Abstract
The UL56 gene product of herpes simplex virus (HSV) has been shown to play an important role in viral pathogenicity. However, the properties and functions of the UL56 protein are little understood. We raised rabbit polyclonal antisera specific for the UL56 protein of HSV type 2 (HSV-2) and examined its expression and properties. The gene product was identified as three polypeptides with apparent molecular masses ranging from 32 to 35 kDa in HSV-2-infected cells, and at least one species was phosphorylated. Studies of their origins showed that the UL56 protein of HSV-2 is also translated from the upstream in-frame methionine codon that is not present in the HSV-1 genome. Synthesis was first detected at 6 h postinfection and was not abolished by the viral DNA synthesis inhibitor phosphonoacetic acid. Indirect immunofluorescence studies revealed that the UL56 protein localized to both the Golgi apparatus and cytoplasmic vesicles in HSV-2-infected and single UL56-expressing cells. Deletion mutant analysis showed that the C-terminal hydrophobic region of the protein was required for association with the cytoplasmic membrane and that the N-terminal proline-rich region was important for its translocation to the Golgi apparatus and cytoplasmic vesicles. Moreover, the results of protease digestion assays and sucrose gradient fractionation strongly suggested that UL56 is a tail-anchored type II membrane protein associated with lipid rafts. We thus hypothesized that the UL56 protein, as a tail-anchored type II membrane protein, may be involved in vesicular trafficking in HSV-2-infected cells.
Herpes simplex virus (HSV) is a large, enveloped DNA virus with a genome encoding at least 74 different genes (15, 34), of which approximately half are not essential for viral replication in cell culture (33). However, these dispensable gene products are thought to be important for viral growth and spread in natural hosts.
The UL56 gene is located in the right end of the unique region of the HSV genome and has no homolog in other herpesviruses (1, 14, 30, 45), suggesting that this gene product may play a unique role in HSV infection. According to nucleotide sequences, the HSV type 2 (HSV-2) UL56 gene is predicted to encode a protein of 235 amino acids containing a C-terminal hydrophobic domain (20, 29). Although the UL56 gene is not required for growth in vitro, lack of the UL56 gene product abrogates the pathogenicity of HSV-1 in vivo (2, 3, 32, 36, 37, 39). It has also been shown that HSV-1 UL56 protein associates with virions (21) and that the C-terminal hydrophobic region of the UL56 protein is important for the pathogenicity of HSV-1 (19). However, the precise function of this protein remains unknown.
Both the HSV-1 and HSV-2 UL56 proteins possess a C-terminal 17-amino-acid stretch of uncharged or hydrophobic amino acids that are bracketed by several charged amino acids and are predicted to be anchored in the membrane, although there is no indication of an N-terminal hydrophobic signal sequence (29). These are features of tail-anchored type II membrane proteins (26). The members of this class of nonconformist membrane proteins have no signal sequence, but instead possess a hydrophobic segment near the C terminus that orients them with their N termini in the cytoplasm.
In the present study, we expressed the HSV-2 UL56 open reading frame in bacteria as a glutathione-S-transferase (GST) fusion protein and prepared an HSV-2 UL56-specific antiserum in rabbits to characterize the UL56 protein. Our observations suggested that UL56 protein was a tail-anchored type II membrane protein targeted to the Golgi component and cytoplasmic vesicles in both infected and transfected cells. Furthermore, we found that a significant amount of UL56 associated with lipid rafts, Triton X-100-insoluble membrane microdomains (43) that have received much attention as potential regulators and organizing centers for signal transduction and membrane traffic pathways (10, 43). Lipid rafts are enriched in cholesterol and glycosphingolipids, and because of the high melting temperature of their sphingolipids, rafts are resistant to Triton X-100 solubilization at low temperatures (9). Although little is known about the role of these microdomains, it has been suggested that lipid rafts perform a key role in the sorting of certain membrane proteins in polarized epithelial cells (48). Taken together with the fact that many tail-anchored type II membrane proteins are involved in vesicular transport, there is a possibility that the UL56 protein is involved in vesicular transport in HSV-infected cells.
MATERIALS AND METHODS
Cells and viruses.
Vero cells, a stable line of African green monkey kidney cells, were grown in Eagle's minimal essential medium (MEM) supplemented with 5% calf serum, 100 IU of penicillin per ml and 100 μg of streptomycin per ml. Cos-1 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Wild type HSV-1 strain KOS and HSV-2 strain 186 were used in this study. Viruses were propagated and titers were determined on Vero cells. Vero cells were infected at a multiplicity of infection (MOI) 3 of PFU per cell.
DNA manipulation.
The UL56 open reading frame is located between nucleotide positions 117075 and 117782 of the HSV-2 genome (15). The UL56 coding sequences were cloned by PCR amplification from HSV-2 HindIII fragment D (47) by using UL56f (CCGGAATTCATGGCGTTGGGGGCTGGGCAT) as the forward primer and UL56r (ATCACTCGAGTTACCGCCAAAGGAAGGCCAA) as the reverse primer. EcoRI and XhoI sites were incorporated into the forward and reverse primers, respectively, to facilitate cloning. The PCR product was digested with EcoRI and XhoI and cloned into pGEX-4T-1 (Amersham Pharmacia) to give plasmid pGEX-UL56. The expression of the GST-UL56 fusion protein is regulated by the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lac operator and the tac promoter. Translation is expected to terminate at the UL56 stop codon. Plasmid pGEX-UL56 was transformed into Escherichia coli strain XL1-Blue, which, following induction with IPTG, expressed large quantities of the GST-UL56 fusion protein.
The expression plasmid pcDNA-UL56 was constructed for expression of the UL56 gene in cultured cells. To construct pcDNA-UL56, the UL56 coding sequence was amplified by PCR with the primers UL56fHind (CCGAAGCTTATGGCGTTGGGGGCTGGGCAT) and UL56r from pGEX-UL56. A HindIII site was incorporated into UL56fHind. The PCR product was digested with HindIII and XhoI and cloned into pcDNA3.1(+).
The expression plasmid pcDNA-UL56-10M was constructed for expression of the UL56 gene from 30 bp upstream of UL56 open reading frame. To construct this plasmid, UL56 coding sequence was amplified by PCR with the primers UL56f-10M (CCGAAGCTTATGCAGGCTTCTGAGCCGCGA) and UL56r from the HSV-2 genome. A HindIII site was incorporated into UL56f-10M. The PCR product was digested with HindIII and XhoI and cloned into pcDNA3.1(+).
Construction of deletion mutants of UL56 protein.
Plasmids pcDNA-UL56F1, pcDNA-UL56F2, pcDNA-UL56F3, pcDNA-UL56F4, pcDNA-UL56R1, pcDNA-UL56R2, pcDNA-UL56R3, pcDNA-UL56R4, pcDNA-UL56R5, and pcDNA-UL56R6 were constructed for expression of N-terminal and C-terminal deletion mutants of the UL56 protein. The forward PCR primer for the C-terminal deletion plasmids was UL56fHind. The reverse primers were UL56r1 (CGCCTCGAGTTAATGACGACACGTCCCGCCACA), UL56r2 (CGCCTCGAGTTACGTGCGCGAGCTTTGCACGCG), UL56r3 (CGCCTCGAGTTAGGCGTAGGTGGGGGGGCGATC), UL56r4 (CGCCTCGAGTTAGCCTCCCACGCCAGGGTGGGC), UL56r5 (CGCCTCGAGTTACTCCACAAACAACCCCCCTGG), and UL56r6 (CGCCTCGAGTTACCGGCGCCGCGCCCGTCGTTC). An XhoI site was incorporated into the reverse primers. The forward primers for N-terminal deletion mutants were UL56f1 (CGAAGCTTATGGACGCCCCGCCCCCATACGAG), UL56f2 (CGAAGCTTATGTTTGTCGTTATTGATATCGAC), UL56f3 (CGAAGCTTATGACGTCGCCGGTTGGGCTTGTT), and UL56f4 (CGAAGCTTATGCGTGGCCGCTCGCGCCGCGCC). A HindIII site was incorporated into the forward primers. The reverse primer was UL56r. The PCR products were ligated into pcDNA3.1(+).
