Abstract
The late genes of SV40 are not expressed at significant levels until after the onset of viral DNA replication. We previously identified two hormone response elements (HREs) in the late promoter that contribute to this delay. Mutants defective in these HREs overexpress late RNA at early, but not late, times after transfection of CV-1PD cells. Overexpression of nuclear receptors (NRs) that recognize these HREs leads to repression of the late promoter in a sequence-specific and titratable manner, resulting in a delay in late gene expression. These observations led to a model in which the late promoter is repressed at early times after infection by NRs, with this repression being relieved by titration of these repressors through simian virus 40 (SV40) genome replication to high copy number. Here, we tested this model in the context of the viral life cycle. SV40 genomes containing mutations in either or both HREs that significantly reduce NR binding without altering the coding of any proteins were constructed. Competition for replication between mutant and wild-type viruses in low-multiplicity coinfections indicated that the +1 HRE offered a significant selective advantage to the virus within a few cycles of infection in African green monkey kidney cell lines CV-1, CV-1P, TC-7, MA-134, and Vero but not in CV-1PD′ cells. Interestingly, the +55 HRE offered a selective disadvantage in MA-134 cells but had no effect in CV-1, CV-1P, TC-7, Vero, and CV-1PD′ cells. Thus, we conclude that these HREs are biologically important to the virus, but in a cell type-specific manner.
Simian virus 40 (SV40) is a member of the primate polyomavirus family. Its small 5.2-kbp double-stranded DNA genome codes for two sets of proteins: the regulatory proteins, large T antigen and small t antigen, and the structural proteins, leader protein 1 (LP1, also called agnoprotein) (27) and virion protein 1 (VP1), VP2, and VP3 (23). The genes encoding the regulatory and structural proteins are separated and controlled by a bidirectional promoter-regulatory region that spans ∼400 bp and that can be bound by numerous factors (30). Expression of these two sets of genes is temporally regulated. The regulatory genes are expressed at early times in the lytic cycle of infection. Their products interact with numerous cellular regulatory factors (2, 44), resulting in the infected cell being pushed into an unchecked proliferation loop. Large T antigen also directly plays multiple roles in transcriptional control and replication of the viral genome. The expression of the structural genes is delayed until after the onset of viral DNA replication (1).
How does SV40 manage this early-to-late switch in gene expression when both sets of genes are controlled from the same bidirectional promoter-regulatory region? Two non-mutually exclusive mechanisms contribute to this temporal regulation. The replication-independent mechanism involves transactivation of the late promoter by large T antigen (7, 21, 31). Evidence that transactivation is, in part, independent of DNA replication includes large T antigen's ability to activate late gene expression in the presence of inhibitors of DNA replication (7, 43), in the absence of SV40's origin of DNA replication (Ori) (7, 11, 32), and in a cell-free transcription system (13). This mechanism likely involves protein-protein interactions between large T antigen and factors that bind SV40's 21-bp repeat region (21), the transcription factor TEF-1 (6, 11, 12, 33), components of the general transcription machinery (14, 24, 26, 28), transcriptional coactivators (3, 17), and transcriptional corepressors (41).
The replication-dependent mechanism of the early-to-late switch probably involves the titration of cellular factors that act as sequence-specific repressors of the late promoter (8, 46). Previous work from our laboratory showed that at least some of these cellular repressors consist of members of the nuclear receptor superfamily (NRs) that bind hormone response elements (HREs) located both surrounding the major site of transcription initiation from the late promoter (+1 HRE) and approximately 55 bp downstream of this site (+55 HRE) (46). These +1 and +55 site HREs are composed of direct repeats similar to the canonical HRE half-site sequence, 5′-AGGTCA-3′, separated by 4 (DR4) and 2 (DR2) bp, respectively. The +1 HRE can be bound by retinoic X receptor α (RXRα) in combination with thyroid hormone receptor α1 (T3Rα1) (15, 48, 49) or liver X receptor α (LXRα) (41), estrogen-related receptor α1 (ERRα1) (29, 46, 49), COUP-TF1, and COUP-TF2 (48). The +55 HRE can be bound by COUP-TF1 and COUP-TF2 (48), ERRα1 (29, 46), testis receptor 2 (TR2) (48), and TR4 (36).
Overexpression of COUP-TF1 or T3Rα1 in African green monkey kidney (AGMK) cell line CV-1PD leads to a delay in the early-to-late switch in SV40 gene expression in an HRE-dependent manner (48, 49). Interestingly, the delay caused by T3Rα1 can be relieved by addition of the T3R ligand, T3 (49). On the basis of these data, we hypothesized that the binding of cellular NRs to these HREs represses transcription of the SV40 late genes through active and/or passive mechanisms until replication of viral DNA to high copy number leads to their being titrated away. Furthermore, SV40 may modulate its life cycle in response to endogenous and exogenous hormone signals via these HREs (46, 48, 49).
The experiments described above were performed by transient transfection with viral DNAs containing a mutation in the VP1-coding region to prevent complications in the interpretation of data due to production of virions and subsequent second cycles of infection. Unanswered was the question of whether these HREs are biologically important to the virus in the context of its natural life cycle. In this report we show that virions containing mutations in these HREs are viable; however, the presence of these HREs has a positive, negative, or no effect on virus replication in a cell type-specific manner.
MATERIALS AND METHODS
Cells.
Cells were grown and maintained at 37°C in a 5% CO2 atmosphere. AGMK cell lines CV-1 (obtained from the American Type Culture Collection), CV-1P (clonal derivative of CV-1 originally obtained from S. Kit's laboratory), CV-1PD (C. Cole laboratory derivative of CV-1P), CV-1PD′ (Mertz laboratory derivative of CV-1PD), MA-134 (obtained from Janet Butel), TC-7 (obtained from Janet Butel), and Vero (obtained from Bill Sugden) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 40 U of penicillin and streptomycin/ml and 5% fetal bovine serum (FBS; HyClone). COS M6 cells (obtained from the American Type Culture Collection) were grown as described above, except that they were supplemented with 10% FBS.
Oligonucleotides, plasmids, and antisera.
Oligonucleotides (Fig. 1) and primers used in this study were synthesized by Integrated DNA Technologies. Double-stranded oligonucleotides were prepared by following standard protocols (45).
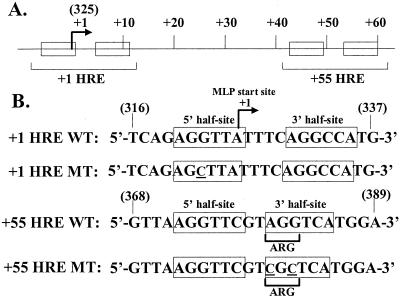
FIG. 1.
Locations and sequences of the HREs present in the SV40 late promoter. (A) Diagram showing locations of the HREs within the SV40 major late promoter (MLP). The +1 and +55 HREs consist of two directly repeated hexanucleotide half-site elements (boxes). The SV40 nucleotides are numbered relative to the site of transcription initiation from the SV40 MLP (arrow), which corresponds to nt 325 in the SV numbering system (9). (B) Sequences of the wild-type and mutant double-stranded HRE-containing oligonucleotides used in this study. Only the strand corresponding to the viral late RNA is shown. The bases altered in the mutants are underlined and in small capitals. The numbers in parentheses indicate the SV40 nucleotides to which the oligonucleotides correspond in the SV numbering system.
