Abstract
A reverse transcription-PCR (RT-PCR) was established to amplify a 379-bp cDNA fragment (nucleotides 747 to 1126, coding for amino acids 241 to 367) of the VP6 gene of group A rotaviruses associated with subgroup (SG) specificity. Thirty-eight human rotavirus strains characterized with SG-specific monoclonal antibodies were subjected to VP6-specific RT-PCR, and PCR amplicons were used for sequencing. Nucleic acid sequencing and phylogenetic analysis of the VP6 amplicons revealed two clusters, or genogroups. Two genetic lineages were distinguished within genogroup I, consisting of strains serologically characterized as SG I, and three genetic lineages were distinguished within genogroup II, composed of strains serologically characterized as SG II, SG I + II, and SG non-I, non-II. Subgrouping of rotaviruses by means of serological methods may result in strains not being assigned the correct SG or in a failure of strains to subgroup. Molecular characterization of the SG-defining region of VP6 provided evidence for independent segregation of the rotavirus genes encoding VP4, VP6, and VP7.
Rotaviruses are triple-layered particles of the Reoviridae family which are classified into groups (A to E) and subgroups (SG) according to the presence of epitopes on the middle-layer protein VP6 (16). VP6 is a trimeric protein that interacts with the inner-layer protein VP2 and with the two outer-layer proteins VP7 and VP4 (20). Most human rotavirus infections are caused by rotaviruses of group A. Within this group, SG I, II, I + II, and non-I, non-II have been defined according to the presence or absence of two distinct epitopes reactive with one, both, or neither of the monoclonal antibodies (MAbs) 255/60 and 631/9 (11). Subgrouping enzyme-linked immunosorbent assays (ELISAs) have been used extensively in epidemiological studies. The rotaviruses most commonly found in humans belong to SG II (2, 4, 8, 14, 19), while SG I is common among animal rotaviruses (18, 23).
Previous studies have mapped SG I specificity to amino acid (aa) position 305 and the region between positions 296 and 299 and SG II specificity to residue 315 (18, 23). It is still unclear whether SG specificity is determined by linear or conformational epitopes, although there is some evidence that the epitopes recognized by SG-specific MAbs are conformational and are present only in the trimeric form of VP6 (9). Amino acid substitutions that are quite distant in the linear molecule may affect the conformation of the epitopes.
Serological characterization of the surface proteins VP7 and VP4 has been shown to be unreliable due to antigenic drift through the accumulation of point mutations (7, 12, 21). It is possible that VP6 will undergo similar antigenic changes, resulting in poor reactivity in serological subgrouping assays.
The aims of this work were to determine the SG of rotavirus strains on the basis of cDNA sequences of a fragment of the VP6 genes, to investigate correlations between molecular and serological subgrouping methods, and to study the variability of the VP6 gene within and among the different SG. This would also allow researchers to investigate at a molecular level whether and to what extent VP6 genes are involved in reassortment events shown to occur in nature during cocirculation of different human and animal rotavirus strains (14, 25, 26).
MATERIALS AND METHODS
Strains.
Thirty-eight rotavirus strains previously characterized by serological assays as SG I (8 strains), SG II (16 strains), SG non-I, non-II (8 strains), or SG I + II (6 strains) were selected from a collection of human rotavirus isolates obtained in the United Kingdom during a 4-year (1995 to 1999) surveillance study (13, 14). Five strains of unusual G and P genotype combinations for which subgroup serology was not available were also included (one strain each of G8 P[8], G9 P[6], and G1 P[9] and two strains of G9 P[8]).
