Abstract
Virion release of hepatitis B virus (HBV) from hepatocytes is a tightly regulated event. It is a dogma that only the mature HBV genome is preferentially allowed to export from the intracellular compartment (J. Summers and W. S. Mason, Cell 29:403-415, 1982). Recently, an “immature secretion” phenotype of a highly frequent naturally occurring HBV variant containing a leucine residue at amino acid 97 of the core protein was identified. Unlike wild-type HBV, this variant secretes almost equal amounts of mature and immature genomes. This phenomenon is not caused by any instability of core particles or by any deficiency in viral reverse transcription (T. T. Yuan, P. C. Tai, and C. Shih, J. Virol. 73:10122-10128, 1999). In this study, our kinetic analysis of virion secretion of the mutant F97L (phenylalanine to leucine) indicates that the secretion of its immature genome does not occur earlier than that of its mature genome. In addition, the secretion kinetics of the mature genomes are comparable between the wild-type HBV and the mutant F97L. Therefore, the immature secretion phenomenon of mutant F97L is not caused by premature secretion or more efficient secretion. Previously, we hypothesized that the immature secretion phenotype is probably caused by the aberrant interaction between its mutant core and wild-type envelope proteins. Here, we further demonstrated that a pre-S1 envelope mutation at position 119, changing an alanine (A) to a phenylalanine (F), can offset the immature secretion phenotype of the mutant I97L (isoleucine to leucine) and successfully restore the wild-type-like selective export of the mature genome of the double mutant pre-S1-A119F/core-I97L.
Hepatitis B virus (HBV) is an enveloped DNA virus belonging to the family Hepadnaviridae. As in many other viruses, virion release of HBV is a regulated process. It is well established that mature genomes of hepadnaviruses, which contain relaxed circular DNA, are preferentially secreted. By contrast, immature genomes, which contain nascent single-stranded linear DNA, are preferentially retained within the hepatocytes (13, 37). It was hypothesized that a genome maturation signal can become more pronounced as replication takes place inside the core particles. Recognition of the genome maturation signal by the envelopment machinery would then lead to virion release. Under this hypothesis, core particles are not a passive structural entity. Instead, they can serve as an active signal transducer to communicate between the genomic DNA replication and the envelopment machinery.
HBV encodes three closely related envelope proteins: small (S), middle (M), and large (L) (16, 28, 36). While the M protein is dispensable for virion secretion, both S and L envelope proteins are required (2, 43). Functional characterization via mutagenesis of the S and L genes has mapped two envelope regions essential for virion secretion (4, 22, 23). One region is localized in an internal domain of the small S protein (amino acids 35 to 80). The other region within the large envelope protein is localized in the C terminus of the pre-S1 domain (amino acids 107 to 119 of the adr subtype) and extends into the adjacent amino acids 1 to 5 of the pre-S2 domain (4, 22, 23). Although these two regions were found to be essential for virion secretion by a genetic approach, their exact roles in virion secretion were unclear. An assay of in vitro binding between core particles and synthetic envelope peptides suggests that these two envelope regions probably directly bind to the core particles (29). An artificial synthetic peptide of non-HBV origin, identified from a random peptide library screening, was able to bind the tip of the spike region of the core particles and was found to be inhibitory to virion secretion (9). To date, it remains unclear if a direct core-envelope interaction in vivo is indeed important for HBV virion secretion.
We reported previously an abnormal virion secretion phenotype dubbed “immature secretion.” This phenotype is characterized by nonselective and excessive secretion of virion particles containing immature genomes with single-stranded DNA. A frequent naturally occurring mutation, changing either isoleucine (I) or phenylalanine (F) to a leucine (L) at position 97 in the HBV core protein, is both necessary and sufficient for this rather surprising phenotype (45, 46). The immature secretion phenotype is not created as a consequence of any intracellular defect in genome maturation per se (46) or from any alteration in the stability of the core particles (45). Furthermore, another frequent core mutation, P130T (proline to threonine), can be compensatory for the immature secretion phenotype of the core mutant I97L (47). In addition to immature secretion, both artificial and frequent naturally occurring mutations of the core protein were found to result in a low-secretion phenotype (20, 21). The mechanism(s) behind these virion secretion phenotypes remains unclear. Given the fact that neither the immature secretion variant I97L nor the low-secretion variants exhibited any intracellular phenotype in viral DNA replication, we hypothesized that these secretion-defective phenotypes could be caused by aberrant core-envelope interaction rather than by aberrant core-polymerase interaction (46).
Here, we demonstrate the importance and specificity of the core-envelope interaction in vivo in the control of HBV virion secretion by creating a pre-S1 mutant, A119F, that can suppress the immature secretion phenotype of core variant I97L.
MATERIALS AND METHODS
DNA transfection of the human hepatoma HepG2 cell line was carried out by the calcium phosphate or FuGENE 6 protocol (Roche, Indianapolis, Ind.). Cell culture conditions, preparation of HBV viral DNA, Southern blot analysis, and gradient centrifugation analysis of virion secretion were as detailed elsewhere (45). Western blot analysis using a monoclonal antibody against the pre-S1 domain (16) was as described by Tai et al. (38).