Plasmid pSVS(WT) contains SV40 wild-type strain WT830 cloned into a pBR322-based vector via their EcoRI sites (20). Plasmid pSV4540(pm322C), containing the +1 HRE mutant (+1 HRE MT) (Fig. 1) in a wild-type SV40 background, was created by recombination of pSV4508(pm322C) (35) with pSVS. Plasmid pSV4541(pm380C,382C), containing the +55 HRE mutant (+55 HRE MT) (Fig. 1), was created by PCR site-directed mutagenesis of pSVS using a primer incorporating the +55 HRE mutations (Fig. 1). Plasmid pSV4542(pm322C,380C,382C), containing both the +1 HRE and +55 HRE mutations (+1/+55 HRE DMT), was created by recombination of plasmids pSV4540 and pSV4541. All mutations were confirmed by DNA sequence analysis. Plasmid pcDNA3.1-hERRα1, containing the 423-amino-acid human ERRα1-coding region expressed from a cytomegalovirus promoter, was obtained from Eric Ariazi; its construction is described elsewhere (R. J. Kraus, E. A. Ariazi, M. L. Farrell, and J. E. Mertz, submitted for publication).
A rabbit polyclonal antiserum against the region of ERRα1 comprising amino acids 49 to 66 was obtained from Eric Ariazi.
Growth and titering of virus stocks.
The viral sequences present in pSVS, pSV4540, pSV4541, and pSV4542 were excised from their cloning vector by incubation with EcoRI and ligated to form monomer circles of wild-type SV40 and mutants +1 HRE MT, +55 HRE MT, and +1/+55 HRE DMT, respectively. These DNAs were transfected into 100-mm-diameter tissue culture dishes containing ∼70% confluent CV-1PD′ cells with LT-1 transfection reagent (PanVera). The infected cells were grown at 37°C in DMEM containing 5% FBS. The medium was changed after 5 days. The cells were harvested when all exhibited virus-induced cytopathic effect (∼10 to 12 days) and collected by centrifugation for 5 min at 300 × g. The supernatant was removed, and the cells were resuspended in 100 μl of virus carrier medium (VCM; DMEM, 10 mM Tris [pH 7.5], 2% FBS) per 100-mm dish of cells. The cells were frozen and thawed three times and extracted with 1/5 volume of chloroform to release the virus from the cells. After centrifugation in a microcentrifuge at maximum speed for 5 min to separate the phases, the virus-containing supernatant was collected and stored in aliquots at −80°C. Virus stocks were titered by a standard agar overlay plaque assay procedure as previously described (38) with TC-7 cells in place of CV-1P cells.
Preparation of whole-cell and nuclear extracts.
Nuclear extracts of CV-1 and CV-1PD′ cells were prepared essentially according to Zuo's modification (48) of the method of Dignam et al. (16). Briefly, cells were washed twice with phosphate-buffered saline (PBS), scraped off the dish, collected by centrifugation at 1,000 × g for 10 min, resuspended in PBS, pelleted again, resuspended in a 5× packed-cell volume of buffer A (10 mM HEPES [pH 7.4], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT]), incubated on ice for 10 min, pelleted again, and resuspended in a 2× packed-cell volume of buffer A. These cells were then lysed by 10 strokes of a glass Dounce homogenizer (A-type pestle). The nuclei were collected by centrifugation at 25,000 × g at 4°C for 30 min, resuspended in 2.5 ml of buffer C (20 mM HEPES [pH 7.4], 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride [PMSF])/109 cells, and homogenized by five strokes of a glass Dounce homogenizer (B-type pestle). The lysate was then rocked at 4°C for 30 min and dialyzed against 50 volumes of buffer D (20 mM HEPES [pH 7.4], 20% glycerol, 0.1 M KCl, 2 mM EDTA, 6 mM MgCl2, 0.5 mM DTT, 0.5 mM PMSF] for 4 h at 4°C. The dialysate was cleared by centrifugation at 25,000 × g at 4°C for 20 min. The supernatant was aliquoted and stored at −80°C.
Whole-cell extracts from COS M6 cells overexpressing ERRα1 were prepared as follows. Dishes (100 mm in diameter) of COS M6 cells in 5 ml of medium were transfected by addition of a mixture containing 3 μg of pcDNA3.1-hERRα1, 300 μl of DMEM, and 18 μl of LT-1 transfection reagent (PanVera) and incubation at 37°C for 24 h. Five milliliters of fresh medium was added, and incubation was continued for an additional 24 h. The cells were then washed three times with PBS, scraped off the dishes, collected by centrifugation at 300 × g for 10 min, washed with cold PBS, resuspended in 100 μl of lysis buffer (50 mM Tris-HCl [pH 7.5], 0.5 M NaCl, 1 mM EDTA, 10% glycerol, 1 mM DTT, 1× protease inhibitor cocktail [4-{2-aminoethyl}benzenesulfonyl fluoride, trans-epoxysuccinyl-l-leucylamido{4-guanidino}butane, bestatin, leupeptin, aprotinin, and EDTA {Sigma}]) per five dishes of cells, and treated with three cycles of freezing and thawing. The resulting extracts were then centrifuged at maximum speed at 4°C in a microcentrifuge for 5 min to clear away cellular debris, and the supernatant was aliquoted and stored at −80°C.
Electrophoretic gel mobility shift assays (EMSAs).
The sequences of the double-stranded synthetic oligonucleotides used as probes and competitors are shown in Fig. 1B. Twenty picomoles of these DNAs was end labeled by incubation for 45 min at 37°C with 50 μCi of [γ-32P]ATP (Amersham Pharmacia) and 10 U of T4 kinase (New England Biolabs). The radiolabeled DNAs were separated from unincorporated nucleotides by size chromatography with G-25 columns (Boehringer Mannheim).
The EMSA reaction mixtures (total volume, 20 μl) contained either 2 μg of CV-1 cell nuclear extract (Fig. 2A) or 5 μg of COS M6 whole-cell extract overexpressing human estrogen-related receptor α1 (hERRα1) (Fig. 2B) in a solution containing 10 mM HEPES (pH 7.9), 10% glycerol, 2% Ficoll, 100 mM NaCl, 1 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 4 μg bovine serum albumin, and 100 ng of MspI-cut pBR322 DNA (34). After preincubation on ice for 10 min with or without competitor oligonucleotide as indicated (Fig. 2B), the indicated (Fig. 2B) radiolabeled probe was added and incubation was continued for another 5 min on ice. Where indicated (Fig. 2B) hERRα1 antiserum was then added, and incubation was continued at room temperature for an additional 5 min prior to electrophoresis in a native 5% polyacrylamide (29:1, acrylamide/bisacrylamide; Bio-Rad) gel at 150 V and 4°C for 2 h in 0.5× Tris-borate-EDTA running buffer. The gel was then transferred to Whatman paper and dried. The radiolabel was visualized with a PhosphorImager (Molecular Dynamics) and quantified with ImageQuant, version 5.1, software (Molecular Dynamics).
