Abstract
The replicative, cytopathic, and antigenic properties of simian immunodeficiency virus (SIV) variants influence its replication efficiency in vivo. To further define the viral properties and determinants that may be important for high-level replication in vivo and progression to AIDS, we compared a minimally pathogenic SIVmne molecular clone with two highly pathogenic variants cloned from late stages of infection. Both variants had evolved greater infectivity than the parental clone due to mutations in nef. Interestingly, a pol determinant in one of the highly pathogenic variants also contributed to its increased infectivity. Furthermore, because replication in vivo may also be influenced by the ability of a virus to evade the cellular immune response of the host, we examined whether the variants were more capable of downregulating surface expression of class I major histocompatibility complex (MHC). Decreased MHC class I expression was not observed in cells infected with any of the viruses. Furthermore, the Nef proteins of the highly pathogenic variants only slightly reduced surface MHC class I expression in transfected cells, although they efficiently downregulated CD4. Together, these data demonstrate that mutations which can enhance viral infectivity, as well as CD4 downregulation, may be important for efficient replication of SIV in the host. However, Nef-mediated reduction of MHC class I expression does not appear to be critical for the increased in vivo replicative ability of highly pathogenic late variants.
Human immunodeficiency virus (HIV) undergoes significant and continuous genetic change throughout the course of infection, resulting in the evolution and selection of variant viruses with distinct characteristics that may be important for persistence and disease (55). Remarkably, HIV replicates continuously throughout the course of infection (33, 57, 71), despite the specific immune responses of the host against the infecting virus. Furthermore, recent studies have shown that the extent of HIV replication is predictive of the rate of progression to AIDS (48, 49, 53, 54). Moreover, several phenotypic characteristics of HIV type 1 (HIV-1) variants, including tropism, coreceptor usage, replication rate, cytopathicity, and syncytium-inducing ability, have been correlated with different stages of infection and disease, suggesting that particular characteristics are acquired by variants for persistence, efficient replication, and disease progression (4, 12, 16, 17, 19, 23-25, 28, 63, 69, 70). Specifically, efficient replication in T cells, greater cytopathicity, and syncytium-inducing ability correlate with progression to AIDS. However, which characteristics enable variant viruses to continuously replicate in the host at high levels is difficult to address because of the absence of a suitable animal model for studying HIV pathogenesis. Thus, the full complement of viral determinants that contribute to pathogenicity are not well understood and could perhaps be best addressed with the simian immunodeficiency virus (SIV)-macaque model.
SIV infection of macaques is a primary animal model for studying HIV pathogenesis (reviewed in reference 75). Although most studies with the SIV-macaque model have focused on defining the molecular determinants of virulence by using highly pathogenic strains and clones of SIV, the model has also provided a system in which to study genetic and phenotypic changes in a primate lentivirus as they relate to the development of AIDS (reviewed in reference 72). Importantly, like variants of HIV-1 that emerge during infection of humans, variant viruses that evolve in macaques infected with macrophage-tropic, minimally cytopathic, non-syncytium-inducing virus derived from Macaca nemestrina (SIVmneCL8) are rapidly replicating, are highly cytopathic, and may be syncytium inducing in culture (31, 40, 56, 60, 61). They also have increased ability to escape from host neutralizing antibody responses (11, 61). Furthermore, molecular clones of these variants replicate more efficiently and cause disease in vivo, demonstrating that emerging viral variants are more pathogenic and drive disease progression (38-40, 61). Thus, the SIV model may be a useful system to dissect which mutations that evolve in the virus influence replication in the host and subsequent disease.
Early studies with a variety of HIV-1 variants demonstrated that the envelope surface protein (env-SU) gene encodes the primary determinant for changes in tropism, replication, and cytopathicity (reviewed in references 45 and 50). Surprisingly, the Env-SU region of SIVmne has not been found to be the major determinant influencing in vitro replication and cytopathicity, although it affects syncytium induction, tropism, and sensitivity to neutralizing antibodies (40, 61). Instead, mutations in other regions of the virus, including those located in the env transmembrane and gag-pol coding regions, confer changes in replication and cytopathicity (39, 40). Together, these data suggest that viral fitness is influenced by mutations selected in multiple determinants.
Nef is a critical factor for high-level replication of both HIV and SIV in the host. The inability of a SIV mutant with nef deleted to replicate efficiently and cause disease in macaques first marked the importance of this gene for pathogenesis (36). Additionally, a cohort of long-term HIV survivors with low viral loads have been shown to harbor only nef deletion viruses, further demonstrating nef's importance for replication and disease (21, 42, 46). However, while these studies establish a significant role for nef in modulating viral replication, other studies have shown that nef deletion SIV can still cause disease in newborn, and occasionally adult, macaques (5, 6, 18). Thus, the exact role of nef in disease remains unclear.
Several functions have been ascribed to the Nef protein that may have relevance to its ability to promote efficient viral replication and, therefore, disease progression in vivo. For example, Nef enhances viral replication and increases virion infectivity by CD4-dependent and -independent mechanisms (1, 13, 14, 30, 43, 51, 59, 66). It may also affect viral replication by altering cell signaling mechanisms that regulate chemokine and interleukin-2 (IL-2) production from infected macrophages and T cells, respectively (3, 7, 58, 62, 65, 68, 73). Furthermore, Nef mediates downregulation of surface CD4 and major histocompatibility complex (MHC) class I expression (26, 64). Decreased surface expression of CD4 may enhance virion infectivity, alter T-cell receptor signaling, and impair immune functions of CD4+ T cells (43, 59, 65). Downregulation of MHC class I may assist virus replication in vivo by protecting infected cells from CD8+ cytotoxic T-lymphocyte (CTL)-mediated killing (15). Interestingly, nef variants are selected with disease progression, suggesting that particular activities may be associated with replication and persistence (31, 41). However, despite these intriguing observations, the importance and relevance of each of these activities for efficient viral replication in vivo remain unclear.
To further delineate the properties and determinants of viruses that may be critical for SIV replication in vivo and progression to AIDS, we compared a minimally pathogenic, parental SIVmne clone (SIVmneCL8) with two highly pathogenic variant viruses (SIVmne170 and SIVmne027) cloned from late stages of infection (31, 38-40). We found that nef alleles from the variants enhanced viral infectivity, and they effectively downregulated surface expression of CD4 but not MHC class I. Furthermore, a determinant in the pol region of the late variant SIVmne027 also conferred greater infectivity. These data suggest that mutations selected during the course of infection that enhance infectivity and CD4 downregulation may be critical for efficient viral replication in vivo. However, Nef-mediated MHC class I downregulation may play a limited role as an immune escape mechanism for SIV.
MATERIALS AND METHODS
Viruses.
