Abstract
The pseudorabies virus (PrV) homolog of the tegument protein encoded by the UL48 gene of herpes simplex virus type 1 (HSV-1) was identified by using a monospecific rabbit antiserum against a bacterial fusion protein. UL48-related polypeptides of 53, 55, and 57 kDa were detected in Western blots of infected cells and purified virions. Immunofluorescence studies demonstrated that the PrV UL48 protein is predominantly localized in the cytoplasm but is also found in the nuclei of infected cells. Moreover, it is a constituent of extracellular virus particles but is absent from primary enveloped perinuclear virions. In noncomplementing cells, a UL48-negative PrV mutant (PrV-ΔUL48) exhibited delayed growth and significantly reduced plaque sizes and virus titers, deficiencies which were corrected in UL48-expressing cells. RNA analyses indicated that, like its HSV-1 homolog, the PrV UL48 protein is involved in regulation of immediate-early gene expression. However, the most salient effect of the UL48 gene deletion was a severe defect in virion morphogenesis. Late after infection, electron microscopy of cells infected with PrV-ΔUL48 revealed retention of newly formed nucleocapsids in the cytoplasm, whereas enveloped intracytoplasmic or extracellular complete virions were only rarely observed. In contrast, capsidless particles were produced and released in great amounts. Remarkably, the intracytoplasmic capsids were labeled with antibodies against the UL36 and UL37 tegument proteins, whereas the capsidless particles were labeled with antisera directed against the UL46, UL47, and UL49 tegument proteins. These findings suggested that the UL48 protein is involved in linking capsid and future envelope-associated tegument proteins during virion formation. Thus, like its HSV-1 homolog, the UL48 protein of PrV functions in at least two different steps of the viral life cycle. The drastic inhibition of virion formation in the absence of the PrV UL48 protein indicates that it plays an important role in virion morphogenesis prior to secondary envelopment of intracytoplasmic nucleocapsids. However, the UL48 gene of PrV is not absolutely essential, and concomitant deletion of the adjacent tegument protein gene UL49 also did not abolish virus replication in cell culture.
Pseudorabies virus (PrV, suid herpesvirus 1), the causative agent of Aujeszky's disease of pigs (41), has been classified as a member of the Varicellovirus genus within the Alphaherpesvirinae subfamily of Herpesviridae (49). Although the DNA sequence of the ca. 150-kbp genome of PrV has not yet been elucidated completely, gene content and arrangement appear to be widely similar to those of other alphaherpesviruses including herpes simplex virus type 1 (HSV-1) (38), and therefore the gene nomenclature established for HSV-1 has been adopted (41).
PrV virions exhibit the typical morphology of herpesvirus particles (26). The nucleoprotein core containing the DNA genome is enclosed in an icosahedral capsid shell. The capsid is surrounded by a protein layer named the tegument and a lipid membrane of cellular origin containing glycosylated and nonglycosylated virus-encoded proteins (41, 49). Nucleocapsids, which are formed in the nucleus of the host cell, are released by budding at the inner nuclear membrane, thereby acquiring a primary envelope (48). As demonstrated for several herpesviruses, the primary enveloped virions are de-enveloped at the outer nuclear membrane and nucleocapsids are released into the cytoplasm. Final tegumentation and envelopment then take place by budding of capsids into trans-Golgi-derived vesicles (reviewed in reference 42).
In the past, the tegument has been considered an amorphous bulk of numerous proteins, but recent studies indicate an at least partly ordered structure (10, 60). For HSV-1, more than 15 different viral gene products are considered to be components of the tegument of mature extracellular virions (42, 48). Among them, the tegument proteins encoded by UL36 and UL37 have been shown to play important roles during virion morphogenesis of PrV and HSV-1 (14, 15, 31, 34). A direct association of the UL36 protein with the pentons of nucleocapsids in HSV-1 particles was found (60), and a physical interaction between the UL36 and UL37 proteins in PrV-infected cells (31) as well as in human cytomegalovirus (3; M. E. Harmon and W. Gibson, Proc. Am. Soc. Virol., abstr. W35-4, 1996) was shown. Apparently, the UL36 and UL37 gene products, which are conserved throughout all herpesvirus subfamilies, form the innermost layer of the tegument. In contrast, the other tegument proteins of alphaherpesviruses possess no significantly conserved homologs in the other herpesvirus subfamilies, and many of them, such as the protein kinases encoded by the UL13 and US3 genes, the viral host shutoff (vhs) protein encoded by the UL41 gene, and the UL46, UL47, and UL49 gene products, were shown to be nonessential for propagation of HSV-1 in cultured cells (47, 48; G. Elliott and A. Whiteley, Abstr. 26th Int. Herpesvirus Workshop, abstr. 8.09, 2001). For PrV it could be demonstrated that the US3, UL13, and UL49 homologs are also dispensable for replication (13, 16, 33, 55; W. Fuchs, B. G. Klupp, H. Granzow, A. Mundt, C. Hengartner, L. W. Enquist, and T. C. Mettenleiter, submitted for publication). The UL49 gene product of PrV may be located in the tegument close to the envelope, since it was shown to interact specifically with the cytoplasmic domains of the envelope glycoproteins gE and gM (Fuchs et al., submitted). The presence of either gE or gM is required for virion incorporation of the UL49 protein, but, whereas the absence of gE and gM is deleterious to virion morphogenesis (5, 6), the absence of the UL49 protein does not affect it detectably (13). Therefore, other virion glycoprotein-tegument protein interactions have to be present to ensure proper secondary envelopment.
Sequencing the PrV genome region adjacent to the UL49 gene identified homologs of the HSV-1 tegument protein genes UL48, UL47, and UL46 (7; Fuchs et al., submitted), but the respective proteins have been neither identified nor functionally characterized. The UL48 gene product of HSV-1, designated VP16, vmw65, or alpha trans-inducing factor, is considered to be essential for virus replication (45, 48, 57). It has primarily been characterized by its capability to transactivate expression of viral immediate-early genes by interaction with cellular transcription factors (2, 8, 52), a function that could also be demonstrated for the homologous gene products of bovine herpesvirus 1 (BHV-1) (43) and varicella-zoster virus (VZV) (44). However, the transactivating function of UL48 apparently is not required for expression of viral genes, since naked herpesvirus DNA is infectious after transfection into appropriate target cells (25, 50). In addition, the UL48 gene product of HSV-1 has been shown to modulate the activity of the vhs protein, thereby preventing rapid degradation of viral mRNA (36).
HSV-1 UL48 deletion mutants also exhibited a striking defect in virion formation, involving impaired DNA encapsidation in the nucleus and a failure to envelope intracytoplasmic capsids and to release mature virions (45, 57). Consequently, no viable progeny virus was detectable from noncomplementing cells infected with HSV-1 UL48 null mutants. Interestingly, neither VZV nor Marek's disease virus, alphaherpesviruses which, in contrast to PrV and HSV-1, are strictly cell associated in vitro, requires its UL48 homolog for productive replication in cell culture (12, 18). To investigate a functional role for PrV UL48 in virion morphogenesis and egress, we identified the PrV UL48 protein, constructed a PrV UL48 deletion mutant, and characterized its in vitro growth in noncomplementing and in UL48-expressing cells. The PrV UL48 protein was also analyzed for its influence on expression of the major viral immediate-early gene. Furthermore, a PrV mutant simultaneously lacking the nonessential UL49 gene (13) and UL48 was isolated and analyzed.
MATERIALS AND METHODS
Viruses and cells.