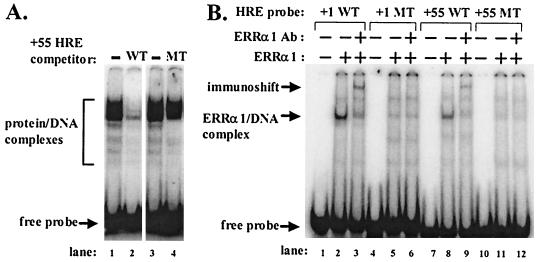
FIG. 2.
NRs bind to the wild-type but not the mutant +1 and +55 HREs. (A) The +55 HRE MT oligonucleotide is defective in binding proteins present in nuclear extract made from CV-1 cells. Shown are results of EMSAs with 2 μg of CV-1 cell nuclear extract incubated with 0.02 pmol of radiolabeled double-stranded probe DNA corresponding to sequences surrounding the wild-type +55 HRE after preincubation with no competitor (lanes 1 and 3) or a 25-fold molar excess of cold wild-type (lane 2) or mutant (lane 4) oligonucleotides as the competitor. Protein-DNA complexes and free probe are indicated. (B) EMSAs showing failure of +1 HRE MT and +55 HRE MT oligonucleotides to bind ERRα1. ERRα1, produced by transfection of pcDNA3.1-hERRα1 into COS M6 cells, was incubated with 0.5 pmol of radiolabeled double-stranded oligonucleotides corresponding to the indicated HRE probe DNAs (lanes 2, 5, 8, and 11). Lanes 1, 4, 7, and 10, probe DNA only; lanes 3, 6, 9, and 12, ERRα1-containing whole-cell extract preincubated with an ERRα1-specific antiserum after incubation with the indicated probe DNAs. Arrows, ERRα1-DNA complexes, immunoshifted complexes, and free probes. Ab, antibody.
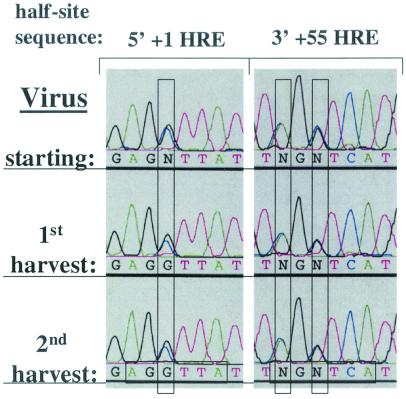
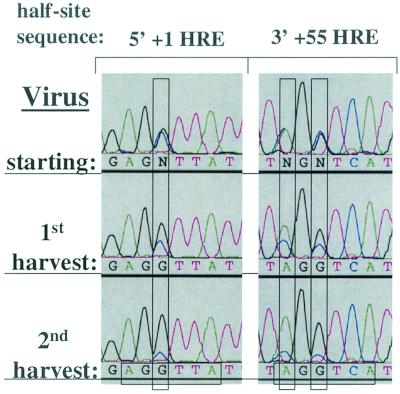
FIG. 5.
Wild-type virus has no replication advantage over the double HRE mutant virus in CV-1PD′ cells. Shown are sequence electropherograms of the +1 HRE 5′ half-site and the +55 HRE 3′ half-site regions of PCR products generated from viral DNA recovered from the initial virus stock (starting) used for infection of CV-1PD′ cells with WT and +1/+55 HRE DMT in an initially equal, low-MOI coinfection and from the first and second harvests following sequential infection and incubation at 37°C for 10 days in DMEM containing 2% FBS. Virus competition experiments were performed in triplicate. The electropherograms are representative of the data obtained from one set of experiments. For the significance of colors and of the vertical and horizontal rectangles, see the Fig. 3 legend.
Virus competition assays.
CV-1, CV-1P, CV-1PD′, MA-134, TC-7, and Vero cells were grown in 35-mm-diameter tissue culture dishes. After reaching confluence, monolayers were coinfected by incubation at 37°C for 90 min with approximately 0.025 PFU each of wild-type and mutant virus/cell in 200 μl of VCM per 35-mm dish. Afterward, unabsorbed virus was removed by aspiration of the infection medium and by washing the monolayers twice with PBS. The cells were then incubated at 37°C in DMEM supplemented with 2% FBS. The medium was changed after 5 days, and incubation continued for an additional 5 days. The cells were then harvested as described above for virus, except for resuspension in 200 μl of VCM per 35-mm dish prior to the freeze/thaw cycles. The virus stocks recovered from the coinfections were diluted in VCM to a multiplicity of infection (MOI) of ∼0.05 PFU/cell, and the protocol was repeated for a second, third and, in some cases, fourth round of infection of cell monolayers.
To determine the relative amounts of the wild-type and mutant virus present in the starting infection mixtures and virus harvests, 75 μl of virus was digested with proteinase K by the addition of 20 μl of 5× PK buffer (1 M Tris [pH 7.5], 125 mM EDTA, 1.5 M NaCl, 10% [wt/vol] sodium dodecyl sulfate) and 5 μl of proteinase K (0.75 μg/μl) (Roche), incubation for 2 h at 37°C, and extraction with an equal volume of phenol-chloroform-isoamyl alcohol (25:25:1 [vol/vol/vol]). The aqueous phase containing the viral DNA was precipitated with ethanol. The DNA pellet was washed with 70% ethanol and resuspended in 20 μl of H2O. One microliter of viral DNA was amplified by PCR in a 100-μl reaction mixture containing 2 pmol each of SV40-specific primers corresponding to SV40 nucleotides (nt) 446 to 423 and 4924 to 4947, 0.2 mM deoxynucleoside triphosphate mixture (Amersham Pharmacia), 3 mM MgCl2, 10× PCR buffer (Promega), and 5 U of Taq DNA polymerase (Promega). The PCR mixtures were subjected to 25 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min. After purification with a PCR clean-up kit (Qiagen), the PCR products were quantified by spectrophotometry. One hundred nanograms of PCR product was sequenced in a 20-μl reaction mixture containing 10 pmol of an SV40 primer corresponding to SV40 nt 446 to 423, 2 μl of Big Dye terminator sequencing mixture (Perkin-Elmer), and 6 μl of 2.5× sequencing buffer (400 mM Tris-HCl [pH 8.3], 10 mM MgCl2). Sequencing reactions were carried out for 25 cycles at 94°C for 15 s, 50°C for 15 s, and 60°C for 2 min. The sequencing products were cleaned up with an AutoSeq G-50 column (Amersham Pharmacia), concentrated by drying, and resuspended in 5 μl of sequencer loading buffer (5 mM EDTA [pH 8.0], 10 mg of blue dextran/ml, 80% deionized formamide). The sequence was determined with an Applied Biosystems 373 DNA sequencer, and the electropherograms were generated with Applied Biosystems Prism, version 2.1.1, software.