FuGENE 6 protocol.
Transfections with FuGENE 6 reagent were performed according to the manufacturer's procedure. Briefly, 2 μg of plasmid DNA was mixed with 9 μl of FuGENE 6 reagent and 180 μl of serum-free medium. After a 1-h incubation at room temperature, the mixture was added to a 10-cm dish with 50 to 70% confluence. Medium was changed 2 days posttransfection.
Mutagenesis.
The QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) was used to introduce mutations in HBV monomers of the adr subtype carrying either the wild-type (WT) core or the I97L core gene. Briefly, plasmids were amplified by Pfu Turbo DNA polymerase using two complementary oligonucleotides (see Table 1), carrying a mutation in the pre-S1 gene. After amplification, the PCR product was treated with DpnI endonuclease, which digested the input plasmid (the parental plasmid isolated from Escherichia coli is methylated and therefore susceptible to DpnI). The amplified DNA was then transformed into competent E. coli cells, and colonies were screened by DNA sequencing with the primer AS180 (5′GGGGTCCTAGGAATCCTGATG3′) (nucleotides 170 to 190; numbering according to the ayw subtype [12]). The mutant HBV monomers were then dimerized in tandem to mimic the circular genetic configuration of HBV (45). Using these procedures, we created a total of 12 pre-S1 mutants: six in the WT core gene context (see Fig. 3) and another six in the I97L mutant core gene context (see Fig. 4).
TABLE 1.
Nucleotide sequences of the primers used for mutagenesis in the pre-S1 region
| Pre-S1 mutanta | Oligonucleotide sequence (5′ to 3′)b | Change(s) in polymerase spacer region |
|---|---|---|
| I108F (S) | CAG CCT ACT CCC TTC TCT CCA CCT CTA | H288L |
| I108F (AS) | TAG AGG TGG AGA GAA GGG AGT AGG CTG | |
| L112F (S) | C ATC TCT CCA CCT TTC AGA GAC AGT CAT C | S292F and K293Q |
| L112F (AS) | G ATG ACT GTC TCT GAA AGG TGG AGA GAT G | |
| L112I (S) | C ATC TCT CCA CCT ATA AGA GAC AGT CAT C | S292Y |
| L112I (AS) | G ATG ACT GTC TCT TAT AGG TGG AGA GAT G | |
| A119F (S) | GAC AGT CAT CCT CAG TTC ATG CAG TGG AAC TCC | G299V |
| A119F (AS) | GGA GTT CCA CTG CAT GAA CTG AGG ATG ACT GTC | |
| A119I (S) | GAC AGT CAT CCT CAG ATC ATG CAG TGG AAC TCC | G299D |
| A119I (AS) | GGA GTT CCA CTG CAT GAT CTG AGG ATG ACT GTC | |
| A119L (S) | GAC AGT CAT CCT CAG CTC ATG CAG TGG AAC TCC | G299A |
| A119L (AS) | GGA GTT CCA CTG CAT GAG CTG AGG ATG ACT GTC |
S, sense; AS, antisense.
The mutated codons are underlined, and the mutation sites are in bold.
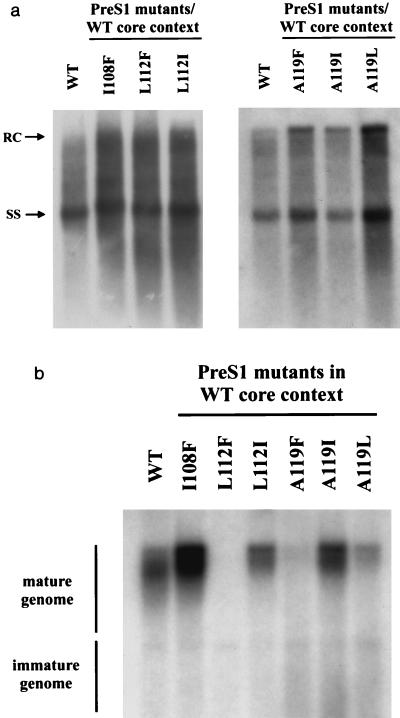
FIG. 3.
Functional analysis of the six pre-S1 mutations in the same WT core gene context via assays for both intracellular HBV DNA replication (a) and extracellular virion secretion (b). The low-secretion phenotype was observed in both mutants L112F and A119F. Briefly, 2 μg of each plasmid DNA was transfected into a human hepatoma cell line HepG2 using the FuGENE 6 reagent and protocol (Roche). Intracellular core particles were harvested 5 days posttransfection, and the core particle-associated HBV DNA was analyzed by Southern blot with a 3.1-kb HBV double-stranded DNA probe. The virion secretion assay was as described for Fig. 1. The full-length RC-form DNA at 4.0 kb and the SS-form DNA at 1.5 kb are indicated. The tandem dimer of WT HBV DNA was included as a control.