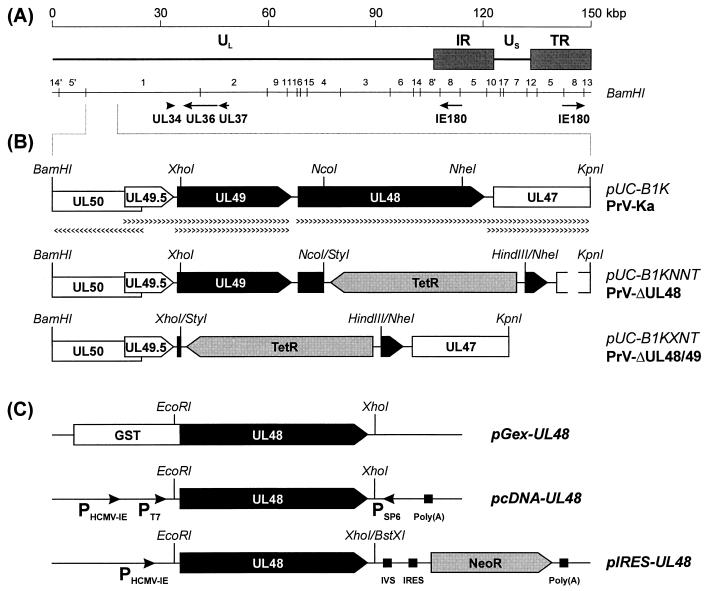
Virus mutants were generated by manipulation of the recently described full-length clone pPrV-K1 (21) of the laboratory strain PrV-Ka (30). Virus was propagated in rabbit kidney (RK13) cells, which were grown in minimum essential medium (Life Technologies) supplemented with 10% fetal calf serum. UL48-expressing cell lines were isolated after calcium phosphate-mediated transfection (24) of RK13 cells with plasmid pIRES-UL48 (Fig. 1C). After 48 h the transfected cells were trypsinized and seeded into microtiter plates with medium containing 500 μg of Geneticin (Life Technologies)/ml. Resistant cell colonies were tested for constitutive UL48 expression by indirect immunofluorescence tests. One positive cell clone, named RK-UL48, was used for trans-complementation studies with UL48-negative PrV mutants.
FIG. 1.
Generation of plasmids and virus mutants. (A) Schematic map of the PrV genome, which consists of a unique long (UL) region and a unique short (US) region, which is bracketed by inverted-repeat sequences (IR and TR). The positions of BamHI restriction fragments and of genes relevant for this study are indicated. (B) Enlarged map of a plasmid-cloned DNA fragment of PrV-Ka (pUC-B1K) including relevant restriction sites. Transcriptional organization of genes UL50 to UL47 (pointed rectangles) is also shown. Deletion mutants PrV-ΔUL48 and PrV-ΔUL48/49 contain the tetracycline resistance gene (encoding TetR), since they were generated by mutagenesis of an infectious clone of PrV-Ka with plasmid pUC-B1KNNT or pUCB1KXNT in E. coli. (C) A UL48-GST fusion protein was expressed from pGex-UL48, whereas pcDNA-UL48 was used for in vitro transcription from the bacteriophage T7 and SP6 promoters (PT7, PSP6) and subsequent in vitro translation. Plasmid pIRES-UL48 contains a bicistronic transcription unit of the UL48 gene and the neomycin resistance gene (NeoR), which are separated by an intron (IVS) and an internal ribosomal entry site (IRES) and which are flanked by the human cytomegalovirus immediate-early promoter (PHCMV-IE) and a polyadenylation signal [Poly(A)].
Construction of UL48 expression plasmids.
The complete UL48 open reading frame (ORF) was amplified from cloned PrV DNA by PCR using Pfx DNA polymerase (Life Technologies). For that purpose primers PUL48-F (GAGAATTCGTGAGGATGCGCGACGAGG; initiation codon underlined) and PUL48-R (GTGCTCGAGCGCCGCAGATCCGACC) were deduced from the recently determined DNA sequence (GenBank accession no. AJ437285). They contained artificial EcoRI and XhoI sites (italics) to facilitate insertion into appropriately cleaved expression vectors pGEX-4T-1 (Amersham Pharmacia) and pcDNA3 (Invitrogen). For cloning UL48 into plasmid pIRES1neo (Clontech) doubly digested with EcoRI and BstXI, the noncompatible single-stranded overhangs were blunt ended by incubation with Klenow polymerase. The resulting plasmids, pGEX-UL48 and pIRES-UL48 (Fig. 1C), were used for prokaryotic protein expression and for generation of cell line RK-UL48, respectively. The T7 promoter of pcDNA-UL48 (Fig. 1C) was used for in vitro transcription and translation of the cloned ORF (TNT coupled reticulocyte lysate system; Promega).
Preparation of a UL48-specific rabbit antiserum.
After transformation of Escherichia coli strain DH5α (Amersham Pharmacia) with pGEX-UL48, the complete UL48 ORF, preceded by 6 nucleotides of originally noncoding PrV DNA, was expressed as a 72-kDa fusion protein with glutathione S-transferase (GST; Fig. 1C). This fusion protein was purified (21), emulsified in mineral oil, and injected intramuscularly into a rabbit four times at 4-week intervals (100 μg of protein per dose) essentially as described previously (31). Sera collected before and 3 weeks after each immunization were analyzed.
Western blot analysis.
For Western blotting, RK13 cells were infected at a multiplicity of infection (MOI) of 5 and incubated on ice for 1 h. Then the inoculum was replaced by prewarmed medium, and incubation was continued at 37°C for 1 to 20 h. Samples of infected and noninfected cells, as well as PrV virions, were prepared as described previously (17), separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, and electrotransferred to nitrocellulose membranes (Schleicher & Schuell). Blots were blocked with 5% low-fat milk in phosphate-buffered saline (PBS) and incubated for 1 h with diluted rabbit sera against the UL34 (1:105) (32), UL48 (1:105), UL49 (1:105) (6), and UL49.5 (1:5,000) (29) gene products of PrV. The bound antibody was detected with peroxidase-conjugated secondary antibodies (Dianova) and visualized by chemiluminescence (Super Signal; Pierce) recorded on X-ray films.
Indirect immunofluorescence and confocal microscopy.
RK13 cells grown on coverslips were infected for 20 h at a MOI of ca. 0.001 with PrV-Ka. Thereafter, they were fixed for 10 min with a 1:1 mixture of methanol and acetone and subsequently incubated for 1 h each with anti-UL48 serum (dilution, 1:500) and Alexa 488-conjugated secondary antibodies (Molecular Probes). Slides were washed repeatedly with PBS after each step. Fluorescence was preserved with a 9:1 mixture of glycerol and PBS containing 25 mg of 1,4-diazabicyclooctane per ml and 1 μg of propidium iodide per ml for chromatin counterstaining. The slides were analyzed with a confocal laser scan microscope (LSM 510; Zeiss).
Generation of PrV UL48 deletion mutants.
For generation of a PrV UL48 deletion mutant, a 3,605-bp BamHI-KpnI fragment of the PrV genome was cloned in pUC-19 (pUC-B1K; Fig. 1B). For deletion of UL48 (codons 58 to 365) and for concomitant deletion of UL49 (codon 4 to end) a 928-bp NcoI-NheI fragment and a 1,899-bp XhoI-NheI fragment, respectively, were removed from pUC-B1K and replaced by the tetracycline resistance gene, which had been isolated as a 1,340-bp HindIII-StyI fragment from plasmid pBR322 (Fig. 1B). In all experiments, blunt DNA ends were generated by treatment with Klenow polymerase prior to ligation of noncompatible restriction fragments. From the resulting plasmids, pUC-B1KNNT and pUC-B1KXNT (Fig. 1B), the mutated virus genes were amplified by PCR with PrV-specific primers PUL48-F and PUL48-R (see above) or PUL49-F (CCCACTCGCTCGCCATGTCCAG) and PUL48-R. The isolated PCR products were used for RecE- and RecT-mediated mutagenesis (58) of a full-length clone of the PrV genome (pPrV-K1) (21) in E. coli. Recombinant bacteria were selected and propagated in medium containing 30 μg of chloramphenicol/ml and 10 μg of tetracycline/ml. To confirm the expected deletions, DNA of double-resistant clones was characterized by blot hybridization and sequencing. Since the bacterial vector insertion at the gG gene locus of pPrV-K1 and its derivatives adversely affects virus replication, the respective DNA sequences were removed by cotransfection of RK13 cells with the cloned PrV genome and a plasmid containing the authentic gG gene (21). Finally, the obtained virus recombinants, PrV-ΔUL48 and PrV-ΔUL48/49 (Fig. 1B), were plaque purified and further characterized.
RNA analyses.
RK13 and RK-UL48 cells were infected with PrV-Ka or PrV-ΔUL48 at a MOI of 20 and incubated for 3 or 6 h at 37°C in the presence or absence of cycloheximide (100 μg/ml). Total RNA was prepared, separated in agarose gels, transferred to nylon membranes, and hybridized as described previously (23), except that due to the high G+C content of the PrV genome blots were washed at 78°C. For identification of the UL48 mRNA, a radiolabeled antisense cRNA was transcribed from pcDNA-UL48 (Fig. 1C) with SP6 RNA polymerase (SP6/T7 transcription kit; Roche). 32P-labeled (Rediprime II system; Amersham Pharmacia) plasmid-cloned BamHI fragment 8 of the PrV genome (Fig. 1A) was used to detect the mRNA of major immediate-early protein IE180.