RESULTS
Rationale for selection of HRE mutations.
A previously characterized mutant, pm322C, contains a substitution mutation at SV40 nt 322 of a cytidine for a guanosine within the 5′ half-site of the +1 HRE at SV40 nt −3 relative to the transcription initiation site at nt 325 (Fig. 1). The pm322C mutation greatly reduces binding by T3Rα1/RXRα, LXRα/RXRα, ERRα1, COUP-TF1, and COUP-TF-2 to the +1 HRE (28, 41, 48, 49) (Fig. 2B; data not shown). Approximately 70 to 80% of SV40's late transcripts are synthesized from the nt 325 initiation site (22). Most of the remaining transcripts are synthesized from minor initiation sites located farther upstream and processed via excision of an intron that includes nt 322 (22). Thus, almost all of the SV40 late mRNAs synthesized from virus containing this mutation should be identical in sequence to the ones made from wild-type SV40. Therefore, any differences between the mutant and wild type are unlikely to be posttranscriptional in nature.
The +1 HRE extensively overlaps the initiator (Inr) element of the major late promoter. Fortunately, the pm322C mutation lies outside of the bases that define the Inr and has no cis-acting effects on the functionality of the basal elements of this promoter (35). Thus, any effects of this mutation on replication of the mutant are likely due to the inability of NRs to bind the +1 HRE.
The +55 HRE lies within the coding region for a late gene product, the agnoprotein (also called LP1). The agnoprotein may play roles in virion assembly (27, 39, 40), cell-to-cell spread of virus particles (37, 42), and nuclear localization of VP1 (10). The +55 HRE mutant used in our previously published studies contained the LS26 mutation (4). This mutation alters the coding of the agnoprotein as well as the sequence of the +55 HRE.
To study the effects of the +55 HRE by itself, we needed a mutant that significantly reduced binding by NRs yet still encoded wild-type agnoprotein. We accomplished this by incorporating two base substitutions: guanosine to cytidine at SV40 nt 382 to inactivate the +55 HRE and adenosine to cytidine at SV40 nt 380 to maintain an arginine codon (Fig. 1B). A competition EMSA confirmed that an oligonucleotide containing these two base changes in the +55 HRE is unable to compete for binding most of the multiple factors present in a nuclear extract of CV-1 cells that bind a radiolabeled probe DNA corresponding to the +55 HRE wild-type sequence (Fig. 2A, lane 4 versus 1 to 3). Thus, these two base substitution mutations greatly reduce the binding of multiple factors present in CV-1 cell nuclear extract that specifically recognize the +55 HRE.
To test the ability of this +55 HRE double mutation to reduce binding by a specific NR known to recognize the wild-type +55 HRE sequence, we synthesized ERRα1 in large-T-antigen-containing COS M6 cells by transfection with pcDNA3.1-hERRα1, an expression plasmid containing an SV40 Ori. As expected, whole-cell extract prepared from these COS M6 cells contained factors that formed a protein-DNA complex on the +55 HRE WT probe but not on the +55 HRE MT probe (Fig. 2B, lane 8 versus 11, respectively). The fact that incubation with an ERRα1-specific antiserum led to a shift in the mobility of much of this complex (Fig. 2B, lane 9 versus 8) confirmed that the complex contained ERRα1. Similar results were obtained with COS M6 cells overexpressing COUP-TF1 (data not shown), the NR representing the majority of the +55 HRE-binding activity present in CV-1PD cells (48). Thus, this +55 HRE double mutation greatly reduces binding by most of the +55 HRE-binding activity present in AGMK cell lines. Fortunately, it does so without affecting replication of SV40 in some cell lines (see below). Thus, this +55 HRE double mutation probably does not have significant effects on posttranscriptional events despite mapping within the coding region of the late mRNAs. Therefore, an SV40 mutant containing these mutations should enable us to examine the importance of this HRE in the context of the whole SV40 genome.
SV40 genomes containing HRE mutations are viable.
Plasmids pSV4540 and pSV4541 contain these +1 HRE and +55 HRE mutations, and plasmid pSV4542 contains both of the HRE mutations, crossed into pSVS(WT). The viral genomes were excised from their cloning vector by treatment with EcoRI, ligated to form monomer circles, and transfected into cells of AGMK cell line CV-1PD′. After incubation for 12 days at 37°C, most of the cells on the dishes exhibited extensive evidence of SV40 cytopathic effect. The cells were harvested and used to prepare stocks of virus. The titers of these virus stocks ranged from ∼108 to 109 PFU/ml as determined by standard plaque assays with TC-7 cells (38). Plaque morphologies of the mutants were essentially indistinguishable from those of wild-type SV40 (data not shown). Thus, the mutants all appear to be viable. We refer to the virus stocks made from these plasmids as +1 HRE MT, +55 HRE MT, +1/+55 HRE DMT, and WT, respectively.
To confirm that the mutants were truly viable rather than the result of revertants arising during growth of the virus stocks, viral DNA was isolated from each virus stock and their promoter-regulatory regions (i.e., SV40 nt 4924 to 446) were amplified by PCR and sequenced. As expected, all of the HRE mutations were retained without second-site mutations being observed within this sequenced region at levels detectable by this assay (18). Digestion of the viral DNA with HindIII followed by electrophoresis in agarose gels also produced no evidence of novel deletions, duplications, or insertions (data not shown). We conclude from these data that virus with mutations in one or both of these HREs is viable and grows nearly as well, if not as well, as the WT in CV-1PD′ cells.
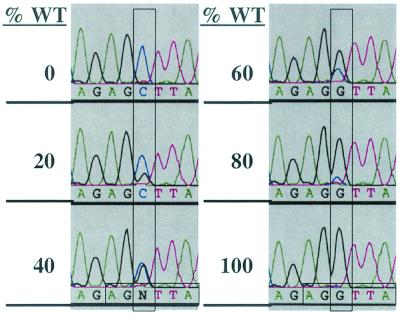
Given the excellent growth of the mutants, any differences in their phenotypes from that of WT would likely be subtle. Therefore, we developed a replication competition assay that would enable us to observe even small effects of the HREs on viral replication. This assay involves performing DNA sequence analysis of PCR products generated from the viral DNAs present in the virus stocks. To determine the sensitivity of this assay, we mixed together WT and +1/+55 HRE DMT virus stocks at various known ratios, recovered the viral DNA from these mixtures, amplified the promoter-regulatory region by PCR, and sequenced the PCR products. The resulting sequence electropherograms demonstrate that the ratio of peaks within the sequence of the 5′ +1 HRE half-site for guanosine versus cytidine at the site of the mutation accurately reflects the ratio of wild-type to mutant virus present in each mixture (Fig. 3). Similar results were obtained at the sequence of the +55 HRE (data not shown). Furthermore, although the peak of a specific nucleotide at one position differed in size from the peak for the same nucleotide at another position, this difference was very consistent (Fig. 3 to 7). It is due to context effects of neighboring bases (Perkin-Elmer product literature). Repeated analyses of the same virus mixtures showed a variation as measured by eye in the ratios of the heights and areas under the peaks at a given position in the sequence of at most a few percent (see, e.g., starting sample in Fig. 4 versus 5; data not shown). We conclude that this assay enables one to determine the relative ratios of genotypes present within a mixture containing two genetically known types of SV40.