Plasmid pcDNA-UL56KPN, which encodes a UL56 protein that lacks amino acids 69 to 110, was constructed to express the internal deletion UL56 protein mutant. The N-terminal region, which is encoded between nucleotides 1 and 196 of the UL56 open reading frame, was amplified by PCR with primers UL56fHind and UL56kpnD (CGCCGGTACCGGAAGAAGCCGGTGAAAC). The C-terminal region, which is encoded between nucleotides 331 and 708 of the UL56 open reading frame, was amplified by PCR with primers UL56kpnR (TGTGGGTACCCCCCTGTTTCTACCGGAA) and UL56r. A KpnI site was incorporated into primers UL56kpnD and UL56kpnR. The PCR product of the N-terminal region was digested with HindIII and KpnI and cloned into pcDNA3.1(+) to give pcDNA-UL56KPN-D. The PCR product of the C-terminal region was digested with KpnI and XhoI and cloned into pcDNA-UL56KPN-D to give pcDNA-UL56KPN.
Plasmid pEGFP-UL56Ct1 was constructed to express the fusion of the enhanced green fluorescence protein (EGFP) and the C-terminal hydrophobic region and flanking charged residues of the UL56 protein. To construct this plasmid, nucleotides 643 to 708 of the UL56 gene were amplified by PCR with primers UL56Cterm1 (TCGAAGATCTCGTCATGCGGTTTTTGGG) and UL56r. A BglII site was incorporated into UL56Cterm1. The PCR product was digested with BglII and XhoI and cloned into pEGFP-C1 (Clontech).
Plasmid transfection.
Cells were transfected by using the Lipofectamine reagent according to the protocols recommended by the supplier (Gibco-BRL).
Preparation of polyclonal antisera.
Antisera were produced in two rabbits by immunization with emulsion containing approximately 0.6 mg of E. coli-expressed GST-UL56 fusion protein, which was purified with the PrepCell system (Bio-Rad), in Freund's complete adjuvant. Rabbits were immunized by subcutaneous injection into the shaven back; 0.6 mg of purified protein in the same adjuvant was used for each of three subsequent booster injections given at 2-week intervals after the primary injection. Two weeks after the last immunization, blood was collected from the heart.
Western blotting.
Mock-infected and HSV-2-infected or -transfected cell lysates were electrophoretically separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels and electrically transferred to Immobilon polyvinylidene difluoride membranes (Millipore). Nonspecific protein binding was blocked by treating membranes at 4°C overnight with phosphate-buffered saline (PBS) containing 0.05% Tween 20 and 3% bovine serum albumin. The membranes were washed twice with PBS containing 0.05% Tween 20 and incubated at 37°C for 1 h with either a 1:5,000 dilution of anti-UL56 serum, a 1:5,000 dilution of anti-UL34 serum (42), a 1:500 dilution of anti-γ-adaptin monoclonal antibody (BD-Transduction), a 1:500 dilution of anti-LAMP-1 monoclonal antibody (BD-Transduction), or a 1:1,000 dilution of anti-caveolin 1 monoclonal antibody (BD-Transduction) in PBS containing 0.05% Tween 20. After washing three times with PBS containing 0.05% Tween 20, the membranes were incubated with a 1:1,000 dilution of either peroxidase-labeled anti-rabbit or anti-mouse immunoglobulin second antibody (New England Biolabs) at 37°C for 1 h. The membranes were then washed three times with PBS containing 0.05% Tween 20, treated with either the ECL or the ECL Plus Western blotting detection system (Amersham Pharmacia), and exposed to Hyperfilm-ECL (Amersham Pharmacia).
Indirect immunofluorescence.
Cells grown on cover slips in 35-mm dishes were subjected to indirect immunofluorescence. Mock-infected or HSV-2-infected or -transfected cells were washed with PBS and fixed in either cold acetone for 5 min or 4% paraformaldehyde at 4°C for 45 min. After fixation with paraformaldehyde, cells were permeabilized with 0.05% Triton X-100 in PBS at 4°C for 45 min. HSV-2-infected cells were treated with 20% goat serum for 1 h at room temperature to block any nonspecific antibody reaction. Primary antibodies were used at a dilution of 1:200 (anti-UL56), 1:100 (anti-EEA1) (BD-Transduction), or 1:50 (anti Golgi-58K-protein) (Sigma). After 30 min at 37°C, cover slips were washed thoroughly in PBS and labeled with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin G (IgG) (MBL) or tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-mouse IgG (Sigma) for 30 min at 37°C. After rinsing again with PBS, the cover slips were quickly mounted onto glass slides with Perma Fluor (Immunon) and subsequently analyzed with a Zeiss LAM510 laser scanning microscope.
Metabolic labeling.
For [35S]methionine labeling, after two washes with methionine-free MEM, mock-infected or HSV-2-infected Vero cells were incubated for 30 min at 37°C in methionine-free MEM containing 5% calf serum and labeled with 50 μCi of methionine per ml. For [32P]orthophosphate labeling, mock-infected or HSV-2-infected Vero cells were labeled with 1 mCi of [32P]orthophosphate per ml. Cells were labeled from 6 to 18 h postinfection (p.i.) or labeled for 30 min from 9 h p.i. and then chased in complete medium for 150 min.
Immunoprecipitation.
At the end of the chase, the cells were quickly chilled by being placed on ice and treated with ice-cold PBS. Immunoprecipitation was performed under denaturing conditions. In brief, the radiolabeled cells were lysed in ice-cold radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 7.4], 1% NP-40, 0.1% SDS, 0.1% deoxycholate, 0.15 M NaCl, 1 mM EDTA) containing a protease inhibitor cocktail (Sigma). The lysates were precleared with protein A-Sepharose 6MB (Amersham Pharmacia) and immunoprecipitated with anti-UL56 serum and protein A-Sepharose 6MB. The Sepharose beads were washed in RIPA buffer seven times to remove nonspecific adsorbed proteins. Immunoprecipitates were eluted from the beads by boiling in 2× SDS sample buffer (125 mM Tris-HCl [pH 6.8], 20% glycerol, 4% SDS, 10% 2-mercaptoethanol). Then antigens were separated by SDS-polyacrylamide gel electrophoresis (PAGE). After the gels were fixed with 10% acetic acid and 10% methanol and dried, protein bands were visualized with the Fujix Bio-Imaging Analyzer BAS2000 system (Fuji Photo Film Co., Ltd., Tokyo, Japan).
Alkaline floatation.
Membrane anchoring was examined by the alkaline floatation method. Cells were scraped into reticulocyte standard buffer (RSB; 10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 1.5 mM MgCl2) containing a protease inhibitor cocktail (Sigma). Cells were homogenized with 20 strokes with a Dounce homogenizer, and then an equal volume of either RSB, RSB containing 0.2 M Na2CO3 (pH 11.0), or RSB containing 2% Triton X-100 was added to aliquots of cell lysates. The mixtures were incubated for 30 min on ice and then centrifuged at 38,000 rpm in a Beckman TLS55 rotor for 30 min at 4°C. At the end of centrifugation, the supernatant was carefully removed from the pellet. After the pH of the Na2CO3 supernatant was adjusted to between 7 and 8, samples were separated by SDS-PAGE and analyzed by Western blotting with anti-UL56 serum. The amount of proteins present in each fraction was then quantitated with an ATTO Lane Analyzer, version 3.
Sucrose gradient fractionation.