One-step growth analysis and determination of plaque sizes.
Confluent monolayers of RK13 and RK-UL48 cells were infected at a MOI of 2 with PrV-Ka or with the described deletion mutants grown on noncomplementing cells and incubated on ice for 1 h. Then, prewarmed medium was added, and incubation was continued at 37°C. After 1 h, nonpenetrated virus was inactivated by low-pH treatment (40), and 3, 6, 9, 12, 24, and 48 h after the temperature shift cells were scraped into the medium and lysed by freezing (−70°C) and thawing (37°C). The mean progeny virus titers of two independent experiments were determined by plaque assays on RK-UL48 cells overlaid with semisolid minimum essential medium containing 5% fetal calf serum and 6 g of methylcellulose/liter. For determination of plaque sizes, infected RK13 and RK-UL48 cells were incubated for 48 h under semisolid medium. Plaques were visualized by indirect immunofluorescence reaction of a gC-specific monoclonal antibody (35), and average diameters of 30 plaques per virus and cell line, as well as standard deviations, were calculated.
Electron microscopy.
RK13 and RK-UL48 cells were infected at an MOI of 1 with PrV-Ka or phenotypically complemented PrV-ΔUL48 which had been propagated on RK-UL48 cells. After 14 h of incubation at 37°C, fixation and embedding for routine microscopy and for intracellular immunolabeling of viral proteins were done as described previously (32). The reactions of monospecific rabbit sera against the UL31 protein, which is a constituent of primary enveloped but not of mature virions (21), or the UL36 (31), UL37 (34), UL46 (M. Kopp et al., submitted), UL47 (M. Kopp et al., submitted), UL48, and UL49 (6) tegument proteins of mature virions were visualized with 10-nm gold-tagged secondary antispecies antibodies (GAR 10; British BioCell International). The counterstained ultrathin sections were examined with an electron microscope (Tecnai 12, 400T; Philips).
RESULTS
Characterization of the UL48 gene product of PrV.
The recently determined DNA sequence of a 2,200-bp fragment of the PrV-Ka genome (accession no. AJ437285) contains two adjacent ORFs which correspond to the UL48 and UL49 genes of HSV-1 (38). The deduced products of both PrV genes exhibited significant homologies to the respective proteins encoded by HSV-1 and other alphaherpesviruses (Fuchs et al., submitted), whereas no related gene products of beta- and gammaherpesviruses were found. Like the UL49 gene of PrV (Fuchs et al., submitted), UL48 is preceded by a putative TATA box element, but, unlike UL49, UL48 is not immediately followed by the polyadenylation consensus sequence AATAAA. Since the next polyadenylation signal was found downstream of the UL46 gene of PrV (7), UL48 is most likely part of a coterminally transcribed gene cluster (Fig. 1B) which includes UL47 and UL46. This was confirmed by Northern blot analyses of total cellular RNA harvested 6 h after infection with PrV-Ka. By hybridization with a labeled antisense cRNA of UL48 a single viral transcript of ca. 5.6 kb was detected, which approximately fitted the expected size (data not shown).
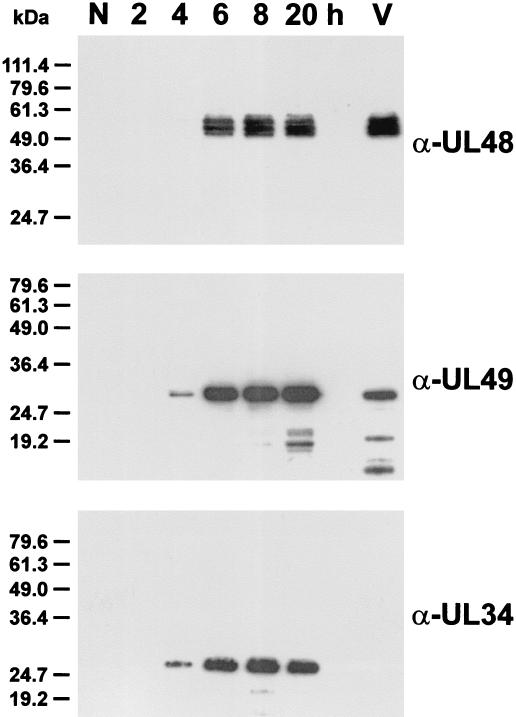
For identification of the 413-amino-acid UL48 protein, the entire ORF was fused in frame to the GST gene in plasmid pGex-UL48 (Fig. 1C) and expressed in E. coli, and the isolated 72-kDa fusion protein was used for immunization of a rabbit. In Western blot analyses of PrV-infected cells the obtained antiserum specifically reacted with at least three polypeptides exhibiting apparent masses of 53, 55, and 57 kDa (Fig. 2, α-UL48). These proteins were not detected in noninfected cells (Fig. 2, N) and also did not react with the respective preimmune serum (not shown). The identified viral gene products appeared significantly larger than 45.5 kDa as calculated from the deduced amino acid sequence encoded by the UL48 ORF. After in vitro transcription and translation of pcDNA-UL48 (Fig. 1C), electrophoretic migration of the products indicated molecular masses of 51 and 55 kDa (data not shown). Thus, the apparent masses of the UL48 proteins are partly caused by aberrant mobility or by posttranslational modifications which do not require a functional endoplasmic reticulum or Golgi apparatus.
FIG. 2.
Identification of the UL48 gene products of PrV. Lysates of noninfected RK13 cells (N), of cells harvested 2 to 20 h after infection with PrV-Ka, and of gradient-purified virions (V) were separated in SDS-polyacrylamide gels and transferred to nitrocellulose membranes. The blots were incubated with monospecific rabbit antisera against the UL48 (α-UL48), UL49 (α-UL49), and UL34 (α-UL34) proteins. Antibody binding was visualized by chemiluminescence reactions of peroxidase-conjugated secondary antibodies.
The processing of the UL48 gene product seems to be independent of other viral proteins, since RK13-derived cell lines which were stably transfected with plasmid pIRES-UL48 (Fig. 1C) expressed UL48 proteins with a size similar to that of UL48 proteins detectable in infected cells (see Fig. 5; α-UL48). However, the cells were considerably affected by constitutive expression of UL48, and the cell division rates of all analyzed clones were drastically reduced compared to that of the parental RK13 cells. Furthermore, although the RK-UL48 cell clones contained the viral ORF within a bicistronic transcription unit together with the neomycin resistance gene (Fig. 1C) and were maintained in the presence of Geneticin, the proportion of UL48-expressing cells declined rapidly upon passaging (results not shown).
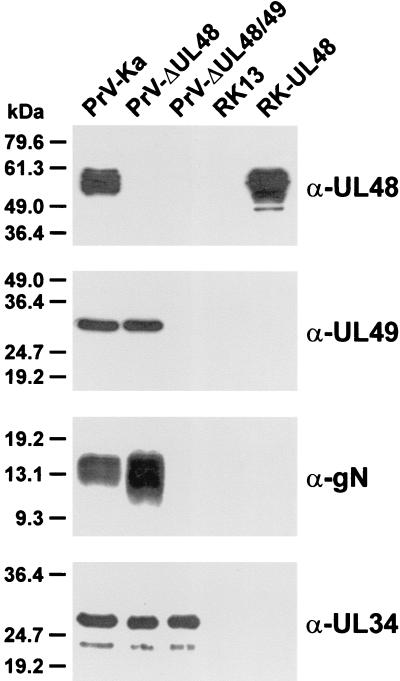
FIG. 5.
Protein expression of PrV recombinants. RK13 cells infected at an MOI of 5 with PrV-Ka, PrV-ΔUL48, or PrV-ΔUL48/49 were incubated for 20 h at 37°C. Western blots of infected RK13, and noninfected RK13 and RK-UL48 cells were probed with monospecific antisera against viral tegument proteins (α-UL48 and α-UL49), against the glycoprotein encoded by UL49.5 (α-gN), and against the UL34 protein, which is absent from mature virions (α-UL34).
In Western blot analyses of normal RK13 cells increasing amounts of the newly synthesized UL48 protein were detected from 6 to 20 h after infection with PrV-Ka (Fig. 2, α-UL48). Thus, like its HSV-1 homolog (48), the UL48 gene of PrV appears to be expressed as a late gene. Similar to what was found for the tegument protein encoded by UL49 (Fuchs et al., submitted), multiple forms of the UL48 protein could be detected in gradient-purified virions of PrV-Ka (Fig. 2, lane V). The purity of the virion preparation was verified by the absence of the UL34 protein (Fig. 2, α-UL34), which is a component of primary enveloped but not of mature virus particles (32).