FIG. 3.
Verification of sensitivity of the assay used for determination of relative genotypic ratios of viruses in mixed stocks. Shown are sequencing electropherograms of the 5′ +1 HRE half-site region of PCR products generated from viral DNA recovered from various mixtures of stocks of WT and +1/+55 HRE DMT virus. Percentages of WT virus present in the mixture are given on the left. Vertical rectangles, bases within the electropherograms that differ between the mutant and wild type; horizontal rectangles (bottom), half-site sequences. Black peak, wild-type guanosine base; blue peak, mutant cytidine base.
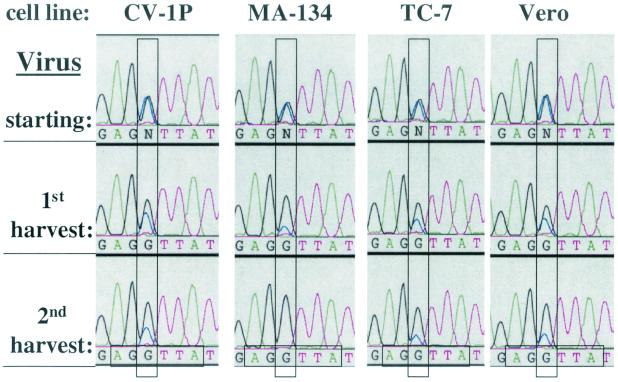
FIG. 7.
Cell lines differ in the extent of replication advantage conferred by the +1 HRE. Shown are sequence electropherograms of the +1 HRE 5′ half-site regions of PCR products generated from viral DNA recovered from the initial virus stock (starting) used for infection of CV-1P, MA-134, TC-7, and Vero cells with WT and +1 HRE MT in an initially equal, low-MOI coinfection and from the first and second harvests following sequential infection and incubation at 37°C for 10 days in DMEM containing 2% FBS. Virus competition experiments were performed in triplicate. The electropherograms are representative of the data obtained from one set of experiments. For the significance of colors and of the vertical and horizontal rectangles, see the Fig. 3 legend.
FIG. 4.
Wild-type virus has a replication advantage over the double-HRE-mutant virus in CV-1 cells. Shown are sequence electropherograms of the +1 HRE 5′ half-site and the +55 HRE 3′ half-site regions of PCR products generated from viral DNA recovered from the initial virus stock (starting) used for infection of CV-1 cells with WT and +1/+55 HRE DMT in an initially equal, low-MOI coinfection and from the first and second harvests following sequential infection and incubation at 37°C for 10 days in DMEM containing 2% FBS. Virus competition experiments were performed in triplicate. The electropherograms are representative of the data obtained from one set of experiments. For the significance of colors and of the vertical and horizontal rectangles, see the Fig. 3 legend.
WT outreplicates +1/+55 HRE DMT in CV-1 cells.
We first used this assay to examine the relative rates of replication of wild-type versus double-HRE-mutant virus. Equivalent PFU of WT and +1/+55 HRE DMT virus were mixed together at a total MOI of ∼0.05 PFU/cell and used to coinfect monolayers of CV-1 cells that had just reached confluence. We chose to use low-MOI coinfection to insure that most infected cells would initially harbor only one infectious genotype of virus, thereby avoiding replication by complementation. The cells were subsequently incubated at 37°C for 10 days, a time at which most cells exhibited virus-induced cytopathic effect. Since the SV40 life cycle under these conditions typically takes ∼60 h (5), we assume that the virus infection spread from ∼5% of the cells to all of them over three or four cycles of infection. Although most cells were likely coinfected by the last cycle, superinfection inhibition probably prevented late-infecting virus from replicating significantly. The cells were harvested, virus was extracted from them, and fresh monolayers of cells were infected at an MOI of ∼0.05 PFU/cell with the virus mixture obtained from the first-round harvest. Once again, the infected cells were incubated at 37°C for 10 days and harvested and virus was extracted. The viral DNAs present in the initial infecting virus stock, first-round harvest, and second-round harvest were processed as described above to determine genotypic ratios. As expected, approximately equivalent amounts of wild-type and mutant genotypes were present in the starting mixture of virus (Fig. 4). However, the wild-type genotype gradually predominated over the mutant genotype, accounting for at least 80% by the time of the second harvest (Fig. 4). Thus, wild-type HREs offer a selective advantage to the virus for replication under the conditions used in this experiment.
WT does not outreplicate +1/+55 HRE DMT in CV-1PD′ cells.
Is the wild-type replication advantage observed above a general phenomenon or cell type specific? To begin to address this question, we repeated the above experiment, but this time with CV-1PD′ cells, the cells originally used to grow the virus stocks. CV-1PD′ cells are a derivative of CV-1 cells that have passed sequentially through the laboratories of S. Kit, P. Berg, C. Cole, and J. Mertz over the past 3 decades, with noted changes in phenotype sometimes selected by cell cloning and regrowth. In contrast to the results obtained with the CV-1 cells, the approximately equimolar ratios of wild-type and mutant genotypes present in the starting mixture were maintained throughout the first and second serial passages (Fig. 5). Thus, we conclude that the replication advantage offered by the HREs is cell type specific. Importantly, we also conclude from this lack of an effect of the mutations in CV-1PD′ cells that the advantage offered by the HREs is not due to a trivial cis-acting defect such as the +1 HRE mutation affecting a basal element of the promoter (e.g., the Inr); the downstream +55 HRE mutation affecting transcription elongation, pre-mRNA processing, stability of the mRNA, or synthesis of agnoprotein; or either mutation affecting viral DNA replication.
+1 HRE, not +55 HRE, provides a replication advantage in CV-1 cells.
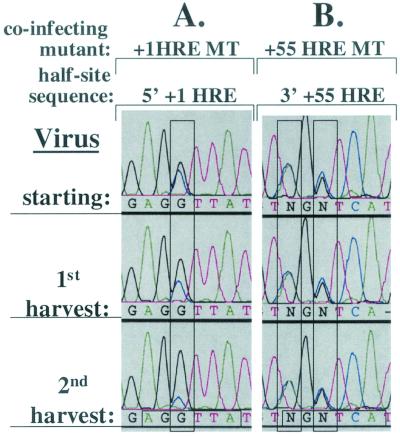
To determine whether one or both of these HREs is involved in the replication advantage observed in CV-1 cells (Fig. 4), we repeated the coinfection competition experiment with CV-1 cells but with +1 HRE MT or +55 HRE MT virus in place of +1/+55 HRE DMT virus. As observed with +1/+55 HRE DMT, +1 HRE MT accounted for only ∼20% of the virus by the time of the second harvest (Fig. 6A). On the other hand, the ratio of WT to +55 HRE MT remained the same throughout 20 days of replication in CV-1 cells (Fig. 6B). Thus, we conclude that the +1 HRE is responsible for the difference in replication of WT in CV-1 versus CV-1PD′ cells but that the +55 HRE contributes little or nothing to WT's replication advantage in CV-1 cells.