The molecular cloning and in vitro phenotypes of SIVmneCL8, SIVmne170, and SIVmne027 have previously been described in detail (31, 37-40, 56, 60, 61). Importantly, SIVmneCL8 is representative of variants found in the early stages of infection, and SIVmne170 and SIVmne027 are representative of variants found at later stages of infection following inoculation with SIVmneCL8. SIVmne170 was cloned from late stages of infection when CD4+ cell counts were low and there were clinical signs of AIDS following inoculation with SIVmneCL8. SIVmne027 was cloned from lymph node tissue of a macaque with declining CD4+ cell counts and early signs of AIDS. This animal was inoculated with the SIVmne isolate from which SIVmneCL8 was cloned (8, 31). The in vivo pathogenicities of SIVmne170 and SIVmne027 have been recently described (38). Compared to SIVmneCL8, SIVmne170 and SIVmne027 cause rapid disease progression and replicate to significantly higher levels in the host. In vitro, all viruses are dualtropic for T cells and macrophages and use the same coreceptors (CCR5 and Bob/GPR15) for entry. However, SIVmne170 and SIVmne027 replicate more efficiently in peripheral blood mononuclear cells than SIVmneCL8, and they are more cytopathic for the CD4+ T-cell population than SIVmneCL8. Finally, neither SIVmne170 nor SIVmne027 is sensitive to neutralizing antibodies against SIVmneCL8 or serum from macaques challenged with these variants.
Plasmid constructs.
Chimeric viruses containing regions of the highly pathogenic proviral clones, SIVmne170 and SIVmne027, in the background of the minimally pathogenic clone, SIVmneCL8, were constructed using conserved restriction sites (31, 38-40). The restriction site positions are numbered according to the sequence of SIVmneCL8 (GenBank accession no. M32741). To construct clone 8/027gag, the portion of gag between DraIII (position 851) and NsiI (position 1886) that includes the gag capsid (CA) and nucleocapsid (NC) sequences was excised from pMneCL8 and replaced with the homologous sequences from pSIVmne027. To construct 8/027pol, the region between NsiI (position 1886) and HpaI (position 3868) that includes the protease (PR) and reverse transcriptase (RT) sequences was removed from pMneCL8 and replaced with the homologous sequences from pSIVMne027. For the clone 8/027vif, the region between HpaI (position 3868) and BstBI (position 5343) that contains the coding region of integrase (IN) and most of vif was excised from pMneCL8 and replaced with the homologous sequences from pSIVmne027. For each of these clones, multiple steps were required to introduce the indicated regions of pSIVmne027 into pMneCL8. To construct the nef chimeric viruses, 8/170nef and 8/027nef, pMneCL8 was digested with NheI (position 8216) and SphI (located in the polylinker of pUC18) to remove a piece of the overlapping envelope transmembrane protein-coding region and the entire nef coding region and 3′ long terminal repeat (LTR). These sequences were replaced with the nef-3′ LTR sequences from either pSIVmne170 or pSIVmne027. As previously described, the LTR sequences are highly conserved. Mutations are not present in known elements important for transcription (39, 40). Partial sequencing and restriction site mapping of the chimeric viruses confirmed the presence of the variant sequences within the SIVmneCL8 provirus. Chimeric viruses 170/8 and 027/8 were previously described (39, 40).
For transient-transfection experiments, the nef genes from SIVmneCL8, SIVmne170, and SIVmne027 were cloned into the expression plasmid pIRES2-EGFP (Clontech, Palo Alto, Calif.) to coexpress Nef and green fluorescent protein (GFP) from a single message. Each nef gene was obtained from the proviral clones by PCR amplification using DNA primers that flank the translational start and stop codons of nef. To facilitate cloning, EcoRI and SalI sites were incorporated into the forward (Nef-ST-RI, 5′-GACTGAATTCACCTACCTACAATATGG-3′ [positions 8531 to 8557]) and reverse (Nef-End-SalI, 5′-GTCCCTGTCGACTCAGCTAGTTTCCTTC-3′ [positions 9330 to 9357]) primers, respectively. (The underlined sequences are the EcoRI and SalI sites.) PCR amplification was carried out in 50-μl reaction mixtures by using Taq plus long enzyme and buffer conditions recommended by the manufacturer (Stratagene, La Jolla, Calif.) and each primer at a final concentration of 250 nM. PCR was performed using the following conditions: denaturation at 94°C for 5 min followed by 35 cycles of a three-step amplification reaction (denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 1 min). An additional extension of 5 min at 72°C followed the final cycle. A sample of each reaction mixture was examined on a 0.8% agarose gel to verify amplification of the expected 826-bp products. The products were digested with EcoRI and SalI and inserted into the multiple cloning site of the pIRES2-EGFP vector. The sequence of each PCR-amplified nef allele was determined to verify that it was identical to the sequence in the provirus from which it was cloned. The nef gene from HIV NL4-3 was cloned into the vector by using a similar strategy but with PCR primers based on the HIV NL4-3 sequence (HIV-NEF2-RI, 5′-GCTTGGAAAGGAATTCGCTATAAGATG G-3′ [positions 8843 to 8870] and HIV-NEF3-SALI, 5′-CTGGAAAGTCGACAGCGGAAAGTCCCTTG-3′ [positions 9502 to 9530]).
Infectivity assay.
To examine the infectivities of SIVmneCL8, SIVmne170, SIVmne027, and chimeric clones, stocks of infectious virus were produced by transient transfection of 293T cells. Two hundred fifty thousand 293T cells were plated into wells of six-well plates and cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated (56°C for 30 min) fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin per ml and 100 μg of streptomycin per ml (DMEM complete). The following day the cells were transfected by the FuGene6 method (Roche, Indianapolis, Ind.), using 2 μg of each proviral clone. After overnight incubation, the cells were washed once with phosphate-buffered saline (PBS), and fresh DMEM complete was added to each culture. At 48 h posttransfection, the supernatants were harvested, passed through a 0.2-μm-pore-size syringe filter (Corning Inc., Corning, N.Y.), and immediately used for infection or stored at −80°C. To determine the virus titer of each stock, serial dilutions of supernatants were tested for infectious virus by using sMAGI cells as described by Chackerian et al. (10). The amount of SIV p27gag antigen in each virus stock was determined by antigen enzyme-linked immunosorbent assay according to the protocol of the manufacturer (Coulter-Immunotech, Miami, Fla.). Infectivity was reported as the number of blue (infected) cells per nanogram of SIV p27gag.
Infection of PBMCs.
Peripheral blood mononuclear cells (PBMCs) were isolated from pig-tailed macaque (M. nemestrina) blood by Ficoll-Hypaque centrifugation as previously described (60). Animals were negative for SIV and simian type D retrovirus. Cells were stimulated with 10 μg of phytohemagglutinin (Difco Laboratories, Detroit, Mich.) per ml and 50 U of IL-2 (Roche, Indianapolis, Ind.) per ml in RPMI complete medium for 3 days. Cells were then concentrated by centrifugation and washed once with RPMI complete, and duplicate cultures of 2 × 106 cells were infected with the SIVmne variants at a multiplicity of infection (MOI) of 0.001 in 1 ml of RPMI complete. The following day, the cells were pelleted by centrifugation, washed twice with PBS to remove residual cell-free virions, and resuspended in 3 ml of RPMI complete plus 50 U of IL-2 per ml. Every 3 days, 2 ml of supernatant was removed from each culture and replaced with fresh RPMI complete plus 50 U IL-2 per ml. Supernatants were stored at −70°C until they were assayed for p27gag by SIV p27gag antigen enzyme-linked immunosorbent assay.