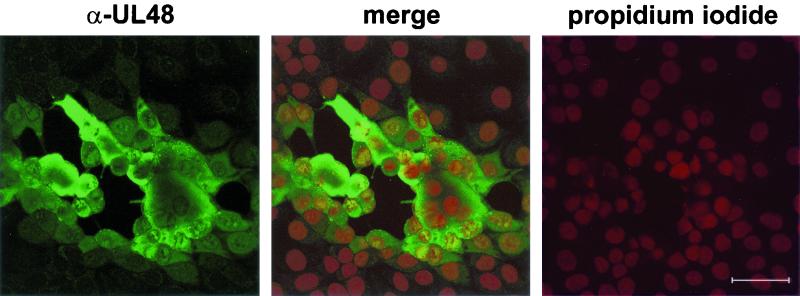
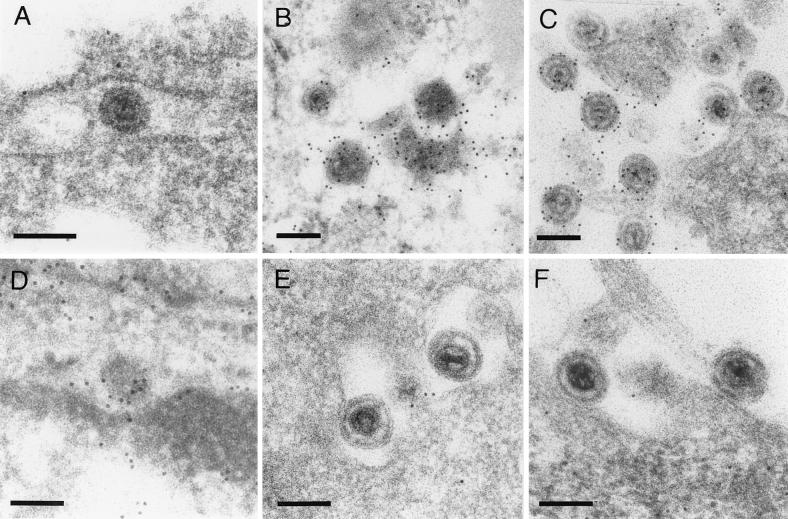
To determine the intracellular distribution of the UL48 gene product, RK13 cells were fixed 20 h after infection with PrV-Ka and reactions of the monospecific antiserum were analyzed by confocal immunofluorescence microscopy (Fig. 3). In infected cells the UL48 protein (green fluorescence) was detectable by a diffuse staining in the cytoplasm as well as in a speckled pattern in the nuclei, which were visualized by chromatin counterstaining with propidium iodide (red fluorescence). However, fluorescence intensities in the nuclei were mostly lower than those in the cytoplasm. Immunoelectron-microscopic studies demonstrated that the UL48 protein is a component of enveloped cytoplasmic (Fig. 4 B) and extracellular PrV virions (Fig. 4C). However, like the tegument protein encoded by UL49 (32), the UL48 gene product was not detectable in primary enveloped virus particles within the perinuclear space (Fig. 4A). In contrast, the UL31 protein (21) was detected in perinuclear virions (Fig. 4D) but not in mature virions (Fig. 4E and F).
FIG. 3.
Subcellular distribution of the UL48 protein. RK13 cells were fixed 20 h after infection with PrV-Ka with a 1:1 mixture of methanol and acetone and incubated with UL48-specific rabbit serum (α-UL48). Separate and merged fluorescence of Alexa 488-conjugated anti-rabbit antibodies (green) and of propidium iodide-stained chromatin (red) was analyzed in a confocal laser scan microscope. Bar, 50 μm.
FIG. 4.
Virion localization of the UL48 protein. RK13 cells were fixed and embedded 14 h after infection with PrV-Ka (MOI, 1). Ultrathin sections were subsequently incubated with anti-UL48 (A to C) or anti-UL31 serum (D to F), and 10-nm gold-tagged secondary anti-rabbit antibodies and counterstained. The UL48 protein was not detectable in primary enveloped virus particles in the perinuclear space (A) but was present in enveloped cytoplasmic (B) and released virus particles (C). In contrast, the UL31 protein is found only in the perinuclear space (D), not in mature virions in the cytoplasm (E) or in the extracellular space (F). Bars, 150 nm.
Deletion of the UL48 and UL49 genes from the PrV genome.
Viral deletion mutants were generated by RecE- and RecT-mediated mutagenesis (58) of a previously described infectious full-length plasmid clone of PrV-Ka (pPrV-K1) in E. coli (21). By this procedure the tetracycline resistance gene was inserted instead of viral DNA fragments ranging either from codon 58 to 365 of UL48 or from codon 4 of UL49 to codon 365 of UL48 (Fig. 1B). Since replication of pPrV-K1 and its derivatives in eukaryotic cells is adversely affected by the F plasmid insertion, these sequences were removed after mutagenesis in E. coli as described previously (21). Thus, as confirmed by DNA restriction analyses, Southern blot hybridization, and sequencing of relevant genome fragments (results not shown), the virus recombinants PrV-ΔUL48 and PrV-ΔUL48/49 differ from the parental strain, PrV-Ka, only at the modified loci (Fig. 1B).
To analyze the effects of the gene deletions on viral protein expression, Western blots of infected RK13 cell lysates were probed with monospecific rabbit antisera against the UL48, UL49, UL49.5, and UL34 gene products (Fig. 5). In cells infected with PrV-ΔUL48 neither the full-length nor truncated forms of the UL48 protein were detectable (Fig. 5, α-UL48), whereas the amounts of all other tested viral gene products were comparable to those obtained with wild-type PrV-Ka. As expected, PrV-ΔUL48/49 failed to produce the UL48 and UL49 proteins (Fig. 5). However, the double mutant also did not express detectable amounts of the UL49.5 gene product, gN (Fig. 5, α-gN), although the ORF was not mutated (Fig. 1B). This has also been observed in a PrV UL49 deletion mutant (Fuchs et al., submitted) and may be due to instability of the UL49.5 mRNA. In PrV-Ka and PrV-ΔUL48, UL49.5 forms a 3′-coterminal transcription unit together with UL49 (Fig. 1B), whereas in PrV-ΔUL48/49 the UL49 ORF and the common polyadenylation signal were deleted. Although the next polyadenylation signal (of UL48 to UL46) may then be used, the expected UL49.5 transcripts would be enlarged and modified by the bacterial antibiotic resistance gene (Fig. 1B). As a control, the UL34 gene product was found at levels similar to those in cells infected with PrV-Ka or with PrV-ΔUL48 (Fig. 5, α-UL34).
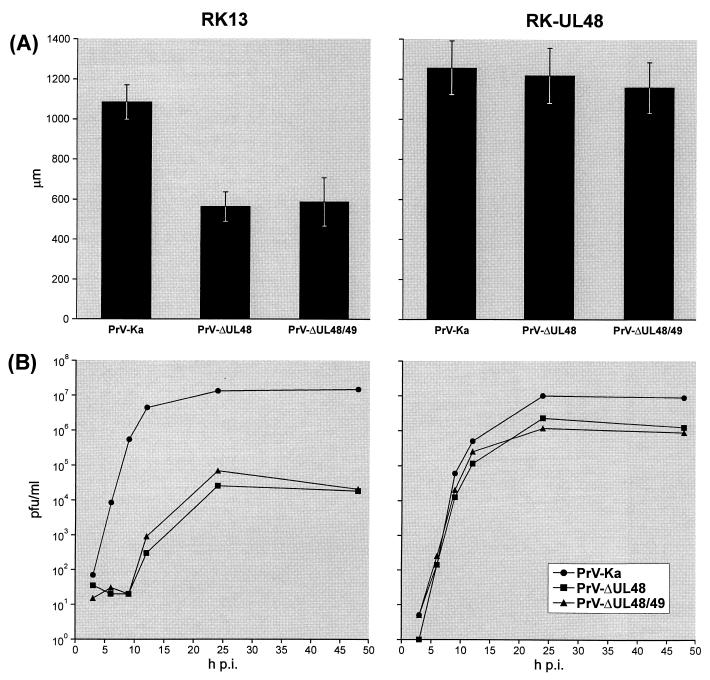
As already known, the gN and UL49 genes of PrV are not required for virus replication in cultured cells (13, 28). Obviously, the UL48 protein is also not strictly essential, since PrV-ΔUL48 could be isolated on noncomplementing RK13 cells after transfection with the modified full-length clone of the PrV genome. However, the plaque sizes of PrV-ΔUL48 and PrV-ΔUL48/49 were significantly smaller than that of PrV-Ka (Fig. 6A) or of deletion mutant PrV-ΔUL49 (Fuchs et al., submitted). Furthermore, one-step growth kinetics in RK13 cells showed that the UL48 deletion mutants exhibit a delayed onset of replication and that their maximum virus titers are reduced nearly 1,000-fold compared to those of PrV-Ka (Fig. 6B). These defects correlated with the deletion of UL48, since the phenotypes of PrV-ΔUL48 and PrV-ΔUL48/49 were almost identical. In RK-UL48 cells (Fig. 5, α-UL48), the growth defects of PrV-ΔUL48 and of PrV-ΔUL48/49 could partially be compensated (Fig. 6). The pronounced instability of the trans-complementing cell line with respect to UL48 expression (see above) might explain the observations that plaques of the UL48 mutants and double mutants were still slightly smaller and that titers remained ca. 10-fold lower than that of PrV-Ka. However, the observed delay in the onset of replication of both analyzed mutants in RK13 cells was fully restored by the UL48 protein provided by RK-UL48 cells (Fig. 6B).
