinfection (Fig. 2C). However, by day 9 p.i., γHV68Δ9473-infected animals had at least a 10-fold reduction in splenic viral titers compared to wt γHV68-infected animals (mean splenic titers [log10] ± the standard error of the mean [SEM]: wt, 2.820 ± 0.092; γHV68Δ9473, 1.700 ± 0.000), with none of the γHV68Δ9473-infected animals having a detectable viral titer at this time (Fig. 2C). Animals infected i.n. with γHV68Δ9473 exhibited a similar phenotype, with lung titers comparable between wt- and γHV68Δ9473-infected animals at day 4 p.i. and a nearly 10-fold reduction in mean lung titers by day 6 p.i. in γHV68Δ9473-infected mice compared to wt γHV68-infected mice (mean lung titers [log10] ± SEM: wt, 4.562 ± 0.145; γHV68Δ9473, 3.732 ± 0.109) (Fig. 2D). By day 9 p.i., there was no detectable virus in either wt γHV68- or γHV68Δ9473-infected lungs (data not shown).
γHV68Δ9473 establishes and reactivates from latency comparably to wt γHV68 following i.p. infection.
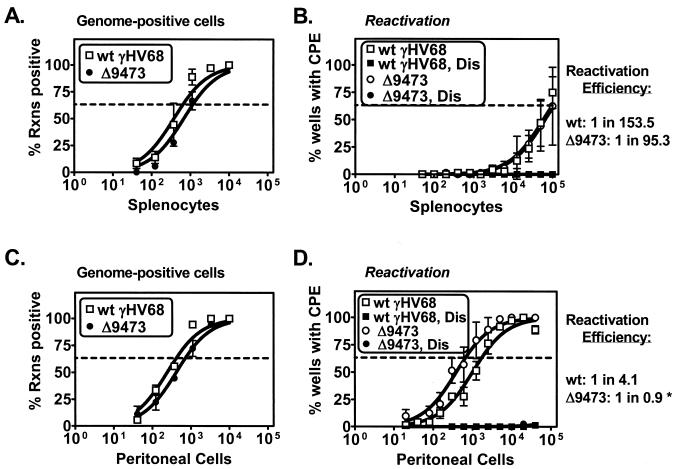
Given that the M2, M3, and viral tRNA-like genes have previously been identified as candidate latency-associated transcripts (7, 18, 28, 33, 46), we hypothesized that γHV68Δ9473 would have a significant defect in latency. We first analyzed latency and reactivation following i.p. infection, a route of infection that may bypass some of the initial stages in the infection process (e.g., trafficking of latently infected cells from the site of infection) (51). To analyze latency and reactivation, we analyzed two parameters: (i) the frequency of γHV68 genome-positive cells with a single-copy sensitivity limiting-dilution PCR assay (51, 52) and (ii) the frequency of spontaneous ex vivo reactivation with a limiting-dilution coculture assay. Surprisingly, animals infected with γHV68Δ9473 had nearly wt frequencies of viral genome-positive cells in both splenocytes and peritoneal cells at early times (days 17 to 18 p.i.) with, at most, a twofold decrease in the frequency of viral genome-positive splenocytes (γHV68 genome-positive splenocytes: wt γHV68, 1 in 644; γHV68Δ9473, 1 in 1,156) (Fig. 3A and C). In addition, animals infected with γHV68Δ9473 had a wt frequency of ex vivo reactivation in splenocytes and a threefold increase in the frequency of reactivation in peritoneal cells (Fig. 3B and D). Mechanically disrupted samples plated in parallel were uniformly negative, demonstrating that these samples did not contain any preformed infectious virus (Dis samples in Fig. 3B and D).
FIG. 3.
γHV68Δ9473 establishes and reactivates from latency comparably to wt γHV68 at early times following i.p. infection. C57BL/6 mice were infected with 106 PFU of wt γHV68 or γHV68Δ9473 by i.p. injection. Splenocytes (A and B) and peritoneal cells (C and D) were harvested on days 17 and 18 p.i. and analyzed for the frequency of viral genome-positive cells (A and C) or ex vivo reactivation (B and D). Reactivation efficiency (defined as the fraction of γHV68 genome-positive cells that reactivated from latency) is indicated to the right of each set of data. The asterisk denotes that although reactivation efficiency cannot be higher than 1 in 1, these assays are limited in the ability to resolve two- to threefold differences. Frequency analysis was based on the Poisson distribution with the horizontal dashed line at 63.2%. Mechanically disrupted (Dis) cells were plated in parallel to identify the presence of preformed infectious virus. The data shown represent three independent experiments with cells pooled from three to five mice per experiment per group. Curve fit lines were derived by nonlinear-regression analysis, and the symbols represent mean percentages of wells positive for virus (viral DNA or a CPE) ± SEM. There were no statistically significant differences between wt γHV68 and γHV68Δ9473 in the frequency of viral genome-positive cells or reactivation or reactivation efficiency at this time. Rxns, reactions.