Flow cytometric analysis of infected and transfected cells.
CEMx174 cells (5 × 105) were infected at an MOI of 1 with SIVmneCL8, SIVmne170, or SIVmne027. Viruses used for infection were generated by transfection of CEMx174 cells as previously described (40). At sequential time points postinfection, viable cells were harvested and examined for surface CD4 and MHC class I expression by two-color fluorescence-activated cell sorting (FACS) analysis. The antibodies used for staining were anti-human CD4-fluorescein isothiocyanate (RPA-T4; BD Pharmingen, San Diego, Calif.) and anti-HLA-ABC-phycoerythrin (PE) (W6/32; Dako, Carpinteria, Calif.). Cells were fixed in 1% paraformaldehyde. and 10,000 gated cells were analyzed for surface CD4 and MHC class I expression with a Becton Dickinson FACS analyzer.
Downregulation of CD4, CD3, and MHC class I by the variant nef alleles expressed from the pIRES2-EGFP vector was examined in the Jurkat E6-1 cell line. Jurkat cells were transfected with 20 μg of plasmid DNA by electroporation using a Bio-Rad Gene Pulser apparatus equipped with a capacitance extender unit (250 V, 960 μF). Cells were then reseeded into RPMI complete medium and analyzed at 24 to 36 h posttransfection, when GFP expression was determined to be highest. Cells were harvested and stained with anti-CD4-PE (RPA-T4), anti-HLA-ABC-PE, or anti-CD3-PE (HIT3a; BD Pharmingen), fixed with 1% paraformaldehyde in PBS, and analyzed by two-color FACS. Fifty thousand gated cells were acquired for analysis, and the levels of CD4, MHC class I, and CD3 expression in the GFP-positive cell population were examined.
RESULTS
Infectivity of SIVmne variants.
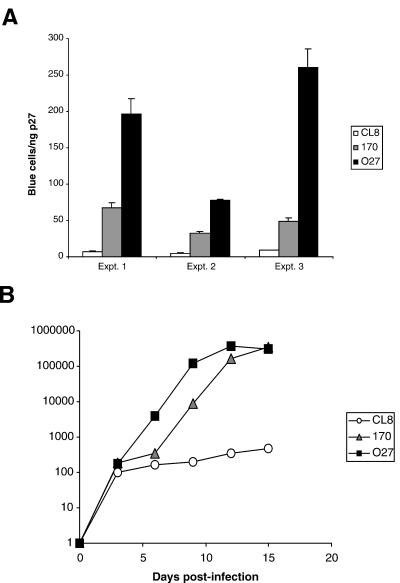
To determine whether the highly pathogenic variants, SIVmne170 and SIVmne027, cloned from late stages of infection were more infectious than the parental virus, SIVmneCL8, we compared their infectivities by using the sMAGI assay (10), which detects single cycles of infection. In each of the three independent experiments shown, both SIVmne170 and SIVmne027 demonstrated greater numbers of blue cells per nanogram of SIV p27gag antigen (Fig. 1A). Five- to 10-fold and 15- to 30-fold more blue cells were counted in sMAGI cell cultures infected with SIVmne170 and SIVmne027, respectively, than in those infected with SIVmneCL8. The total amount of cell-free p27gag antigen produced from the 293T cells transfected with each provirus was similar (data not shown), suggesting that equivalent amounts of virus were released over a 24-h period. Thus, the differences in infectivity of these viruses were likely not determined by transcriptional or posttranscriptional events leading to viral protein expression. Additionally, PBMCs infected with either SIVmne170 or SIVmne027 produced higher levels of SIV p27gag antigen than did those infected with SIVmneCL8 (Fig. 1B), indicating that the increase in infectivity of the variants was associated with enhanced replicative ability. Together, these data demonstrate that highly pathogenic late-stage variant viruses acquire mutations that enhance replication by increasing virion infectivity.
FIG. 1.
Infectivity and replication of SIVmne variant viruses. (A) Infectivity of SIVmne variants. Dilutions of virus stocks were used to infect sMAGI cells as described in Materials and Methods. Infectivity is reported as the mean number of blue cells per nanogram of SIV p27gag antigen ± the standard error of the mean. Results from 3 of 10 independent experiments are shown. (B) Replication of SIVmne variants in pig-tailed macaque PBMCs. Duplicate cultures of cells were infected at an MOI of 0.001 with each virus. Data are representative of those from three independent experiments.
CD4 and MHC class I downregulation on infected cells.
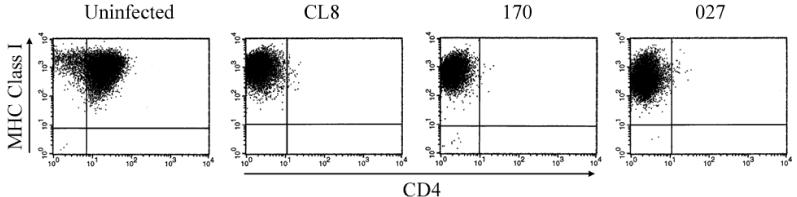
To determine whether there were changes in the ability to decrease surface expression of either CD4 or MHC class I, we infected CEMx174 cells with the variant viruses and examined CD4 and MHC class I surface expression at sequential time points postinfection by FACS analysis. Decreased CD4 surface expression was observed within 2 days of infection, although MHC class I levels were high and indistinguishable from those in the uninfected control cells (data not shown). By 3 weeks postinfection, when the cells had become chronically infected, the early virus, SIVmneCL8, and each of the late variants, SIVMne170 and SIVmne027, had completely downregulated CD4 surface expression (Fig. 2). Even at this late stage of infection, no difference in MHC class I expression was observed in cells infected with any of the viruses compared with the uninfected control cells, although the infected cells produced high levels of cell-free p27gag and contained at least one proviral genome per cell as determined by quantitative Taqman PCR (data not shown).
FIG. 2.
CD4 and MHC class I downregulation in CEMx174 cells infected with SIVmne variant viruses. Levels of CD4 and MHC class I surface expression were determined by FACS at 3 weeks postinfection with the indicated viruses. Data are representative of those from three independent experiments.
Because MHC class I downregulation by the SIVmne variants may occur only in monkey and not in human cells, we examined whether the variant viruses could downmodulate MHC class I surface expression in the sMAGI indicator cells, which were derived from a rhesus macaque mammary tumor cell line (10). Although the majority of the cells stained blue following infection at an MOI of 1, MHC class I levels in the infected cell cultures remained similar to those in the uninfected cell cultures (data not shown). Together, these data suggest that the ability to downmodulate MHC class I is not a characteristic of highly pathogenic SIV variants that emerge late in infection.
Determinants of infectivity.