Subgrouping of rotaviruses into SG I, II, I + II, and non-I, non-II was performed by ELISA using 96-well microtiter plates coated with rabbit polyclonal anti-rotavirus VP6 antibodies (provided by DAKO Diagnostics Ltd., Ely, United Kingdom) with minor modifications to a published procedure (3). MAbs 255/60 and 631/9 (10, 11), specific for SG I and SG II, respectively, were used for subgrouping ELISAs. Briefly, plates were blocked with 100 μl of 0.1 M phosphate-buffered saline (PBS) (pH 7.4) containing 5% (wt/vol) skim milk powder and 0.05% (vol/vol) Tween 20 (5% SMP-PBST) for 2 h at 37°C. After aspiration of the blocking solution, 75 μl of 2.5% SMP-PBST was added to all the wells, followed by 25 μl of a 10% fecal suspension (2 wells/sample); three negative controls were included in each plate. Plates were incubated at 4°C for 16 to 18 h and washed five times with PBST. One hundred microliters of a SG I- or SG II-specific MAb diluted 1:1,000 in 2.5% SMP-PBST was added to the appropriate wells. Plates were incubated for 2.5 h at 37°C and then washed five times with PBST, and 100 μl of a 1:10,000 dilution of a commercial horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (Sigma, Dorset, United Kingdom) was added to each well. Plates were incubated for 1.5 h at 37°C and then washed five times with PBST. One hundred microliters of tetramethylbenzidine (0.1 mg/ml)-H2O2 (0.014%) (Sigma, Dorset) in 0.05 M phosphate-citrate buffer (pH 6) was added to each well, and the reaction was stopped after 10 min by addition of 50 μl of 2 M H2SO4. Optical density was measured at 450 nm (OD450). The cutoff value was calculated as the mean of the OD450 values of the rotavirus-negative fecal sample controls for each plate plus 3 standard deviations. A sample was considered to be reactive for one or both of the two MAbs if the OD450 was above the cutoff value. SG I or SG II was assigned when the OD450 value obtained with one of the MAbs was at least twice the OD450 value obtained with the other MAb. A sample was classified as SG non-I, non-II if it was not reactive with either MAb; it was classified as SG I + II if it was reactive with both MAbs but the OD450 ratio between the two was <2.
Primers were designed to amplify a 379-bp region (nucleotides [nt] 747 to 1126, coding for aa 241 to 367) of the VP6 gene; they were VP6-F (sense) (5′ GACGGVGCRACTACATGGT 3′) (nt 747 to 766) and VP6-R (antisense) (5′ GTCCAATTCATNCCTGGTGG 3′) (nt 1126 to 1106).
Reverse transcription was performed after guanidinium isothiocyanate-silica nucleic acid extraction (5) using random priming with hexamers (Amersham-Pharmacia Biotech) (5, 13), and the cDNA was used in the VP6-specific PCR. The PCR was optimized for use with the LightCycler (Roche, Molecular Biochemicals) real-time PCR thermal cycler.
Amplicons obtained were purified by using a commercial spin column (QIAquick; Qiagen) and sequenced by using primers VP6-F and VP6-R. Nucleic acid sequences were derived by using an automated sequencer (CEQ2000; Beckman-Coulter).
Phylogenetic analyses were performed by using the Clustal and neighbor-joining methods and were confirmed by the bootstrapping and maximum parsimony methods (using Megalign, DNAstar [Lasergene, Madison, Wis.], and Bionumerics [Applied Maths, Kortrijk, Belgium]).
RESULTS
PCR.
A rotavirus group A-specific real-time PCR which amplifies a 379-bp fragment of the VP6 gene of rotavirus strains of different subgroups was optimized. The melting temperatures (Tm) of the VP6 amplicons ranged between 83.0 and 86.0°C. No correlation was found between amplicon Tm and serologically defined SG specificity (data not shown).
Sequence analysis.