FIG. 6.
The +1 HRE, not the +55 HRE, provides the replication advantage in CV-1 cells. Shown are sequence electropherograms of the +1 HRE 5′ half-site and the +55 HRE 3′ half-site regions of PCR products generated from viral DNA recovered from the initial virus stock (starting) used for infection of CV-1 cells with WT and +1 HRE MT (A) or WT and +55 HRE MT (B) in an initially equal, low-MOI coinfection and from the first and second harvests following sequential infection and incubation at 37°C for 10 days in DMEM containing 2% FBS. Virus competition experiments were performed in triplicate. The electropherograms are representative of the data obtained from one set of experiments. For the significance of colors and of the vertical and horizontal rectangles, see the Fig. 3 legend.
Cell lines differ in the extent of replication advantage conferred by the +1 HRE.
Is the replication advantage offered by the +1 HRE common or rare among cell lines? To address this question, we also performed coinfection competition experiments with WT and the +1 HRE MT virus using CV-1P, MA-134, TC-7, and Vero cells, four permissive cell lines independently derived from the kidneys of different African green monkeys. By the time of the second harvest, the wild-type genotype predominated over the mutant genotype in all four of these cell lines (Fig. 7). Thus, the presence of the wild-type +1 HRE offers a replication advantage to SV40 in these cell lines as well. However, the rate at which WT outreplicated +1 HRE MT differed among the cell lines, with the mutant genotype already being undetectable by our assay (at most 5%) in MA-134 cells by the time of the second harvest (Fig. 7). Therefore, we conclude that cell lines differ in the degree to which the +1 HRE confers a selective advantage for replication.
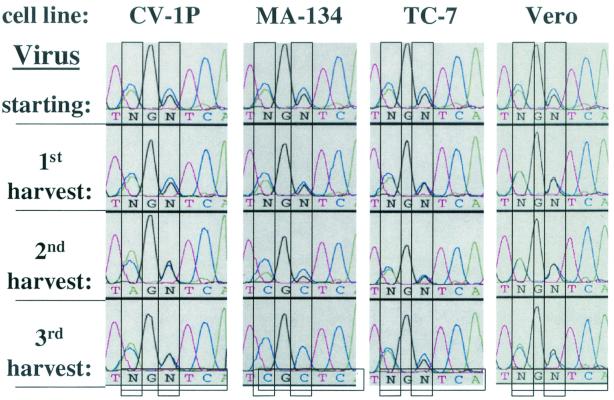
The +55 HRE confers a replication disadvantage in MA-134 cells but not in CV-1P, TC-7, or Vero cells.
Although the presence of the +55 HRE conferred no obvious effect on replication in CV-1 and CV-1PD′ cells, might it, too, confer effects in some other cell context? To address this question, we performed coinfection competition experiments with WT and +55 HRE MT virus using CV-1P, MA-134, TC-7, and Vero cells. By the time of the third harvest, i.e., three passages of 10 days each in culture, there were no significant changes in the genotypic ratios in CV-1P, TC-7, or Vero cells (Fig. 8). On the other hand, the mutant virus almost completely outreplicated the wild type in MA-134 cells by the time of the third harvest (Fig. 8). Thus, the presence of the +55 HRE confers a negative effect on the rate of replication of SV40 in MA-134 cells but has little or no effect in the other cell lines tested. Taking this together with the above findings, we conclude that the +1 and +55 HREs can, indeed, affect the replication of SV40, with the direction and degree of the effect differing with both the specific HRE and the cell line in which the virus is grown.
FIG. 8.
The +55 HRE confers a selective disadvantage for replication in MA-134 cells, but not in CV-1P, TC-7, or Vero cells. Shown are sequence electropherograms of the +55 HRE 3′ half-site regions of PCR products generated from viral DNA recovered from the initial virus stock (starting) used for infection of CV-1P, MA-134, TC-7, and Vero cells with WT and +1 HRE MT in an initially equal, low-MOI coinfection and from the first and second harvests following sequential infection and incubation at 37°C for 10 days in DMEM containing 2% FBS. Virus competition experiments were performed in triplicate. The electropherograms shown here are representative of the data obtained from one set of experiments. For the significance of colors and of the vertical and horizontal rectangles, see the Fig. 3 legend.
DISCUSSION
The central question addressed here and elsewhere (18) was whether the HREs present within the SV40 late promoter contribute to the life cycle of the virus. We showed that SV40 genomes containing mutations in the +1 and +55 HREs which inactivate binding by known and unknown factors recognizing the wild-type HREs (Fig. 2A) (18) are viable (data not shown) and that the associated viruses grow to high titers (data not shown) and make phenotypically normal plaques (data not shown). Also, because the +1 and +55 HRE mutations do not affect growth in CV-1PD′ cells (Fig. 5), the mutant viruses probably do not possess cis-acting defects in basal elements of the promoter, pre-mRNA processing, mRNA stability, synthesis of agnoprotein, or viral DNA replication. Rather, the growth characteristics of the mutants likely result from inherent differences among the cell types in trans-acting factors that directly or indirectly bind these HREs. Interestingly, the presence of the +1 HRE offers a selective advantage for replication of SV40 in CV-1, CV-1P, MA-134, TC-7, and Vero cells (Fig. 6 and 7) but not in CV-1PD′ cells (Fig. 5). The rate associated with this replication advantage varies and is greatest in MA-134 cells (Fig. 7). The presence of the +55 HRE confers no replication advantage in CV-1, CV-1PD′, CV-1P, TC-7, and Vero cells (Fig. 5, 6, and 8) but confers a replication disadvantage in MA-134 cells (Fig. 8). Taken together, these findings show that these HREs are biologically important to SV40 in the context of a viral infection, with the effects being dependent on both the specific HRE and the cell line in which the virus is growing.
The cellular environment may determine effects of HREs.
Our finding that the HREs of SV40 can have a positive, negative, or no effect on replication in a cell type-specific manner suggests that these cells must differ in factors that interact directly or indirectly with the HREs. The most obvious difference could be in the types and amounts of the NRs that interact with the HREs. We examined by immunoblot analysis the levels in CV-1 versus CV-1PD′ nuclear extracts of five NRs known to recognize the +1 HRE. No significant differences were observed (18). Since these five NRs represent only a subset of NRs from this large superfamily capable of recognizing the +1 HRE (25), we also used EMSAs to look for differences in CV-1 versus CV-1PD′ nuclear extract in the multiple protein complexes that form on a radiolabeled +1 HRE probe. Again, no significant differences were noted (18). Also, addition to cell culture media of ligands for the NRs known to bind the +1 HRE had no effect on the outcome of WT versus +1 HRE MT virus competition experiments in MA-134 cells (18). These experiments do not rule out the hypothesis that cell type differences in the NR population determine the replication advantage since the biologically relevant NR(s) might not have been detected in these assays. An alternative, non-mutually exclusive hypothesis is that the important differences are in the posttranslationally modified states of the NRs and/or cofactors (e.g., corepressor and coactivator complexes) with which these NRs interact to perform their functional activities. Additional experiments are needed to test these hypotheses.