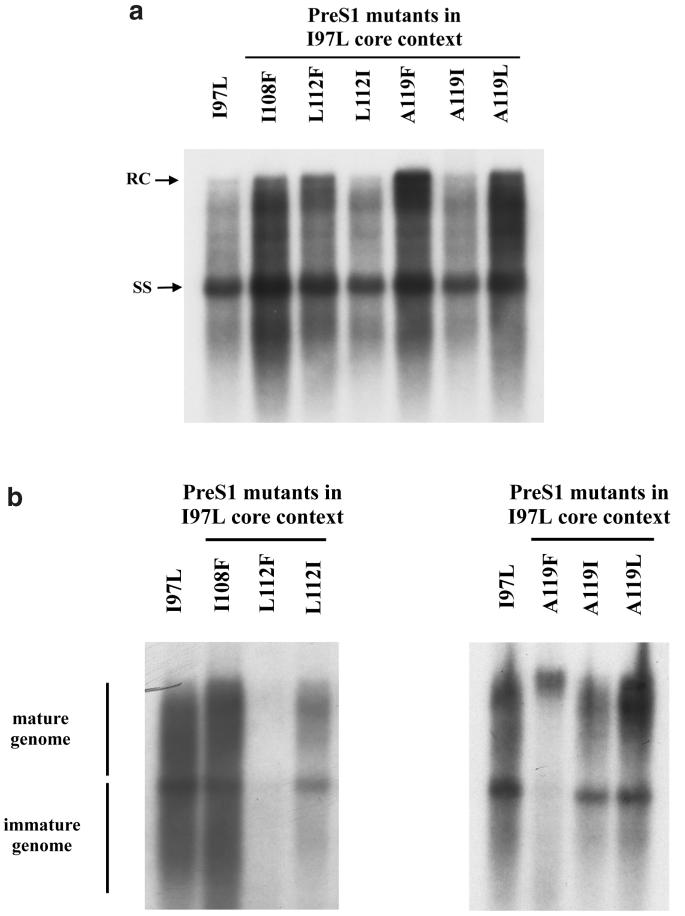
FIG. 4.
Functional analysis of the six pre-S1 mutations in the same mutant I97L core gene context via assays for both intracellular HBV DNA replication (a) and extracellular virion secretion (b). While the double mutant L112F/I97L continues to exhibit a low-secretion phenotype, the double mutant A119F/I97L behaves like a WT HBV, with a highly selective virion secretion of mature genome. The assays were as described for Fig. 1 and 3. The tandem dimer of core mutant I97L was included as a control.
RESULTS
As described in the introduction, it was previously demonstrated that the loss of selectivity in HBV virion secretion associated with the variants F97L and I97L is not caused by any intracellular defect in viral DNA synthesis or instability of core particles (45, 46; data not shown). Instead, it could be caused by an aberrant interaction between core and envelope proteins. Such an aberrant interaction could, for example, lead to premature secretion of immature genome.
Kinetic analysis of virion secretion.
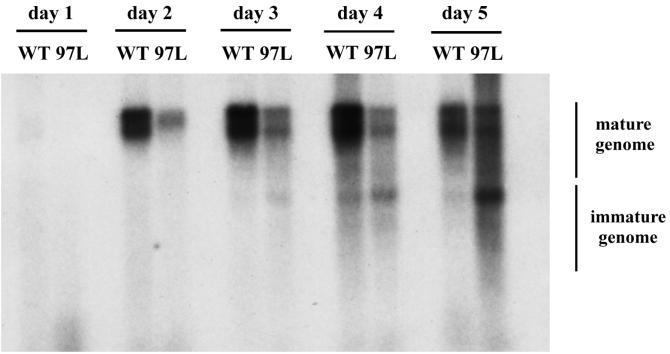
To investigate the mechanism of immature secretion, we compared the kinetics of virion secretion between the immature and the mature genomes in mutant F97L as well as the secretion of mature genomes between the WT and the mutant F97L (Fig. 1). For this purpose, cell culture media were collected from day 1 to day 5 posttransfection and viral particles were isolated via cesium chloride gradient centrifugation (see the Fig. 1 legend). In the case of variant F97L, the immature genome was detectable on day 3 whereas the mature genome appeared on day 2 posttransfection. Furthermore, there is no kinetic difference in the secretion of mature genomes between WT HBV and mutant F97L, both being detectable on day 2. In summary, the variant F97L does not secrete its mature or immature virions any earlier or faster than the WT HBV.
FIG. 1.
Comparison of virion secretion kinetics of WT HBV and core variant F97L via gradient centrifugation analysis. Cell culture media were collected from day 1 to day 5 posttransfection. After centrifugation through a 20% sucrose cushion, the resuspended pellets of HBV particles were separated by isopycnic centrifugation through a cesium chloride gradient (20 to 50%). The fractions corresponding to the enveloped Dane particles (fractions 10, 12, and 14, density = 1.2 g/cm3) were pooled before DNA analysis by Southern blotting with a 3.1-kb HBV double-stranded-DNA probe.
