Abstract
Mammalian hepatitis B viruses encode an essential regulatory protein, termed X, which may also be implicated in liver cancer development associated with chronic infection. X protein, also referred to as HBx in human virus and WHx in woodchuck virus, has been reported to bind to a number of cellular proteins, including the DDB1 subunit of the damaged DNA-binding (DDB) complex. Our previous work provided genetic evidence for the importance of WHx-DDB1 interaction in both the activity of the X protein and establishment of viral infection in woodchucks. In the present study, a direct action of DDB1 on the X protein is documented. Physical interaction between the two proteins leads to an increase in X protein stability. This effect results from protection of the viral protein from proteasome-mediated degradation. Protection of WHx is overcome in the presence DDB2, the second subunit of the DDB heterodimer. In keeping with observations reported for HBx, DDB2 was found to directly bind to WHx. Nonetheless, the counteracting effect of DDB2 on X stabilization requires DDB2-DDB1 interaction. Taken together, these findings substantiate the physical and functional connection between the X protein and the DDB1-DDB2 heterodimer, leading to the regulation of the pool of the viral protein.
Chronic infection by human hepatitis B virus (HBV) causes severe liver pathologies, including cirrhosis and liver cancer (4). Due to the lack of fully effective and cost-effective antiviral treatments, the number of persistent HBV carriers is still increasing and exceeds 350 million worldwide (for reviews, see references 3 and 23).
HBV is a member of the family Hepadnaviridae, which includes the hepatitis viruses of the woodchuck, ground squirrel, Pekin duck, and heron (38). These DNA viruses possess a common replication mode in which the pregenomic viral transcript is reverse transcribed by the viral polymerase within neosynthesized viral capsids (27). Mammalian hepadnavirus genomes contain a unique regulatory gene termed X (also referred to as HBx in HBV and WHx in woodchuck hepatitis virus), which codes for an ∼17-kDa protein. Establishment of hepadnaviral infection requires functional X protein (6, 49). However, the protein is dispensable for the late steps of the viral life cycle, that is, from the reverse-transcription step up to secretion of enveloped virions (49). Thus, the mechanism of action of X during viral infection remains to be elucidated. In vitro, HBx protein has two well-documented activities, namely, modest and promiscuous transactivation of transcription (reviewed in references 11 and 35) and proapoptosis (reviewed in references 2 and 24), although the relevance of these activities to the role of X during infection has yet to be established. Both activities are genetically linked (5) and conserved by WHx in the heterologous context of human cells (8, 43). This suggests a common and conserved mechanism of host cell protein recruitment by X proteins from different hepadnaviruses, despite their rather low level of identity (50%) at the amino acid level. Among human proteins reported to physically interact with HBx, we identified UV-DDB, now renamed DDB1, as a protein conserving interactions with X proteins across viral species (44). Using well-defined WHx substitution mutants variously affected in affinity for DDB1, we further showed that correct X-DDB1 interaction is a required upstream step in X activities, including transactivation, proapoptosis, and function in woodchuck infection (42, 43). A recent independent study also revealed a strict correlation between DDB1-binding proficiency and proapoptotic activity in a series of HBx substitution mutants (21). However, the specific function of X-DDB1 interaction remains to be elucidated.
DDB1 is a ubiquitous 127-kDa protein which was initially characterized on the basis of its specific binding to UV-damaged DNA (1). The corresponding damaged DNA-binding (Ddb) activity is defective in some of the group E patients suffering from the genetic disease xeroderma pigmentosum (7). Therefore, a role of DDB1 in nucleotide excision repair of DNA was strongly suspected. It turned out that in Ddb− patients, DDB1 is not affected, whereas mutations occur in a 48-kDa protein termed DDB2 (17, 29), which forms a heterodimeric complex with DDB1 (18). DDB2 has recently been implicated in global genomic repair (45), namely, the mechanism which processes lesions in nontranscribed DNA. Interestingly DDB2 is a UV-inducible nuclear protein, whereas the absence of nuclear localization signals in DDB1 argues for a cytoplasmic location (28). One proposed role for DDB2 is DDB1 nuclear import by a piggyback mechanism. Indeed, wild-type DDB2 induces DDB1 nuclear accumulation, whereas the naturally occurring DDB2 mutants fail to stimulate DDB1 nuclear import (39). In addition to its Ddb activity, the DDB complex has been shown to bind the E2F1 transcription factor and to stimulate its transactivation activity (15, 39). The appearance of DDB1 well before DDB2 in evolution (48) predicts conserved DDB1 functions which are independent of its heterodimeric association with DDB2. This raised the possibility that a DDB2-independent action of DDB1 is involved in X function.
In the present study, we establish a direct functional connection of X with the DDB heterodimer. DDB1 markedly increases X stability, whereas DDB2 relieves this effect. We further show that physical interactions occurring within the tripartite X-DDB complex mediate these effects through the modulation of proteasome-dependent degradation of X.
MATERIALS AND METHODS
Plasmid constructs.
The WHx-expressing plasmids have been described previously (43). The HBx expression plasmid was created by ligating the BglII insert (HA-X gene) from the yeast pACT2-HBx vector (44) into the BamHI site of pCT152, an episomal vector modified from pCEP4 (Invitrogen) (43). The HBx 66-to-101 coding region was excised from pDS103 (43) by Asp718-BamHI digestion. Ligation of this fragment along with a BamHI-SalI insert of the human β-catenin open reading frame into pcDNA3 digested with Asp718-XhoI created an expression construct of β-catenin N-terminally fused to the DDB1-binding domain of HBx. The initial wild-type DDB2 construct (12) was a generous gift of S. Linn. To obtain N-terminally tagged DDB1 and DDB2 proteins, a c-myc tag coding sequence was cloned in frame with the DDB1 and DDB2 inserts of pcDNA3 constructs. The naturally occurring 2RO and 82TO DDB2 mutants (29) were artificially recreated by site-directed mutagenesis using the QuickChange kit (Stratagene). The entire open reading frames of the obtained mutants were sequenced to ensure that no additional mutation was introduced by the mutagenesis procedure. Expression constructs of E2F and of V protein from simian virus 5 were kindly provided by J. Seeler and R. A. Lamb.