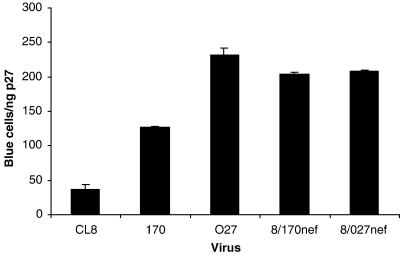
Variant nef alleles of HIV-1 and SIV have been shown to differ in their ability to enhance viral infectivity (1, 9, 31). To determine whether the nef alleles of the late variants from SIVmne170 and SIVmne027 contained mutations that increased viral infectivity compared to nef of the early virus, SIVmneCL8, we created chimeric viruses that contained the nef coding regions from either SIVmne170 (8/170nef) or SIVmne027 (8/027nef) but were isogenic with respect to SIVmneCL8. The infectivity of each chimeric virus was five- to sixfold greater than that of the wild-type early virus, SIVmneCL8 (Fig. 3), demonstrating that both late variants evolved increased infectivity due to mutations in nef. Interestingly, 8/170nef was slightly more infectious than wild-type SIVmne170, suggesting that mutations in Nef alone could account for the enhanced infectivity of this virus. By contrast, 8/027nef was less infectious than wild-type SIVmne027, indicating that in addition to nef, another determinant may also enhance its infectivity.
FIG. 3.
Infectivity of nef chimeric viruses. The nef coding regions of SIVmne170 and SIVmne027 were inserted into the parental provirus, SIVmneCL8, as described in Materials and Methods. Infectivity was determined by infection of sMAGI cells and is reported as the mean number of blue cells per nanogram of p27gag antigen ± the standard error of the mean. Infections are representative of three independent experiments.
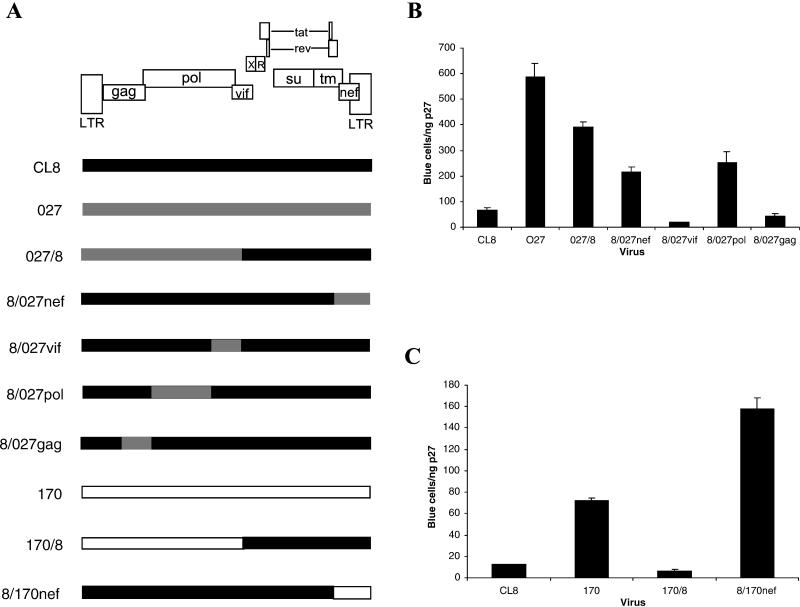
To address whether other viral determinants may contribute to the infectivity of SIVmne027, but not SIVmne170, we constructed additional chimeric viruses (Fig. 4A), and tested their infectivity with the sMAGI single-cycle infection assay (Fig. 4B and C). Interestingly, the chimeric virus 027/8, which contains the 5′ half of SIVmne027 and 3′ half of SIVmneCL8, was as infectious as the 8/027nef chimera, demonstrating an infectivity determinant in the 5′ half of SIVmne027. To identify which region increased infectivity, the 5′ half of SIVmne027 was further subdivided and the individual regions were introduced into the SIVmneCL8 provirus. The resulting chimeric viruses harbored the gag (8/027gag), PR-RT (8/027pol), or IN-vif (8/027vif) region of SIVmne027 but were isogenic with respect to SIVmneCL8. Of these chimeras, only 8/027pol showed an increase in viral infectivity compared to wild-type SIVmneCL8. The three- to fourfold enhancement in infectivity by the pol determinant was equal to that of the nef gene from SIVmne027 (8/027nef). By contrast, a chimeric virus containing the 5′ half of SIVmne170 and the 3′ half of SIVmneCL8 (170/8) was only as infectious as SIVmneCL8, suggesting that the pol infectivity mutation was unique to SIVmne027 (Fig. 4C). Together, these data demonstrate that highly pathogenic viruses that evolve during infection contain mutations in nef which enhance infectivity. Interestingly, mutations in pol can also enhance infectivity.
FIG. 4.
Identification of a 5′ infectivity determinant in SIVmne027. (A) Schematic diagram of chimeras between SIVmneCL8 and SIVmne027 or between SIVmneCL8 and SIVmne170. (B) Infectivity of SIVmneCL8/SIVmne027 chimeras. (C) Infectivity of SIVmneCL8/SIVmne170 chimeric viruses. Infectivity was determined as described in Materials and Methods and is shown as the mean number of blue cells per nanogram of p27gag antigen ± the standard error of the mean. The results are representative of those from three independent experiments.
Nef-mediated CD4 and MHC class I downregulation.
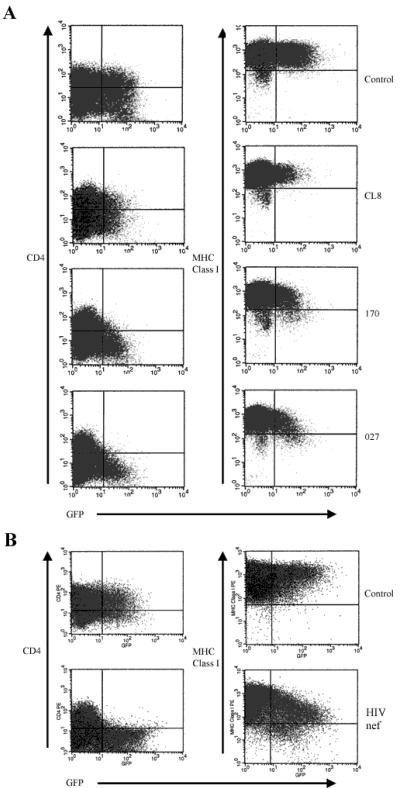
Nef activities such as CD4 downregulation may be masked by other viral proteins during infection (20). To determine whether Nef from the late variant SIVmne clones downregulated either CD4 or MHC class I more efficiently than Nef of SIVmneCL8, we transfected Jurkat cells with bicistronic Nef-GFP expression vectors and examined CD4 and MHC class I expression in the GFP-positive population by FACS analysis (Fig 5A). Compared to cells transfected with the vector without nef sequences (control), expression of the SIVmneCL8 Nef (CL8) did not reduce the percentage of GFP-positive cells expressing CD4 (37.6% vs. 39.4%). Furthermore, there was no difference in the percentage of GFP-positive cells expressing high levels of MHC class I (99.0% for the control versus 99.4% for SIVmneCL8 Nef) or in the mean fluorescence intensity (MFI). In contrast, expression of nef alleles from the late variant viruses, SIVmne170 and SIVmne027, resulted in 75 and 90% decreases in the number of GFP-positive CD4+ cells, respectively. Interestingly, while the level of Nef expression was sufficient for CD4 downregulation, neither late variant Nef efficiently decreased surface MHC class I expression. Although the percentage of GFP-positive cells expressing moderate levels of MHC class I was greater for cells expressing SIVmne170 Nef (12%) and SIVmne027 Nef (17%) than for the control (1%), there was no decrease in the MFI of MHC class I for cells expressing the SIVmne170 nef allele relative to the control and only a 1.2-fold decrease in the MFI of MHC class I for cells expressing the SIVmne027 nef allele. Additionally, each SIVmne nef allele had a limited ability to decrease CD3 expression (data not shown). As a control for both CD4 and MHC class I downregulation, the nef gene from HIV-1 NL4-3 was expressed from the bicistronic vector. As previously shown (2), NL4-3 Nef downregulated both CD4 and MHC class I surface expression (Fig. 5B). The percentage of CD4+ GFP-positive cells decreased by 90%, and while the percentage of GFP-positive cells expressing high levels of MHC class I decreased by only 16%, the MFI of the GFP-positive, MHC class I-positive population was reduced by fourfold relative to the control. This indicated that there was no defect in the Jurkat cell line used that would affect Nef's ability to decrease either CD4 or MHC class I. Thus, these data demonstrate that SIVmne Nef evolves to efficiently downmodulate CD4 but not MHC class I. Furthermore, they are in agreement with the infection experiments (Fig. 2). Thus, Nef-mediated downmodulation of MHC class I may not be an important immune escape mechanism for late-stage SIVmne variants.