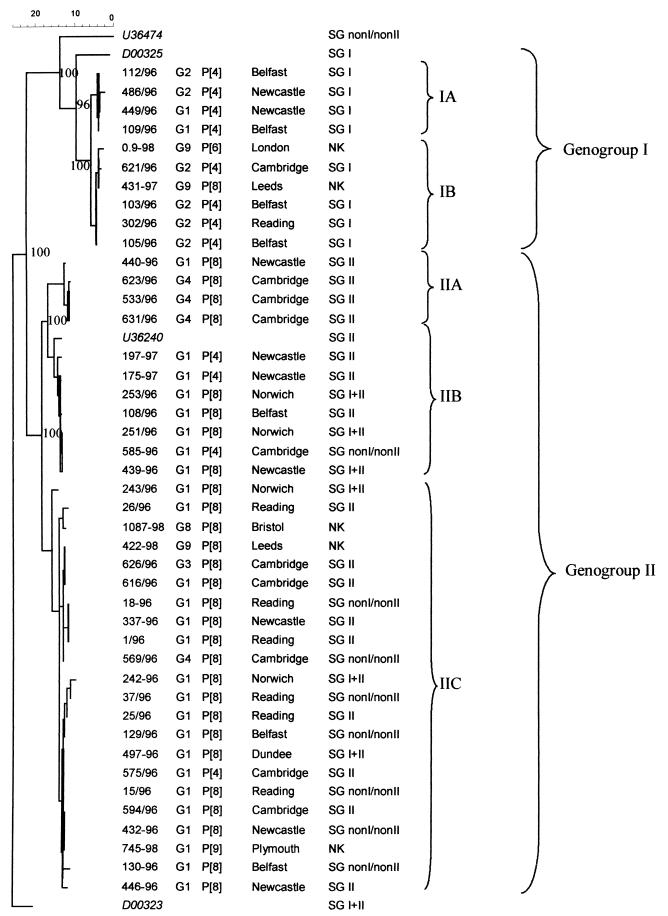
Phylogenetic analyses of the nucleic acid sequences of VP6 amplicons obtained from these strains showed that all of the strains determined as SG I by ELISA clustered together into two genetic lineages, IA and IB (genogroup I) (Fig. 1). All the other strains (SG II, SG I + II, and SG non-I, non-II) formed a second cluster (genogroup II) (Fig. 1). Three genetic lineages, IIA, IIB, and IIC, were distinguishable within genogroup II, but no correlations were found between serologically determined SG and genetic lineage (Fig. 1; Table 1).
FIG. 1.
Phylogenetic tree constructed from 379-bp cDNA fragments of the VP6 genes of 43 human rotavirus strains by using the Clustal and neighbor-joining methods. For each strain, the strain designation and year of isolation, G and P types, geographical location, and serologically determined SG are shown. Significant bootstrap values (>90%) are shown at the branching point of each cluster. Accession numbers of sequences of prototype strains of SG I, SG II, SG I + II, and SG non-I, non-II obtained from GenBank are given in italics.
TABLE 1.
Subgroup assignment of 43 human rotavirus strains according to sequencing and phylogenetic analysis versus subgrouping ELISAs
| Sample identifier | G and P types | OD450 ratioa
|
SG by ELISA | Genetic lineage | |
|---|---|---|---|---|---|
| SG I/SG II | SG II/SG I | ||||
| 112-96 | G2 P[4] | 4.6 | SG I | IA | |
| 486-96 | G2 P[4] | 4.9 | SG I | IA | |
| 449-96 | G1 P[4] | 4.3 | SG I | IA | |
| 109-96 | G1 P[4] | 2.4 | SG I | IA | |
| 0.9-98 | G9 P[6] | NT | NT | NK | IB |
| 621-96 | G2 P[4] | 9.2 | SG I | IB | |
| 431-97 | G9 P[8] | NT | NT | NK | IB |
| 103-96 | G2 P[4] | 4.5 | SG I | IB | |
| 302-96 | G2 P[4] | 3.5 | SG I | IB | |
| 105-96 | G2 P[4] | 4.1 | SG I | IB | |
| 440-96 | G1 P[8] | 3.5 | SG II | IIA | |
| 623-96 | G4 P[8] | 7.3 | SG II | IIA | |
| 533-96 | G4 P[8] | 5.0 | SG II | IIA | |
| 631-96 | G4 P[8] | 4.7 | SG II | IIA | |
| 197-97 | G1 P[4] | 3.3 | SG II | IIB | |
| 175-97 | G1 P[4] | 2.9 | SG II | IIB | |
| 253-96 | G1 P[8] | 1.3 | SG I + II | IIB | |
| 108-96 | G1 P[8] | 3.0 | SG II | IIB | |
| 251-96 | G1 P[8] | 1.7 | SG I + II | IIB | |
| 585-96 | G1 P[4] | NR | NR | SG non-I, non-II | IIB |
| 439-96 | G1 P[8] | 1.8 | SG I + II | IIB | |
| 243-96 | G1 P[8] | 1.4 | SG I + II | IIC | |
| 26-96 | G1 P[8] | 4.2 | SG II | IIC | |
| 1087-98 | G8 P[8] | NT | NT | NK | IIC |
| 422-98 | G9 P[8] | NT | NT | NK | IIC |
| 626-96 | G3 P[8] | 4.8 | SG II | IIC | |
| 616-96 | G1 P[8] | 5.6 | SG II | IIC | |
| 18-96 | G1 P[8] | NR | NR | SG non-I, non-II | IIC |
| 337-96 | G1 P[8] | 2.8 | SG II | IIC | |
| 1-96 | G1 P[8] | 2.8 | SG II | IIC | |
| 569-96 | G4 P[8] | NR | NR | SG non-I, non-II | IIC |
| 242-96 | G1 P[8] | 1.6 | SG I + II | IIC | |
| 37-96 | G1 P[8] | NR | NR | SG non-I, non-II | IIC |
| 25-96 | G1 P[8] | 2.7 | SG II | IIC | |
| 129-96 | G1 P[8] | NR | NR | SG non-I, non-II | IIC |
| 497-96 | G1 P[8] | 1.8 | SG I + II | IIC | |
| 575-96 | G1 P[4] | 5.2 | SG II | IIC | |
| 15-96 | G1 P[8] | NR | NR | SG non-I, non-II | IIC |
| 594-96 | G1 P[8] | 2.3 | SG II | IIC | |
| 432-96 | G1 P[8] | NR | NR | SG non-I, non-II | IIC |
| 745-98 | G1 P[9] | NT | NT | NK | IIC |
| 130-96 | G1 P[8] | NR | NR | SG non-I, non-II | IIC |
| 446-96 | G1 P[8] | 3.4 | SG II | IIC | |
NR, nonreactive; NT, not tested; NK, not known.
Analysis of the deduced amino acid sequences showed conserved amino acid substitutions distinguishing the strains in genogroups I and II at residues 305 (Ala→Asn); 310 (Asn→Leu), 315 (Glu→Leu), 339 (Ser→Asn), 342 (Met→Leu), and 348 (Ser→Ala). In the strains that were identified as SG non-I, non-II or SG I + II by serology, the residues at these positions were identical to those of SG II strains (Table 2).