HSV-2-infected Vero cells were washed twice in ice-cold PBS and once in 25 mM MES (morpholinepropanesulfonic acid)-150 mM NaCl, pH 6.5 (MBS). The cells were then resuspended in 2 ml of 1% Triton X-100 in MBS supplemented with a protease inhibitor cocktail (Sigma) and incubated at 4°C for 20 min. The solubilized cells were homogenized with 10 strokes of a Dounce homogenizer, and 1.25 ml of the homogenate was added to an equal volume of 80% (wt/vol) sucrose in MBS. The solubilized cells (in 40% sucrose) were overlaid successively with 5 ml of 30% sucrose and 3.5 ml of 5% sucrose. After centrifugation at 240,000 × g in a Hitachi P40ST rotor for 18 h, 0.5-ml fractions were collected, carefully so not to disrupt the pellet, from the bottom of the gradient (designated fractions 1 [bottom] through 22 [top]) and immediately supplemented with the protease inhibitor cocktail. The pellet was resuspended in 0.5 ml of MBS and designated fraction P. These fractions were then separated by SDS-PAGE and analyzed by Western blotting with anti-UL56 serum. The amount of proteins present in each fraction was then quantitated with an ATTO Lane Analyzer, version 3.
Protease digestion assay.
HSV-2-infected cells, isolated virions, or pcDNA-UL56 transfected cells were scraped into RSB supplemented with protease inhibitor cocktail to prevent proteolysis by cellular proteases. After the cells were homogenized by 10 strokes with a glass homogenizer, proteinase K (TaKaRa) was added to cell homogenates in the presence or absence of 1% Triton X-100. After incubation for 30 min at room temperature, phenylmethylsulfonyl fluoride was added to each sample to a final concentration of 2 mM to inhibit further proteolysis. Samples were then separated by SDS-PAGE and analyzed by Western blotting with anti-UL56 serum.
Transmission immunoelectron microscopy and immunolabeling.
For transmission immunoelectron microscopy, HSV-2-infected Vero cells were fixed in periodate-lysine-paraformaldehyde fixative for 30 min at room temperature. The samples were then scraped and dehydrated through a graded series of ethanol and embedded in Lowicryl K4M (Polyscience, Inc.). Thin sections were cut and mounted on 2% collodion-coated nickel grids. Postembedding labeling of ultrathin sections was performed after blocking of surfaces with 5% normal goat serum in PBS containing 1% bovine serum albumin for 1 h at room temperature and 2 h of incubation at room temperature with anit-UL56 serum diluted in PBS containing 1% bovine serum albumin. Diluted gold-tagged goat anti-rabbit IgG antibodies (British BioCell International, Cambridge, United Kingdom) were added for 1 h at room temperature, and excess antibodies were removed by washing. Ultrathin immunolabeled sections were stained with Leynol's lead citrate and uranyl acetate (2%) and examined at 100 kV in an H-7100 (Hitachi) transmission electron microscope.
RESULTS
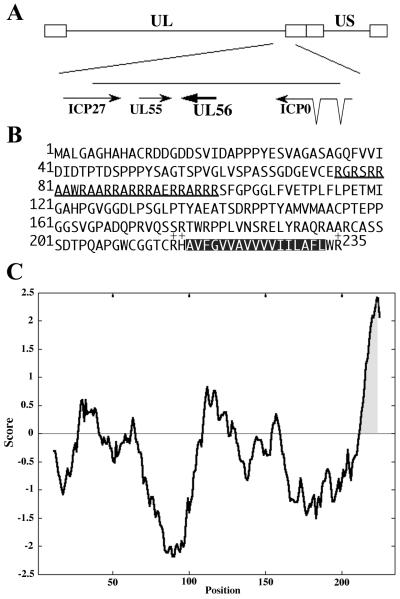
Hydrophobicity plot of UL56.
The UL56 protein contains three domains that would be useful for identifying the function of the protein. The first is a proline-rich region at the N terminus of the protein that is negatively charged. The second is an arginine-rich region (amino acids 76 to 100). In the HSV-2 UL56 protein, this region contains 60% arginine and 28% alanine residues, and alanine residues occur at three or four other positions. The third is a C-terminal hydrophobic region (amino acids 217 to 233). A Kyte and Doolittle hydrophobicity plot of UL56 revealed that the C-terminal hydrophobic region reached the threshold value of 1.6, which is generally used as a criterion for prediction of transmembrane domains (Fig. 1C). Programs which predict putative transmembrane domains according to the algorithms of Klein et al. (23) or Kyte and Doolittle (27) indeed classified UL56 as an integral membrane protein with a single transmembrane domain at its C terminus, but no N-terminal cleavable signal sequence was observed. These are the features of tail-anchored type II membrane proteins.
FIG. 1.
(A) Schematic representation of the HSV-2 genome, showing the UL56 gene. The HSV genome is shown with its unique long (UL) and unique short (US) regions. The arrows indicate the open reading frames. (B) Amino acid sequence of the HSV-2 UL56 open reading frame. The arginine-rich region is underlined. The putative transmembrane domain is shown as a hatched box, and the surrounding basic residues are indicated (+). (C) Hydrophobicity plot of UL56 using the algorithm described by Kyte and Doolittle (27). The window size used is 21 amino acids. The predicted transmembrane domain is highlighted.
Preparation and specificity of HSV-2 UL56 antiserum.
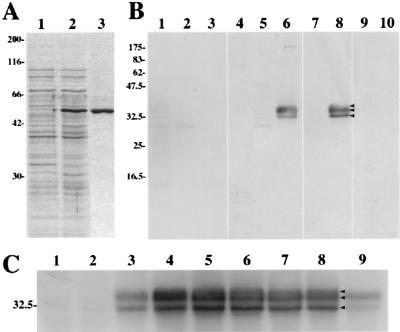
To characterize the UL56 gene product, we first generated anti-UL56 rabbit sera by using a recombinant HSV-2 UL56 fusion protein as the antigen. For this purpose, plasmid pGEX-UL56 was constructed as described in Materials and Methods, and the UL56 fusion protein was expressed in E. coli by treatment with IPTG and then purified with the PrepCell system (Bio-Rad) (Fig. 2A). Using Western blotting analysis to examine the reactivity and specificity of the antisera (Fig. 2B), we found that one of the antisera reacted strongly with 35-, 34-, and 32-kDa proteins in HSV-2-infected Vero cell lysates. Sometimes the 31- and 30-kDa proteins were also detectable (data not shown). The antiserum recognized those proteins in HSV-2-infected cell lysates (Fig. 2B, lane 6), but not in mock-infected or HSV-1-infected cell extracts (Fig. 2B, lanes 4 and 5). They were also not recognized by preimmune serum (Fig. 2B, lanes 1 to 3). The reactivity of this antiserum was clearly eliminated by preadsorption with E. coli lysate expressing the GST-UL56 fusion protein (Fig. 2B, lanes 9 and 10), but there was no significant change in the reactivity after adsorption with the lysate of control E. coli (Fig. 2B, lanes 7 and 8). The results suggest that these 35-, 34-, and 32-kDa proteins are UL56 gene products.
FIG. 2.