FIG. 5.
Nef-mediated CD4 and MHC class I downregulation in Jurkat cells. CD4 and MHC class I downregulation by SIV nef variants (A) or the HIV-1 NL4-3 nef (B) is shown. The pIRES2-EGFP vector without nef (control) or containing the nef genes from SIVmneCL8 (CL8), SIVmne170 (170), SIVmne027 (027), or HIV-1 NL4-3 (HIV nef) was transiently transfected into Jurkat cells by electroporation. CD4 and MHC class I surface expression in the GFP-positive cell populations were monitored by FACS.
Comparison of the variant Nef sequences.
To determine which mutations might be important for changing the functional activity of SIVmne Nef, we compared the predicted Nef amino acid sequences of the late variant SIVmne clones, SIVmne170 and SIVmne027, with that of SIVmneCL8 (Fig. 6). Overall, the variant Nefs were highly conserved (94% identical) with that of SIVmneCL8. Tyrosine residues at positions 223 and 226 and the aspartic acid residue at position 155 that had been previously shown to be important for MHC class I downregulation in mutagenesis studies using the SIVmac239 nef gene were conserved among these viruses (67). Furthermore, amino acids residues P73, A74, and D204, which were shown to be important for CD4 downmodulation, were also conserved between the variant Nef proteins (34, 35, 44). Mutations that were conserved between the two late variants occurred at four positions in the central region of Nef (I87M, N95D, I111M, and M144I). Of these mutations, only three (D95 in the acidic domain, M111, and I144) are also found in the Nef protein of SIVmac239. Interestingly, the asparagine-to-aspartic acid and isoleucine-to-methionine mutations at amino acid positions 95 and 111, respectively, have been found to be selected within 4 to 8 weeks following infection with SIVmneCL8 (31), suggesting their importance early during in vivo replication. By contrast, the isoleucine-to-methionine mutation at amino acid position 87 typically appeared later in the course of infection. These data implicate the N95D and I111 M mutations in Nef-mediated downregulation of CD4 and enhanced infectivity.
FIG. 6.
Comparison of the predicted Nef sequences of the SIVmne variants. Differences in the Nef protein sequences of SIVmne170 and SIVmne027 are shown relative to that of SIVmneCL8. The amino acid position number is indicated above the sequence. The amino acid sequence is shown in single-letter code. Residues in the variants that are identical to those in SIVmneCL8 are represented by dots. Specific amino acid differences are shown.
DISCUSSION
Using molecular clones of SIVmne from different stages of infection and disease, we previously demonstrated that variants of SIV which evolve during an infection have increased pathogenicity and drive disease progression (38). The data support a model in which variant viruses acquire mutations that increasingly enhance their fitness for replication in the host. Such mutations could affect the intrinsic replicative ability of the viruses and/or immune escape mechanisms. Interestingly, changes in the envelope surface protein that confer neutralization resistance are among the earliest mutations found in variant viruses isolated from macaques inoculated with the minimally pathogenic virus SIVmneCL8. These mutations enhance viral replication in vivo but not in vitro (11, 38, 61). However, enhancement of pathogenicity appears to require mutations in other genetic loci, which together increase the replicative and cytopathic properties of the virus. Indeed, we have mapped replicative and cytopathic determinants to gag and env transmembrane coding sequences (39, 40). Here, we provide evidence for the selection of nef and pol mutations that enhance viral infectivity. Furthermore, the Nef protein also evolves a greater ability to downmodulate CD4 from the surface of cells, but downregulation of MHC class I molecules is minimal. These data demonstrate that selected mutations which enhance viral replication and potentially disrupt the function of infected CD4+ T cells may be critical for pathogenicity of the variant viruses. The lack of extensive downregulation of MHC class I by late variant viruses suggests that this function may not be important for efficient viral replication in the host at late stages of infection and disease. Successful avoidance of the host immune response and disease progression may therefore primarily depend upon genetic variation, efficient virus replication, and the ability to disrupt CD4+ T-helper cell viability and function.
A recent study by Carl et al. has shown functional changes in HIV-1 Nef as infected individuals progressed to AIDS, suggesting that particular properties of Nef may be associated with different stages of infection (9). Interestingly, with progression to AIDS, Nef either maintains or evolves enhanced ability to downregulate CD4 and stimulate virus replication, but it becomes inefficient at downmodulating MHC class I expression. Furthermore, in ex vivo human lymphoid tissue cultures, Nef-mediated CD4 downregulation correlates with increased CD4+ T-cell depletion and enhanced viral replication (29). In addition, site-directed mutations in the SIVmac239 nef, which abolish the ability to downregulate CD4 and enhance infectivity, attenuate replication in macaques (35). Our studies on late variant SIVmne nef alleles are in agreement with these data. Importantly, we demonstrate by using variant viruses of known pathogenicity that a nef allele with limited activity acquires the ability to downmodulate CD4 and increase viral infectivity but not an efficient ability to downregulate MHC class I. These data may be interpreted to suggest that the ability to downregulate MHC class I is not significantly enhanced because late variants are adapted to replication in an immunocompromised environment where there is no pressure to escape from the host's CTL response. However, both late variant viruses, SIVmne170 and SIVmne027, are rapidly replicating and highly cytopathic for macaque CD4+ T cells, and they replicate efficiently and cause accelerated disease in naive macaque hosts compared to the early virus, SIVmneCL8 (38). Even during the postacute stages of infection with these variant viruses, the steady-state viral load is significantly higher that that found in SIVmneCL8-infected macaques, indicating that viral replication is poorly controlled by the host immune response. Thus, an explanation for why there may not be selection for nef mutations that enhance MHC class I downregulation is that viruses with increased inherent replicative and cytopathic properties sufficiently disrupt and destroy CD4+ T-helper responses required for priming and maintaining virus-specific immune responses. Virally induced loss of CD4+ T-helper responses against HIV or SIV could establish a state of tolerance to the virus (47). Also, CTL responses may be evaded by virus-induced Fas ligand (CD95L) expression in infected cells (27, 74).