TABLE 2.
Deduced amino acid sequence of the VP6 region spanning positions 281 to 350
| Samplea | Geno- group | SGb by ELISA | VP6 sequencec at position:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 281 | 291 296-299 | 301 305 | 311 315 | 321 | 331 | 341 | |||
| U36240 | II | II | IARNFDTIRL | SFQLMRPPNM | TPAVNALFPQ | AQPFQHHATV | GLTLRIESAV | CESVLADANE | TLLANVTAMR |
| 251-96 | II | I + II | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| 439-96 | II | I + II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 253-96 | II | I + II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 197-97 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 175-97 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 108-96 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 585-96 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 440-96 | II | II | V......... | L......... | .......... | .......... | .......... | .......... | .........V |
| 631-96 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 623-96 | II | II | .......... | L......... | .......... | .......... | .......... | .......... | .........V |
| 533-96 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 745-98 | II | NK | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 422-98 | II | NK | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 497-96 | II | I + II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 243-96 | II | I + II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 242-96 | II | I + II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 616-96 | II | II | V......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 626-96 | II | II | V......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 25-96 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 594-96 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 337-96 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 575-96 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 569-96 | II | nI, nII | .......... | .......... | .......... | .......... | .......... | .......S.. | .........V |
| 37-96 | II | nI, nII | .......... | .......... | .......... | .......... | .......... | .......S.. | .........V |
| 130-96 | II | nI, nII | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 15-96 | II | nI, nII | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 432-96 | II | nI, nII | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 18-96 | II | nI, nII | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 129-96 | II | nI, nII | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 449-96 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 112-96 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 109-96 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| 26-96 | II | II | .......... | .......... | .......... | .......... | .......... | .......... | .........V |
| D00325 | I | I | V......... | .......... | ....A....N | ....E..... | .......... | ........S. | .M.......V |
| 0.9-98 | I | NK | V......... | .......... | ....A....N | ....E..... | .......... | ........S. | .M.......V |
| 431-96 | I | I | V......... | .......... | ....A....N | ....E..... | .......... | ........S. | .M.......V |
| 621-96 | I | I | V......... | .......... | ....A....N | ....E..... | ....K..... | ........S. | .M.......V |
| 103-96 | I | I | V......... | .......... | ....A....N | ....E..... | ....K..... | ........S. | .M.......V |
| 302-96 | I | I | V......... | .......... | ....A....N | ....E..... | ....K..... | ........S. | .M.......V |