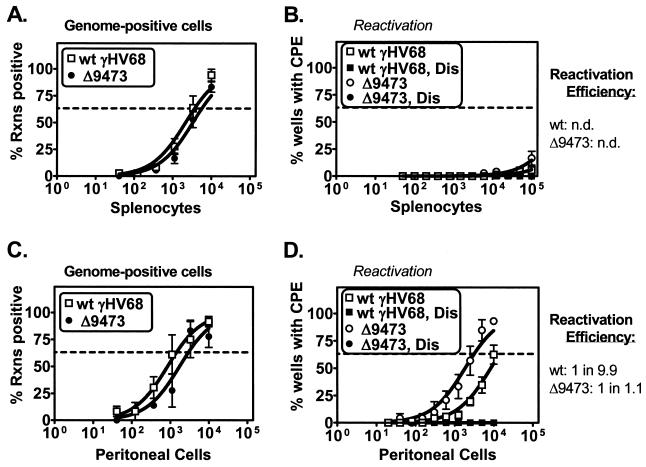
To extend the analysis of viral latency, we tested whether γHV68Δ9473 exhibited any defect in latent infection at later times after i.p. infection. γHV68Δ9473-infected animals harvested at late times (days 42 to 48 p.i.) had wt frequencies of viral genome-positive cells in both splenocytes and peritoneal cells with no more than a twofold decrease relative to wt virus-infected animals (Fig. 4A and C). At this late time, neither wt γHV68- nor γHV68Δ9473-infected splenocytes had any detectable level of ex vivo reactivation (Fig. 4B). In contrast, peritoneal cells from animals infected with γHV68Δ9473 had a ca. fivefold increase in the frequency of ex vivo reactivation relative to wt-infected animals (frequency of reactivation: wt γHV68, 1 in 14,387; γHV68Δ9473, 1 in 3,165) (Fig. 4D) and a ca. 10-fold increase in reactivation efficiency (reactivation efficiency: wt γHV68, 1 in 9.9; γHV68Δ9473, 1 in 1.1).
FIG. 4.
γHV68Δ9473 maintains latency and has increased reactivation efficiency from peritoneal cells relative to that of wt γHV68 at late times following i.p. infection. C57BL/6 mice were infected with 106 PFU of wt γHV68 or γHV68Δ9473 by i.p. injection, and splenocytes (A and B) and peritoneal cells (C and D) were harvested between days 42 and 48 postinfection. Cells were analyzed for the frequency of viral genome-positive cells (A and C) or ex vivo reactivation (B and D). Reactivation efficiency (defined as the fraction of γHV68 genome-positive cells that reactivated from latency) is indicated to the right of each set of data. Frequency analysis was based on the Poisson distribution with the horizontal dashed line at 63.2%. Mechanically disrupted (Dis) cells were plated in parallel to identify the presence of preformed infectious virus. The data shown represent three independent experiments with cells pooled from three to five mice per experiment per group. Curve fit lines were derived by nonlinear-regression analysis, and the symbols represent mean percentages of wells positive for virus (viral DNA or a CPE) ± SEM. There were no statistically significant differences between wt γHV68 and γHV68Δ9473 in the frequency of viral genome-positive cells or reactivation or in reactivation efficiency. n.d., not determined. Rxns, reactions.
To confirm that virus that reactivated from γHV68Δ9473-infected mice was not a revertant or wt γHV68, virus was harvested from the ex vivo reactivation assay. Reactivation isolates for both wt γHV68 and γHV68Δ9473 were harvested from splenocytes and peritoneal cells at day 17 p.i. and from peritoneal cells at day 42 p.i. Isolates were harvested as clonal reactivation events (where less than 63.2% of the wells were positive for CPE) and amplified on NIH 3T12 monolayers, and the structure of the viral genome was analyzed by digestion with NotI and NsiI as described above (Fig. 1). On the basis of an analysis of 13 reactivation isolates from two independent experiments (day 17, 8 isolates; day 42, 5 isolates), γHV68Δ9473 reactivation isolates were indistinguishable from parental γHV68Δ9473 (data not shown). A parallel analysis demonstrated that wt γHV68 isolates were identical to parental wt γHV68 (data not shown). These data indicate that the M1, M2, M3, and M4 genes and the eight viral tRNA-like genes are dispensable for the establishment and maintenance of latency in vivo and reactivation from latency following i.p. infection.
γHV68Δ9473 has a specific defect in splenic latency that is not observed in peritoneal cells following i.n. infection.
Given the size and nature of the deletion in γHV68Δ9473, we anticipated that γHV68Δ9473 would have alterations in latency and reactivation. Previous analyses of B-cell-deficient animals infected with wt γHV68 identified a defect in splenic latency following i.n. infection but not following i.p. infection (51). The latter result indicated that the route of infection may, in some cases, influence the requirements for establishment of latency and, further, that infection at a mucosal site might serve as a more stringent test for potential defects in viral mutants.