The mutations in SIVmne Nef that enhance infectivity and CD4 downmodulation are distinct from those amino acids previously shown to be important by mutagenesis studies of SIVmac239 Nef (34, 35, 44). P73, A74, and D204 are all highly conserved in the SIVmne variants. However, similarly to Heidecker et al. (31), we note several mutations in the central domain of SIVmne Nef, particularly N95D and I111 M, which correlate with an increase in infectivity as well as CD4 downmodulation. Furthermore, Swigut et al. have shown that changing tyrosine 223 to phenylalanine in SIVmac 239 Nef specifically disrupts the ability to downregulate MHC class I without altering other known Nef functions, including CD4 downmodulation and infectivity (67). In vivo, there is a strong selective pressure for restoring the tyrosine residue at position 223 (52). Although the reversion of Y223 coincides with the decrease in primary viremia and likely appearance of anti-SIV CTLs, if Y223 has other important functions related to virus replication, it would be selected rapidly, similar to the case for functionally significant mutations in the envelope transmembrane protein (32, 40). By comparison, Y223 is highly conserved among the SIVmne clones. Thus, changes at amino acid residues other than those previously identified by site-directed mutagenesis may modulate Nef's ability to downmodulate MHC class I or CD4 and enhance infectivity. Finally, it is intriguing that mutations in P73, A74, and D204 of SIVmac239 Nef attenuate viral replication in vivo, despite having no effect on MHC class I downregulation (35). This provides further evidence that Nef-mediated MHC class I downregulation is not sufficient for the positive effects of Nef on SIV virulence. Nevertheless, it will be important to determine whether variant viruses cloned from intermediate asymptomatic stages of infections initiated with the minimally pathogenic SIVmneCL8 virus evolve greater ability to downregulate MHC class I. Furthermore, in vivo studies will be necessary to prove whether the Nef D95 and M111 mutations contribute to the enhanced replicative ability of SIVmne variants.
In addition to Nef, mutations selected in other determinants may enhance infectivity. We demonstrated that SIVmne027 has an infectivity determinant in pol that is not present in either the minimally pathogenic SIVmneCL8 or the highly pathogenic SIVmne170. At this time it is unclear how a mutation in pol enhances infectivity. A comparison of the sequences from the SIVmne variants identifies only three amino acid residues within the determinant that are unique to SIVmne027 (39). One mutation is located in the amino-terminal region of PR and the other two mutations are in the RNase H domain of RT. It is important to note that the activity of the SIVmneCL8 RT is similar to that of the SIVmac239 RT (22); therefore, it is unlikely that the mutations in SIVmne027 RT have arisen simply because the SIVmne RT has suboptimal activity. Additionally, the SIVmne170 variant virus is highly pathogenic (38), but the pol region does not enhance infectivity. Further investigation will be necessary to identify the important amino acid residues and mechanism of how the determinant increases infectivity and to determine if it functions independent of Nef. Intriguingly, we previously demonstrated that the SIVmne027 virus has the ability to replicate in unstimulated PBMCs (39), and the determinant important for this phenotype maps to the 5′ region of the virus. Furthermore, our preliminary data also demonstrate that SIVmne027 is highly adapted to replication in either macrophage-resting-T-cell or dendritic-cell-resting-T-cell cocultures compared to either SIVmne170 or SIVmneCL8 (J. T. Kimata and P. G. Patel, unpublished data). We hypothesize that this determinant may confer a selective advantage upon SIVmne027 for replication in lymph nodes, the tissue from which it was directly cloned. In vivo studies will be required to establish the determinant's importance for infection and pathogenesis.
Acknowledgments
We thank Paul Zhou and Jon Allan for their comments and criticism and Stephanie Fargo for assistance with the FACS analysis. Macaque blood was provided by the Washington Regional Primate Research Center. pNL4-3 was obtained from Malcolm Martin through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This work was supported by NIH grant AI47725 to J.T.K. J.E.B. was supported, in part, by an NIH training grant in viral pathogenesis (AI07522).
REFERENCES
- 1.Aiken, C., and D. Trono. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J. Virol. 69:5048-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akari, H., S. Arold, T. Fukumori, T. Okazaki, K. Strebel, and A. Adachi. 2000. Nef-induced major histocompatibility complex class I down-regulation is functionally dissociated from its virion incorporation, enhancement of viral infectivity, and CD4 down-regulation. J. Virol. 74:2907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, L., Z. Du, M. Rosenzweig, J. U. Jung, and R. C. Desrosiers. 1997. A role for natural simian immunodeficiency virus and human immunodeficiency virus type 1 nef alleles in lymphocyte activation. J. Virol. 71:6094-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Åsjö, B., L. Morfeldt-Manson, J. Albert, G. Biberfeld, A. Karlsson, K. Lidman, and E. M. Fenyö. 1986. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet ii:660-662. [PubMed]
- 5.Baba, T. W., Y. S. Jeong, D. Pennick, R. Bronson, M. F. Greene, and R. M. Ruprecht. 1995. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science 267:1820-1825. [DOI] [PubMed] [Google Scholar]
- 6.Baba, T. W., V. Liska, A. H. Khimani, N. B. Ray, P. J. Dailey, D. Penninck, R. Bronson, M. F. Greene, H. M. McClure, L. N. Martin, and R. M. Ruprecht. 1999. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat. Med. 5:194-203. [DOI] [PubMed] [Google Scholar]
- 7.Bell, I., C. Ashman, J. Maughan, E. Hooker, F. Cook, and T. A. Reinhart. 1998. Association of simian immunodeficiency virus Nef with the T-cell receptor (TCR) zeta chain leads to TCR down-modulation. J. Gen. Virol. 79:2717-2727. [DOI] [PubMed] [Google Scholar]
- 8.Benveniste, R. E., W. R. Morton, E. A. Clark, C.-C. Tsai, H. D. Ochs, J. M. Ward, L. Kuller, W. B. Knott, R. W. Hill, M. J. Gale, and M. E. Thouless. 1988. Inoculation of baboons and macaques with simian immunodeficiency virus/mne, a primate lentivirus closely related to human immunodeficiency virus type 2. J. Virol. 62:2091-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carl, S., T. C. Greenough, M. Krumbiegel, M. Greenberg, J. Skowronski, J. L. Sullivan, and F. Kirchhoff. 2001. Modulation of different human immunodeficiency virus type 1 Nef functions during progression to AIDS. J. Virol. 75:3657-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chackerian, B., N. L. Haigwood, and J. Overbaugh. 1995. Characterization of a CD4-expressing macaque cell line that can detect virus after a single replication cycle and can be infected by diverse simian immunodeficiency virus isolates. Virology 213:386-394. [DOI] [PubMed] [Google Scholar]