FIG. 6.
Growth properties of PrV-Ka and deletion mutants PrV-ΔUL48 and PrV-ΔUL48/49 in noncomplementing (RK13) and UL48-expressing (RK-UL48) cells. (A) Average diameters of 30 single plaques per virus mutant and cell line were determined after incubation of infected-cell monolayers under semisolid medium for 48 h. Standard deviations are indicated. (B) For analysis of one-step growth kinetics cells were harvested together with the supernatants 3, 6, 9, 12, 24, and 48 h after synchronized infection at a MOI of 2. Titers were determined by plaque assays of RK-UL48 cells. The average results of two independent experiments are shown. p.i., postinfection.
The UL48 gene product of PrV is required for efficient expression of the major immediate-early gene
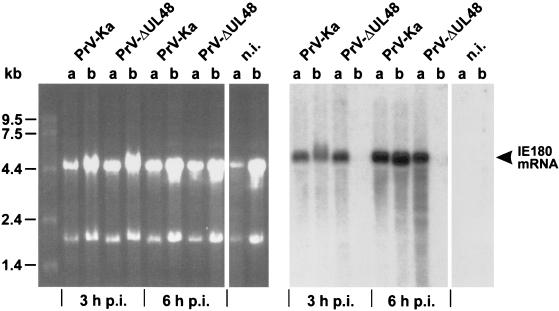
The UL48 homologs of different alphaherpesviruses, such as HSV-1, VZV, and BHV-1 were shown to encode structural proteins which augment expression of viral immediate-early (α) genes in newly infected host cells (2, 43, 44). Furthermore, the UL48 protein of HSV-1 was shown to stabilize viral mRNAs by modulation of the vhs activity of the UL41 tegument protein (36). To test whether the UL48 protein of PrV has similar properties, comparative Northern blot analyses of infected RK-UL48 (Fig. 7, lanes a) and RK13 cells (Fig. 7, lanes b) were performed. From the time when virus was added, the cells were incubated in the presence of cycloheximide to avoid any influence of newly synthesized viral proteins on transcription. Total cellular RNA was analyzed 3 and 6 h after infection with either PrV-Ka or PrV-ΔUL48 and hybridized with 32P-labeled BamHI fragment 8, which contains the major immediate-early gene of PrV (Fig. 1A) (11, 54). In RK13 and RK-UL48 cells infected at high MOI with PrV-Ka, an accumulation of the ca. 5.2-kb immediate-early mRNA was detected irrespective of cellular UL48 expression (Fig. 7). This finding indicates that, if the UL48 gene product of PrV influences immediate-early expression at all, its amount in the infecting virions is sufficient for maximal α-gene expression. In contrast, in RK13 cells infected with PrV-ΔUL48 which had previously been passaged in noncomplementing cells, IE180 mRNA was not detectable (Fig. 7, lanes b), but wild-type-like levels of this transcript were found in similarly infected RK-UL48 cells (Fig. 7, lanes a). This striking difference was not influenced by the total amounts of RNA used for blotting, as can be seen from the ethidium bromide-stained agarose gel (Fig. 7). Thus, like its homologs, the UL48 protein of PrV increases expression and/or steady-state levels of viral immediate-early RNA and thereby might accelerate the onset of virus replication.
FIG. 7.
Induction of immediate-early gene expression by the UL48 protein of PrV. RK-UL48 (lanes a) and normal RK13 cells (lanes b) were infected with either PrV-Ka or PrV-ΔUL48 and incubated in the presence of cycloheximide for 3 or 6 h. Total RNA of infected and noninfected (n.i.) cells was separated in a denaturing agarose gel (left) and transferred to a nylon membrane. The blot was hybridized with a probe specific for the major immediate-early transcript of PrV (IE180 mRNA; right).
The UL48 protein of PrV is required for efficient formation of mature virus particles.
Although the virus particles obtained after propagation of PrV-ΔUL48 in RK-UL48 cells contained the UL48 protein, the phenotypically complemented virus did not grow to higher titers in normal RK13 cells than the noncomplemented UL48 deletion mutant (results not shown). This finding indicated that, besides the influence on the level of immediate-early transcripts by virion-associated UL48, the newly synthesized UL48 gene product may also have an important function during the late phase of virus replication. To characterize this additional function, RK13 cells were fixed 14 h after infection with phenotypically complemented PrV-ΔUL48 and investigated by electron microscopy (Fig. 8). These studies indicated that capsid formation and DNA encapsidation in the nucleus were not detectably affected in the absence of the UL48 protein (Fig. 8A). Moreover, nucleocapsids were efficiently released from the nucleus by primary envelopment at the inner nuclear membrane (Fig. 8C and D) and de-envelopment at the outer nuclear membrane (Fig. 8D) as previously described for wild-type PrV (26). No perinuclear accumulation of primary enveloped virions could be observed. However, large numbers of unenveloped nucleocapsids were detected in the cytoplasm (Fig. 8B and E), and secondary envelopment was rarely detected. Instead, budding of tegument-like electron-dense material into cytoplasmic vesicles (Fig. 8E), which resulted in the release of high numbers of capsidless particles into the extracellular space (Fig. 8F and G), was observed. Electron-microscopic analyses of RK13 cells infected with PrV-ΔUL48/49 revealed a similar impairment of virus egress (data not shown). These defects were caused by the UL48 deletion, since electron microscopy of RK-UL48 cells infected with PrV-ΔUL48 showed numerous enveloped capsid-containing virions in the cytoplasm and in the extracellular space (results not shown).
FIG. 8.
Virion morphogenesis of PrV-ΔUL48. For electron microscopy, RK13 cells were fixed and embedded 14 h after infection with phenotypically complemented PrV-ΔUL48 (MOI, 1). The counterstained ultrathin sections showed all stages of intranuclear capsid maturation (A), as well as primary envelopment (C) and de-envelopment (D). However, nonenveloped nucleocapsids accumulated in the cytoplasm (B and E), and numerous capsidless particles were formed (E, arrows) and released from the cells (F and G). Bars, 500 nm (A), 2 μm (B), 200 nm (C and D), and 500 nm (E to G).