Experimental design to look for an intermolecular suppressor mutation.
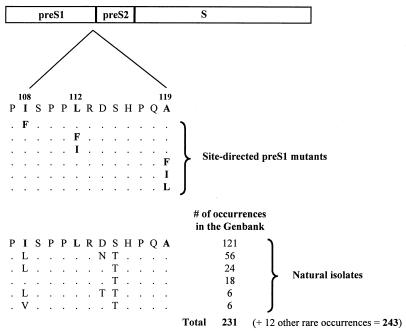
If an aberrant core-envelope interaction is indeed responsible for the immature virion secretion phenotype created by the core mutation I97L, one might be able to reverse the mutant phenotype by creating a suppressor mutation at the putative core-interacting domains of the envelope proteins (e.g., amino acids 107 to 119 of the pre-S1 domain). However, unless we could create a feasible number of mutations at the right positions with the right amino acid substitutions, we would not be able to handle the astronomical number of arbitrary mutations within amino acids 107 to 119 (1320 possibilities). We reasoned that since amino acid 97 of HBV core protein is always a hydrophobic residue (isoleucine in the WT adr subtype, phenylalanine in the WT ayw subtype, and a leucine in mutant 97L), it is most likely that this amino acid will interact with another hydrophobic residue(s) in the envelope proteins. Using this rationale, we created six mutations at positions 108, 112, and 119, changing from one hydrophobic residue to another (Fig. 2). The remaining 10 amino acids within pre-S1 107 to 119 are not as hydrophobic and thus were not tested further.
FIG. 2.
Site-directed missense mutations were introduced into the pre-S1 domain of the large envelope protein at positions 108, 112, and 119 (HBV subtype adr). The amino acid sequences from positions 107 to 119 of the pre-S1 domain of natural HBV isolates are highly conserved. For example, only 2 of 243 occurrences available in GenBank are different from the consensus residue at position 119 (data not shown). Functional consequences of these pre-S1 mutations on virion secretion were analyzed in WT (Fig. 3) and mutant (Fig. 4) core gene contexts.
It was shown previously that an excessive amount of the L envelope protein could interfere with small envelope protein secretion (7, 26, 27). Furthermore, as mentioned earlier, proper levels of the S and L envelope proteins have been shown to be required for virion secretion (2, 43). If we chose a strategy to analyze the aforementioned pre-S1 mutations by complementation assay between an L envelope-deficient HBV and an expression vector containing various mutant L envelope genes under a heterologous enhancer/promoter, we would then need to consider the complication of improper stoichiometry between envelope proteins for virion secretion. To preserve the natural stoichiometry between the L, M, and S envelope proteins, these six pre-S1 mutations were analyzed in the context of a tandem dimer configuration. As a control for the effect of pre-S1 mutations in the context of core mutant I97L, we have also examined the effect of these six pre-S1 mutations in the WT core gene context. It should also be noted that, because of the overlapping open reading frames, these pre-S1 mutations overlap with the spacer region of the polymerase protein. The spacer region of polymerase is well known to be very tolerant to drastic mutations, including insertions and deletions of short peptides (32, 38). To be on the cautious side, whenever possible, we designed pre-S1 mutations by minimizing the number of concurrent amino acid changes within the spacer region of the polymerase (Table 1).
PreS1 mutations in the context of the WT core gene.
As shown in Fig. 3a, the intracellular viral DNA replication of all six pre-S1 mutants (I108F, L112F, L112I, A119F, A119I, and A119L), in the context of the WT core gene, appears to be more or less similar. Except in pre-S1 mutants A119F and A119I, stronger overall replication was observed. Similar steady-state levels of HBV large envelope proteins were observed between WT and all the mutant viruses by Western blot analysis using a monoclonal antibody specific for the pre-S1 domain (data not shown) (16). Virion secretion analysis of these mutants revealed several interesting features. While mutants I108F, L112I, and A119I behaved like WT HBV, with a normal level of virion secretion, mutants L112F and A119F exhibited a low-secretion phenotype, with almost undetectable extracellular HBV DNA (Fig. 3b). In addition, mutant A119L displayed a reduced level of virion secretion, with very weak signals of extracellular HBV DNA.
PreS1 mutations in the context of the mutant I97L core gene.