The use of HREs to regulate early and late gene expression may be advantageous to SV40's survival. In its natural host, SV40 may use HREs to modulate progeny virion production so as to produce enough virus to insure transmission yet not enough to cause host pathogenesis or elicit a vigorous host immune response. This hypothesis could explain the opposing effects on replication of the +1 and +55 HREs in MA-134 cells (Fig. 7 and 8, respectively). Alternatively, SV40 may replicate efficiently only in cell types expressing NRs favorable to the proper temporal regulation of early and late gene expression. Thus, with different cell types within a host each expressing a unique array of NRs, HREs may contribute to the tissue tropism of SV40.
Mechanisms by which HREs affect SV40 replication.
We previously observed that SV40 plasmids containing mutations in the +1 or +55 HREs that significantly reduce binding by NRs overexpress late genes at early times posttransfection (46-49). These observations led us to hypothesize that the SV40 late-promoter HREs are binding sites for cellular NRs that act as repressors, delaying late gene expression until titrated away by viral DNA replication to high copy number (46, 48). Our observation here that the +1 HRE mutation has a negative effect on virus replication relative to that of WT in some cell types (Fig. 7) is consistent with this hypothesis and indicates that late-gene regulation through this element is biologically important to the virus. In data to be presented elsewhere (19) we show that the reason for the disadvantage of +1 HRE MT in MA-134 cells is that it accumulates higher levels of viral late RNA and lower levels of viral early RNA at very early times postinfection relative to WT. This lower accumulation of viral early RNA by +1 HRE MT results in lower accumulation of large T antigen, replicated viral DNA, and, subsequently, production of virions.
Interestingly, in contrast to mutation of the +1 HRE, mutation of the +55 HRE has a positive effect on replication of SV40 in MA-134 cells (Fig. 8). How might this be when, presumably, both HREs are binding sites for NRs that can act as repressors of the late promoter? One hypothesis is that the +55 HRE, being a DR2 rather than a DR4, binds different NRs, some of which may be highly abundant and, thus, not readily titrated away by viral DNA replication to high copy number. This prolonged repression of the late promoter in WT SV40 may result in underexpression of the late genes relative to that in +55 HRE MT, leading to a lag in the production of virions. Alternatively, some of the NRs binding the +55 HRE in MA-134 cells may interact with corepressors that are refractory to large T antigen's replication-independent transactivation of the late promoter (41). Experiments to test these and other hypotheses to explain the mechanism behind the replication disadvantage conferred by the +55 HRE in MA-134 cells have yet to be done.
In summary, we showed here that the SV40 HREs are biologically important to the virus as they can affect replication in a cell type-dependent manner.
Acknowledgments
We thank Janet Butel, Chuck Cole, and Bill Sugden for cell lines and Eric Ariazi for plasmid pcDNA3.1-hERRα1 and the ERRα1-specific antiserum. We also thank Paul Lambert, Bill Sugden, and members of our laboratory for helpful advice, discussions, and suggestions relating to the manuscript and Jackie Perrigoue for technical assistance.
This research was supported by U.S. Public Health Service grants CA22443, CA07175, and CA09135 from the National Cancer Institute.
REFERENCES
- 1.Acheson, N. H. 1982. Lytic cycle of SV40 and polyoma virus, p. 125-204. In J. Tooze (ed.), DNA tumor viruses, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Ali, S. H., and J. A. DeCaprio. 2001. Cellular transformation by SV40 large T-antigen: interaction with host proteins. Semin. Cancer Biol. 11:15-23. [DOI] [PubMed] [Google Scholar]
- 3.Avantaggiati, M. L., M. Carbone, A. Graessmann, Y. Nakatani, B. Howard, and A. S. Levine. 1996. The SV40 large T antigen and adenovirus E1a oncoproteins interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 15:2236-2248. [PMC free article] [PubMed] [Google Scholar]
- 4.Ayer, D. E., and W. S. Dynan. 1988. Simian virus 40 major late promoter: a novel tripartite structure that includes intragenic sequences. Mol. Cell. Biol. 8:2021-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barkan, A., and J. E. Mertz. 1981. DNA sequence analysis of simian virus 40 mutants with deletions mapping in the leader region of the late viral mRNAs: mutants with deletions similar in size and position exhibit varied phenotypes. J. Virol. 37:730-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, L. C., D. B. Smith, I. Davidson, J.-J. Hwang, E. Fanning, and A. G. Wildeman. 1996. Interaction between T antigen and TEA domain of the factor TEF-1 derepresses simian virus 40 late promoter in vitro: identification of T-antigen domains important for transcription control. J. Virol. 70:1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brady, J., J. B. Bolen, M. Radonovich, N. Salzman, and G. Khoury. 1984. Stimulation of simian virus 40 late gene expression by simian virus 40 tumor antigen. Proc. Natl. Acad. Sci. USA 81:2040-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brady, J., and G. Khoury. 1985. Trans-activation of the simian virus 40 late transcription unit by T-antigen. Mol. Cell. Biol. 5:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchman, A. R., L. Burnett, and P. Berg. 1982. The SV40 nucleotide sequence, p. 799-841. In J. Tooze (ed.), DNA tumor viruses, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Carswell, S., and J. C. Alwine. 1986. Simian virus 40 agnoprotein facilitates perinuclear-nuclear localization of VP1, the major capsid protein. J. Virol. 60:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casaz, P., P. W. Rice, C. N. Cole, and U. Hansen. 1995. A TEF-1-independent mechanism for activation of the simian virus 40 (SV40) late promoter by mutant SV40 large T antigens. J. Virol. 69:3501-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casaz, P., R. Sundseth, and U. Hansen. 1991. Trans-activation of the simian virus 40 late promoter by large T antigen requires binding sites for the cellular transcription factor TEF-1. J. Virol. 65:6535-6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulombe, J., L. Berger, D. B. Smith, R. K. Hehl, and A. G. Wildeman. 1992. Activation of simian virus 40 transcription in vitro by T antigen. J. Virol. 66:4591-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damania, B., and J. C. Alwine. 1996. TAF-like function of SV40 large T antigen. Genes Dev. 10:1369-1381. [DOI] [PubMed] [Google Scholar]
- 15.Desvergne, B., and T. Favez. 1997. The major transcription initiation site of the SV40 late promoter is a potent thyroid hormone response element. Nucleic Acids Res. 25:1774-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckner, R., J. W. Ludlow, N. L. Lill, E. Oldread, Z. Arany, N. Modjtahedi, J. A. DeCaprio, D. M. Livingston, and J. A. Morgan. 1996. Association of p300 and CBP with simian virus 40 large T antigen. Mol. Cell. Biol. 16:3454-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrell, M. L. 2001. Effects of hormone response elements on the SV40 life cycle. Ph.D. thesis. University of Wisconsin, Madison.