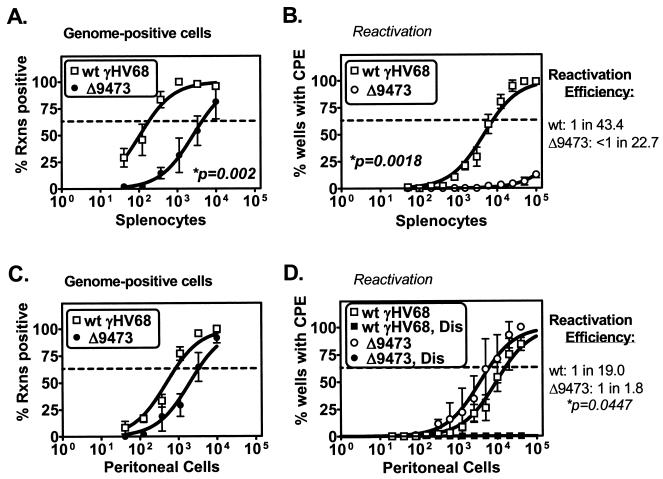
To assess whether γHV68Δ9473 exhibited alterations in latency following infection by a mucosal route, we infected animals with wt γHV68 or γHV68Δ9473 by i.n. injection. First, we analyzed latency in splenocytes and peritoneal cells at an early time (day 17) postinfection. Notably, animals infected with γHV68Δ9473 had a 24-fold decrease in the frequency of viral genome-positive splenocytes relative to wt γHV68-infected animals (γHV68 genome-positive splenocytes: wt γHV68, 1 in 183; γHV68Δ9473, 1 in 4,397) (Fig. 5A). γHV68Δ9473-infected splenocytes also exhibited a severe defect in ex vivo reactivation (a ≥13-fold decrease) relative to wt-infected splenocytes (frequency of reactivation: wt γHV68, 1 in 7,938; γHV68Δ9473, <1 in 100,000) (Fig. 5B). γHV68Δ9473-infected spleens also had greatly diminished splenomegaly compared to wt γHV68-infected spleens upon gross inspection (data not shown).
FIG. 5.
γHV68Δ9473 has a defect in splenic latency, not observed in peritoneal cells, relative to wt γHV68 at early times following i.n. infection. C57BL/6 mice were infected with 4 × 105 PFU of wt γHV68 or γHV68Δ9473 by i.n. injection, and splenocytes (A and B) and peritoneal cells (C and D) were harvested at day 17 postinfection. Cells were analyzed for the frequency of viral genome-positive cells (A and C) or ex vivo reactivation (B and D). For clarity, disrupted samples are not included in reactivation data on splenocytes (Fig. 5B); these samples were uniformly negative. Reactivation efficiency (defined as the fraction of γHV68 genome-positive cells that reactivated from latency) is indicated to the right of each set of data. Frequency analysis was based on the Poisson distribution with the horizontal dashed line at 63.2%. Mechanically disrupted (Dis) cells were plated in parallel to identify the presence of preformed infectious virus. The data shown represent four independent experiments, with cells pooled from three to five mice per experiment per group. Curve fit lines were derived by nonlinear-regression analysis, and symbols represent mean percentages of wells positive for virus (viral DNA or a CPE) ± SEM. Statistically significant differences in γHV68Δ9473 latency were observed in the frequency of viral genome-positive splenocytes, reactivation from splenocytes, and efficiency of reactivation from peritoneal cells, as indicated. Rxns, reactions.
In contrast to these defects in splenic latency, latent γHV68Δ9473 infection of peritoneal cells was relatively unaffected at this early time. At day 17 postinfection, the frequency of viral genome-positive peritoneal cells in γHV68Δ9473-infected animals was decreased fourfold relative to that in wt γHV68-infected mice (γHV68 genome-positive splenocytes: wt γHV68, 1 in 845; γHV68Δ9473, 1 in 3,368) (Fig. 5C) and the frequency of ex vivo reactivation in peritoneal cells was modestly increased (about twofold) (Fig. 5D), resulting in an overall increased reactivation efficiency for γHV68Δ9473 within peritoneal cells (reactivation efficiency: wt γHV68, 1 in 19.0; γHV68Δ9473, 1 in 1.8). On the basis of the pronounced defect of γHV68Δ9473 in splenic latency and reactivation, not observed in peritoneal cells, there appear to be tissue-specific genetic requirements for γHV68 latency and reactivation. Furthermore, the defect of γHV68Δ9473 in splenic latency following i.n., but not i.p., infection demonstrates that the route of infection influences the viral genes required for γHV68 latency and reactivation.
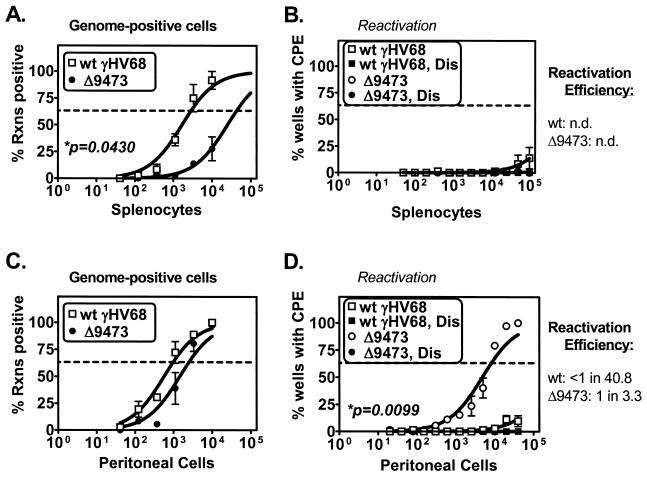
γHV68Δ9473-infected mice also exhibited severe alterations in latency and reactivation at late times following i.n. infection (days 42 to 48 p.i.). γHV68Δ9473-infected animals had a 15-fold decrease in the frequency of viral genome-positive splenocytes compared to wt γHV68-infected animals (γHV68 genome-positive splenocytes: wt γHV68, 1 in 2,884; γHV68Δ9473, 1 in 42,817) (Fig. 6A). At this time, neither wt- nor γHV68Δ9473-infected splenocytes exhibited any detectable ex vivo reactivation (Fig. 6B). Within peritoneal cells, there was a threefold decrease in the frequency of γHV68Δ9473 viral genome-positive peritoneal cells relative to the frequency of wt γHV68-infected animals (γHV68 genome-positive peritoneal cells: wt γHV68, 1 in 980; γHV68Δ9473, 1 in 2,510) (Fig. 6C) and a significant (≥12-fold) increase in the frequency of ex vivo reactivation in γHV68Δ9473-infected peritoneal cells (frequency of reactivation: wt γHV68, <1 in 40,000; γHV68Δ9473, 1 in 8,215) (Fig. 6D). As a result, γHV68Δ9473 had a greatly increased reactivation efficiency relative to that of wt γHV68 within peritoneal cells at 6 weeks postinfection (reactivation efficiency: wt γHV68, <1 in 40.8; γHV68Δ9473, 1 in 3.3).