Cell culture and transfection procedures.
Chang cells, CCL-13 in the American Type Culture Collection, were grown at 37°C with a 5% CO2 atmosphere in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum containing penicillin (100 U/ml), streptomycin (100 μg/ml), and glutamine (2 mM). The day before transfection, cells were seeded at 0.8 × 106 per 6-cm-diameter dish or at 0.45 × 106 per well of six-well plates. Transient transfections were performed by either the standard calcium phosphate method or the lipid-based method using Lipofectamine-Plus (Life Technologies) according to the manufacturer's instructions. In each transfection experiment, the amount of DNA was kept constant by addition of the relevant empty expression vector. Total proteins were extracted 36 h posttransfection for calcium phosphate precipitation and 24 h posttransfection for the lipid method. For proteasome inhibition studies, cells were incubated for 5 h in medium containing 50 μM benzyloxycarbonyl-Leu-Leu-Leu aldehyde (MG132) before lysis.
Immunoprecipitation.
Cells were washed with phosphate-buffered saline and lysed in SD buffer (50 mM Tris-HCl [pH 8], 0.15 M NaCl, 1% NP-40) in the presence of protease inhibitors. For immunoprecipitation of chimeric proteins carrying the immunoglobulin G (IgG)-binding domain of protein A (ZZ tag) (43), cell extracts (approximately 1 mg of protein) were incubated with rabbit IgG-Sepharose beads (Amersham Pharmacia Biotech) for 15 h at 4°C. The beads were washed twice in SD buffer and twice in SD buffer containing 1 M NaCl. Proteins were recovered from the beads by elution with 0.1 M glycine-HCl buffer (pH 2.5) containing 0.15 M NaCl, followed by neutralization with 1.5 M Tris-HCl (pH 8.8). For classical immunoprecipitation, cell extracts (approximately 1 mg of protein) were incubated with 5 μl of rabbit polyclonal anti-WHx antiserum (a generous gift of W. S. Mason) for 2 h at 4°C under gentle agitation. Protein A-Sepharose beads (Amersham Pharmacia Biotech) previously equilibrated in lysis buffer were then added for an 18-h incubation at 4°C. The beads were washed four times in lysis buffer and twice in 10 mM Tris-HCl (pH 7.5)-0.1% Triton X-100 and resuspended in gel-loading buffer (60 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 100 mM dithiothreitol, 0.1% bromophenol blue, 10% glycerol). The immunoprecipitates were eluted from the beads by incubation at 95°C for 5 min. The eluted proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE).
Immunoblotting.
After a washing step in phosphate-buffered saline, the cells were lysed either in SDS-PAGE loading buffer or in M-PER (mammalian protein extraction reagent; Pierce) according to the manufacturer's instructions. The cell extracts were separated by SDS-PAGE and transferred to nitrocellulose membranes (Hybond C extra; Amersham). Proteins were revealed by chemiluminescence using the Western-Star detection kit (Tropix) according to the manufacturer's instructions. Mouse monoclonal anti-human c-myc 9E10 (PharMingen) simultaneously revealed transfected myc-DDB1 and myc-DDB2. Endogenous DDB1 was revealed with a purified polyclonal rabbit antibody, kindly provided by A. Nairn. WHx proteins were revealed with either polyclonal rabbit anti-WHx antibody or monoclonal anti-hemagglutinin (HA) antibody 16B12 (BabCo), also used for the detection of HA-HBx. Endogenous p53 was revealed with the monoclonal anti-P53 antibody DO-1 (Santa Cruz Biotechnology). Transfected and endogenous β-catenins were revealed with the mouse monoclonal anti-β-catenin antibody clone 14 (BD Biosciences).
Determination of X half-life.
Twenty-four hours posttransfection by the Lipofectamine-Plus method, cells were starved for 30 min in methionine- and cysteine-free minimal essential medium (ICN). The cells were then labeled for 15 min in fresh methionine- and cysteine-free minimal essential medium containing 0.2 mCi of a [35S]methionine-[35S]cysteine mixture (TRAN 35S Label; ICN). The labeling medium was removed, and the cells were lysed following various chase periods performed by further incubation in medium containing serum. The cell lysates were processed for immunoprecipitation as described above.
RESULTS
X protein half-life is increased by its interaction with DDB1.
One previously reported feature of X protein is its rapid turnover (37). Since DDB1 behaves as a key upstream element in the control of X activity (42, 43), we reasoned that it might be involved in the control of X degradation. To address this hypothesis, we transiently transfected an expression construct of WHx into Chang cells and we first monitored the decay of the viral protein in pulse-chase labeling experiments.