- 19.Farrell, M. L., and J. E. Mertz. Hormone response element in SV40 late promoter directly affects synthesis of early as well as late viral RNAs. Virology, in press. [DOI] [PubMed]
- 20.Fromm, M., and P. Berg. 1982. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J. Mol. Appl. Genet. 1:457-481. [PubMed] [Google Scholar]
- 21.Gilinger, G., and J. C. Alwine. 1993. Transcriptional activation by simian virus 40 large T antigen: requirements for simple promoter structures containing either TATA or initiator elements with variable upstream factor binding sites. J. Virol. 67:6682-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Good, P. J., R. C. Welch, W.-S. Ryu, and J. E. Mertz. 1988. The late spliced 19S and 16S RNAs of simian virus 40 can be synthesized from a common pool of transcripts. J. Virol. 62:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffin, B. E. 1982. Structure and genomic organization of SV40 and polyoma virus, p. 61-124. In J. Tooze (ed.), DNA tumor viruses, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Gruda, M. C., J. M. Zabolotny, J. H. Xiao, I. Davidson, and J. C. Alwine. 1993. Transcriptional activation by simian virus 40 large T antigen: interactions with multiple components of the transcription complex. Mol. Cell. Biol. 13:961-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinemeyer, T., E. Wingender, I. Reuter, H. Hermjakob, A. E. Kel, O. V. Kel, E. V. Ignatieva, E. A. Ananko, O. A. Podkolodnaya, F. A. Kolpakov, N. L. Podkolodny, and N. A. Kolchanov. 1998. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 26:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herbig, U., K. Weisshart, P. Taneja, and E. Fanning. 1999. Interaction of the transcription factor TFIID with simian virus 40 (SV40) large T antigen interferes with replication of SV40 DNA in vitro. J. Virol. 73:1099-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jay, G., S. Nomura, C. W. Anderson, and G. Khoury. 1981. Identification of the SV40 agnogene product: a DNA binding protein. Nature 291:346-349. [DOI] [PubMed] [Google Scholar]
- 28.Johnston, S. D., X.-M. Yu, and J. E. Mertz. 1996. The major transcriptional transactivation domain of simian virus 40 large T antigen associates nonconcurrently with multiple components of the transcriptional preinitiation complex. J. Virol. 70:1191-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnston, S. D., X. Liu, F. Zuo, T. L. Eisenbraun, S. R. Wiley, R. J. Kraus, and J. E. Mertz. 1997. Estrogen-related receptor α1 functionally binds as a monomer to extended half-site sequences including ones contained within estrogen-response elements. Mol. Endocrinol. 11:342-352. [DOI] [PubMed] [Google Scholar]
- 30.Jones, N. C., P. W. J. Rigby, and E. B. Ziff. 1988. Trans-acting protein factors and the regulation of eukaryotic transcription: lessons from studies on DNA tumor viruses. Genes Dev. 2:267-281. [DOI] [PubMed] [Google Scholar]
- 31.Keller, J. M., and J. C. Alwine. 1984. Activation of the SV40 late promoter: direct effects of T antigen in the absence of viral DNA replication. Cell 36:381-389. [DOI] [PubMed] [Google Scholar]
- 32.Keller, J. M., and J. C. Alwine. 1985. Analysis of an activatable promoter: sequences in the simian virus 40 late promoter required for T-antigen-mediated trans-activation. Mol. Cell. Biol. 5:1859-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly, J. J., and A. G. Wildeman. 1991. Role of the SV40 enhancer in the early to late shift in viral transcription. Nucleic Acids Res. 19:6799-6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein-Hitpass, L., S. Y. Tsai, G. L. Greene, J. H. Clark, M.-J. Tsai, and B. W. O'Malley. 1989. Specific binding of estrogen receptor to the estrogen response element. Mol. Cell. Biol. 9:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraus, R. J., E. E. Murray, S. R. Wiley, N. M. Zink, K. Loritz, G. W. Gelembiuk, and J. E. Mertz. 1996. Experimentally determined weight matrix definitions of the initiator and TBP binding site elements of promoters. Nucleic Acids Res. 24:1531-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, H.-J., Y. Lee, J. P. H. Burbach, and C. Chang. 1995. Suppression of gene expression on the simian virus 40 major late promoter by human TR4 orphan receptor. J. Biol. Chem. 270:30129-30133. [DOI] [PubMed] [Google Scholar]
- 37.Mertz, J. E., and P. Berg. 1974. Viable deletion mutants of simian virus 40: selective isolation by means of a restriction endonuclease from Haemophilus parainfluenzae. Proc. Natl. Acad. Sci. USA 71:4879-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mertz, J. E., and P. Berg. 1974. Defective simian virus 40 genomes: isolation and growth of individual clones. Virology 62:112-124. [DOI] [PubMed] [Google Scholar]
- 39.Mertz, J. E., A. Murphy, and A. Barkan. 1983. Mutants deleted in the agnogene of simian virus 40 define a new complementation group. J. Virol. 45:36-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng, S.-C., J. E. Mertz, S. Sanden-Will, and M. Bina. 1985. Simian virus 40 maturation in cells harboring mutants deleted in the agnogene. J. Biol. Chem. 260:1127-1132. [PubMed] [Google Scholar]
- 41.O'Reilly, G. H. 2000. Regulation of the SV40 late promoter by nuclear receptors and large T antigen. Ph.D. thesis. University of Wisconsin, Madison.
- 42.Resnick, J., and T. Shenk. 1986. Simian virus 40 agnoprotein facilitates normal nuclear location of the major capsid polypeptide and cell-to-cell spread of virus. J. Virol. 60:1098-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenthal, L. J., and M. Brown. 1977. The control of SV40 transcription during a lytic infection: late RNA synthesis in the presence of inhibitors of DNA replication. Nucleic Acids Res. 4:551-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rundell, K., and R. Parakati. 2001. The role of the SV40 T-antigen in cell growth promotion and transformation. Semin. Cancer Biol. 11:5-13. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Wiley, S. R., R. J. Kraus, F. Zuo, E. E. Murray, K. Loritz, and J. E. Mertz. 1993. SV40 early-to-late switch involves titration of cellular transcriptional repressors. Genes Dev. 7:2206-2219. [DOI] [PubMed] [Google Scholar]
- 47.Zuo, F. 1995. Regulation of the SV40 late promoter by members of the steroid/thyroid hormone receptor superfamily. Ph.D. thesis. University of Wisconsin, Madison.
- 48.Zuo, F., and J. E. Mertz. 1995. Simian virus 40 late gene expression is regulated by members of the steroid/thyroid hormone receptor superfamily. Proc. Natl. Acad. Sci. USA 92:8586-8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuo, F., R. J. Kraus, T. Gulick, D. D. Moore, and J. E. Mertz. 1997. Direct modulation of simian virus 40 late gene expression by thyroid hormone and its receptor. J. Virol. 71:427-436. [DOI] [PMC free article] [PubMed] [Google Scholar]