FIG. 6.
γHV68Δ9473 has a defect in splenic latency and increased reactivation from peritoneal cells relative to wt γHV68 at late times following i.n. infection. C57BL/6 mice were infected with 4 × 105 PFU of wt γHV68 or γHV68Δ9473 by i.n. injection, and splenocytes (A and B) and peritoneal cells (C and D) were harvested between day 42 and 46 postinfection. Cells were analyzed for the frequency of viral genome-positive cells (A and C) or ex vivo reactivation (B and D). Reactivation efficiency (defined as the fraction of γHV68 genome-positive cells that reactivated from latency) is indicated to the right of each set of data. Frequency analysis was based on the Poisson distribution with the horizontal dashed line at 63.2%. Mechanically disrupted (Dis) cells were plated in parallel to identify the presence of preformed infectious virus. The data shown represent three independent experiments with cells pooled from three to five mice per experiment per group. Curve fit lines were derived by nonlinear-regression analysis, and symbols represent mean percentages of wells positive for virus (viral DNA or a CPE) ± SEM. Statistically significant differences with γHV68Δ9473 were observed in the frequency of viral genome-positive splenocytes and reactivation from peritoneal cells, as indicated. n.d., not determined. Rxns, reactions.
To further validate these findings, Southern blot analysis of reactivation isolates (days 17 and 42 p.i.) showed that reactivation isolates from γHV68Δ9473-infected mice were genotypically identical to parental γHV68Δ9473 (data not shown; day 17, eight isolates; day 42, two isolates). Curiously, DNA from one additional γHV68Δ9473 isolate harvested at day 42 p.i. from peritoneal cells hybridized with a smaller fragment than parental γHV68Δ9473 DNA, consistent with deletion of an additional 0.7 to 1.6 kb of the γHV68 sequence. Whether this deletion species represents a low-level subspecies within the γHV68Δ9473 viral stock or whether this isolate arose during infection in vivo is unknown. The genomic structure of this isolate has not been further defined.
DISCUSSION
Here we have described the identification and characterization of a large spontaneous deletion mutant of γHV68, that we have named γHV68Δ9473. This virus lacks at least 12 different gene products, including the M1, M2, M3, and M4 genes and the eight viral tRNA-like genes. Among these genes, expression of M2, M3, and the viral tRNAs has been detected during latent infection, with M2 and M3 being detected in at least two independent screens (7, 18, 28, 33, 46). Given the size and nature of this deletion, we initially hypothesized that this virus would have a severe defect in the ability to establish a latent infection in vivo. While γHV68Δ9473 does exhibit alterations in latency and reactivation, these phenotypes are tissue specific and route of infection dependent. Furthermore, analysis of γHV68Δ9473 latency following i.p. infection revealed only mild alterations in latency and reactivation (Fig. 3 and 4).
Overall, our findings are consistent with those recently reported for γHV76, another gammaherpesvirus isolated at the same time as γHV68 (5, 22). Recently, Macrae et al. have shown that γHV76 has a deletion nearly identical to that observed in γHV68Δ9473, with both viruses lacking approximately 9.5 kb of the γHV68 unique coding sequence (22). These investigators demonstrated that γHV76 exhibited a defect in acute replication in the lung, as well as an increased inflammatory infiltrate in the lung. In addition, γHV76-infected animals had decreased splenomegaly and decreased establishment of splenic latency following i.n. infection (22). The authors also provided preliminary evidence that γHV76 might exhibit a long-term defect in persistent infection in the lung (22). On the basis of these results and construction of a marker rescue, the authors concluded that the M1-through-M4 locus is important for acute pathogenesis during γHV68 infection, including host evasion and the development of splenic pathology. Our results are consistent with these observations and extend these observations to a quantitative analysis of latency and reactivation comparing different tissues and routes of infection, at early and late times during infection.
Tissue-specific genetic requirements for γHV68 latency and reactivation.