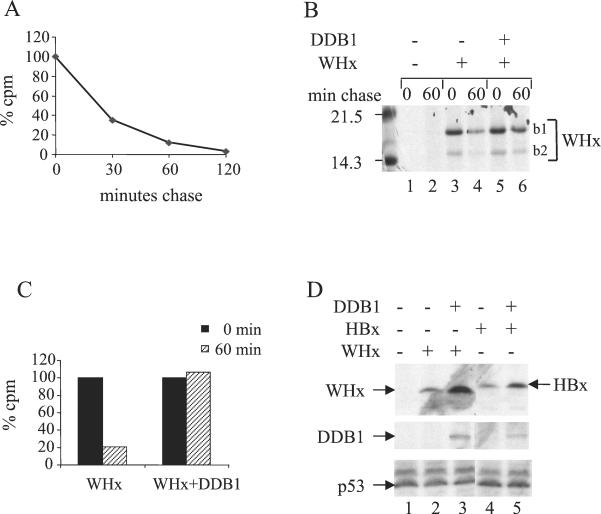
In agreement with the results obtained in other experimental systems (9, 37), we observed that WHx was rapidly degraded, with <40% of the labeled pool remaining after a 30-min chase period (Fig. 1A). Coexpression of DDB1 dramatically increased the half-life of the protein, since no measurable decay of WHx was observed after a 1-h chase (Fig. 1B and C).
FIG. 1.
DDB1 specifically modulates X protein stability. (A) Decay kinetics of WHx protein. An episomal construct of HA-WHx (0.2 μg) was transfected into Chang cells, and 24 h posttransfection, the cells were labeled for 15 min with a [35S]methionine-[35S]cysteine mixture. At the indicated times of chase with normal medium, the cells were lysed, and the cell lysates were processed for immunoprecipitation with anti-WHx polyclonal antibody. Proteins were separated by SDS-15% PAGE. Signals were quantified using a phosphorimager. The data are presented as the percentage of WHx signal obtained at the beginning of chase. (B and C) Comparison of WHx decay in the absence (−) or presence (+) of ectopic DDB1. Cells were transfected with 0.2 μg of HA-WHx construct together with 0.6 μg of either empty pcDNA3 or pcDNA3 expressing DDB1. Pulse-chase labeling and immunoprecipitation were performed as described for panel A. (B) Autoradiogram of the gel. Note that two WHx products were immunoprecipitated. The upper band (b1) corresponds to full-length HA-WHx. The lower band (b2) migrates at the size expected for nontagged WHx produced by internal translation initiation at the normal AUG codon of the protein. (C) Quantification of the signals shown in panel B. (D) Effect of DDB1 expression on the steady-state level of X protein. Chang cells were transfected using Ca2PO4 precipitates of 0.1 μg of X-expressing constructs with 0.5 μg of either empty vector or myc-tagged DDB1 construct. The cell lysates were separated by SDS-PAGE followed by Western blot analysis using monoclonal anti-HA antibody to reveal WHx and HBx (top gel), monoclonal anti-myc antibody to reveal DDB1 (middle gel), and monoclonal anti-p53 (bottom gel).
At the steady-state level, WHx abundance strongly increased when the protein was coexpressed with DDB1 (Fig. 1D). A similar effect was observed with HBx, the X protein encoded by HBV (Fig. 1D, compare lanes 4 and 5). The latter observation excluded any peculiarity of WHx due to its heterologous expression in human cells. In contrast, the abundance of another short-lived protein, p53, was not affected by overexpression of DDB1 (Fig. 1D). Taken together, the observations shown in Fig. 1 indicate that DDB1 specifically up-regulates X protein stability.
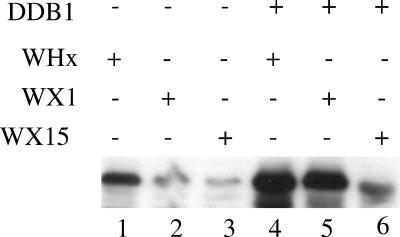
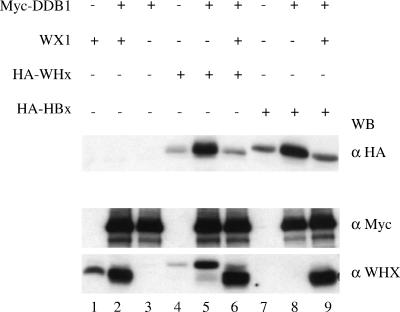
Whether stabilization of X by DDB1 involved the physical interaction between the two proteins was investigated using two previously characterized WHx mutants: WX1 (V72D), which shows gain of affinity to DDB1, and WX15 (N86D and Q95H), which shows strong loss of affinity to DDB1 (43). As shown in Fig. 2, DDB1 expression strongly increased accumulation of both wild-type WHx and WX1, whereas it barely affected WX15 accumulation. In addition, when expressed in trans, the WX1 gain-of-binding mutant prevented DDB1-mediated accumulation of both wild-type WHx and wild-type HBx (Fig. 3, compare lanes 5 and 6 to lanes 8 and 9).
FIG. 2.
Stabilization of X protein requires its efficient binding to DDB1. Cells were transfected with the indicated combinations of expression constructs (WHx, WX1, and WX15, 0.2 μg; empty vector and DDB1, 0.6 μg). The cell extracts were analyzed by Western blotting using polyclonal anti-WHx antibody. +, present; −, absent.
FIG. 3.
DDB1-mediated stabilization of wild-type X is inhibited by trans expression of gain-of-binding WX1 mutant. Cells were transfected with the indicated combinations of expression constructs (HA-WHx and HA-HBx, 40 ng; WX1, 2 μg; myc-DDB1, 0.2 μg), and the cell extracts were analyzed by Western blotting using the indicated antibodies. Note that anti-WHx (α WHX) reveals both the HA-WHx target protein and the nontagged WX1 used as a competitor. +, present; −, absent.
Taken together, the results shown in Fig. 2 and 3 indicate that stabilization of X requires its efficient binding to DDB1 and that occupancy of DDB1 at its X-binding site represents a way of controlling the pool of X protein.
Fusion of the DDB1-binding domain of X to an irrelevant short-lived protein confers DDB1-mediated stabilization.