- 11.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng-Mayer, C., D. Seto, M. Tateno, and J. A. Levy. 1988. Biologic features of HIV-1 that correlate with virulence in the host. Science 240:80-82. [DOI] [PubMed] [Google Scholar]
- 13.Chowers, M. Y., M. W. Pandori, C. A. Spina, D. D. Richman, and J. C. Guatelli. 1995. The growth advantage conferred by HIV-1 nef is determined at the level of viral DNA formation and is independent of CD4 downregulation. Virology 212:451-457. [DOI] [PubMed] [Google Scholar]
- 14.Chowers, M. Y., C. A. Spina, T. J. Kwoh, N. J. Fitch, D. D. Richman, and J. C. Guatelli. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J. Virol. 68:2906-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins, K. L., B. K. Chen, S. A. Kalams, B. D. Walker, and D. Baltimore. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397-401. [DOI] [PubMed] [Google Scholar]
- 16.Connor, R. I., and D. D. Ho. 1994. Human immunodeficiency virus type 1 variants with increased replicative capacity develop during the asymptomatic stage before disease progression. J. Virol. 68:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connor, R. I., H. Mohri, Y. Cao, and D. D. Ho. 1993. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J. Virol. 67:1772-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connor, R. I., D. C. Montefiori, J. M. Binley, J. P. Moore, S. Bonhoeffer, A. Gettie, E. A. Fenamore, K. E. Sheridan, D. D. Ho, P. J. Dailey, and P. A. Marx. 1998. Temporal analyses of virus replication, immune responses, and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J. Virol. 72:7501-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crise, B., L. Buonocore, and J. K. Rose. 1990. CD4 is retained in the endoplasmic reticulum by the human immunodeficiency virus type 1 glycoprotein precursor. J. Virol. 64:5585-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deacon, N. J., A. Tsykin, A. Solomon, K. Smith, M. Ludford-Menting, D. J. Hooker, D. A. McPhee, A. L. Greenway, A. Ellett, C. Chatfield, et al. 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270:988-991. [DOI] [PubMed] [Google Scholar]
- 22.Diamond, T. L., J. Kimata, and B. Kim. 2001. Identification of a simian immunodeficiency virus reverse transcriptase variant with enhanced replicational fidelity in the late stage of viral infection. J. Biol. Chem. 276:23624-23631. [DOI] [PubMed] [Google Scholar]
- 23.Fenyö, E. M., J. Albert, and B. Åsjö. 1989. Replicative capacity, cytopathic effects and cell tropism of HIV. AIDS 3(Suppl.):S5-S12. [DOI] [PubMed] [Google Scholar]
- 24.Fenyö, E. M., L. Morfeldt-Manson, F. Chiodi, B. Lind, A. von Gegerfelt, J. Albert, E. Olausson, and B. Åsjö. 1988. Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J. Virol. 62:4414-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiore, J. R., A. Bjorndal, K. A. Peipke, M. Di Stefano, G. Angarano, G. Pastore, H. Gaines, E. M. Fenyo, and J. Albert. 1994. The biological phenotype of HIV-1 is usually retained during and after sexual transmission. Virology 204:297-303. [DOI] [PubMed] [Google Scholar]
- 26.Garcia, J. V., and A. D. Miller. 1991. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature 350:508-511. [DOI] [PubMed] [Google Scholar]
- 27.Geleziunas, R., W. Xu, K. Takeda, H. Ichijo, and W. C. Greene. 2001. HIV-1 Nef inhibits ASK1-dependent death signalling providing a potential mechanism for protecting the infected host cell. Nature 410:834-838. [DOI] [PubMed] [Google Scholar]
- 28.Glushakova, S., J.-C. Grivel, W. Fitzgerald, A. Sylwester, J. Zimmerberg, and L. B. Margolis. 1998. Evidence for the HIV-1 phenotype switch as a causal factor in acquired immunodeficiency. Nat. Med. 4:346-349. [DOI] [PubMed] [Google Scholar]
- 29.Glushakova, S., J. Munch, S. Carl, T. C. Greenough, J. L. Sullivan, L. Margolis, and F. Kirchhoff. 2001. CD4 down-modulation by human immunodeficiency virus type 1 Nef correlates with the efficiency of viral replication and with CD4+ T-cell depletion in human lymphoid tissue ex vivo. J. Virol. 75:10113-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldsmith, M. A., M. T. Warmerdam, R. E. Atchison, M. D. Miller, and W. C. Greene. 1995. Dissociation of the CD4 downregulation and viral infectivity enhancement functions of human immunodeficiency virus type 1 Nef. J. Virol. 69:4112-4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heidecker, G., H. Munoz, P. Lloyd, D. Hodge, F. W. Ruscetti, W. R. Morton, S. Hu, and R. E. Benveniste. 1998. Macaques infected with cloned simian immunodeficiency virus show recurring nef gene alterations. Virology 249:260-274. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch, V. M., P. Edmondson, M. Murphy-Corb, B. Arbeille, P. R. Johnson, and J. I. Mullins. 1989. SIV adaptation to human cells. Nature 341:572-573. [DOI] [PubMed] [Google Scholar]
- 33.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 34.Iafrate, A. J., S. Bronson, and J. Skowronski. 1997. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J. 16:673-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iafrate, A. J., S. Carl, S. Bronson, C. Stahl-Hennig, T. Swigut, J. Skowronski, and F. Kirchhoff. 2000. Disrupting surfaces of Nef required for downregulation of CD4 and for enhancement of virion infectivity attenuates simian immunodeficiency virus replication in vivo. J. Virol. 74:9836-9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kestler, H. W., 3rd, D. J. Ringler, K. Mori, D. L. Panicali, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65:651-662. [DOI] [PubMed] [Google Scholar]
- 37.Kimata, J. T., J. J. Gosink, L. M. Rudensey, V. N. KewalRamani, D. R. Littman, and J. Overbaugh. 1999. Coreceptor specificity of temporal variants of SIVMne. J. Virol. 73:1655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimata, J. T., L. Kuller, D. B. Anderson, P. Dailey, and J. Overbaugh. 1999. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat. Med. 5:535-541. [DOI] [PubMed] [Google Scholar]
- 39.Kimata, J. T., A. Mozaffarian, and J. Overbaugh. 1998. A lymph node-derived cytopathic simian immunodeficiency virus Mne variant replicates in nonstimulated peripheral blood mononuclear cells. J. Virol. 72:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimata, J. T., and J. Overbaugh. 1997. The cytopathicity of a simian immunodeficiency virus Mne variant is determined by mutations in Gag and Env. J. Virol. 71:7629-7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirchhoff, F., P. J. Easterbrook, N. Douglas, M. Troop, T. C. Greenough, J. Weber, S. Carl, J. L. Sullivan, and R. S. Daniels. 1999. Sequence variations in human immunodeficiency virus type 1 Nef are associated with different stages of disease. J. Virol. 73:5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirchhoff, F., T. C. Greenough, D. B. Brettler, J. L. Sullivan, and R. C. Desrosiers. 1995. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 332:228-232. [DOI] [PubMed] [Google Scholar]