With the identification of tissue-specific defects in γHV68Δ9473 latency, there are now at least four examples of differential regulation of γHV68 latency between splenocytes and peritoneal cells. First, there are distinct cellular and genetic requirements for latency in peritoneal cells and splenocytes, as revealed by either infection of B6 mice with γHV68Δ9473 or wt γHV68 infection of B-cell-deficient mice (μMT/μMT) (51). In both cases, there are specific defects in splenic latency but not in peritoneal cell latency. Second, peritoneal cells and splenocytes have different transcriptional programs during latency in B-cell-deficient mice (46), indicating that, even in the absence of B cells as a latent reservoir, latent infection in peritoneal cells and splenocytes is fundamentally different. Differences in viral gene expression have also been observed between the mesenteric lymph nodes and spleen in BALB/c mice (28). Third, latency reservoirs are regulated by different immune effectors (S. A. Tibbetts, L. F. van Dyk, S. H. Speck, and H. W. Virgin IV, submitted for publication). Gamma interferon is important for controlling persistence in peritoneal cells, but not in splenocytes, at late times (day 42), and perforin is more important for controlling reactivation in splenocytes than in peritoneal cells. Fourth, wt γHV68 has a higher efficiency of reactivation from peritoneal cells than from splenocytes. One example of this difference is at the peak of latency following i.p. infection (day 17), where only ∼1 in 150 γHV68 genome-positive splenocytes shows reactivation from latency compared to the ∼1 in 4 γHV68 genome-positive peritoneal cells that show reactivation from latency (Fig. 3). Even at the peak of latency following i.n. infection, only ∼1 in 43.4 splenocytes shows reactivation from latency (Fig. 5). This poor reactivation efficiency within splenocytes raises the question of whether all γHV68 genome-positive splenocytes are truly latently infected. This is less of a concern in peritoneal cells, since virtually all peritoneal cells can reactivate from latency, especially in certain mutants (Fig. 3D and 4D).
What is the basis of this tissue-specific regulation? A lead hypothesis is that the cell types that are latently infected significantly differ between different tissues. While γHV68 establishes a latent infection in multiple cell types, including B cells, macrophages, and dendritic cells (17, 39, 52), no one has compared the latently infected cell types present in splenocytes and peritoneal cells or determined how the composition of the latent reservoir changes during infection. A second hypothesis is that the local environment of the tissue significantly influences γHV68 latency. The spleen is a lymphoid organ packed with lymphocytes, and it is an environment that places latently infected cells in close physical proximity to potentially γHV68-reactive lymphocytes. In stark contrast, peritoneal cells reside in a diffuse space with no defined architecture. Interestingly, while the peritoneum is not a lymphoid tissue, latently infected mice do show a long-term increase in the number of T cells present in the peritoneum (52).
Tissue-specific differences in latency have also been observed in EBV-infected individuals. In the peripheral blood, latently EBV-infected B cells have extremely limited transcription (27). This is in contrast to latency in the tonsils, where multiple transcriptional programs are observed, as well as ongoing lytic replication (1, 2, 20, 27). While the basis for these differences is unknown, it has been proposed that exposure to antigen at a mucosal site such as the tonsils might lead to alterations in the latently infected cell (40). Alternatively, it is interesting to speculate that cell trafficking and tissue-specific cues for homing to mucosal and nonmucosal surfaces may provide one molecular mechanism by which a latently infected cell could be cued to undergo activation within specific tissues (13).
Route of infection-dependent latency and reactivation phenotypes.
There is increasing evidence that the route of infection also influences the cellular and molecular requirements for latency and reactivation. Infection with viral mutants (γHV68Δ9473 and the γHV68-M2.Stop) or wt virus infection of a specific host mutant (B-cell-deficient mice) has identified a defect in splenic latency following i.n. infection but not following i.p. infection (19, 51). These data indicate that B cells are an essential cell type for the establishment of splenic latency following i.n. infection. Consistent with this hypothesis, transfer of B cells into B-cell-deficient mice restores splenic latency (37), indicating that B cells can traffic γHV68 to the spleen.
It is unclear why B cells do not play an equivalent role in the establishment of splenic latency following i.p. infection. It is possible that i.p. infection bypasses the need for B-cell-dependent trafficking either through the infection of a different cell type (e.g., macrophages) or through direct spreading of virus particles to the spleen. Whatever the explanation is, it is clear from this analysis that i.n. infection provides additional requirements for the establishment of latency beyond those observed with i.p. infection. Furthermore, the defect of γHV68 mutants following i.n. infection but not following i.p. infection supports the hypothesis that γHV68 may naturally be transmitted by a mucosal route of infection. At this time, the natural route of transmission is unknown and the only documented case of γHV68 transmission between animals occurred when a mother ate her infected pups (6).
Genetic basis for γHV68Δ9473 phenotypes.