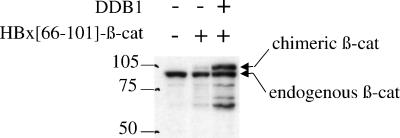
Besides X, DDB1 binds to several proteins, including the V protein of simian virus 5 (20) and the cellular transcription factor E2F1, the latter interaction occurring in the context of the DDB complex (15, 39). However, none of these proteins was stabilized when coexpressed with DDB1 (data not shown). This suggested that the nature of the interaction with DDB1, rather than interaction by itself, led to the stabilization effect. To address this point, we fused the HBx region encompassing residues 66 to 101, which includes the DDB1-binding site of X (44), to β-catenin, a known short-lived protein (32, 36). As shown in Fig. 4, ectopic expression of DDB1 led to a strong increase in the steady-state level of the chimeric β-catenin protein. In contrast, the turnover of β galactosidase, a known stable protein, did not change upon fusion with the DDB1-binding domain of X (data not shown).
FIG. 4.
β-Catenin fused to the DDB1-binding domain of HBx is sensitive to DDB1-mediated stabilization. Cells were cotransfected with 0.1 μg of an expression construct of the human β-catenin N-terminally fused to the 66-to-101 region of HBx (HBx[66-101]-β-cat) and 0.5 μg of empty vector or vector expressing DDB1. The cell extracts were subjected to Western blot analysis using a monoclonal anti-β-catenin antibody, which reveals both endogenous β-catenin and chimeric β-catenin. +, present; −, absent.
Therefore, the DDB1 stabilization effect can be conferred on an irrelevant unstable protein provided that it expresses the DDB1-binding X epitope.
DDB complex formation interferes with DDB1-mediated X stabilization.
As mentioned above, although DDB1 is predicted to bear DDB2-independent activities, it has so far been functionally characterized only in the context of the DDB heterodimer. We therefore investigated whether DDB1-mediated stabilization of X protein was functionally connected to DDB complex formation by comparing X accumulation when expressed alone or in combination with either a single DDB subunit or both subunits.
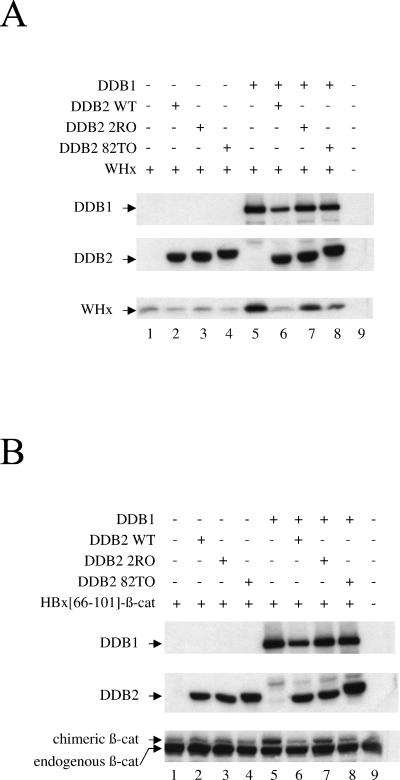
In our experiments, we included two naturally occurring DDB2 mutants isolated from patients suffering from genetic deficiency in DNA repair (xeroderma pigmentosum group E). Both the 2RO and 82TO DDB2 mutants (consisting of an R273H and a K244E substitution, respectively) are compromised in the formation of a functional DDB complex in the nucleus (17, 39). 2RO is deficient in DDB1 binding, whereas 82TO still interacts with DDB1 and the origin of its functional defect remains unclear (39).
The results in Fig. 5A show that the basal level of X protein was not affected by overexpression of the DDB2 subunit alone (compare lanes 2 to 4 to lane 1). In contrast, DDB1-mediated stabilization of X did not occur when DDB1 was expressed along with wild-type DDB2 (Fig. 5A, compare lanes 5 and 6). Under the same conditions, the 2RO mutant had only slight effect on X accumulation (Fig. 5A, lane 7), whereas the 82TO mutant showed a phenotype intermediate between wild-type DDB2 and the 2RO mutant (Fig. 5A, compare lane 8 to lanes 6 and 7). As shown in Fig. 5B, β-catenin fused to the HBx DDB1-binding tag behaved similarly to full-length WHx. Together, the results shown in Fig. 5 strongly suggest that, depending on its ability to efficiently interact with DDB1, DDB2 impairs stabilization of X by DDB1.
FIG. 5.
Effect of DDB subunits on X stability. (A) Cells were transfected with an expression construct of full-length WHx (40 ng), along with constructs encoding DDB1 (0.2 μg) and wild-type (WT) or mutant (2RO and 82TO) DDB2 (2 μg), as indicated. The cell extracts were analyzed by Western blotting using monoclonal anti-HA antibody to reveal HA-tagged WHx protein (bottom gel) and monoclonal anti-myc antibody to reveal myc-tagged DDB1 (top gel) and myc-tagged DDB2 (middle gel). +, present; −, absent. (B) Cells were transfected and analyzed as described for panel A except that full-length WHx was replaced by the chimeric HBx(66-101)-β-catenin protein, which was revealed by monoclonal anti β-catenin antibody.