(A) Induction of the GST-UL56 fusion protein. E. coli cells harboring pGEX-UL56 were grown in the absence (lane 1) or presence (lane 2) of IPTG. The fusion protein was purified with the PrepCell system (Bio-Rad). Proteins were separated by SDS-PAGE and stained with Coomassie brilliant blue. (B) Reactivity and specificity of rabbit polyclonal antiserum raised against the GST-UL56 fusion protein. Vero cells were mock infected (lanes 1 and 4) or infected with HSV-1 (lanes 2, 5, 7, and 9) or HSV-2 (lanes 3, 6, 8, and 10) and harvested at 15 h p.i. Samples were separated by SDS-PAGE and analyzed by Western blotting with preimmune serum (lanes 1 to 3), anti-UL56 serum (lanes 4 to 6), anti-UL56 serum preadsorbed with E. coli lysate (lanes 7 and 8), or anti-UL56 serum preadsorbed with E. coli lysate expressing the UL56 fusion protein (lanes 9 and 10). (C) Production of the UL56 protein in HSV-2-infected Vero cells. Vero cells were mock infected (lane 1) or infected with HSV-2 (lanes 2 to 9) at an MOI of 3 PFU per cell. The cells were cultured in the absence or presence of phosphonoacetic acid (lane 9) and harvested at 3 (lane 2), 6 (lane 3), 9 (lane 4), 12 (lane 5), 15 (lanes 6 and 9), 18 (lane 7), and 24 (lane 8) h p.i. Samples were separated by SDS-PAGE and then analyzed by Western blotting with anti-UL56 serum. Positions of molecular mass markers (in kilodaltons) are indicated on the left.
Production of UL56 in HSV-2-infected cells.
All three species of UL56 gene product were detected by 6 h p.i. (Fig. 2C, lane 3) and gradually increased in amounts until 15 h p.i. The UL56 proteins were detectable even in the presence of 300 μg of phosphonoacetic acid, an inhibitor of viral DNA synthesis (Fig. 2C, lane 9), per ml. These results suggest that the UL56 protein is a late gene product whose expression is regulated as a γ1 gene.
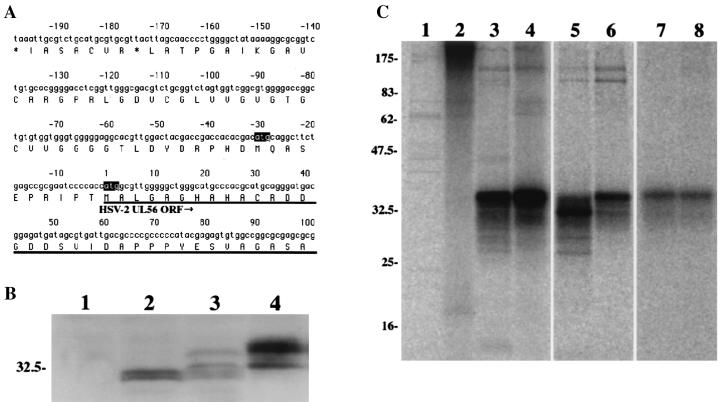
In HSV-2-infected cells, the UL56 gene products were observed as at least three species of 35, 34, and 32 kDa (Fig. 2C and Fig. 3B, lane 4). However, only two species of 31 and 30 kDa were detected in wild-type-UL56-transfected cells (Fig. 3B, lane 2). The HSV-2 UL56 gene possesses an in-frame ATG 10 codons upstream of the start site, which could be an alternative translation initiation site (Fig. 3A). This in-frame upstream ATG has fewer consensus matches to the Kozak sequence and is not observed in the HSV-1 genome. To determine whether translation of the UL56 protein is initiated from the upstream ATG in HSV-2-infected cells, we compared the molecular masses of the UL56 proteins in single UL56-expressing cells. In pcDNA-UL56-10M-transfected cells, the UL56 protein was observed as four species, and their molecular masses were 34, 32, 31, and 30 kDa (Fig. 3B, lane 3). These results suggest that both ATG codons are active for translation in UL56-10M-transfected cells.
FIG. 3.
(A) HSV-2 DNA sequence upstream of the UL56 open reading frame (ORF). Proposed encoded amino acid sequences are indicated in the single-letter code. The UL56 open reading frame is underlined, and the in-frame ATG codons of the UL56 open reading frame are highlighted. (B) UL56 protein differences between singly expressed and HSV-2-infected cells. Control Vero cells (lane 1), wild-type UL56-expressing cells (lane 2), UL56-10M-expressing cells (lane 3), or HSV-2-infected cells (lane 4) were separated by SDS-PAGE and analyzed by Western blotting with anti-UL56 serum. Positions of molecular mass markers are indicated at the left (in kilodaltons). (C) Detection of UL56 protein by immunoprecipitation. Vero cells were mock infected (lanes 1 and 2) or infected with HSV-2 (lanes 3 and 4) at an MOI of 3 PFU per cell. The cells were labeled with [35S]methionine (lanes 1 and 3) or [32P]orthophosphate (lanes 2 and 4) from 6 h p.i to 18 h p.i. In the pulse-chase experiment, HSV-2-infected cells were labeled with [35S] methionine (lanes 5 and 6) or [32P]orthophosphate (lanes 7 and 8) for 30 min at 9 h p.i. and chased for 0 (lanes 3 and 5) or 150 min (lanes 4 and 6).
To investigate the posttranslational modification of UL56 protein in infected cells, radioisotope labeling experiments were done. The 32P- and 35S-labeled proteins were immunoprecipitated with anti-UL56 antiserum from infected cell lysates and separated by SDS-PAGE (Fig. 3C). When cells were labeled from 6 to 18 h p.i., 32P-labeled proteins with molecular masses ranging from 30 to 35 kDa were immunoprecipitated by the anti-UL56 antiserum; a 35-kDa species was detected as a major and broad band. None of these proteins were immunoprecipitated from mock-infected cells (Fig. 3C, lanes 1 to 4). When cells were pulse labeled with [35S]methionine for 30 min at 9 h p.i., a 32-kDa species was immunoprecipitated as a major protein and the 35-kDa species was also easily detectable (Fig. 3C, lane 5). After a 150-min chase, the amount of the 32-kDa protein decreased markedly, while the intensity of the 35-kDa band increased (Fig. 3C, lane 6). However, when cells were labeled with [32P]orthophosphate, the 35-kDa species was predominantly detected, even immediately after the pulse (Fig. 3C, lanes 7 and 8). Although several additional bands were observed in these immunoprecipitation experiments, it remains unclear whether they were unmodified precursors or degradation products. These results indicate that the UL56 gene product is phosphorylated in infected cells.
Intercellular distribution of UL56 protein.
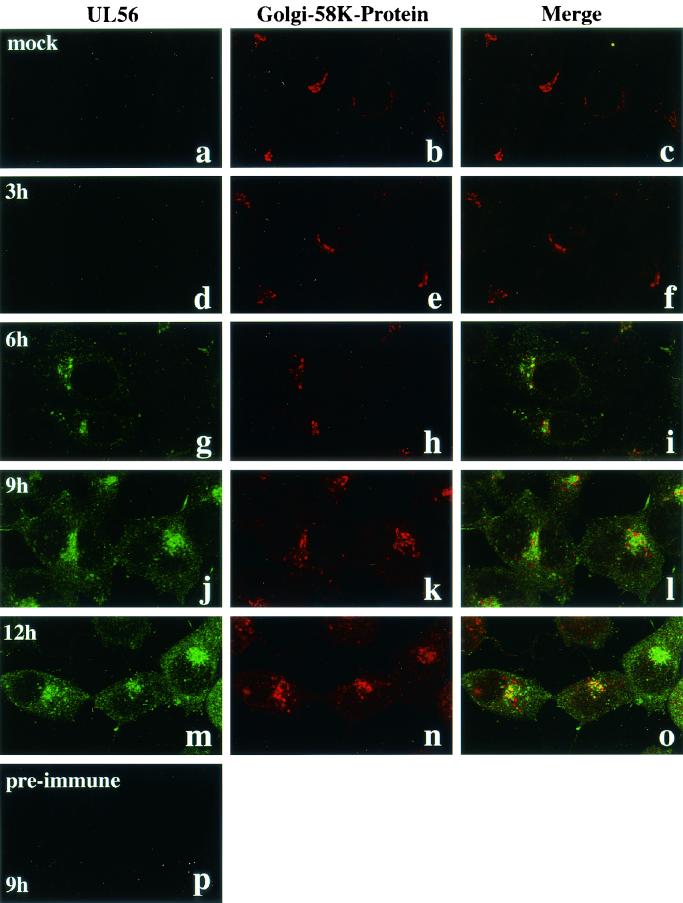
To identify the cellular compartment containing UL56, double-labeling immunofluorescence microscopy was done. HSV-2-infected Vero cells were fixed with cold acetone at various times after infection and reacted with anti-UL56 rabbit serum and anti-Golgi-58K-protein mouse monoclonal antibody. No specific staining was seen in mock-infected cells with anti-UL56 serum (Fig. 4a) and in HSV-2-infected cells with preimmune serum (Fig. 4p). The UL56 protein was first detected at 6 h p.i. and predominantly colocalized with the Golgi-58K-protein (Fig. 4g to i). At 9 h p.i., UL56 staining was partially dispersed in the cytoplasm, where UL56 stained a fine speckled pattern and the intensity of cytoplasmic fluorescence of UL56 gradually increased (Fig. 4j and m). Golgi-58K-protein localized to a compact perinuclear element until 6 h p.i.(Fig. 4h), but became gradually dispersed thereafter (Fig. 4k and n).