Previously, single-gene mutations have been characterized in the M1, M2, and M3 genes. Analysis of γHV68Δ9473 has recapitulated each of the reported phenotypes for single-gene mutations as follows (Table 1). First, disruption of the M1 gene results in an increased efficiency of reactivation from latency in peritoneal cells after i.p. infection (11), a phenotype also observed with γHV68Δ9473 (Fig. 4D). Second, disruption of the M2 gene results in decreased viral titers in the spleen after i.p. infection (also seen in γHV68Δ9473; Fig. 2C) and a decreased frequency of viral genome-positive cells in the spleen after i.n. infection (seen in γHV68Δ9473; Fig. 5A) (19). At this time, the phenotype of an M3 mutant remains controversial. A lacZ insertional mutation in the M3 gene has recently been reported to result in decreased splenic latency after i.n. infection (8). However, this latency phenotype has not been observed upon analysis of an M3 mutant virus containing a translational stop codon near the 5′ end of the M3 gene that ablates the expression of a functional M3 protein (van Berkel et al., unpublished data). lacZ mutations within the M1 and M2 genes and gene 4 have each resulted in compound phenotypes not observed upon analysis of deletions or stop codon insertions (11, 19) (S. B. Kapadia, S. H. Speck, and H. W. Virgin IV, unpublished data). Based on this observation and the similarity of a stop codon mutation within the M2 gene with the M3.LacZ phenotype, alterations in M3.LacZ may be due to alterations in the neighboring M2 gene and not to disruption of M3. Notably, the deletion present in γHV68Δ9473 also removes all eight viral tRNA-like genes, whose role in infection is unknown. No γHV68 mutants have been characterize that selectively disrupt the viral tRNA genes, and it is unclear whether deletion of these noncoding RNAs contributes to the observed phenotypes of γHV68Δ9473.
TABLE 1.
Phenotypes observed upon infection with γHV68Δ9743
| γHV68Δ9743 phenotype | Tissue | Time: (days) p.i. | Route of infection | Candidate genea |
|---|---|---|---|---|
| Reactivation, 5× increase | Peritoneal cells | 42 | i.p. | M1 (M1Δ511) (ref. 11) |
| Acute-phase titer, 10× decrease | Spleen | 9 | i.p. | M2 (M2.Stop) (ref. 19) |
| Latent load and reactivation, 24× and >13× decreases | Spleen | 17 | i.n. | M2 (M2.Stop) (ref. 19) |
| Acute-phase titer, 10× decrease | Lung | 6 | i.n. | ? |
| Latent load, 15× decrease | Spleen | 42 | i.n. | ? |
| Reactivation, >12× increase | Peritoneal cells | 42 | i.n. | M1?b |
Candidate genes are based on specific gene mutations that have given similar phenotypes. ref., reference.
While M1 mutants exhibit increased reactivation from latency in peritoneal cells following i.p. infection (11), it is unknown whether these viruses show increased reactivation following i.n. infection.
Analysis of γHV68Δ9473 has also identified three phenotypes not yet identified by single-gene mutations (Table 1). These include (i) decreased acute-phase viral titers in the lung after i.n. infection (also seen with γHV76 [22]), (ii) decreased frequency of viral genome-positive splenocytes at late times in wt immunocompetent mice, and (iii) increased reactivation in peritoneal cells following i.n. infection. It is important to emphasize that since a marker rescue of γHV68Δ9473 has not been generated and characterized, we have not unambiguously mapped the observed phenotypes to the deletion at the left end of the viral genome. One of the goals of future studies is to identify the genetic basis of these phenotypes, whether they result from single-gene mutations (e.g., M4), combinatorial gene effects, or disruption of noncoding elements (e.g., a latency-associated origin of replication or cis elements required for regulation of latency-associated transcription). On the basis of the hyperreactivation phenotype of M1 mutants following i.p. infection (11), we hypothesize that M1 mutants might also hyperreactivate following i.n. infection.
Notably, γHV68Δ9473 is the first mutant or experimental condition identified in the γHV68 system that confers a decreased splenic latent viral genome load at late times in wt mice. The nature of this defect is unknown and, curiously, appears to be route of infection dependent. These large deletion viruses, such as γHV68Δ9473 and γHV76, may also have a more global defect in long-term infection, with preliminary evidence indicating that γHV76 may exhibit a defect in long-term persistent infection in the lung (22).
Implications of γHV68Δ9473 for γHV68 passage and mutagenesis.
There is a history of spontaneous deletions and genomic rearrangements within the herpesviruses. These include the identification of herpesvirus saimiri deletions that lack oncogenic activity in primates and EBV deletion genomes that have mapped genes involved in transformation and lytic replication (21, 41). Spontaneous deletions have also been observed in common laboratory strains, including the identification of a 13-kb sequence that is present in human cytomegalovirus clinical isolates but absent in laboratory strains (10).
While analysis of γHV68Δ9473 has provided important information regarding γHV68 latency, the generation of a large spontaneous deletion virus during passage urges caution when working with γHV68. First, the appearance of this spontaneous deletion mutant during tissue culture passage of γHV68 suggests that γHV76 may have arisen during in vitro passage of a γHV68 isolate. Second, identification of γHV76 suggests that even the current wt γHV68 may lack genes that are present in related viruses in the wild.
The generation of a spontaneous deletion during γHV68 mutagenesis stresses the need for rigor when generating and analyzing γHV68 recombinants. Generation of γHV68Δ9473 was not an isolated incident; we have observed the generation of large deletion mutants at the left end of the γHV68 genome in at least two other independent screens for γHV68 mutants (M. A. Jacoby, S. B. Kapadia, H. W. Virgin IV, and S. H. Speck, unpublished data). Based on the propensity of the γHV68 genome to undergo rearrangements at the left end of the viral genome, it is imperative that all γHV68 mutants be assessed for intact genomic structure, especially at the left end of the viral genome. Furthermore, these deletion mutants can also be present as a low-frequency contaminant in viral stocks (11). These contaminants can only be detected by Southern blot analysis with NotI-NsiI restriction digestion, as shown in Fig. 1. Importantly, this is not simply an academic concern because γHV68Δ9473 has at least one gain-of-function phenotype: hyperreactivation in peritoneal cells (Fig. 4D and 6D). Given these concerns, we routinely screen viral mutant stocks, as well as reactivation isolates, with NotI-NsiI digestion and Southern blotting as described in Fig. 1.