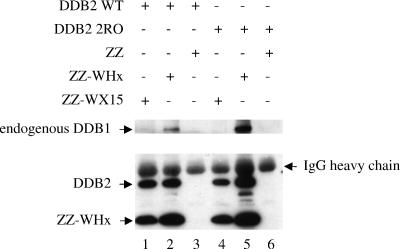
However, Nag et al. recently reported a previously unsuspected direct interaction between the HBx protein and a DDB2 subunit (26). Therefore, the possibility that the 2RO mutant was deficient in the binding of both X and DDB1 remained to be investigated. To this end, we performed coimmunoprecipitation experiments using myc-tagged DDB2 and X protein fused to the IgG-binding domain ZZ tag (43) (Fig. 6). We used two isoforms of DDB2, either the wild-type protein or the DDB1-binding-deficient mutant 2RO, as well as two isoforms of WHx, either the wild-type protein or the DDB1-binding-deficient mutant WX15.
FIG. 6.
WHx interacts with DDB2 independently of DDB1 binding. Cells were transfected with the indicated ZZ constructs (1 μg) together with the indicated DDB2 construct (0.5 μg). Cell lysates were processed for immunoprecipitation with rabbit IgG-coupled Sepharose beads, which specifically retain the ZZ IgG-binding tag. The retained proteins were eluted from the beads and were subjected to Western blot analysis using a monoclonal anti-myc antibody to reveal myc-DDB2 and ZZ-fused WHx and WX15 proteins (bottom gel). Part of the coupled IgG is released from the beads during the elution process, leading to a visible signal corresponding to the IgG heavy chain. Endogenous DDB1 was detected using a polyclonal anti-DDB1 antibody (top gel). WT, wild type; +, present; −, absent.
As expected, endogenous DDB1 coimmunoprecipitated with wild-type WHx (Fig. 6, lanes 2 and 5) but not with WX15 (Fig. 6, lanes 1 and 4). Regardless of its DDB1-binding status, DDB2 efficiently coimmunoprecipitated with wild-type WHx (Fig. 6, lanes 2 and 5), as well as with the WX15 mutant (Fig. 6, lanes 1 and 4). These observations, therefore, show that DDB2 interaction is conserved by WHx, extending previous observations made with HBx (26). Since WX15 still interacted with DDB2, these results further suggest that on the X molecule, the DDB2-binding site is likely distinct from the DDB1-binding site.
In summary, the DDB2 mutant 2RO, which shows loss of binding to DDB1 (39), was inactive in relieving X stabilization by DDB1 (Fig. 5), although it efficiently bound to WHx (Fig. 6). Taken together, these data strongly suggest that DDB2 acts on X stability in an indirect way involving the formation of the DDB heterodimer, in contrast to DDB1, which appears to directly protect X from the degradation process.
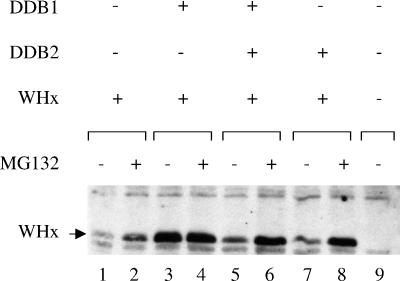
DDB1 protects X protein from proteasome-dependent degradation.
Earlier studies have reported that the short half-life of the X protein reflects its rapid degradation by the proteasome pathway (13, 16, 41). We therefore used the MG132 proteasome inhibitor to gain insight into the mechanisms used by DDB1 and the DDB heterodimer to modulate X protein stability (Fig. 7). MG132 treatment strongly increased the amount of intact X protein when it was expressed alone (Fig. 7, lanes 1 and 2) or in combination either with DDB2 (Fig. 7, lanes 7 and 8) or with DDB1 and DDB2 (Fig. 7, lanes 5 and 6). In contrast, when the X protein was coexpressed with DDB1 alone, the pool of intact X protein reached a maximal level which was not further increased upon MG132 treatment (Fig. 7, lanes 3 and 4).
FIG. 7.
DDB1 prevents proteasome-dependent degradation of X protein. Cells were transfected by the Ca2PO4 procedure using 0.1 μg of the HA-WHx construct, 0.5 μg of the DDB1 construct, and 4 μg of the DDB2 construct in the indicated combinations. For each DNA combination, a single precipitate was formed and split on two dishes; 36 h posttransfection, the medium was replaced by normal medium or medium containing 50 μM MG132, and the cells were further incubated for 5 h before lysis. The cell lysates were analyzed by Western blotting using monoclonal anti-HA antibody. +, present; −, absent.
We conclude from these observations that in the presence of an excess amount of DDB1 subunit, the proteasome pathway can no longer operate on X protein, whereas further addition of the DDB2 subunit restores X targeting to proteasome-mediated degradation.
DISCUSSION
In this study, we demonstrate that the turnover rate of X protein is regulated by a network of protein-protein interactions involving the host cell DDB complex. This is the first report of a mechanism operating on X activity through the modulation of its stability.
A novel mechanism controlling X activity by the regulation of its steady-state level.
Earlier, we showed that ectopic expression of DDB1 increases both the transactivation and proapoptotic activities of X protein (43). Here, we show that DDB1 enhances the steady-state levels of both the WHx and HBx proteins and that this stabilization effect requires efficient X-DDB1 interaction. Therefore, our present findings give insight into a molecular mechanism by which DDB1 up-regulates X protein activity.
Stabilization of the p53 transcription factor induced by the corepressor Sin3a (47) and stabilization of the yeast transcription factor Crz1p induced by the response regulator transcription factor Skn7p (46) provide related examples of specific stabilization effects mediated by physical interactions. In both cases, the prolonged half-life of the stabilized protein might ensure its efficient recruitment into a particular functional pathway by the other partner. In other examples, the stabilized protein is the target of a chaperone, which assists it in acquisition of a specific folding. Thus, proper binding of Hsp90 chaperone to its target proteins, which include steroid hormone nuclear receptors and protein kinases, is essential for both their ability to transduce signal and their stability (33). That DDB1 does not modify the steady-state level of all its binding partners argues against a general chaperone function of DDB1. However, the DDB1-mediated stabilization conferred on β-catenin protein by the fused DDB1-binding domain of X might be indicative of a specific chaperone-like effect of DDB1. Such a cis-dominant effect is indeed observed with the ligand-binding domain of estrogen receptor, which confers Hsp90-mediated hormone dependence on a fused irrelevant protein, a property widely used to obtain conditionally active proteins (34).