| 105-96 | I | I | V......... | .......... | ....A....N | ....E..... | ....K..... | ........S. | .M.......V |
| 431-97 | I | NK | V......... | .......... | ..S.A....N | ....E..... | .......... | ........S. | .M.......V |
| 486-96 | I | I | V......... | .......... | ....A....N | ....E..... | .......... | ........S. | .M.......V |
| 112-96 | I | I | V......... | .......... | ....A....N | ....E..... | .......... | ........S. | .M.......V |
| D00323 | I + II | V......... | .......... | .......... | .......... | ......D... | .......S.. | .M.......V | |
| U36474 | nI, nII | V......... | .......... | ....T....N | ....E..... | ......D..I | ........S. | .M.......V | |
Italicized designations are accession numbers of sequences of prototype strains of the different SGs obtained from GenBank.
NK, not known; nI, nII, SG non-I, non-II.
Amino acid residues previously identified as defining SG I and SG II reactivities are boldfaced. Dots indicate consensus with sequence of prototype SG II strain. Residues that differ from this consensus are shown.
Analysis of the distribution of G and P genotypes revealed likely reassortment events involving the VP6 gene among strains of genogroups I and II (Table 3).
TABLE 3.
Distribution of G and P genotypes among strains of genogroups I and IIa
| Genotype | Possible parental strains | No. of strains
|
||
|---|---|---|---|---|
| Genogroup I | Genogroup II | Total | ||
| G1 P[4] | G1 P[8] | 2b | 4c | 6 |
| G2 P[4] | ||||
| G1 P[8] | 21 | 21 | ||
| G2 P[4] | 6 | 6 | ||
| G3 P[8] | 1 | 1 | ||
| G4 P[8] | 4 | 4 | ||
| G9 P[6] | 1 | |||
| G9 P[8] | G1 P[8], G3 P[8], or G4 P[8] | 1d | 1e | 2 |
| G9 P[6] | ||||
| G1 P[9] | G1 P[8] | 1c | 1 | |
| G? P[9] | ||||
| G8 P[8] | G8 P? | 1e | 1 | |
| G1 P[8], G3 P[8], or G4 P[8] | ||||
| Total | 10 | 33 | 43 | |
Putative reassortant strains are boldfaced. Strains with genotypes suggestive of an animal origin are italicized.
Gene 6 is provided by the parental strain carrying the gene encoding P[4].
Gene 6 is provided by the parental strain carrying the gene encoding G1.
Gene 6 is provided by the parental strain carrying the gene encoding G9.
Gene 6 is provided by the parental strain carrying the gene encoding P[8].
DISCUSSION
The ability to amplify a 379-bp region of the VP6 gene which encompassed the region previously defined as encoding the SG-specific epitopes allowed the molecular subgrouping of rotaviruses according to diversity within the VP6 gene.
The unreliability of serological methods for the characterization of VP7 and VP4 into G and P types, respectively, is a well-recognized problem (7, 12, 21). This originates from an accumulation of point mutations which leads to amino acid changes on the epitopes recognized by the type-specific MAbs. Many strains cannot be serotyped due to the limited availability of MAbs specific for all the variants, which has resulted in the gradual replacement of serotyping by molecular typing methods. Therefore, it could be postulated that the existence of mutants is not restricted to the VP7 and VP4 molecules but may also apply to other structural proteins such as VP6.
A total of 20 SG-specific amino acids have been identified, and studies using site-directed mutagenesis (23) or recombination (18) of VP6 genes have shown that substitutions at positions 296 to 299, 305, 306, 308, and 315 are capable of changing reactivities to the SG I- and SG II-specific MAbs. It has been proposed that an Ala residue at position 305 and the region between 296 and 299 contribute to determining reactivity to MAb 255/60 (SG I), whereas a Glu residue at position 315 contributes to reactivity to MAb 631/9 (SG II) (23).