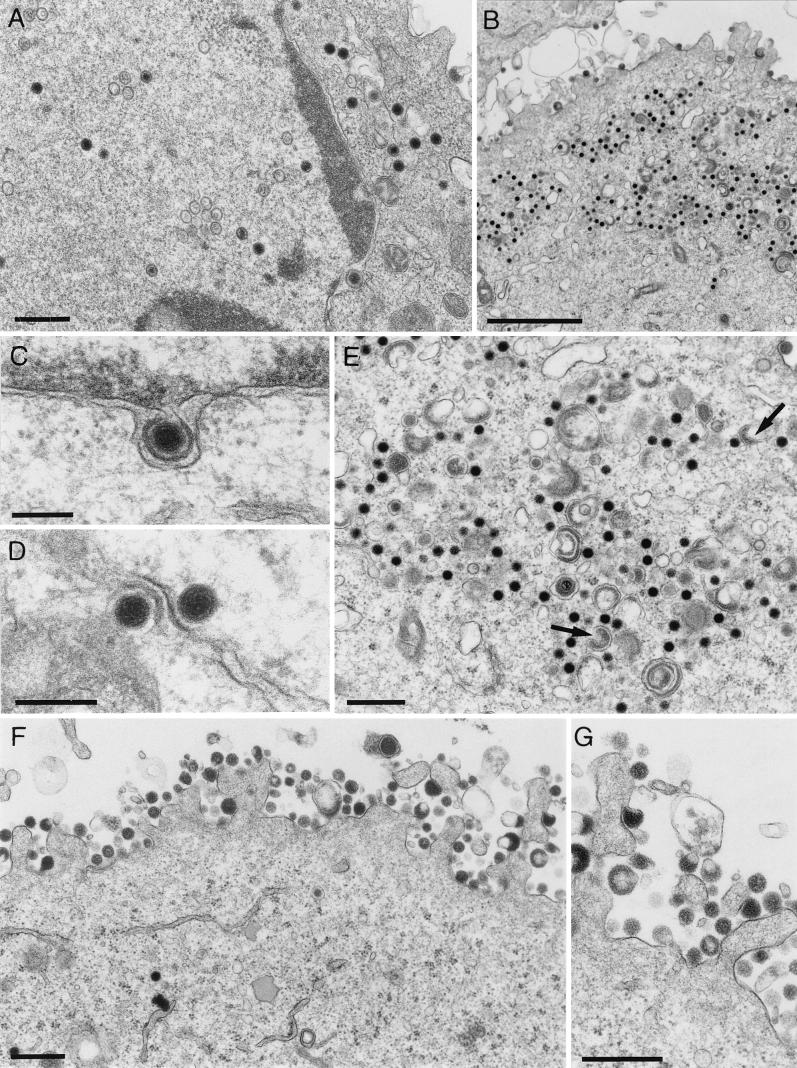
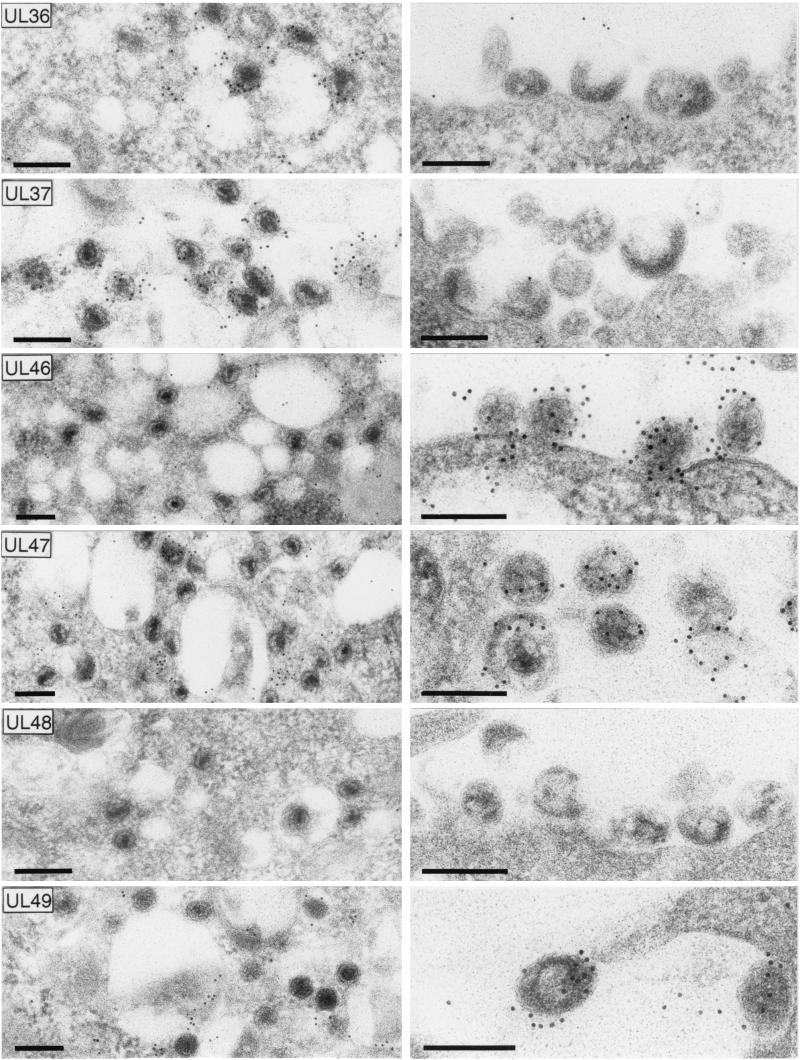
Since tegumentation of nascent herpesvirus particles seems to occur partly by interaction with capsid proteins (3, 31, 34, 42, 60) and partly by interaction with viral envelope proteins at the future budding site (42, 56; Fuchs et al., submitted), it was of particular interest to investigate the distribution of tegument proteins between the cytoplasmic nucleocapsids and the capsidless particles formed by PrV-ΔUL48. For that purpose, ultrathin sections of Lowicryl-embedded infected RK13 cells were incubated with monospecific antisera against six different tegument proteins and gold-tagged anti-rabbit secondary antibodies prior to electron microscopy (Fig. 9). The UL36 and UL37 proteins were primarily found in association with cytoplasmic nucleocapsids of PrV-ΔUL48 (Fig. 9, left panels), as has also been described for wild-type PrV-Ka (31, 34). In contrast, these proteins were not detectable in cytoplasmic (not shown) and extracellular capsidless particles (Fig. 9, right panels). The tegument proteins encoded by UL46, UL47, and UL49 exhibited a reverse distribution. They were present within capsidless particles of PrV-ΔUL48 (Fig. 9, right panels) and were found dispersed in the cytoplasm but were not associated with intracytoplasmic nucleocapsids (Fig. 9, left panels). Capsidless particles that are also occasionally released from wild-type PrV-infected cells contain the same set of tegument proteins but, in addition, carry the UL48 protein (data not shown), which, as expected, is absent from cells infected with the UL48 deletion mutant (Fig. 9). Taken together, these findings indicate that the UL48 protein of PrV plays a central role in morphogenesis of virions.
FIG. 9.
Association of tegument proteins with cytoplasmic nucleocapsids (left panels) and released capsidless particles (right panels) of PrV-ΔUL48. Infected RK13 cells (see Fig. 8) were embedded for immunoelectron microscopy, and ultrathin sections were incubated with monospecific rabbit sera against the UL36, UL37, UL46, UL47, UL48, and UL49 gene products and 10-nm gold-tagged secondary anti-rabbit antibodies. Bars, 200 nm.
DISCUSSION
In the present study the UL48 protein of PrV was identified and functionally characterized by using a monospecific antiserum and specific viral deletion mutants. UL48-homologous genes are present in many alphaherpesvirus genomes. Like the homologous gene products of HSV-1 (VP16) (48) and BHV-1 (43), the PrV UL48 protein is expressed during the late phase of the replication cycle and is incorporated into mature virus particles. Interestingly, after SDS-polyacrylamide gel electrophoresis the molecular masses of all these UL48-homologous proteins appeared more than 10 kDa higher than the values calculated from the deduced amino acid sequences, a difference which might be caused by phosphorylation, as demonstrated for the UL48 gene product of HSV-1 (37). Different phosphorylation could also account for the appearance of several distinct protein bands of the PrV UL48 gene product. Phosphorylated and nonphosphorylated forms of the UL49 tegument proteins of HSV-1 and PrV were shown to be differentially distributed in the cytoplasm and nuclei of infected cells and in mature virions, suggesting different protein functions at different sites (13, 20, 46). In contrast, the multiple forms of the UL48 protein of PrV were detectable in infected cells as well as in purified virus particles. By confocal laser scan and immunoelectron microscopy of PrV-infected cells the UL48 protein was predominantly detectable in the cytoplasm and detectable only to a minor extent in the nucleus, which correlates with absence of the UL48 protein from primary enveloped virions within the perinuclear space.
The intranuclear UL48 protein could function by stimulation of viral immediate-early (α-) gene expression, as reported for the homologous gene products of HSV-1, BHV-1, and VZV (2, 8, 43, 44). Our study shows that the UL48 protein of PrV influences expression and/or steady-state levels of immediate-early RNA, since Northern blot analyses of cells infected with a PrV UL48 null mutant revealed that immediate-early transcripts encoding the ICP4 homolog IE180 (11, 54) were substantially increased by concomitant expression of PrV UL48. This could be a direct transactivating effect since the IE180 promoter of PrV has previously been demonstrated to be inducible by UV-inactivated HSV-1 virions as well as by VP16 expression constructs and since it was also shown to contain putative binding sites for transcription factor Oct-1, which mediates interaction with VP16 (9, 52). However, in disagreement with our results, former infection studies revealed that UV-irradiated PrV, unlike similarly treated HSV-1, failed to stimulate expression of reporter genes controlled by immediate-early promoters of either HSV-1 or PrV (2, 9). Possibly, the trans-inducing activity of the UL48 protein in PrV particles is less pronounced or less stable than that of VP16. On the other hand, the increased amounts of IE180 mRNA could also be due to a modulatory interaction with another regulatory tegument component, the vhs protein, since HSV-1 VP16 has been shown to prevent rapid degradation of viral mRNA (36). A homolog of the vhs gene, UL41, was also identified in the PrV genome (4), but it remains to be investigated whether the gene product is functional and whether it interacts with the UL48 protein.