In conclusion, this study demonstrates that the M1, M2, M3, and M4 genes and the viral tRNA-like genes are not essential for latency and reactivation. Instead, a virus lacking these genes exhibits tissue-specific and route-of-infection-dependent alterations in latency and reactivation. These studies highlight the importance of loss-of-function mutations to characterize the in vivo relevance of latency-associated transcripts and emphasize the emerging complexity of γHV68 latency in vivo.
Acknowledgments
This research was supported by NIH grants CA74730 and HL60090 to H.W.V. and S.H.S.; NIH grants CA43143, CA52004, and CA58524 to S.H.S.; NIH grant AI39616 to H.W.V.; and ACS grant RP6-97-134-01-MBC to H.W.V. E.T.C. was supported by a Lucille P. Markey Pathway predoctoral fellowship and NIH grant CA43143.
We acknowledge helpful discussions with members of the Speck and Virgin laboratories, as well as discussions during joint laboratory meetings with members of the laboratories of David Leib and Lynda Morrison.
REFERENCES
- 1.Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. [DOI] [PubMed] [Google Scholar]
- 2.Babcock, G. J., and D. A. Thorley-Lawson. 2000. Tonsillar memory B cells, latently infected with Epstein-Barr virus, express the restricted pattern of latent genes previously found only in Epstein-Barr virus-associated tumors. Proc. Natl. Acad. Sci. USA 97:12250-12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 4.Bellows, D. S., B. N. Chau, P. Lee, Y. Lazebnik, W. H. Burns, and J. M. Hardwick. 2000. Antiapoptotic herpesvirus Bcl-2 homologs escape caspase-mediated conversion to proapoptotic proteins. J. Virol. 74:5024-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaskovic, D., M. Stancekova, J. Svobodova, and J. Mistrikova. 1980. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 24:468. [PubMed] [Google Scholar]
- 6.Blaskovic, D., D. Stanekova, and J. Rajcani. 1984. Experimental pathogenesis of murine herpesvirus in newborn mice. Acta Virol. 28:225-231. [PubMed] [Google Scholar]
- 7.Bowden, R. J., J. P. Simas, A. J. Davis, and S. Efstathiou. 1997. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J. Gen. Virol. 78:1675-1687. [DOI] [PubMed] [Google Scholar]
- 8.Bridgeman, A., P. G. Stevenson, J. P. Simas, and S. Efstathiou. 2001. A secreted chemokine binding protein encoded by murine gammaherpesvirus-68 is necessary for the establishment of a normal latent load. J. Exp. Med. 194:301-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardin, R. D., J. W. Brooks, S. R. Sarawar, and P. C. Doherty. 1996. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J. Exp. Med. 184:863-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clambey, E. T., H. W. Virgin IV, and S. H. Speck. 2000. Disruption of the murine gammaherpesvirus 68 M1 open reading frame leads to enhanced reactivation from latency. J. Virol. 74:1973-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter, M. A., and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 13.Cyster, J. G. 1999. Chemokines and cell migration in secondary lymphoid organs. Science 286:2098-2102. [DOI] [PubMed] [Google Scholar]
- 14.Doherty, P. C., J. P. Christensen, G. T. Belz, P. G. Stevenson, and M. Y. Sangster. 2001. Dissecting the host response to a gamma-herpesvirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:581-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Efstathiou, S., Y. M. Ho, S. Hall, C. J. Styles, S. D. Scott, and U. A. Gompels. 1990. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. J. Gen. Virol. 71:1365-1372. [DOI] [PubMed] [Google Scholar]
- 16.Efstathiou, S., Y. M. Ho, and A. C. Minson. 1990. Cloning and molecular characterization of the murine herpesvirus 68 genome. J. Gen. Virol. 71:1355-1364. [DOI] [PubMed] [Google Scholar]
- 17.Flano, E., S. M. Husain, J. T. Sample, D. L. Woodland, and M. A. Blackman. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J. Immunol. 165:1074-1081. [DOI] [PubMed] [Google Scholar]
- 18.Husain, S. M., E. J. Usherwood, H. Dyson, C. Coleclough, M. A. Coppola, D. L. Woodland, M. A. Blackman, J. P. Stewart, and J. T. Sample. 1999. Murine gammaherpesvirus M2 gene is latency-associated and its protein a target for CD8+ T lymphocytes. Proc. Natl. Acad. Sci. USA 96:7508-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacoby, M. A., H. W. Virgin IV, and S. H. Speck. 2002. Disruption of the M2 gene of murine gammaherpesvirus 68 alters splenic latency following intranasal, but not intraperitoneal, inoculation. J. Virol. 76:1790-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joseph, A. M., G. J. Babcock, and D. A. Thorley-Lawson. 2000. Cells expressing the Epstein-Barr virus growth program are present in and restricted to the naive B-cell subset of healthy tonsils. J. Virol. 74:9964-9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 22.Macrae, A. I., B. M. Dutia, S. Milligan, D. G. Brownstein, D. J. Allen, J. Mistrikova, A. J. Davison, A. A. Nash, and J. P. Stewart. 2001. Analysis of a novel strain of murine gammaherpesvirus reveals a genomic locus important for acute pathogenesis. J. Virol. 75:5315-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mistrikova, J., H. Raslova, M. Mrmusova, and M. Kudelova. 2000. A murine gammaherpesvirus. Acta Virol. 44:211-226. [PubMed] [Google Scholar]