The epitopes recognized by the SG-specific MAbs are thought to be conformational and therefore present only in the trimeric form of the protein. Therefore, the VP6 region chosen for amplification in our study encompasses the amino acid positions previously implicated in recognition by SG-specific MAbs, and those associated with trimerization (aa 246 to 315) (1, 6), as well as most of those necessary for the formation of double-layered particles (aa 281 to 397) (24). The deduced amino acid sequences of all the strains serologically determined as SG I showed an Ala residue at position 305, which is characteristic of SG I. However, the amino acid sequences of the amplicons obtained from strains serologically determined to be SG II, SG I + II, or SG non-I, non-II were indistinguishable from each other. All these strains had the Glu residue at aa position 315 characteristic of SG II strains. None of the strains serologically determined as SG I + II or SG non-I, non-II showed substitutions at any of the other SG-determining amino acid positions different from those of the SG II strains (Table 2). None of the strains serologically determined as SG non, non-II showed the insertion Pro-Glu at positions 299 and 300 or an amino acid substitution at position 308 (Table 2) previously shown to abolish reactivity with both MAb 255/60 and MAb 631/9 (22).
The genetic similarity among strains subgrouped as SG II, SG I + II, or SG non-I, non-II by using serological methods would suggest either serological cross-reactivity between SG I and SG II or the loss of an SG-determining epitope. However, the cross-reactivity would appear to be unidirectional, as strains of SG II may cross-react with the SG I-specific MAb, but strains of SG I do not appear to cross-react with the anti-SG II MAb. All the strains serologically determined as SG I + II showed greater reactivity with the SG II-specific MAb (Table 1). It is possible that amino acid positions or regions outside those analyzed here may contribute to reactivity of SG II strains with the SG I-specific MAb. An Ala residue at position 172 was suggested to contribute to the formation of the SG I-specific epitope; however, single point mutations at this position showed only low reactivity values in immunoprecipitation assays with the SG I-specific MAb (23), and a chimeric VP6 containing an Ala residue at this position in the presence of substitutions characteristic of SG II at aa positions 305 and 315 did not react with with the SG I-specific MAb (18). Furthermore, the unsatisfactory definition of an SG through a lack of reactivity with either SG-specific MAb does not rule out the possibility that SG non-I, non-II strains are misclassified as a result of insufficient antigen being present in the ELISA. Nevertheless, this is unlikely given the fact that strains failing to react in the subgrouping ELISAs reacted with group A cross-reactive antibodies in diagnostic ELISAs. There is the theoretical possibility that conformationally determined SG specificity might be affected by different variants of VP2 with which VP6 interacts. This possibility awaits exploration.
Only one strain of SG I + II (equine strain F1-14) and two strains of SG non-I, non-II (the murine strain EW and the equine strain H-2) have been found in the sequence databases, and none of these are of human origin (9, 23). Their nucleic acid and amino acid sequences were significantly different from the SG I and SG II strains throughout the VP6 region chosen for amplification in this study.
No true human SG I + II or SG non-I, non-II strains, based on sequence analysis of VP6, were found in this study. SG I + II or SG non-I, non-II strains detected by serological methods in this study differed considerably in nucleotide and amino acid sequences from prototype strains and were more closely related to SG II strains. This may indicate that rotaviruses of SG I + II and SG non-I, non-II are rare in the human population and that the majority of the SG I + II and SG non-I, non-II strains described in seroepidemiological studies may have been misclassified due to poor reactivity of the common SG II strains with the MAbs used.
Molecular methods for subgrouping provide more-accurate information on the diversity of the VP6 genes of rotavirus strains circulating in the human population and may contribute to understanding of the interrelationships among different genes and different proteins.