As shown in Fig. 4a, all six pre-S1 mutants in the context of the mutant core I97L are replication competent, although their respective replication efficiencies are somewhat variable, about twofold. In the virion secretion assay, the double mutant L112F/I97L (Fig. 4b) displayed the same low-secretion phenotype as the single mutant L112F/WT core (Fig. 3b). Most strikingly, the double mutant A119F/I97L behaved like a WT HBV and did not have any detectable virion-associated immature genomes (Fig. 4b). Previously, we measured quantitatively the degree of immature secretion by comparing the signal intensities between the virion-associated mature genomes (relaxed-circle DNA [RC form]) and immature genomes (single-stranded DNA [SS form]). Based on more precise quantitation, the RC/SS ratio for adr-WT HBV in HepG2 cells is usually around 9.30 ± 2.40, and that for mutants F97L and I97L is usually around 0.87 ± 0.18 (45, 46). Here, the RC/SS ratio for the double mutant A119F/I97L, averaged from four independent experiments, is around 5.94 ± 2.60 (data not shown), a value that would be expected from a WT HBV with normal control of virion secretion. While the overall level of virion-associated mature genomes secreted by double mutant A119F/I97L is reduced relative to the WT HBV or the single mutant I97L, it is clearly much higher than the almost undetectable secretion for the single mutant A119F (Fig. 3b). In summary, neither the low-secretion phenotype of the pre-S1 mutant A119F nor the immature secretion phenotype of the core mutant I97L was observed for the WT-like double mutant A119F/I97L.
DISCUSSION
Control of HBV virion secretion has been studied by introducing mutations into the envelope or core genes. In all cases, loss of either viral DNA replication or virion secretion was observed (4, 20, 21). The present study differs from previous ones in several important aspects. First, the core mutation at amino acid 97 is not an artificial mutation. In fact, I97L is the most frequent clinical mutation within the core gene (1, 8, 10, 17, 24) and has been found in chronic HBV carriers worldwide (5, 6, 11, 14, 15, 18, 19, 25, 33-35, 39-42, 44). Second, in contrast to more drastic deletions or insertional mutagenesis, immature secretion is a unique and intriguing phenotype created by a very subtle amino acid change at position 97, from a hydrophobic phenylalanine or isoleucine to another hydrophobic leucine residue (45, 46). Third, immature secretion is a loss-of-specificity phenotype, rather than a loss-of-competency phenotype, as is commonly found in low-secretion core or envelope mutants (4, 20-22). Fourth, to our knowledge, pre-S1 mutant A119F is the first example of an intermolecular compensatory mutation between HBV core and envelope proteins. It lends strong support to the general hypothesis that the regulation of virion secretion is mediated via specific core-envelope interactions.
Premature versus immature virion secretion.
Following the general hypothesis mentioned above, a more specific hypothesis assumes that the core-envelope interaction in mutant 97L is somehow more efficient than in WT HBV. Therefore, mutant 97L would have less intracellular retention time for maturation to occur than WT, thus leading to the secretion of virions containing immature genomes. A comparison of the time course of virion release between WT and mutant F97L is shown in Fig. 1. We detected no kinetic difference in virion release between WT and mutant F97L or between immature genomes and mature genomes. Therefore, premature secretion or more efficient secretion cannot be a reason for the immature secretion phenomenon of mutant F97L. Finally, it should be noted that the results in Fig. 1 do not necessarily rule out the importance of core-envelope interaction in immature virion of mutant 97L. For example, a different, less efficient, core-envelope interaction could lead to the loss of selectivity in virion secretion. Interestingly, it has been shown that the extracellular duck hepatitis B virus core proteins are more dephosphorylated than the core proteins purified from the intracellular origin (31). At present, we have no evidence that the phosphorylation status of the core proteins is different between the WT and mutant F97L or between the mature and immature core particles in the WT or mutant HBV. Further study will be needed to address this issue.
Low-secretion pre-S1 mutant L112F.
The leucine residue at amino acid 112 of the WT pre-S1 domain seems to be important for virion secretion. For example, the mutation changing a leucine to a phenylalanine dramatically reduced the virion secretion, whereas the change to an isoleucine at the same position had only a modest effect on virion secretion. This result is consistent with a previous study using alanine substitutions at positions 113 and 114 (4). Taken together, amino acids 112 to 114 of the pre-S1 domain seem to play an important role in the control of virion secretion. It should be noted that the low-secretion phenotype of our mutant L112F is independent from the contexts of the core gene (Fig. 3b and 4b). It will be interesting to see if the low-secretion pre-S1 variants and the low-secretion core variants are arrested at the same step along their virion morphogenesis pathway (21).
Does low secretion plus immature secretion equal WT secretion?
We have previously identified a naturally occurring intramolecular compensatory mutation, P130T, for the immature secretion phenotype of mutant I97L (47). When the core protein contains both mutations I97L and P130T, the immature secretion phenotype reverts to a WT-like phenotype with normal virion secretion. Similarly, the present study of the double mutant pre-S1-A119F/core-I97L demonstrates the importance of a highly precise intermolecular interaction between core and envelope proteins in virion secretion (Fig. 4b). Although HBV core mutant P130T is a naturally occurring variant (5, 6, 11, 14, 15, 18, 19, 25, 33-35, 39-42, 44), we have not found any naturally occurring pre-S1-A119F variants in the GenBank database (Fig. 2).