Tight connection of X protein with the cellular DDB complex.
Our finding of direct binding of WHx to DDB2, the second subunit of the DDB complex, was quite unexpected. Indeed, simple indirect interaction of DDB2 with WHx via a bridging effect of DDB1 could have been predicted. Notably, Nag et al. (26) described direct interaction between HBx and DDB2. Given that HBx and WHx show only 50% identity at the amino acid level (44), conservation of DDB2 binding most likely reflects a functional constraint relevant to X protein activity in viral infection.
Since the X protein can engage separate interactions with DDB1 and DDB2 and since DDB subunits interact with each other, three pairs of direct interactions have to be envisioned. We have shown a requirement for X-DDB1 interaction in nuclear accumulation of the WHx protein in stable cell lines (43). Shiyanov et al. (39) and Liu et al. (22) reported DDB2-mediated nuclear import of DDB1. Nag et al. (26) showed that in transient transfection experiments, ectopic DDB2 targets HBx to the nucleus through a mechanism dependent on X-DDB2 interaction but independent of DDB1-DDB2 interaction. Finally, data of Forgues et al. (14) suggest the existence of a nuclear export signal in the X protein. Taken together, these observations suggest that DDB2 has to engage separate interactions with DDB1 and with X to import the two proteins into the nucleus and that once in the nucleus, X is retained in this compartment via its interaction with DDB1. In this view, each of the three interactions involving DDB subunits and the X protein plays a role in X compartmentation within the cell. It should also be recalled that the gap in the positive strand of mammalian hepadnavirus genomes is repaired by a host cell-dependent mechanism which has yet to be characterized. As an attractive speculation, the DDB subunits through their physical interaction with X might acquire the function of signaling the viral DNA gap to the host DNA repair machinery.
Regarding the stability of X protein, our present data show that DDB1 may be the important node in the interaction network: DDB1 has to bind to X to protect it from degradation, and DDB2 has to bind to DDB1 to relieve this effect. Of note, our data do not exclude the possibility that X-DDB1 and DDB2-DDB1 interactions are mutually exclusive. In that case, the DDB2 effect would be merely displacement of DDB1-X interaction. However, we do not favor this hypothesis because reciprocal displacement of DDB1-DDB2 interaction by X protein overexpression would lead to inhibition of Ddb activity, which we did not observe (43). Finally, the fact that ectopic expression of DDB2 affects X stability only when coexpressed with ectopic DDB1 indicates that endogenous DDB1 available for binding to both X and DDB2 is limiting. Since DDB1 is known to be abundant (39), this might indicate that a large fraction of the DDB1 pool is recruited for functions other than damaged DNA recognition, as previously suggested (48).
Functional connection of X protein with the proteasome pathway.
Rapid turnover of the X protein has previously been shown to reflect its targeting to proteasome-mediated degradation through ubiquitination (16). Ubiquitination of proteins is a stepwise process in which a given E3 ligase complex specifically recognizes the substrate protein, which subsequently undergoes covalent ubiquitin linkage at specific lysine residues. The latter step is catalyzed by E1 and E2 enzymes (for reviews, see references 19 and 31). DDB1-mediated protection of X by simple masking of the ubiquitination site can be excluded, since the 66-to-100 region of WHx, which includes the DDB1-binding domain, is devoid of lysine residues. Therefore, DDB1 more likely acts on the E3 ligase recognition step. The DDB1-binding domain of X might contain an intrinsic destabilization motif, recognized by E3 ligase in the absence of DDB1 masking. However, the steady-state level of β-galactosidase, a known long-lived protein, did not decrease when the protein is fused with the DDB1-binding domain of either WHx or HBx. In contrast, the DDB1-binding domain of X conferred DDB1-mediated stabilization on the irrelevant short-lived β-catenin protein. In combination, these data suggest that upon interaction with the X domain, DDB1 transmits a signal of conformational change to the entire molecule, which impairs further recognition by E3 ligases.
Remarkably, DDB2 is itself targeted to proteasome-dependent degradation by its direct interaction with cullin 4A (25), a known component of a specific E3 ligase complex (10, 30). The DDB2 mutant 2RO, which is deficient in DDB1 binding (40), is also incompetent in cullin 4A-mediated degradation (25). Therefore, an attractive hypothesis is that DDB2 restores signaling of X to proteasome-mediated degradation through the recruitment of cullin 4A to the X-DDB1-DDB2 complex.
Hu et al. (16) suggested that X might be both a substrate and an inhibitor of proteasome. Together with the present data, the possibility that the X protein in its DDB1-bound state traps essential components of the proteasome pathway is interesting to consider. In our study, coexpression of X with DDB1 did not affect the stability of endogenous short-lived proteins, such as p53 and β-catenin. However, the possibility remains that the stability of still-unknown cellular substrates sharing specific components with X in their processing by the proteasome are indeed affected by X expression.
The role of DDB1-X interaction is unlikely to be restricted to up-regulation of X stability. Indeed, gain of binding to DDB1 in the WX1 mutant is associated with loss of activity both in vitro and in vivo in the viral infection process (42, 43), although the mutant still responds to DDB1-mediated stabilization (this study). In addition, we show here that the DDB1 stabilization effect can be competed out by the WX1 mutant. This observation provides an explanation for the previously described trans-dominant negative effect of this mutant on X activity (43). It also gives further support to the potential therapeutic value of agents able to occupy DDB1 at its X-binding site. Such agents may show a dual inhibitory action: impairing the activity of X protein and inducing its destabilization.