FIG. 4.
Subcellular localization of UL56 protein in HSV-2-infected cells. Confocal microscopic images of UL56 (a, d, g, j, and m; FITC), Golgi-58K-protein (b, e, h, k, and n; TRITC), and merged regions (c, e, i, k, and o) are shown. Cells were fixed with cold acetone. (a to c) Mock-infected cells. HSV-2-infected cells were fixed at 3 (d to f), 6 (g to I), 9 (j to l and p), or 12 (m to o) h p.i. A confocal microscopic image of preimmune serum is also shown (p).
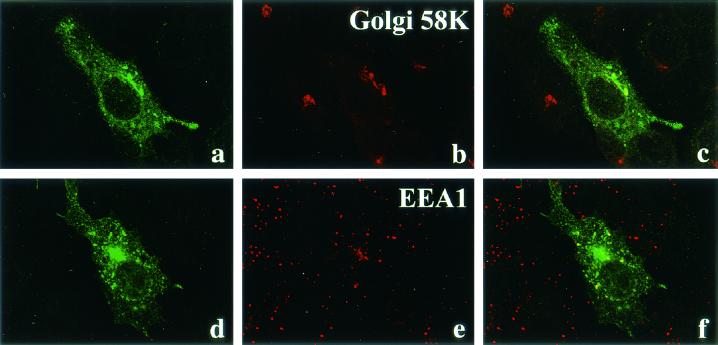
The fragmentation and dispersal of the Golgi apparatus have been described previously in HSV-1-infected cells (11). We found that the colocalization of UL56 with Golgi-58K-protein tended to be obscured with the advance of HSV-2 infection (Fig. 4l and o). In transfected cells, UL56 protein accumulated in a compact perinuclear element, and this perinuclear fluorescence colocalized with Golgi-58K-protein (Fig. 5a to c). As observed with HSV-2-infected cells, fine speckled fluorescence was also detected in the cytoplasm and most colocalized with EEA1, a marker protein of early endosomes (Fig. 5d to f).
FIG. 5.
Immunofluorescence of Vero cells expressing the UL56 protein. At 24 h after transfection, the cells were fixed with cold acetone (a to c) or 4% paraformaldehyde (d to f). After being fixed with cold acetone, cells were double-stained with anti-UL56 rabbit serum (a) and anti-Golgi-58K-protein monoclonal antibody (b). After being fixed with paraformaldehyde, cells were permeabilized with 0.05% Triton X-100 and then double stained with anti-UL56 rabbit serum (d) and anti-EEA1 monoclonal antibody (e).
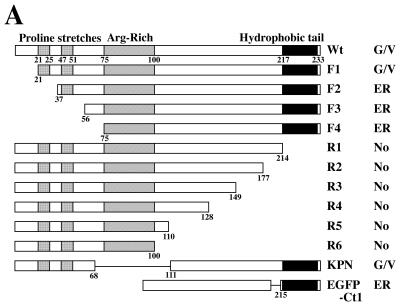
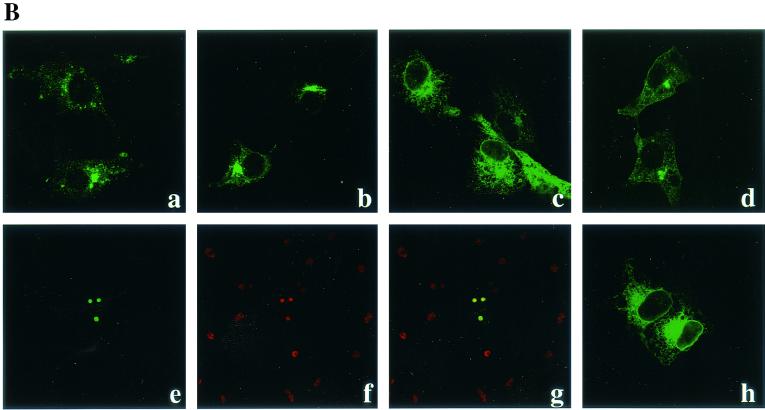
In order to determine the domain responsible for the subcellular localization, we generated a series of UL56 deletion mutants (Fig. 6A) and analyzed their localization in Vero cells. Deletion of the N-terminal 20 amino acids of the UL56 protein did not significantly affect the subcellular localization of the protein (Fig. 6B, b). When the N-terminal 36 amino acids were deleted, the protein UL56F2 was detected in a staining pattern reminiscent of the endoplasmic reticulum (ER) (Fig. 6B, c). The immunofluorescence patterns of other N-terminal deletion mutants such as UL56F3 and F4 were very similar to that of UL56F2 (data not shown). The mutant UL56R1, which lacks the C-terminal hydrophobic region, was detected almost exclusively in the nucleoli (Fig. 6B, e to g), and the other C-terminal deletion mutant proteins also accumulated in the nucleolus (data not shown). The mutant UL56KPN, which lacks the arginine-rich domain spanning amino acids 69 to 110, showed a staining pattern similar to that of wild-type UL56 (Fig. 6B, d). Intrinsic fluorescence of EGFP-Ct1, EGFP fused with the C-terminal hydrophobic region of the UL56 protein, was detected in a staining pattern reminiscent of the ER (Fig. 6B, h). These results suggest that the C-terminal hydrophobic region of the UL56 protein is required for anchoring to cytoplasmic membranes and also that the N-terminal proline-rich region is important for its translocations to the Golgi apparatus and cytoplasmic vesicles.
FIG. 6.
(A) Construction and summary of intracellular localization of UL56 protein and its deletion mutants. Bars represent translated amino acids. Numbers indicate amino acid positions. Abbreviations: No, nucleolus; G/V, Golgi and cytoplasmic vesicles. (B) Subcellular localization of deletion mutants of UL56 protein. Cells were transfected with pcDNA-UL56 (a), pcDNA-UL56F1 (b), pcDNA-UL56F2 (c), pcDNA-UL56KPN (d), pcDNA-UL56R1 (e to g), or pEGFP-UL56Ct1 (h). After 24 h, cells were fixed with cold acetone (a to g) or 4% paraformaldehyde (h) and stained with anti-UL56 serum (a to d). UL56R1-expressing cells were double stained with anti-UL56 serum (e) and anti-nucleolin monoclonal antibody (f) and merged (g). (h) Intrinsic fluorescence of EGFP-UL56Ct1.
Association of UL56 with lipid rafts.