As illustrated in Fig. 5, the fact that the pre-S1-A119F mutation alone exhibits a low-secretion phenotype and the core-I97L mutation alone exhibits an immature secretion phenotype suggests that simply by combining these two mutations, one can generate a WT-like phenotype. However, unlike pre-S1 mutant A119F, another pre-S1 low-secretion mutant, L112F, in combination with core mutant I97L, cannot revert to the WT phenotype. Instead, the double mutant (pre-S1-L112F/core-I97L) continues to exhibit a low- or no-secretion phenotype (Fig. 4b). Furthermore, we have checked several mutations within the putative core-interacting domain of the small envelope protein and have not found any compensatory mutations (unpublished results). Taken together, these data suggest that there is a high degree of specificity in such an intermolecular suppression of immature secretion. Further insights about this putative molecular interaction between HBV core and envelope proteins may be gained when the high-resolution three-dimensional structure of the HBV envelope proteins becomes available.
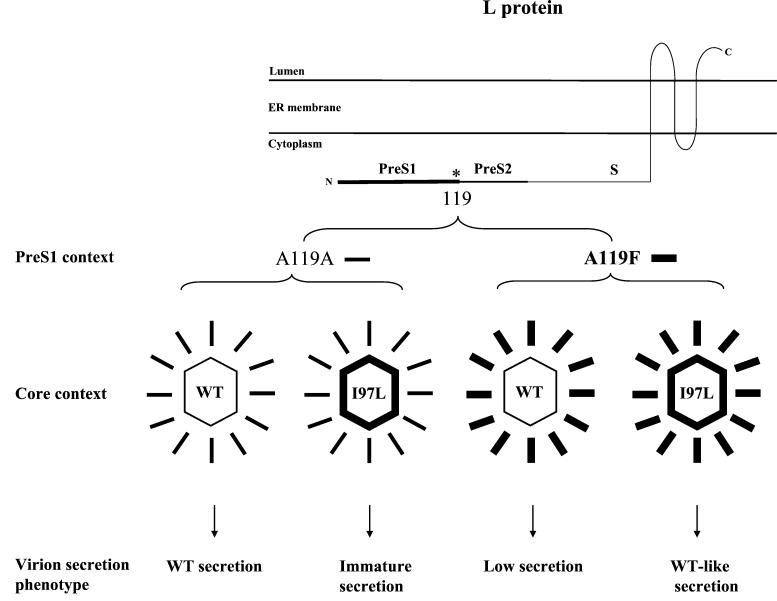
FIG. 5.
Summary of the various HBV virion secretion phenotypes associated with various core and envelope contexts. The N-terminal part of the pre-S1 domain of the large envelope protein has been hypothesized to adopt two distinct membrane topologies: exposed on either the lumen side or the cytoplasmic side of the endoplasmic reticulum (ER) (3, 30). The latter topology was illustrated here to describe its highly specific interactions with the core particles.
Finally, current therapy for hepatitis B patients includes alpha interferon and nucleoside analogues (e.g., lamivudine). Our study on the regulation of HBV virion secretion could identify new targets for future therapeutic interventions.
Acknowledgments
We thank Peggy Newman in C. Shih's laboratory for careful reading of the manuscript.
S.L.P. is a McLaughlin Postdoctoral Fellow. This study is supported in part by NIH grants RO1 CA70336 and CA84217 to C.S.
REFERENCES
- 1.Akarca, U. S., and A. S. F. Lok. 1995. Naturally occurring hepatitis B virus core gene mutations. Hepatology 22:50-60. [PubMed] [Google Scholar]
- 2.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. USA 88:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruss, V., X. Lu, R. Thomssen, and W. H. Gerlich. 1994. Post-translational alterations in trans-membrane topology of the hepatitis B virus large envelope protein. EMBO J. 13:2273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruss, V. 1997. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J. Virol. 71:9350-9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carman, W. F., W. Boner, G. Fattovich, K. Colman, E. S. Dornan, M. Thursz, and S. Hadziyannis. 1997. Hepatitis B virus core protein mutations are concentrated in B cell epitopes in progressive disease and in T helper cell epitopes during clinical remission. J. Infect. Dis. 175:1093-1100. [DOI] [PubMed] [Google Scholar]
- 6.Chang, S.-F., H. J. Netter, M. Bruns, R. Schneider, K. Frolich, and H. Will. 1999. A new hepadnavirus infecting snow geese (Anser caerulescens) produces a significant fraction of virions containing single-stranded DNA. Virology 262:39-54. [DOI] [PubMed] [Google Scholar]
- 7.Chisari, F. V., P. Filippi, A. McLachlan, D. R. Milich, M. Riggs, S. Lee, R. D. Palmiter, C. A. Pinkert, and R. L. Brinster. 1986. Expression of hepatitis B virus large envelope polypeptide inhibits hepatitis B surface antigen secretion in transgenic mice. J. Virol. 60:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuang, W. L., M. Omata, T. Ehata, O. Yokosuka, Y. Ito, F. Imazeki, S. N. Lu, W. Y. Chang, and M. Ohto. 1993. Precore mutations and core clustering mutations in chronic hepatitis B virus infection. Gastroenterology 104:263-271. [DOI] [PubMed] [Google Scholar]
- 9.Dyson, M. R., and K. Murray. 1995. Selection of peptide inhibitors of interactions involved in complex protein assemblies: association of the core and surface antigens of hepatitis B virus. Proc. Natl. Acad. Sci. USA 92:2194-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehata, T., M. Omata, O. Yokosuka, K. Hosoda, and M. Ohto. 1992. Variations in codons 84-101 in the core nucleotide sequence correlate with hepatocellular injury in chronic hepatitis B virus infection. J. Clin. Investig. 89:332-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehata, T., M. Omata, W. L. Chuang, O. Yokosuka, Y. Ito, K. Hosoda, and M. Ohto. 1993. Mutations in core nucleotide sequence of hepatitis B virus correlate with fulminant and severe hepatitis. J. Clin. Investig. 91:1206-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galibert, F., E. Mandart, F. Fitoussi, P. Tiollais, and P. Charnay. 1979. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature 281:646-650. [DOI] [PubMed] [Google Scholar]