Acknowledgments
We thank S. Linn, W. S. Mason, A. C. Nairn, J. Seeler, and R. A. Lamb for providing useful reagents. We are grateful to Y. Wei and C. Labalette for critical reading of the manuscript. We thank C.-A. Renard and M.-L. Michel for helpful discussions. We thank J. Bianchi and C. Legout for technical assistance. This work was performed in the INSERM U163 research unit headed by P. Tiollais.
REFERENCES
- 1.Abramic, M., A. S. Levine, and M. Protic. 1991. Purification of an ultraviolet-inducible, damage-specific DNA-binding protein from primate cells. J. Biol. Chem. 266:22493-22500. [PubMed] [Google Scholar]
- 2.Arbuthnot, P., A. Capovilla, and M. Kew. 2000. Putative role of hepatitis B virus X protein in hepatocarcinogenesis: effects on apoptosis, DNA repair, mitogen-activated protein kinase and JAK/STAT pathways. J. Gastroenterol. Hepatol. 15:357-368. [DOI] [PubMed] [Google Scholar]
- 3.Arbuthnot, P., and M. Kew. 2001. Hepatitis B virus and hepatocellular carcinoma. Int. J. Exp. Pathol. 82:77-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beasley, R. P., L. Y. Hwang, C. C. Lin, and C. S. Chien. 1981. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet 2:1129-1133. [DOI] [PubMed] [Google Scholar]
- 5.Bergametti, F., S. Prigent, B. Luber, A. Benoit, P. Tiollais, A. Sarasin, and C. Transy. 1999. The proapoptotic effect of hepatitis B virus HBx protein correlates with its transactivation activity in stably transfected cell lines. Oncogene 18:2860-2871. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H. S., S. Kaneko, R. Girones, R. W. Anderson, W. E. Hornbuckle, B. C. Tennant, P. J. Cote, J. L. Gerin, R. H. Purcell, and R. H. Miller. 1993. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J. Virol. 67:1218-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu, G., and E. Chang. 1988. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science 242:564-567. [DOI] [PubMed] [Google Scholar]
- 8.Colgrove, R., G. Simon, and D. Ganem. 1989. Transcriptional activation of homologous and heterologous genes by the hepatitis B virus X gene product in cells permissive for viral replication. J. Virol. 63:4019-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dandri, M., J. Petersen, R. J. Stockert, T. M. Harris, and C. E. Rogler. 1998. Metabolic labeling of woodchuck hepatitis B virus X protein in naturally infected hepatocytes reveals a bimodal half-life and association with the nuclear framework. J. Virol. 72:9359-9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 11.Diao, J., R. Garces, and C. D. Richardson. 2001. X protein of hepatitis B virus modulates cytokine and growth factor related signal transduction pathways during the course of viral infections and hepatocarcinogenesis. Cytokine Growth Factor Rev. 12:189-205. [DOI] [PubMed] [Google Scholar]
- 12.Dualan, R., T. Brody, S. Keeney, A. F. Nichols, A. Admon, and S. Linn. 1995. Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics 29:62-69. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, M., L. Runkel, and H. Schaller. 1995. HBx protein of hepatitis B virus interacts with the C-terminal portion of a novel human proteasome alpha-subunit. Virus Genes 10:99-102. [DOI] [PubMed] [Google Scholar]
- 14.Forgues, M., A. J. Marrogi, E. A. Spillare, C. G. Wu, Q. Yang, M. Yoshida, and X. W. Wang. 2001. Interaction of the hepatitis B virus X protein with the Crm1-dependent nuclear export pathway. J. Biol. Chem. 276:22797-22803. [DOI] [PubMed] [Google Scholar]
- 15.Hayes, S., P. Shiyanov, X. Chen, and P. Raychaudhuri. 1998. DDB, a putative DNA repair protein, can function as a transcriptional partner of E2F1. Mol. Cell. Biol. 18:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, Z., Z. Zhang, E. Doo, O. Coux, A. L. Goldberg, and T. J. Liang. 1999. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J. Virol. 73:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang, B. J., S. Toering, U. Francke, and G. Chu. 1998. p48 activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol. Cell. Biol. 18:4391-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keeney, S., G. J. Chang, and S. Linn. 1993. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J. Biol. Chem. 268:21293-21300. [PubMed] [Google Scholar]
- 19.Laney, J. D., and M. Hochstrasser. 1999. Substrate targeting in the ubiquitin system. Cell 97:427-430. [DOI] [PubMed] [Google Scholar]
- 20.Lin, G. Y., R. G. Paterson, C. D. Richardson, and R. A. Lamb. 1998. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology 249:189-200. [DOI] [PubMed] [Google Scholar]
- 21.Lin-Marq, N., S. Bontron, O. Leupin, and M. Strubin. 2001. Hepatitis B virus X protein interferes with cell viability through interaction with the p127-kDa UV-damaged DNA-binding protein. Virology 287:266-274. [DOI] [PubMed] [Google Scholar]
- 22.Liu, W., A. F. Nichols, J. A. Graham, R. Dualan, A. Abbas, and S. Linn. 2000. Nuclear transport of human DDB protein induced by ultraviolet light. J. Biol. Chem. 275:21429-21434. [DOI] [PubMed] [Google Scholar]
- 23.Lok, A. S., E. J. Heathcote, and J. H. Hoofnagle. 2001. Management of hepatitis B: 2000—summary of a workshop. Gastroenterology 120:1828-1853. [DOI] [PubMed] [Google Scholar]