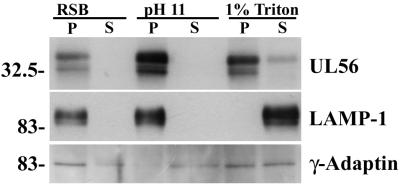
The above results suggested that UL56 was a membrane-associated protein. To investigate the nature of the UL56-membrane interaction, we carried out cell fractionation experiments as described in Materials and Methods (Fig. 7). LAMP-1 and γ-adaptin were used as an integral membrane marker and a peripheral membrane marker, respectively. While most of the γ-adaptin was found in the soluble fraction of high pH, UL56 and LAMP-1 were found almost exclusively in the insoluble fraction. Even when cells were treated with RSB containing 1% Triton X-100, most of the UL56 protein was detected in the insoluble fraction, while LAMP-1 was found predominantly in the soluble fraction. These results suggest that the UL56 protein is an integral membrane protein and may associate with Triton X-100-insoluble structures. It should also be noted that the 35-kDa species of UL56 protein appeared to be selectively released into the soluble fraction by 1% Triton X-100. Thus, we examined the association of UL56 with lipid rafts, which are known as the Triton X-100-insoluble membrane microdomain.
FIG. 7.
Membrane association of UL56 protein. Vero cells were infected with HSV-2 for 12 h and then fractionated as described in Materials and Methods. Samples were separated by SDS-PAGE and analyzed by Western blotting with anti-UL56 serum, anti-γ-adaptin monoclonal antibody, or anti-LAMP-1 monoclonal antibody. P, pellet; S, supernatant. Positions of molecular mass markers (in kilodaltons) are indicated on the left.
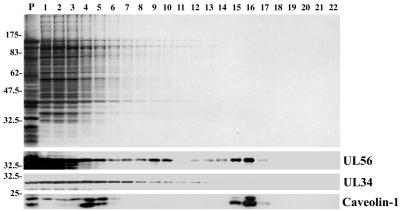
To determine if UL56 is incorporated into the rafts, infected cells were harvested at 12 h p.i. and fractionated as described in Materials and Methods (Fig. 8). It is well established that Triton-insoluble material recovered from the 30%-5% sucrose interface contains both lipid rafts and caveolae (16, 41), and caveolin 1, the major structural protein of caveolae, has been used as a marker of detergent-insoluble lipid fractions. Thus, fractions were analyzed by Western blotting with the anti-UL56, anti-UL34, and anti-caveolin 1 antibodies. Caveolin 1 was detected as peaks at fractions 4 and 16, and the latter corresponded to the 30%-5% sucrose interface from its position (Fig. 8C). About 30% of caveolin 1 was detected at fraction 16, suggesting that fraction 16 contains lipid rafts. Analysis with anti-UL56 serum showed that while UL56 was predominantly found in the loading zone, UL56 protein also made a clear peak at fraction 16. About 10% of UL56 protein was detected at fraction 16 (Fig. 8B). UL34, which is a tail-anchored type II membrane protein encoded by HSV, was detected in the loading zone but not in the 30%-5% sucrose interface. These observations suggest that UL56 protein associated with lipid rafts.
FIG. 8.
Association of UL56 protein with lipid rafts. Cells were solubilized in 1% Triton X-100 and fractionated on a discontinuous sucrose gradient, as described in Materials and Methods. Equal volumes of the recovered fractions were separated by SDS-PAGE and analyzed by silver staining (upper panel) or by Western blotting with anti-UL56 serum, anti-UL34 serum, or anti-caveolin 1 monoclonal antibody. Positions of molecular mass markers (lane P, in kilodaltons) are indicated on the left.
Membrane orientation of UL56.
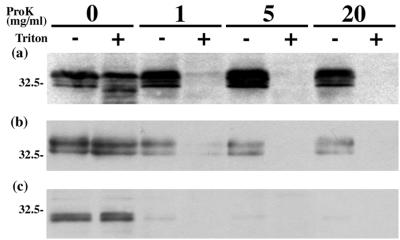
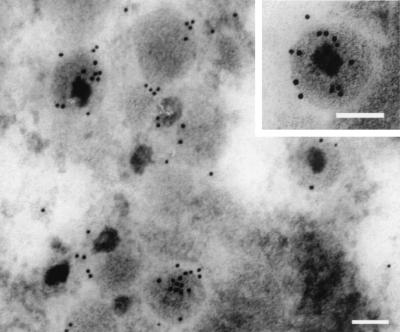
According to the PSORT II prediction (http://psort.nibb.ac.jp), it seems likely that the UL56 protein is a C-terminally anchored type II membrane protein. If so, the N-terminal region of UL56 would be found in the cytoplasm of infected cells and in the tegument of virions. To determine whether UL56 has such an orientation, we performed a protease digestion assay. In this assay, the cytoplasmic parts of membrane proteins are sensitive to protease, but proteins that are localized at the inner side of cytoplasmic vesicles and the envelope are resistant. However, the addition of detergents such as Triton X-100 disrupts cytoplasmic vesicles, making proteins localized within the vesicles sensitive to protease. In HSV-2-infected cells (Fig. 9a) and purified HSV-2 virions (Fig. 9b), all species of UL56 were resistant to proteinase K in the absence of detergent. In singly expressed cells, UL56 was highly sensitive to protease even in the absence of detergent (Fig. 9c). These results suggest that UL56 protein faces the cytoplasm in singly expressed cells. We also examined infected cells by transmission immunoelectron microscopy. As shown in Fig. 10, which shows a cytoplasmic virion, the UL56 signals were mostly detected at the inside of the viral envelope. These observations suggest that the UL56 protein is a tail-anchored type II membrane protein inserted into the viral envelope.
FIG. 9.
Topology of the UL56 protein. HSV-2-infected cells (a), virions (b), and UL56-expressing cells (c) were treated with proteinase K in either the presence (+) or the absence (−) of 1% Triton X-100. The samples were then separated by SDS-PAGE and analyzed by Western blotting with anti-UL56 serum. Positions of molecular mass markers (in kilodaltons) are indicated on the left.
FIG. 10.
Transmission immunoelectron microscopy of HSV-2-infected Vero cells, showing the labeling for UL56 proteins at 12 h p.i. Thin sections were prepared as described in Materials and Methods. Bars, 100 nm.
DISCUSSION
In this study, we identified the UL56 gene product of HSV-2 as virion-associated phosphoproteins with molecular masses ranging from 30 to 35 kDa and suggest that the UL56 protein is a tail-anchored type II membrane protein associating with lipid rafts.
The products of the HSV-2 UL56 open reading frame had a number of isoforms. In infected cells, we identified at least three isoforms having apparent masses of 35, 34, and 32 kDa, and the 31- and 30-kDa species were also sometimes detectable. A possible explanation is a posttranslational modification of the product; we observed phosphorylation of the UL56 protein. Another explanation is that the UL56 open reading frame possesses an additional in-frame initiation site. McGeoch et al. suggested that the HSV-2 UL56 open reading frame possesses an in-frame ATG upstream of the start site (29). In singly expressed cells, the wild-type UL56 was identified as two species of 31 and 30 kDa, while the 34- and 32-kDa species appear to be the products of translation initiated from the upstream ATG. Moreover, in infected cells the primary product of the UL56 open reading frame appeared to be the 32-kDa species.
Kozak has proposed a consensus sequence, PuPyPyPuPyPyAUGG, where Pu is a purine, Py is a pyrimidine, and the AUG initiation codon is in italics, for the initiation of translation in multicellular eukaryotes (24). The sequence around the translation initiation site of the HSV-2 UL56 gene is CCCACCAUGG (PyPyPyPuPyPyAUGG), which is different in only one site from the Kozak sequence. The sequence around the upstream initiation site was CACGACAUGC (PyPuPyPuPuPyAUGC), which was different in four sites from the Kozak sequence. In the case of HSV-1, the sequence around the initiation site is GCAUCCAUGG, which is different in two sites from the Kozak sequence. In the HSV-1 UL56 gene, the important positions flanking the codon were not conserved; that is, the sequence had U in position −3. According to the Kozak sequence, HSV-2 UL56 is predicted to be translated from the downstream initiation site. However, our observations indicate that translation of the HSV-2 UL56 open reading frame is also initiated from the upstream ATG. We have shown in this study that at least the 35-kDa species of UL56 was phosphorylated but have not yet specifically addressed the function of the various isoforms of UL56 protein product.