- 13.Ganem, D. 1996. Hepadnaviridae and their replication, p. 2703-2737. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 14.Gotoh, K., S. Mima, T. Uchida, T. Shikata, K. Yoshizawa, M. Irie, and M. Mizui. 1995. Nucleotide sequence of hepatitis B virus isolated from subjects without serum anti-hepatitis B core antibody. J. Med. Virol. 46:201-206. [DOI] [PubMed] [Google Scholar]
- 15.Gunther, S., W. Paulij, H. Meisel, and H. Will. 1998. Analysis of hepatitis B virus population in interferon-α-treated patients reveals predominant mutations in the C-gene and changing e-antigenicity. Virology 244:146-160. [DOI] [PubMed] [Google Scholar]
- 16.Heermann, K. H., U. Goldmann, W. Schwartz, T. Seyffarth, H. Baumgarten, and W. H. Gerlich. 1984. Large surface proteins of hepatitis B virus containing the pre-S sequence. J. Virol. 52:396-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosono, S., P. C. Tai, W. Wang, M. Ambrose, D. Hwang, T. T. Yuan, B. H. Peng, C. S. Yang, C. S. Lee, and C. Shih. 1995. Core antigen mutations of human hepatitis B virus in hepatomas accumulate in MHC class II-restricted T cell epitopes. Virology 212:151-162. [DOI] [PubMed] [Google Scholar]
- 18.Hur, G. M., Y. I. Lee, D. J. Sun, J. H. Lee, and Y. I. Lee. 1996. Gradual accumulation of mutations in precore/core region of HBV in patients with chronic active hepatitis: implication of clustering changes in a small region of the HBV core region. J. Med. Virol. 48:38-46. [DOI] [PubMed] [Google Scholar]
- 19.Karasawa, T., T. Shirasawa, Y. Okawa, A. Kuramoto, N. Shimada, Y. Aizawa, M. Zeniya, and G. Toda. 1997. Association between frequency of amino acid changes in core region of hepatitis B virus (HBV) and the presence of precore mutation in Japanese HBV carriers. J. Gastroenterol. 32:611-622. [DOI] [PubMed] [Google Scholar]
- 20.Koschel, M., D. Oed, T. Gerelsaikhan, R. Thomssen, and V Bruss. 2000. Hepatitis B virus core gene mutations which block nucleocapsid envelopment. J. Virol. 74:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Pogam, S., T. T. Yuan, G. K. Sahu, S. Chatterjee, and C Shih. 2000. Low-level secretion of human hepatitis B virus virions caused by two independent, naturally occurring mutations (P5T and L60V) in the capsid protein. J. Virol. 74:9099-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and P. Gripon. 1998. Role of the pre-S2 domain of the large envelope protein in hepatitis B virus assembly and infectivity. J. Virol. 72:5573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Löffler-Mary, H., J. Dumortier, C. Klentsch-Zimmer, and R. Prange. 2000. Hepatitis B virus assembly is sensitive to changes in the cytosolic S loop of the envelope proteins. Virology 270:358-367. [DOI] [PubMed] [Google Scholar]
- 24.Minami, M., K. Poussin, M. Kew, T. Okanoue, C. Brechot, and P. Paterlini. 1996. Precore/core mutations of hepatitis B virus in hepatocellular carcinomas developed on noncirrhotic livers. Gastroenterology 111:691-700. [DOI] [PubMed] [Google Scholar]
- 25.Miska, S., S. Gunther, M. Vassilev, H. Meisel, G. Pape, and H. Will. 1993. Heterogeneity of hepatitis B virus C-gene sequences: implications for amplification and sequencing. J. Hepatol. 18:53-61. [DOI] [PubMed] [Google Scholar]
- 26.Ou, J. H., and W. J. Rutter. 1987. Regulation of secretion of the hepatitis B virus major surface antigen by the preS-1 protein. J. Virol. 61:782-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Persing, D. H., H. E. Varmus, and D. Ganem. 1986. Inhibition of secretion of hepatitis B surface antigen by a related presurface polypeptide. Science 234:1388-1391. [DOI] [PubMed] [Google Scholar]
- 28.Peterson, D. L., J. M. Roberts, and G. N. Vyas. 1977. Partial amino acid sequence of two major component polypeptides of hepatitis B surface antigen. Proc. Natl. Acad. Sci. USA 74:1530-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poisson, F., A. Severac, C. Hourioux, A. Goudeau, and P. Roingeard. 1997. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology 228:115-120. [DOI] [PubMed] [Google Scholar]