- 24.Moore, P. S., and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus, p. 2803-2833. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 25.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. 2001. Natural history of murine gamma-herpesvirus infection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parry, C. M., J. P. Simas, V. P. Smith, C. A. Stewart, A. C. Minson, S. Efstathiou, and A. Alcami. 2000. A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J. Exp. Med. 191:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 28.Rochford, R., M. L. Lutzke, R. S. Alfinito, A. Clavo, and R. D. Cardin. 2001. Kinetics of murine gammaherpesvirus 68 gene expression following infection of murine cells in culture and in mice. J. Virol. 75:4955-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roizman, B., and P. E. Pellet. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 30.Roy, D. J., B. C. Ebrahimi, B. M. Dutia, A. A. Nash, and J. P. Stewart. 2000. Murine gammaherpesvirus M11 gene product inhibits apoptosis and is expressed during virus persistence. Arch. Virol. 145:2411-2420. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Simas, J. P., and S. Efstathiou. 1998. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 6:276-282. [DOI] [PubMed] [Google Scholar]
- 33.Simas, J. P., D. Swann, R. Bowden, and S. Efstathiou. 1999. Analysis of murine gammaherpesvirus-68 transcription during lytic and latent infection. J. Gen. Virol. 80:75-82. [DOI] [PubMed] [Google Scholar]
- 34.Speck, S. H., and H. W. Virgin. 1999. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr. Opin. Microbiol. 2:403-409. [DOI] [PubMed] [Google Scholar]
- 35.Stevenson, P. G., S. Efstathiou, P. C. Doherty, and P. J. Lehner. 2000. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. USA 97:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart, J. P. 1999. Of mice and men: murine gammaherpesvirus 68 as a model. Epstein-Barr Virus Rep. 6:31-35. [Google Scholar]
- 37.Stewart, J. P., E. J. Usherwood, A. Ross, H. Dyson, and T. Nash. 1998. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J. Exp. Med. 187:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sunil-Chandra, N. P., S. Efstathiou, J. Arno, and A. A. Nash. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 73:2347-2356. [DOI] [PubMed] [Google Scholar]
- 39.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 40.Thorley-Lawson, D. A. 1999. EBV: schizophrenic but not dysfunctional. Epstein-Barr Virus Rep. 6:158-163. [Google Scholar]
- 41.Trimble, J. J., and R. C. Desrosiers. 1991. Transformation by herpesvirus saimiri. Adv. Cancer Res. 56:335-355. [DOI] [PubMed] [Google Scholar]
- 42.Usherwood, E. J., D. J. Roy, K. Ward, S. L. Surman, B. M. Dutia, M. A. Blackman, J. P. Stewart, and D. L. Woodland. 2000. Control of gammaherpesvirus latency by latent antigen-specific CD8+ T cells. J. Exp. Med. 192:943-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Usherwood, E. J., K. A. Ward, M. A. Blackman, J. P. Stewart, and D. L. Woodland. 2001. Latent antigen vaccination in a model gammaherpesvirus infection. J. Virol. 75:8283-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Berkel, V., J. Barrett, H. L. Tiffany, D. H. Fremont, P. M. Murphy, G. McFadden, S. H. Speck, and H. W. Virgin IV. 2000. Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J. Virol. 74:6741-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Virgin, H. W., IV, R. M. Presti, X.-Y. Li, C. Liu, and S. H. Speck. 1999. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J. Virol. 73:2321-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Virgin, H. W., and S. H. Speck. 1999. Unraveling immunity to gamma-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr. Opin. Immunol. 11:371-379. [DOI] [PubMed] [Google Scholar]
- 48.Wakeling, M. N., D. J. Roy, A. A. Nash, and J. P. Stewart. 2001. Characterization of the murine gammaherpesvirus 68 ORF74 product: a novel oncogenic G protein-coupled receptor. J. Gen. Virol. 82:1187-1197. [DOI] [PubMed] [Google Scholar]
- 49.Wang, G. H., T. L. Garvey, and J. I. Cohen. 1999. The murine gammaherpesvirus-68 M11 protein inhibits Fas- and TNF-induced apoptosis. J. Gen. Virol. 80:2737-2740. [DOI] [PubMed] [Google Scholar]
- 50.Weck, K. E., M. L. Barkon, L. I. Yoo, S. H. Speck, and H. W. Virgin IV. 1996. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J. Virol. 70:6775-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weck, K. E., S. S. Kim, H. W. Virgin IV, and S. H. Speck. 1999. B cells regulate murine gammaherpesvirus 68 latency. J. Virol. 73:4651-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weck, K. E., S. S. Kim, H. W. Virgin IV, and S. H. Speck. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol. 73:3273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]