The UL56 protein was located in the Golgi apparatus as well as in the cytoplasmic vesicles of infected and transfected cells. The results of deletion mutant protein analysis are summarized as follows. (i) The C-terminal hydrophobic region acted as a transmembrane domain and was necessary and sufficient for cytoplasmic localization of UL56 protein. (ii) The N-terminal region (amino acids 20 to 36) was important for targeting of UL56. In this region, however, we have not definitely identified any characteristic motif for its transport from the ER to the Golgi and cytoplasmic vesicles. One tyrosine-based motif, an internalization signal from the plasma membrane to the trans-Golgi network, was identified (22). It is unclear whether this tyrosine motif is involved in the subcellular localization of UL56 protein. (iii) We did not make clear the function of the Arg-rich region in this study. Surprisingly, the C-terminal hydrophobic region-truncated proteins accumulated predominantly in the nucleolus. The nucleolar localization targeting sequences have been identified in various viral proteins such as Tat (40), Rev (13, 25), and human T-cell lymphotropic virus type 1 (HTLV-1) pX (44) and show little homology, but all contain long basic sequences resembling nuclear localization signals. The Arg-rich region of UL56 protein probably acts as a nucleolar localization signal when the C-terminal hydrophobic domain is deleted.
Our data strongly suggest that the UL56 protein is classified in a unique subclass of type II membrane proteins called tail-anchored membrane proteins. Typically in tail-anchored type II membrane proteins, the N-terminal side of the transmembrane domain is more positively charged than the C-terminal side. In the case of HSV-2 UL56 protein, shown in Fig. 1B, the N-terminal side of the transmembrane domain is slightly more positively charged than the C-terminal side. Taken together with the charge of the C-terminal COOH group, the N-terminal side of the transmembrane domain is clearly more positively charged than the C-terminal side. Furthermore, UL56 protein, like typical tail-anchored type II membrane proteins, has no N-terminal cleavable signal sequence but has one transmembrane domain at the C terminus (26).
Tail-anchored membrane proteins have no obvious signal sequence, an N terminus exposed to the cytosol, and a C-terminal membrane anchor. Other examples of proteins in this class include cytochrome b5, Bcl-2, syntaxin, and several proteins involved in vesicular transport (26). At present, the role of UL56 protein in the replication of HSV is unknown. However, we propose the following hypothesis regarding its function. UL56 protein localizes in the Golgi apparatus and the cytoplasmic vesicles, where HSV may gain an envelope (18). The UL34 protein, which is another tail-anchored type II membrane protein encoded by the HSV genome (42), has been shown to be involved in the budding of intranuclear capsids through the inner nuclear membrane into the perinuclear cisterna (35). It seems possible that UL56 plays a role in the secondary envelopment. Envelopment is an essential event for the formation of infectious particles and the lack of viral genes responsible for envelopment must markedly reduce viral growth even in cultured cells. However, as UL56-deficient HSV replicates in cultured cells as efficiently as the wild-type virus, we tend to exclude the possibility that UL56 is directly involved in the process of secondary envelopment.
Another hypothesis is that UL56 may play a role in the transport of viral glycoproteins to the site of secondary envelopment. While lack of the UL56 product does not affect viral replication in most types of cultured cells, it does markedly reduce the neuroinvasiveness of HSV-1 (3, 36, 37, 38, 39). Moreover, Kehm et al. have shown that deletion of the C-terminal hydrophobic region of the UL56 protein abrogates the virulent phenotype (19). Taken together with our observations, it is suggested that UL56 protein is involved in HSV pathogenicity as a tail-anchored type II membrane protein. Miranda-Saksena et al. reported that, in axons of peripheral neurons, viral nucleocapsids coated with tegument proteins are transported separately from envelope glycoproteins and suggested that the final assembly of an enveloped virus occurs at the axon terminus. If this is true, the envelope glycoproteins must be transported as conventional neuronal transport vesicles (31). In the absence of UL56, the vesicles might not be efficiently targeted to the site of assembly, and the virus may thereby fail to obtain an envelope. UL56 has a topology similar to that of proteins involved in vesicular transport (26). Furthermore, UL56 protein associated with lipid rafts, and some of the membrane proteins involved in vesicular transport are known to associate with lipid rafts (12, 28). It thus seems that these observations support our latter hypothesis regarding the role of UL56.
Interestingly, UL56 protein has some significant similarities to the US9 gene product of pseudorabies virus (PRV), another neurotropic alphaherpesvirus. The US9 protein of PRV is a phosphorylated, tail-anchored type II membrane protein ranging from 17 to 20 kDa, which is inserted into the virion envelope and accumulates in the trans-Golgi network (5, 8). Furthermore, PRV US9 null mutants exhibit no obvious phenotype after infection in tissue culture but do exhibit a defect in anterograde transneuronal spread in the rodent nervous system (6, 7). Recently, it was shown that the PRV US9 protein is necessary for axonal localization of viral membrane proteins such as gB and gC (46). Although the HSV-1 US9 protein is also a virion phosphoprotein ranging from 12 to 20 kDa which contains a hydrophobic stretch near the carboxy terminus, earlier studies have shown that it is associated with nucleocapsids in the nuclei of infected cells (17) and may be involved in the regulation of ubiquitin-mediated protein degradation (4). Further studies are required to determine similarities and differences in properties and roles among US9 homologs and HSV UL56.
Acknowledgments
We thank E. Iwata and T. Tsuruguchi for technical assistance.