- 30.Prange, R., and R. E. Streeck. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pugh, J., A. Zweidler, and J. Summers. 1989. Characterization of the major duck hepatitis B virus core particle protein. J. Virol. 63:1371-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radziwill, G., W. Tucker, and H. Schaller. 1990. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J. Virol. 64:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez-Frias, F., M. Buti, R. Jardi, M. Cotrina, L. Viladomiu, R. Esteban, and J. Guardia. 1995. Hepatitis B virus infection: precore mutants and its relation to viral genotypes and core mutations. Hepatology 22:1641-1647. [DOI] [PubMed] [Google Scholar]
- 34.Sterneck, M., S. Gunther, T. Santantonio, L. Fischer, C. E. Broelsch, H. Greten, and H. Will. 1996. Hepatitis B virus genomes of patients with fulminant hepatitis do not share a specific core mutation. Hepatology 24:300-306. [DOI] [PubMed] [Google Scholar]
- 35.Sterneck, M., S. Gunther, J. Gerlach, N. V. Naoumov, T. Santantonio, L. Fischer, X. Rogiers, H. Greten, R. Williams, and H. Will. 1997. Hepatitis B virus sequence changes evolving in liver transplant recipients with fulminant hepatitis. J. Hepatol. 26:754-764. [DOI] [PubMed] [Google Scholar]
- 36.Stibbe, W., and W. H. Gerlich. 1983. Structural relationships between minor and major proteins of hepatitis B surface antigen. J. Virol. 46:626-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis-B like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 38.Tai, P. C., F. M. Suk, W. H. Gerlich, A. R. Neurath, and C. Shih. 2002. Hypermodification of an internally deleted middle-protein (M) of frequent and predominant hepatitis B variants. Virology 292:44-58. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi, K., Y. Akahane, K. Hino, Y. Ohata, and S. Mishiro. 1998. Hepatitis B virus genomic sequence in the circulation of hepatocellular carcinoma patients: comparative analysis of 40 full-length isolates. Arch. Virol. 143:2313-2326. [DOI] [PubMed] [Google Scholar]
- 40.Tsubota, A., H. Kumada, K. Takaki, K. Chayama, M. Kobayashi, M. Kobayashi, Y. Suzuki, S. Saitoh, Y. Arase, N. Murashima, and K. Ikeda. 1998. Deletions on the hepatitis B virus core gene may influence the clinical outcome in hepatitis B e antigen-positive asymptomatic healthy carriers. J. Med. Virol. 56:287-293. [DOI] [PubMed] [Google Scholar]
- 41.Uchida, T., T. T. Aye, S. O. Becher, M. Hirashima, T. Shikata, F. Komine, M. Moriyama, Y. Arakawa, S. Takase, and S. Mima. 1993. Detection of precore/core-mutant hepatitis B virus genome in patients with acute or fulminant hepatitis without serological markers for recent HBV infection. J. Hepatol. 18:369-372. [DOI] [PubMed] [Google Scholar]
- 42.Uchida, T., T. T. Aye, T. Shihata, M. Yano, H. Yatsuhashi, M. Koga, and S. Mima. 1994. Evolution of the hepatitis B virus gene during chronic infection in seven patients. J. Med. Virol. 43:148-154. [DOI] [PubMed] [Google Scholar]
- 43.Ueda, K., T. Tsurimoto, and K. Matsubara. 1991. Three envelope proteins of hepatitis B virus; large S, middle S, and major S proteins needed for the formation of Dane particles. J. Virol. 65:3521-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wakita, T., S. Kakumu, M. Shibata, K. Yoshioka, Y. Ito, T. Shinagawa, T. Ishikawa, M. Takayanagi, and T. Morishima. 1991. Detection of pre-c and core region mutants of hepatitis B virus in chronic hepatitis B virus carriers. J. Clin. Investig. 88:1793-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan, T. T., G. K. Sahu, W. E. Whitehead, R. Greenberg, and C. Shih. 1999. The mechanism of an immature secretion phenotype of a highly frequent naturally occurring missense mutation at codon 97 of human hepatitis B virus core antigen. J. Virol. 73:5731-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan, T. T., P. C. Tai, and C. Shih. 1999. Subtype-independent immature secretion and subtype-dependent replication deficiency of a highly frequent, naturally occurring mutation of human hepatitis B virus core antigen. J. Virol. 73:10122-10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan, T. T., and C. Shih. 2000. A frequent, naturally occurring mutation (P130T) of human hepatitis B virus core antigen is compensatory for immature secretion phenotype of another frequent variant (I97L). J. Virol. 74:4929-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]