In any case, the productive virus replication observed in cells transfected with highly purified virion DNA, or even with cosmids or plasmids carrying the PrV genome (51, 53; this study), clearly demonstrates that the UL48 protein of incoming virions is not required for the onset of viral replication. Consistently, genomic DNA of many other alphaherpesviruses proved to be infectious (25, 50), and an HSV-1 mutant expressing a mutated UL48 protein which cannot activate immediate-early gene transcription was shown to be viable, although progeny virus titers were significantly reduced, especially after infection of cells at low MOI (1).
Compared to parental wild-type PrV-Ka, PrV-ΔUL48 also exhibited drastically reduced virus titers (ca. 1,000-fold), delayed appearance of infectious progeny virus in one-step growth kinetics, and a small-plaque phenotype. All these defects could be largely corrected by propagation in cells which constitutively expressed the PrV UL48 protein. However, the progeny virus titers obtained after high-MOI infection of normal cells with PrV-ΔUL48 that had been grown on complementing cells and that contained the UL48 protein in virus particles were no higher than those obtained with PrV-ΔUL48 grown on nonconplementing cells, which indicated that the input UL48 protein of the infecting virions is not sufficient to fulfil all relevant functions of the UL48 gene product.
Therefore, electron-microscopic studies were performed to analyze the late phase of the viral replication cycle in cell culture after infection with PrV-ΔUL48. Similar investigations of UL48-negative HSV-1 mutants indicated that virion maturation and egress were severely impaired (45, 57), as demonstrated by inefficient capsid formation and DNA encapsidation in the nucleus, retention of enveloped particles in the perinuclear space, and presence of naked nucleocapsids in the cytoplasm (45, 57). Although formation of mature extracellular virions was also blocked in the absence of the PrV UL48 protein, the nuclear stages of virus morphogenesis were not detectably affected. Most of the intranuclear capsids of PrV-ΔUL48 contained DNA, and subsequent transit through the nuclear membrane was efficient, since large amounts of unenveloped nucleocapsids were detectable in the cytoplasm. Furthermore, unlike the UL31 and UL34 proteins, which are involved in egress from the nucleus (21, 32), the UL48 protein was not detectable by immunoelectron microscopy in perinuclear virions of wild-type PrV-Ka but was abundantly detected in enveloped cytoplasmic and extracellular virions.
What is the precise function of the UL48 protein during tegumentation and secondary envelopment of PrV in the cytoplasm? Several PrV deletion mutants, e.g., those lacking the UL37 tegument protein (34), the UL3.5 protein (22), or envelope glycoproteins gE, gI, and gM (5, 6) have defects in secondary envelopment and accumulated nucleocapsids in the cytoplasm. However, the PrV UL37 null mutant exhibited ordered aggregates of nucleocapsids, which carry the UL36 tegument protein (31, 34). In contrast, the intracytoplasmic capsids in PrV-ΔUL48-infected cells were dispersed and contained both UL36 and UL37 proteins. Thus, addition of an inner tegument layer consisting of the UL36 and UL37 proteins to cytoplasmic nucleocapsids apparently does not require the UL48 protein. A PrV gE-, gI-, and gM-negative triple mutant (5), as well as a mutant virus lacking only the gE intracytoplasmic domain and gM (6), formed intracytoplasmic inclusions consisting of nucleocapsids and large amounts of electron-dense material containing the UL49 protein (6) and other tegument components including the UL48 gene product (results not shown). No similar inclusions were detectable with PrV-ΔUL48. Instead, noncomplementing cells infected with PrV-ΔUL48 exhibited another unique feature, which was the presence of high numbers of capsidless particles in cytoplasmic vesicles and in the extracellular space. Moderate amounts of such tegument-containing capsidless particles (which, for HSV-1, have been named L particles) were reproducibly found in cells infected with other herpesviruses (39) besides PrV, but, remarkably, not in cells infected with the PrV triple mutant lacking gE, gI, and gM (5) or a mutant virus lacking only the gE cytoplasmic domain in addition to gM (6). Thus, in the presence of these glycoproteins within the membranes of trans-Golgi-derived vesicles, interactions with viral tegument proteins seem to trigger budding events which are independent of nucleocapsid incorporation (42). In contrast, the UL48 tegument protein of PrV is obviously required to prevent the excessive formation of capsidless particles. This assumption was confirmed by electron microscopy of UL48-expressing cells in which wild-type-like morphogenesis of PrV-ΔUL48 was observed and in which viral particles found in the extracellular space were primarily mature virions (results not shown). Immunoelectron microscopy revealed that the capsidless particles formed by PrV-ΔUL48 contained the major tegument proteins encoded by UL46, UL47, and UL49, whereas the UL36 and UL37 gene products were not detectable. The same proteins and, in addition, the UL48 gene product, were found in the occasionally occurring capsidless particles from wild-type PrV-infected cells (results not shown).
In a proposed model, tegumentation of PrV in the cytoplasm occurs at two different sites (42). The UL36 and UL37 proteins are added to nucleocapsids (31, 34), and the future budding site is formed by interaction of the UL49 gene product and, presumably, other tegument components with cytoplasmic domains of viral glycoproteins gE and gM present in membranes of trans-Golgi-derived vesicles (Fuchs et al., submitted). For the initiation of both processes the UL48 gene product is apparently not required since intracytoplasmic capsids in PrV-ΔUL48-infected normal cells contain the UL36 and UL37 proteins and since capsidless particles contain the UL49 as well as UL46 and UL47 tegument proteins. However, we hypothesize that the UL48 protein may be required for an efficient connection of the two processes. Thus, the UL48 protein might target the semitegumented nucleocapsids to the budding site and prevent premature budding in the absence of capsids. Unfortunately, the molecular details of viral egress are still not clear, and we cannot exclude the possibility that the observed impairment of virus maturation is an indirect consequence of intranuclear effects of the UL48 protein on, e.g., relative steady-state levels of viral mRNAs encoding other proteins. However, it appears more likely that the defects are caused directly by the failure to assemble a functional tegument in the absence of the UL48 tegument component. Although first approaches to identify interactions with other proteins by yeast two-hybrid studies failed, since the UL48 gene product of PrV proved to be a potent transactivator of the reporter constructs used in this system (data not shown), cross-linking experiments indicated that HSV-1 VP16 may contact several envelope glycoproteins (61), and interactions with the UL41, UL46, UL47, and UL49 tegument proteins which affect vhs regulation and α-gene induction were demonstrated (19, 36, 59). These interactions might also be responsible for targeting the UL48 protein to the future budding site.
We also analyzed a PrV UL48/UL49 double-deletion mutant, which additionally was deficient for expression of the UL49.5 gene product (gN). Despite these multiple genetic defects, PrV-ΔUL48/49 was phenotypically indistinguishable from PrV-ΔUL48. Earlier investigations had already shown that single deletions of the UL49.5 or UL49 genes of PrV have little effect on virus replication (13, 28). However, the present results further demonstrate that UL49 does not execute redundant functions which can be compensated by the UL48 protein. This finding is of particular interest, since the UL49 tegument protein of PrV, although nonessential, has been shown to interact with the cytoplasmic domains of glycoproteins E and M (Fuchs et al., submitted). Since the presence of either gE or gM is required for efficient secondary envelopment of PrV (5, 6), it has been speculated that interaction with either the UL49 gene product or another tegument protein might be required for virion assembly at the budding site (42). However, because replication of a PrV UL48 deletion mutant was not further affected by additional deletion of UL49, the UL48 tegument protein is presumably not the alternative partner which can compensate for the UL49 gene product with respect to interaction with gE or gM. Finally, electron-microscopic studies of cells infected with PrV-ΔUL48/49 revealed that the strongly enhanced production of capsidless particles caused by deletion of UL48 was not altered by the additional deletion of UL49 (results not shown), indicating that the observed interactions of the UL49 tegument protein with envelope glycoproteins are not solely responsible for the formation of capsidless particles.