- 24.Murakami, S. 2001. Hepatitis B virus X protein: a multifunctional viral regulator. J. Gastroenterol. 36:651-660. [DOI] [PubMed] [Google Scholar]
- 25.Nag, A., T. Bondar, S. Shiv, and P. Raychaudhuri. 2001. The xeroderma pigmentosum group e gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 21:6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nag, A., A. Datta, K. Yoo, D. Bhattacharyya, A. Chakrabortty, X. Wang, B. L. Slagle, R. H. Costa, and P. Raychaudhuri. 2001. DDB2 induces nuclear accumulation of the hepatitis b virus x protein independently of binding to DDB1. J. Virol. 75:10383-10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nassal, M., and H. Schaller. 1996. Hepatitis B virus replication—an update. J. Viral Hepat. 3:217-226. [DOI] [PubMed] [Google Scholar]
- 28.Nichols, A. F., T. Itoh, J. A. Graham, W. Liu, M. Yamaizumi, and S. Linn. 2000. Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J. Biol. Chem. 275:21422-21428. [DOI] [PubMed] [Google Scholar]
- 29.Nichols, A. F., P. Ong, and S. Linn. 1996. Mutations specific to the xeroderma pigmentosum group E Ddb-phenotype. J. Biol. Chem. 271:24317-24320. [DOI] [PubMed] [Google Scholar]
- 30.Ohta, T., J. J. Michel, A. J. Schottelius, and Y. Xiong. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3:535-541. [DOI] [PubMed] [Google Scholar]
- 31.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 32.Polakis, P. 2001. More than one way to skin a catenin. Cell 105:563-566. [DOI] [PubMed] [Google Scholar]
- 33.Pratt, W. B. 1998. The hsp90-based chaperone system: involvement in signal transduction from a variety of hormone and growth factor receptors. Proc. Soc. Exp. Biol. Med. 217:420-434. [DOI] [PubMed] [Google Scholar]
- 34.Pratt, W. B., and D. O. Toft. 1997. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 18:306-360. [DOI] [PubMed] [Google Scholar]
- 35.Rossner, M. T. 1992. Hepatitis B virus X-gene product: a promiscuous transcriptional activator. J. Med. Virol. 36:101-117. [DOI] [PubMed] [Google Scholar]
- 36.Rubinfeld, B., P. Robbins, M. El-Gamil, I. Albert, E. Porfiri, and P. Polakis. 1997. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science 275:1790-1792. [DOI] [PubMed] [Google Scholar]
- 37.Schek, N., R. Bartenschlager, C. Kuhn, and H. Schaller. 1991. Phosphorylation and rapid turnover of hepatitis B virus X-protein expressed in HepG2 cells from a recombinant vaccinia virus. Oncogene 6:1735-1744. [PubMed] [Google Scholar]
- 38.Schödel, F., R. Sprengel, T. Weimer, D. Fernholz, R. Schneider, and H. Will. 1989. Animal hepatitis B viruses, p. 73-102. In G. Klein (ed.), Advances in viral oncology, vol. 8. Raven Press, New York, N.Y. [Google Scholar]
- 39.Shiyanov, P., S. A. Hayes, M. Donepudi, A. F. Nichols, S. Linn, B. L. Slagle, and P. Raychaudhuri. 1999. The naturally occurring mutants of DDB are impaired in stimulating nuclear import of the p125 subunit and E2F1-activated transcription. Mol. Cell. Biol. 19:4935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiyanov, P., A. Nag, and P. Raychaudhuri. 1999. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J. Biol. Chem. 274:35309-35312. [DOI] [PubMed] [Google Scholar]
- 41.Sirma, H., R. Weil, O. Rosmorduc, S. Urban, A. Israel, D. Kremsdorf, and C. Brechot. 1998. Cytosol is the prime compartment of hepatitis B virus X protein where it colocalizes with the proteasome. Oncogene 16:2051-2063. [DOI] [PubMed] [Google Scholar]
- 42.Sitterlin, D., F. Bergametti, P. Tiollais, B. C. Tennant, and C. Transy. 2000. Correct binding of viral X protein to UVDDB-p127 cellular protein is critical for efficient infection by hepatitis B viruses. Oncogene 19:4427-4431. [DOI] [PubMed] [Google Scholar]
- 43.Sitterlin, D., F. Bergametti, and C. Transy. 2000. UVDDB p127-binding modulates activities and intracellular distribution of hepatitis B virus X protein. Oncogene 19:4417-4426. [DOI] [PubMed] [Google Scholar]
- 44.Sitterlin, D., T. H. Lee, S. Prigent, P. Tiollais, J. S. Butel, and C. Transy. 1997. Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J. Virol. 71:6194-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang, J. Y., B. J. Hwang, J. M. Ford, P. C. Hanawalt, and G. Chu. 2000. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell 5:737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams, K. E., and M. S. Cyert. 2001. The eukaryotic response regulator Skn7p regulates calcineurin signaling through stabilization of Crz1p. EMBO J. 20:3473-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zilfou, J. T., W. H. Hoffman, M. Sank, D. L. George, and M. Murphy. 2001. The corepressor mSin3a interacts with the proline-rich domain of p53 and protects p53 from proteasome-mediated degradation. Mol. Cell. Biol. 21:3974-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zolezzi, F., and S. Linn. 2000. Studies of the murine DDB1 and DDB2 genes. Gene 245:151-159. [DOI] [PubMed] [Google Scholar]
- 49.Zoulim, F., J. Saputelli, and C. Seeger. 1994. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 68:2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]