This work was supported by a grant from the Japan Society for the Promotion of Science (JSPS-RFTF97L00703) and also by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Baumeister, J., B. G. Klupp, and T. C. Mettenleiter. 1995. Pseudorabies virus and equine herpesvirus 1 share a nonessential gene which is absent in other herpesviruses and located adjacent to highly conserved gene cluster. J. Virol. 69:5560-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, Y., J. Hadar, E. Tabor, T. Ben-Hur, I. Raibstein, A. Rosen, and G. Darai. 1986. A sequence in HpaI-P fragment of herpes simplex virus-1 DNA determines intraperitoneal virulence in mice. Virology 149:255-259. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz, C., M. Moyal, A. Rosen-Wölff, G. Darai, and Y. Becker. 1994. Herpes simplex virus type 1 (HSV-1) UL56 gene is involved in viral intraperitoneal pathogenicity to immunocompetent mice. Arch. Virol. 134:73-83. [DOI] [PubMed] [Google Scholar]
- 4.Brandimarti, R., and B. Roizman. 1997. Us9, a stable lysine-less herpes simplex virus 1 protein, is ubiquitinated before packaging into virions and associates with proteasomes. Proc. Natl. Acad. Sci. USA 94:13973-13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brideau, A. D., B. W. Banfield, and L. W. Enquist. 1998. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J. Virol. 72:4560-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brideau, A. D., J. P. Card, and L. W. Enquist. 2000. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. J. Virol. 74:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brideau, A. D., M. G. Eldridge, and L. W. Enquist. 2000. Directional transneuronal infection by pseudorabies virus is dependent on an acidic internalization motif in the Us9 cytoplasmic tail. J. Virol. 74:4549-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brideau, A. D., T. Del Rio, E. J. Wolffe, and L. W. Enquist. 1999. Intracellular trafficking and localization of the pseudorabies virus Us9 type II envelope protein to host and viral membranes. J. Virol. 73:4372-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, D. A., and E. London. 1997. Structure of detergent-resistant membrane domains: does phase separation occur in biological membranes? Biochem. Biophys. Res. Commun. 240:1-7. [DOI] [PubMed] [Google Scholar]
- 10.Brown, D. A., and E. London. 1998. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14:111-136. [DOI] [PubMed] [Google Scholar]
- 11.Campadelli, G., R. Brandimarti, C. Di Lazzaro, P. L. Ward, B. Roizman, and M. R. Torrisi. 1993. Fragmentation and dispersal of Golgi proteins and redistribution of glycoproteins and glycolipids processed through the Golgi apparatus after infection with herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 90:2798-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chamberlain, L. H., R. D. Burgoyne, and G. W. Gould. 2001. SNARE proteins are highly enriched in lipid rafts in PC12 cells: implications for the spatial control of exocytosis. Proc. Natl. Acad. Sci. USA 98:5619-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cochrane, A. W., A. Perkins, and C. A. Rosen. 1990. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J. Virol. 64:881-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davison, A. J., and J. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 15.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fra, A. M., E. Williamson, K. Simons, and R. G. Parton. 1994. Detergent-insoluble glycolipid microdomains in lymphocytes in the absence of caveolae. J. Biol. Chem. 269:30745-30748. [PubMed] [Google Scholar]
- 17.Frame, M. C., D, J. McGeoch, F. J. Rixon, A. C. Orr, and H. S. Marsden. 1986. The 10K virion phosphoprotein encoded by gene US9 from herpes simplex virus type 1. Virology 150:321-332. [DOI] [PubMed] [Google Scholar]
- 18.Harley, C. A., A. Dasgupta, and D. W. Wilson. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 75:1236-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kehm, R., A. Rosen-Wölff, and G. Darai. 1996. Restitution of the UL56 gene expression of HSV-1 HFEM led to restoration of virulent phenotype; deletion of the amino acids 217 to 234 of the UL56 protein abrogates the virulent phenotype. Virus Res. 40:17-31. [DOI] [PubMed] [Google Scholar]
- 20.Kehm, R., E. Lorentzen, A. Rosen-Wölff, and G. Darai. 1994. In vitro expression of UL56 gene of herpes simplex virus type 1; detection of UL56 gene product in infected cells and in virions. Virus Res. 33:55-66. [DOI] [PubMed] [Google Scholar]
- 21.Kehm, R., H. R. Gelderblom., and G. Darai. 1998. Identification of the UL56 protein of herpes simplex virus type 1 within the virion by immuno electron microscopy. Virus Gene 17:49-53. [DOI] [PubMed] [Google Scholar]
- 22.Kirchhausen, T., J. S. Bonifacino, and H. Riezman. 1997. Linking cargo to vesicle formation: receptor tail interactions with coat proteins. Curr. Opin. Cell Biol. 9:488-495. [DOI] [PubMed] [Google Scholar]
- 23.Klein, P., M. Kanehisa, and C. DeLisi. 1985. The detection and classification of membrane-spanning proteins. Biochim. Biophys. Acta 815:468-476. [DOI] [PubMed] [Google Scholar]
- 24.Kozak, M. 1991. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 115:887-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubota, S., H. Siomi, T. Satoh, S. Endo, M. Maki, and M. Hatanaka. 1989. Functional similarity of HIV-I rev and HTLV-I rex proteins: identification of a new nucleolar-targeting signal in rev protein. Biochem. Biophys. Res. Commun. 162:963-970. [DOI] [PubMed] [Google Scholar]
- 26.Kutay, U., E. Hartmann, and T. A. Rapoport. 1993. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 3:72-75. [DOI] [PubMed] [Google Scholar]
- 27.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 28.Lafont, F., P Verkade, T. Galli, C. Wimmer, D. Louvard, and K. Simons. 1999. Raft association of SNAP receptors acting in apical trafficking in Madin-Darby canine kidney cells. Proc. Natl. Acad. Sci. USA 96:3734-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGeoch, D. J., C. Cunningham, G. McIntyre, and A. Dolan. 1991. Comparative sequence analysis of the long repeat region and adjoining parts of the long unique regions in the genome of herpes simplex viruses types 1 and 2. J. Gen. Virol. 72:3057-3075. [DOI] [PubMed] [Google Scholar]
- 30.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 31.Miranda-Saksena, M., P. Armati, R. A. Boadle, D. J. Holland, and A. L. Cunningham. 2000. Anterograde transport of herpes simplex virus type 1 in cultured, dissociated human and rat dorsal root ganglion neurons. J. Virol. 74:1827-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peles, E., H. Rosen, A. Rosen-Wölff, and Y. Becker. 1990. Importance of the HpaI-P sequence for herpes simplex virus-1 replication in the adrenal glands. Arch. Virol. 113:151-163. [DOI] [PubMed] [Google Scholar]
- 33.Roizman, B. 1996. The function of herpes simplex virus genes: a primer for genetic engineering of novel vectors. Proc. Natl. Acad. Sci. USA 93:11307-11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 1043-1107. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 35.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosen, A., F. Ernst, H. G. Koch, H. Gelderblom, G. Darai, J. Hadar, E. Tabor, T. Ben-Hur, and Y. Becker. 1986. Replacement of the deletion in the genome (0.762-0.789 mu) of avirulent HSV-1 HFEM using cloned MluI DNA fragment (0.7615-0.796 mu) of virulent HSV-1 F leads to generation of virulent intratypic recombinant. Virus Res. 5:157-175. [DOI] [PubMed] [Google Scholar]
- 37.Rosen, A., and G. Darai. 1985. Mapping of the deletion in the genome of HSV-1 strain HFEM responsible for its avirulent phenotype. Med. Microbiol. Immunol. 173:329-343. [DOI] [PubMed] [Google Scholar]
- 38.Rosen-Wölff, A., W. Lamadé, C. Berkowitz, Y. Becker, and G. Darai. 1991. Elimination of UL56 gene by insertion of LacZ cassette between nucleotide position 116030 to 121753 of the herpes simplex virus type 1 genome abrogates intraperitoneal pathogenicity in tree shrews and mice. Virus Res. 20:205-221. [DOI] [PubMed] [Google Scholar]
- 39.Rosen-Wöllf, A., and G. Darai. 1991. Identification and mapping of the UL56 gene transcript of herpes simplex virus type I. Virus Res. 19:115-126. [DOI] [PubMed] [Google Scholar]
- 40.Ruben, S., A. Perkins, R. Purcell, K. Joung, R. Sia, R. Burghoff, W. A. Haseltine, and C. A. Rosen. 1989. Structural and functional characterization of human immunodeficiency virus tat protein. J. Virol. 63:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnitzer, J. E., D. P. McIntosh, A. M. Dvorak, J. Liu, and P. Oh. 1995. Separation of caveolae from associated microdomains of GPI-anchored proteins. Science 269:1435-1439. [DOI] [PubMed] [Google Scholar]
- 42.Shiba, C., T. Daikoku, F. Goshima, H. Takakuwa, Y. Yamauchi, O. Koiwai, and Y. Nishiyama. 2000. The UL34 gene product of herpes simplex virus type 2 is a tail-anchored type II membrane protein that is significant for virus envelopment. J. Gen. Virol. 81:2397-2405. [DOI] [PubMed] [Google Scholar]
- 43.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 44.Siomi, H., H. Shida, S. H. Nam, T. Nosaka, M. Maki, and M. Hatanaka. 1988. Sequence requirements for nucleolar localization of human T cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell 55:197-209. [DOI] [PubMed] [Google Scholar]
- 45.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 46.Tomishima, M. J., and L. W. Enquist. 2001. A conserved alpha-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. Cell Biol. 154:741-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsurumi, T., K. Maeno, and Y. Nishiyama. 1986. Molecular cloning of herpes simplex virus type 2 DNA. J. Biochem. 99:981-984. [DOI] [PubMed] [Google Scholar]
- 48.Zegers, M. M., and D. Hoekstra. 1998. Mechanisms and functional features of polarized membrane traffic in epithelial and hepatic cells. Biochem. J. 336:257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]