Although the UL48 and UL49 proteins of PrV are major tegument proteins, mutants with deletions of either one or both are replication competent. However, whereas deletion of UL49 caused no detectable phenotypic alterations, either in vitro or in vivo (13), virus growth was significantly affected in the absence of UL48. As concerns the homologous HSV-1 proteins, HSV-1 UL49 mutants are impaired but viable (47; G. Elliott and A. Whiteley, Abstr. 26th Int. Herpesvirus Workshop, abstr. 8.09, 2001) and a very limited growth of an HSV-1 UL48 null mutant was also observed after compensatory second-site mutations occurred (45). In contrast, the UL48 homolog of VZV was shown to be dispensable for virion formation and productive replication in cell culture (12). In Marek's disease virus UL48 is also nonessential, whereas UL49 appears to be required for productive replication (18). Therefore, although the UL48 and UL49 proteins, like several other tegument and envelope components, are structurally conserved within the Alphaherpesvirinae subfamily of the Herpesviridae (38), the functions of the respective proteins during virion morphogenesis may differ. Whereas viral DNA replication and encapsidation (27), nuclear egress, and the initial tegumentation events leading to addition of the UL36 and UL37 proteins to cytoplasmic nucleocapsids appear to involve proteins conserved in all herpesvirus subfamilies (42), the final steps in virion morphogenesis include less-conserved gene products, with variable, and, at least in some cases, redundant functions. Thus, for a better understanding of the general principles of secondary envelopment and virus release mutants with multiple gene deletions and also gene substitutions between different herpesviruses will be required.
Acknowledgments
Part of this study was supported by a grant from the Deutsche Forschungsgemeinschaft (Me 854/5-1) to T.C.M.
We thank L. W. Enquist, G. A. Smith, and P. Ioannou for providing the plasmids required for BAC cloning and mutagenesis, E. Mundt for help with antiserum preparation, and N. Osterrieder for performing confocal microscopy. The technical assistance and photographic help of C. Ehrlich, P. Meyer, E. Zorn, and H. Stephan are greatly appreciated.
REFERENCES
- 1.Ace, C. I., T. A. McKee, J. M. Ryan, J. M. Cameron, and C. M. Preston. 1989. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J. Virol. 63:2260-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bechtel, J. T., and T. Shenk. 2002. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol. 76:1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthomme, H., B. Jacquemont, and A. Epstein. 1993. The pseudorabies virus host-shutoff homolog gene: nucleotide sequence and comparison with alphaherpesvirus protein counterparts. Virology 193:1028-1032. [DOI] [PubMed] [Google Scholar]
- 5.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bras, F., S. Dezelee, B. Simonet, X. Nguyen, P. Vende, A. Flamand, and M. J. Masse. 1999. The left border of the genomic inversion of pseudorabies virus contains genes homologous to the UL46 and UL47 genes of herpes simplex virus type 1, but no UL45 gene. Virus Res. 60:29-40. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, M. E., and C. M. Preston. 1987. DNA sequences which regulate the expression of the pseudorabies virus major immediate early gene. Virology 157:307-316. [DOI] [PubMed] [Google Scholar]
- 10.Chen, D. H., H. Jiang, M. Lee, F. Liu, and Z. H. Zhou. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10-16. [DOI] [PubMed] [Google Scholar]
- 11.Cheung, A. K. 1989. DNA nucleotide sequence analysis of the immediate-early gene of pseudorabies virus. Nucleic Acids Res. 17:4637-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen, J. I., and K. Seidel. 1994. Varicella-zoster virus (VZV) open reading frame 10 protein, the homolog of the essential herpes simplex virus protein VP16, is dispensable for VZV replication in vitro. J. Virol. 68:7850-7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai, P., G. L. Sexton, J. M. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Wind, N., J. Domen, and A. Berns. 1992. Herpesviruses encode an unusual protein-serine/threonine kinase which is nonessential for growth in cultured cells. J. Virol. 66:5200-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dietz, P., B. G. Klupp, W. Fuchs, B. Köllner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorange, F., B. K. Tischer, J. F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elliott, G., D. O'Reilly, and P. O'Hare. 1996. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology 226:140-145. [DOI] [PubMed] [Google Scholar]
- 21.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchs, W., B. G. Klupp, H. Granzow, H. J. Rziha, and T. C. Mettenleiter. 1996. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J. Virol. 70:3517-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchs, W., and T. C. Mettenleiter. 1996. DNA sequence and transcriptional analysis of the UL1 to UL5 gene cluster of infectious laryngotracheitis virus. J. Gen. Virol. 77:2221-2229. [DOI] [PubMed] [Google Scholar]
- 24.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 25.Graham, F. L., G. Veldhuisen, and N. M. Wilkie. 1973. Infectious herpesvirus DNA. Nat. New Biol. 245:265-266. [DOI] [PubMed] [Google Scholar]
- 26.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 28.Jöns, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jöns, A., H. Granzow, R. Kuchling, and T. C. Mettenleiter. 1996. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J. Virol. 70:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan, A. S., and A. Vatter. 1959. A comparison of herpes simplex and pseudorabies virus. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 31.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 34.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klupp, B. G., and T. C. Mettenleiter. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam, Q., C. A. Smibert, K. E. Koop, C. Lavery, J. P. Capone, S. P. Weinheimer, and J. R. Smiley. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15:2575-2581. [PMC free article] [PubMed] [Google Scholar]
- 37.Lemaster, S., and B. Roizman. 1980. Herpes simplex virus phosphoproteins. II. Characterization of the virion protein kinase and of the polypeptides phosphorylated in the virion. J. Virol. 35:798-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 39.McLauchlan, J., and F. J. Rixon. 1992. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J. Gen. Virol. 73:269-276. [DOI] [PubMed] [Google Scholar]
- 40.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623-625. [DOI] [PubMed] [Google Scholar]
- 41.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 42.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Misra, V., A. C. Bratanich, D. Carpenter, and P. O'Hare. 1994. Protein and DNA elements involved in transactivation of the promoter of the bovine herpesvirus (BHV) 1 IE-1 transcription unit by the BHV α gene trans-inducing factor. J. Virol. 68:4898-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moriuchi, H., M. Moriuchi, S. E. Straus, and J. I. Cohen. 1993. Varicella-zoster virus open reading frame 10 protein, the herpes simplex virus VP16 homolog, transactivates herpesvirus immediate-early gene promoters. J. Virol. 67:2739-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pomeranz, L. E., and J. A. Blaho. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J. Virol. 73:6769-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pomeranz, L. E., and J. A. Blaho. 2000. Assembly of infectious herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 74:10041-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 49.Roizman, B., and P. E. Pellet. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 50.Sheldrick, P., M. Laithier, D. Lando, and M. L. Ryhiner. 1973. Infectious DNA from herpes simplex virus: infectivity of double-stranded and single-stranded molecules. Proc. Natl. Acad. Sci. USA 70:3621-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stern, S., M. Tanaka, and W. Herr. 1989. The Oct-1 homoeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature 341:624-630. [DOI] [PubMed] [Google Scholar]
- 53.van Zijl, M., W. Quint, J. Briaire, T. de Rover, A. Gielkens, and A. Berns. 1988. Regeneration of herpesviruses from molecularly cloned subgenomic fragments. J. Virol. 62:2191-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vlc̆ek, C̆., Z. Kozmík, V. Pac̆es, S. Schirm, and M. Schwyzer. 1990. Pseudorabies virus immediate-early gene overlaps with an oppositely oriented open reading frame: characterization of their promoter and enhancer regions. Virology 179:365-377. [DOI] [PubMed] [Google Scholar]
- 55.Wagenaar, F., J. M. Pol, B. Peeters, A. L. Gielkens, N. de Wind, and T. G. Kimman. 1995. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J. Gen. Virol. 76:1851-1859. [DOI] [PubMed] [Google Scholar]
- 56.Wang, Z. H., M. D. Gershon, O. Lungu, Z. Zhu, S. Mallory, A. M. Arvin, and A. A. Gershon. 2001. Essential role played by the C-terminal domain of glycoprotein I in envelopment of varicella-zoster virus in the trans-Golgi network: interactions of glycoproteins with tegument. J. Virol. 75:323-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123-128. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, Y., D. A. Sirko, and J. L. C. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in αTIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, Q., and R. J. Courtney. 1994. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology 204:590-599. [DOI] [PubMed] [Google Scholar]