Previously we provided evidence for the independent segregation of the genes encoding VP7 and VP4 in reassortant rotavirus strains found in the United Kingdom (14). Rotavirus reassortant strains of genotype G1 P[4] probably originated through reassortment between commonly circulating human rotavirus strains of genotypes G1 P[8] and G2 P[4] (14). Usually G1 P[8] strains are associated with a VP6 of genogroup (SG) II, while G2 P[4] strains are associated with a VP6 of genogroup (SG) I (Tables 1 and 3) (14). The G1 P[4] reassortant strains were found in association with VP6 of either genogroup I or genogroup II (Fig. 1), suggesting that the gene encoding VP6 segregated independently of the genes encoding VP7 and VP4 during reassortment (Table 3). VP6 genes of SG I have been shown to be associated with G9 P[6] strains (15, 22). Analysis of the VP7 and VP4 genes of G9 P[6] and G9 P[8] strains provided evidence for the emergence of G9 P[8] strains through reassortment (14). The detection of G9 P[8] rotavirus strains with VP6 genes of either genogroup I or genogroup II provided further evidence of their reassortant origin and of the independent segregation of the genes encoding VP4, VP6, and VP7. Thus, this study adds to the molecular evidence for the involvement of VP6 in reassortment, strengthening previous reports that have described possible VP6 reassortants on the basis of serologically defined SG (14, 17). Other strains found in the United Kingdom, G1 P[9] and G8 P[8], may also represent reassortants derived from human and animal strains (14). Both these reassortants were found in association with VP6 molecules of genogroup II which are possibly derived from the human parental strains carrying the gene segment encoding P[8]. Study of the epidemiology and characterization of rotavirus strains infecting animals living in close contact with humans is necessary in order to identify possible parental strains associated with human-animal rotavirus reassortment events and their subsequent introduction into the human population.
REFERENCES
- 1.Affranchino, J. L., and S. A. Gonzalez. 1997. Deletion mapping of functional domains in the rotavirus capsid protein VP6. J. Gen. Virol. 78:1949-1955. [DOI] [PubMed] [Google Scholar]
- 2.Arista, S., L. Giovannelli, D. Pistoia, A. Cascio, M. Parea, and G. Gerna. 1990. Electropherotypes, subgroups and serotypes of human rotavirus strains causing gastroenteritis in infants and young children in Palermo, Italy, from 1985 to 1989. Res. Virol. 141:435-448. [DOI] [PubMed] [Google Scholar]
- 3.Beards, G. 1987. Serotyping of rotavirus by NADP-enhanced enzyme immunoassay. J. Virol. Methods 18:77-85. [DOI] [PubMed] [Google Scholar]
- 4.Beards, G. M., U. Desselberger, and T. H. Flewett. 1989. Temporal and geographical distributions of human rotavirus serotypes, 1983 to 1988. J. Clin. Microbiol. 27:2827-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boom, R., C. J. A. Sol, M. M. M. Salismans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. van den Noordaa. 1990. Rapid and simple method for the purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clapp, L. L., and J. T. Patton. 1991. Rotavirus morphogenesis: domains in the major inner capsid protein essential for binding to single-shelled particles and for trimerization. Virology 180:697-708. [DOI] [PubMed] [Google Scholar]
- 7.Coulson, B. S. 1996. VP4 and VP7 typing using monoclonal antibodies. Arch. Virol. Suppl. 12:113-118. [DOI] [PubMed] [Google Scholar]
- 8.Georges-Courbot, M. C., A. M. Beraud, G. M. Beards, A. D. Campbell, J. P. Gonzalez, A. J. Georges, and T. H. Flewett. 1988. Subgroups, serotypes, and electrophoretypes of rotavirus isolated from children in Bangui, Central African Republic. J. Clin. Microbiol. 26:668-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorziglia, M., Y. Hoshino, K. Nishikawa, W. L. Maloy, R. W. Jones, A. Z. Kapikian, and R. M. Chanock. 1988. Comparative sequence analysis of the genomic segment 6 of four rotaviruses each with a different subgroup specificity. J. Gen. Virol. 69:1659-1669. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg, H., V. McAuliffe, J. Valdesuso, R. Wyatt, J. Flores, A. Kalica, Y. Hoshino, and N. Singh. 1983. Serological analysis of the subgroup protein of rotavirus, using monoclonal antibodies. Infect. Immun. 39:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenberg, H. B., J. Valdesuso, K. van Wyke, K. Midthun, M. Walsh, V. McAuliffe, R. G. Wyatt, A. R. Kalica, J. Flores, and Y. Hoshino. 1983. Production and preliminary characterization of monoclonal antibodies directed at two surface proteins of rhesus rotavirus. J. Virol. 47:267-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iturriza Gómara, M., D. Cubitt, U. Desselberger, and J. Gray. 2001. Amino acid substitution within the VP7 protein of G2 rotavirus strains associated with failure to serotype. J. Clin. Microbiol. 39:3796-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iturriza Gómara, M., J. Green, D. W. Brown, U. Desselberger, and J. J. Gray. 1999. Comparison of specific and random priming in the reverse transcriptase polymerase chain reaction for genotyping group A rotaviruses. J. Virol. Methods 78:93-103. [DOI] [PubMed] [Google Scholar]
- 14.Iturriza Gómara, M., B. Isherwood, U. Desselberger, and J. Gray. 2001. Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J. Virol. 75:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain, V., B. K. Das, M. K. Bhan, R. I. Glass, and J. R. Gentsch. 2001. Great diversity of group A rotavirus strains and high prevalence of mixed rotavirus infections in India. J. Clin. Microbiol. 39:3524-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1834. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 17.Krishnan, T., T. N. Naik, and U. Desselberger. 1996. Molecular epidemiology of human rotaviruses: reassortment in vivo as a mechanism for strain diversity? J. Infect. 32:169-170. [DOI] [PubMed] [Google Scholar]
- 18.López, S., R. Espinosa, H. B. Greenberg, and C. F. Arias. 1994. Mapping the subgroup epitopes of rotavirus protein VP6. Virology 204:153-162. [DOI] [PubMed] [Google Scholar]
- 19.Mohammed, K. A., S. M. el Assouli, and Z. M. Banjar. 1994. Human rotavirus subgroups and serotypes in children with acute gastroenteritis in Saudi Arabia from 1988 to 1992. J. Med. Virol. 44:237-242. [DOI] [PubMed] [Google Scholar]
- 20.Prasad, B., and M. Estes. 2000. Electron cryomicroscopy and computer image processing techniques. Methods Mol. Med. 34:9-31. [DOI] [PubMed] [Google Scholar]
- 21.Raj, P., D. O. Matson, B. S. Coulson, R. F. Bishop, K. Taniguchi, S. Urasawa, H. B. Greenberg, and M. K. Estes. 1992. Comparisons of rotavirus VP7-typing monoclonal antibodies by competition binding assay. J. Clin. Microbiol. 30:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramachandran, M., J. R. Gentsch, U. D. Parashar, S. Jin, P. A. Woods, J. L. Holmes, C. D. Kirkwood, R. F. Bishop, H. B. Greenberg, S. Urasawa, G. Gerna, B. S. Coulson, K. Taniguchi, J. S. Bresee, and R. I. Glass. 1998. Detection and characterization of novel rotavirus strains in the United States. J. Clin. Microbiol. 36:3223-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang, B., J. M. Gilbert, S. M. Matsui, and H. B. Greenberg. 1997. Comparison of the rotavirus gene 6 from different species by sequence analysis and localization of subgroup-specific epitopes using site-directed mutagenesis. Virology 237:89-96. [DOI] [PubMed] [Google Scholar]
- 24.Tosser, G., T. Delaunay, E. Kohli, J. Grosclaude, P. Pothier, and J. Cohen. 1994. Topology of bovine rotavirus (RF strain) VP6 epitopes by real-time biospecific interaction analysis. Virology 204:8-16. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe, M., T. Nakagomi, Y. Koshimura, and O Nakagomi. 2001. Direct evidence for genome segment reassortment between concurrently circulating human rotavirus strains. Arch. Virol. 146:557-570. [DOI] [PubMed] [Google Scholar]
- 26.Zao, C.-L., W.-N. Yu, C.-L. Kao, K. Taniguchi, C.-Y. Lee, and C.-N. Lee. 1999. Sequence analysis of VP1 and VP7 genes suggests occurrence of a reassortant of G2 rotavirus responsible for an epidemic of gastroenteritis. J. Gen. Virol. 80:1407-1415. [DOI] [PubMed] [Google Scholar]