- 43.Lama, J., A. Mangasarian, and D. Trono. 1999. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr. Biol. 9:622-631. [DOI] [PubMed] [Google Scholar]
- 44.Lang, S. M., A. J. Iafrate, C. Stahl-Hennig, E. M. Kuhn, T. Nisslein, F. J. Kaup, M. Haupt, G. Hunsmann, J. Skowronski, and F. Kirchhoff. 1997. Association of simian immunodeficiency virus Nef with cellular serine/threonine kinases is dispensable for the development of AIDS in rhesus macaques. Nat. Med. 3:860-865. [DOI] [PubMed] [Google Scholar]
- 45.Levy, J. A. 1993. Pathogenesis of human immunodeficiency virus infection. Microbiol. Rev. 57:183-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mariani, R., F. Kirchhoff, T. C. Greenough, J. L. Sullivan, R. C. Desrosiers, and J. Skowronski. 1996. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J. Virol. 70:7752-7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMichael, A. 1998. T cell responses and viral escape. Cell 93:673-676. [DOI] [PubMed] [Google Scholar]
- 48.Mellors, J. W., L. A. Kingsley, C. R. J. Rinaldo, J. A. Todd, B. S. Hoo, R. P. Kokka, and P. Gupta. 1995. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann. Intern. Med. 122:573-579. [DOI] [PubMed] [Google Scholar]
- 49.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 50.Miedema, F., L. Meyaard, K. M., M. R. Klein, M. T. L. Roos, M. Groenink, R. A. M. Fouchier, A. B. Van't Wout, M. Tersmette, P. T. A. Schellekens, and H. Schuitemaker. 1994. Changing virus-host interactions in the course of HIV-1 infection. Immunol. Rev. 140:35-72. [DOI] [PubMed] [Google Scholar]
- 51.Miller, M. D., M. T. Warmerdam, I. Gaston, W. C. Greene, and M. B. Feinberg. 1994. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J. Exp. Med. 179:101-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munch, J., N. Stolte, D. Fuchs, C. Stahl-Hennig, and F. Kirchhoff. 2001. Efficient class I major histocompatibility complex down-regulation by simian immunodeficiency virus Nef is associated with a strong selective advantage in infected rhesus macaques. J. Virol. 75:10532-10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Brien, T. R., W. A. Blattner, D. Waters, E. Eyster, M. W. Hilgartner, A. R. Cohen, N. Luban, A. Hatzakis, L. M. Aledort, P. S. Rosenberg, W. J. Miley, B. L. Kroner, and J. J. Goedert. 1996. Serum HIV-1 RNA levels and time to development of AIDS in the multicenter hemophilia cohort study. JAMA 276:105-110. [PubMed] [Google Scholar]
- 54.O'Brien, W. A., P. M. Hartigan, D. Martin, J. Esinhart, A. Hill, S. Benoit, M. Rubin, M. S. Simberkoff, and J. D. Hamilton. 1996. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. N. Engl. J. Med. 334:426-431. [DOI] [PubMed] [Google Scholar]
- 55.Overbaugh, J., and C. R. Bangham. 2001. Selection forces and constraints on retroviral sequence variation. Science 292:1106-1109. [DOI] [PubMed] [Google Scholar]
- 56.Overbaugh, J., L. M. Rudensey, M. D. Papenhausen, R. E. Benveniste, and W. R. Morton. 1991. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J. Virol. 65:7025-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piatak, M., Jr., M. S. Saag, L. C. Yang, S. J. Clark, J. C. Kappes, K. C. Luk, B. H. Hahn, G. M. Shaw, and J. D. Lifson. 1993. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science 259:1749-1754. [DOI] [PubMed] [Google Scholar]
- 58.Remkema, G. H., and K. Saksela. 2000. Interactions of HIV-1 NEF with cellular signal transducing proteins. Front. Biosci. 5:D268-D283. [DOI] [PubMed] [Google Scholar]
- 59.Ross, T. M., A. E. Oran, and B. R. Cullen. 1999. Inhibition of HIV-1 progeny virion release by cell-surface CD4 is relieved by expression of the viral Nef protein. Curr. Biol. 9:613-621. [DOI] [PubMed] [Google Scholar]
- 60.Rudensey, L. M., J. T. Kimata, R. E. Benveniste, and J. Overbaugh. 1995. Progression to AIDS in macaques is associated with changes in the replication, tropism, and cytopathic properties of the simian immunodeficiency virus variant population. Virology 207:528-542. [DOI] [PubMed] [Google Scholar]
- 61.Rudensey, L. M., J. T. Kimata, E. M. Long, B. Chackerian, and J. Overbaugh. 1998. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J. Virol. 72:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schrager, J. A., and J. W. Marsh. 1999. HIV-1 Nef increases T cell activation in a stimulus-dependent manner. Proc. Natl. Acad. Sci. USA 96:8167-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. Y. de Goede, R. P. van Steenwijk, J. M. A. Lange, J. K. M. E. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwartz, O., V. Marechal, S. Le Gall, F. Lemonnier, and J. M. Heard. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2:338-342. [DOI] [PubMed] [Google Scholar]
- 65.Skowronski, J., D. Parks, and R. Mariani. 1993. Altered T cell activation and development in transgenic mice expressing the HIV-1 nef gene. EMBO J. 12:703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spina, C. A., T. J. Kwoh, M. Y. Chowers, J. C. Guatelli, and D. D. Richman. 1994. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J. Exp. Med. 179:115-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Swigut, T., A. J. Iafrate, J. Muench, F. Kirchhoff, and J. Skowronski. 2000. Simian and human immunodeficiency virus Nef proteins use different surfaces to downregulate class I major histocompatibility complex antigen expression. J. Virol. 74:5691-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swingler, S., A. Mann, J. Jacque, B. Brichacek, V. G. Sasseville, K. Williams, A. A. Lackner, E. N. Janoff, R. Wang, D. Fisher, and M. Stevenson. 1999. HIV-1 Nef mediates lymphocyte chemotaxis and activation by infected macrophages. Nat. Med. 5:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tersmette, M., R. A. Gruters, F. De Wolf, R. E. Y. De Goede, J. M. A. Lange, P. T. A. Schellekens, J. Goudsmit, H. G. Huisman, and F. Miedema. 1989. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J. Virol. 63:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tersmette, M., J. M. A. Lange, R. E. Y. DeGoede, F. DeWolf, J. K. M. Eeftink-Schattenkerk, P. T. A. Schellekens, R. A. Coutinho, J. G. Huisman, J. Goudsmit, and F. Miedeme. 1989. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet i:983-985. [DOI] [PubMed]
- 71.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 72.Whetter, L. E., I. C. Ojukwu, F. J. Novembre, and S. Dewhurst. 1999. Pathogenesis of simian immunodeficiency virus infection. J. Gen. Virol. 80:1557-1568. [DOI] [PubMed] [Google Scholar]
- 73.Wu, Y., and J. W. Marsh. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503-1506. [DOI] [PubMed] [Google Scholar]
- 74.Xu, X. N., B. Laffert, G. R. Screaton, M. Kraft, D. Wolf, W. Kolanus, J. Mongkolsapay, A. J. McMichael, and A. S. Baur. 1999. Induction of Fas ligand expression by HIV involves the interaction of Nef with the T cell receptor zeta chain. J. Exp. Med. 189:1489-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zack, J. A., and J. Overbaugh. 1994. SIV and HIV pathogenesis in animal systems. AIDS 8(Suppl. 1):S43-S52. [Google Scholar]