Abstract
The complete genome of bovine herpesvirus 1 (BoHV-1) strain V155 has been cloned as a bacterial artificial chromosome (BAC). Following electroporation into Escherichia coli strain DH10B, the BoHV-1 BAC was stably propagated over multiple generations of its host. BAC DNA recovered from DH10B cells and transfected into bovine cells produced a cytopathic effect which was indistinguishable from that of the parent virus. Analysis of the replication kinetics of the viral progeny indicated that insertion of the BAC vector into the thymidine kinase gene did not affect viral replication. Specific manipulation of the BAC was demonstrated by deleting the gene encoding glycoprotein E by homologous recombination in DH10B cells facilitated by GET recombination. These studies illustrate that the propagation and manipulation of herpesviruses in bacterial systems will allow for rapid and accurate characterization of BoHV-1 genes. In turn, this will allow for the full utilization of BoHV-1 as a vaccine vector.
Bovine herpesvirus 1 (BoHV-1) is a member of the family Herpesviridae and has been placed within the subfamily Alphaherpesvirinae. The herpesviruses are characterized by large double-stranded DNA genomes ranging in size from 80 to 250 kbp (21). The genome of the Cooper strain of BoHV-1 has been completely sequenced. The genome is 135,301 bp in length and contains an estimated 73 genes (GenBank accession number AJ004801).
BoHV-1 is a major pathogen of cattle throughout the world. BoHV-1 is responsible for severe respiratory, reproductive, neonatal, and dermal disease in cattle. One of the most important clinical syndromes associated with BoHV-1 infection is the multifactorial disease bovine respiratory disease (BRD). BRD-related economic losses to the cattle industries of North America have been estimated at $1 billion (U.S.) (32). It has been demonstrated that BoHV-1 vaccination can prevent the development of BRD.
On the basis of clinical signs and restriction endonuclease profiles of genomic DNA, BoHV-1 has been divided into three subtypes: 1.1, 1.2a, and 1.2b (6, 14). Subtypes 1.1 and 1.2a have been associated with severe disease and with infections of the fetus resulting in abortion. Subtype 1.2b does not cause abortion and is the only subtype found in Australia (25).
To date there has been no genetic information to support the subtyping of BoHV-1. Rijsewijk et al. (19) reported two monoclonal antibodies which reacted only with glycoprotein C (gC) from type 1.1 viruses. The importance of antigenic differences in gC to the BoHV-1-related clinical syndromes has not been determined. Earlier, Nadin-Davis et al. (15) reported sequence variations at the 3′ end of glycoprotein B gene, as subtype 1.1 viruses had fewer G/C repeats than did type 1.2 viruses in this region.
There are many reports in the literature describing the development of recombinant BoHV-1 (rBoHV-1) as a vaccine vector (2, 33). All of these reports have utilized conventional techniques based on eukaryotic homologous recombination, and while these techniques have been successful generally, only poor efficiencies have been achieved. These approaches have utilized homologous recombination of BoHV-1 genes in bovine cells for reduced virulence, such as deletion of the gene encoding thymidine kinase (TK) (11, 26), or for the construction of marker vaccines through deletion of antigenic markers (10, 30). Homologous recombination has also been used to insert gene expression cassettes into the genome of BoHV-1 to deliver antigens from other viruses and, more recently, immunomodulating molecules (12, 17, 18, 22, 29). None of these approaches has fully utilized the potential of rBoHV-1 as a vaccine vector. The large size of the BoHV-1 genome should enable the delivery of multiple genes in a single rBoHV-1; however, current technologies do not facilitate the efficient construction of these viruses.
Recently there have been a number of reports describing the propagation of infectious herpesvirus genomes in bacteria as bacterial artificial chromosomes (BACs). The BAC is stably propagated in the bacterial host, and subsequent transfection of a BAC DNA into virus-permissive cells results in the reconstitution of infectious virus. To date, the genomes of murine cytomegalovirus (13), human cytomegalovirus (3), herpes simplex virus type 1 (9, 28), pseudorabies virus (24), Epstein-Barr virus (5), and Marek's disease virus serotype 1 (MDV-1) (23) have been successfully propagated as infectious BAC clones. The manipulation of herpesvirus genomes is more efficient when using the BAC technology, as it is no longer dependent on either the growth of viruses in eukaryotic cells or homologous recombination in eukaryotic cells. Further, any manipulations made can be fully characterized prior to the reconstitution of infectious virus, and as a result the reconstituted virus requires no further purification. Finally, there is also the advantage of making genetic changes that inhibit viral replication. Previously, these types of modifications which resulted in either partial or complete inhibition of viral replication could not be characterized, as the virus could not be isolated or grown in culture without complementing cell lines.
Due to the large size of the BACs, it is not possible to manipulate these clones with standard cloning techniques. However, increased utilization of BACs to generate accurate disease models in animal transgenics has resulted in the development of methods for efficient and specific BAC manipulation. One such mutagenesis system is the GET system, which utilizes transient expression of recE and recT to enable homologous recombination in the commonly used laboratory Escherichia coli strains. Recently the GET system has been modified to allow for BAC modification in the BAC host E. coli strain DH10B (16). GET recombination has also proven suitable for the manipulation of a BAC which contains the MDV-1 genome (23).
To enable the efficient evaluation of BoHV-1 as a vaccine vector, a BAC which contains the complete genome of BoHV-1 strain V155 has been constructed. Infectious BoHV-1 virions were recovered following transfection of this BAC into bovine cells. Further, the BAC was stable after multiple generations in the bacterial host and passage in bovine cells. The GET recombination system was also utilized to demonstrate that the infectious BoHV-1 BAC could be manipulated via homologous recombination in the bacterial host. The advantages of the BAC approach for construction of recombinant herpesviruses were demonstrated by deleting the gene for gE. Previous attempts to isolate a gE gene-negative mutant of the BoHV-1 strain V155 using conventional approaches were not successful. However, by using the BoHV-1 BAC and GET recombination the gE gene was successfully deleted. These studies confirm the advantages of using BAC technology for the development of herpesvirus-vectored vaccines. The utilization of BAC technology to help define the genetic elements responsible for the different clinical signs observed between proposed BoHV-1 subtypes is also discussed.
MATERIALS AND METHODS
Virus and cells.
BoHV-1 strain V155 (27) was propagated in CRIB-1 cells (ATCC CRL-11883), a pestivirus-resistant derivative of Madin-Darby bovine kidney cells (7). The CRIB-1 cells were maintained in Hanks' minimal essential medium containing antibiotics and antimycotics, nonessential amino acids, glutaMAX, 25 mM HEPES, and 5% donor calf serum at 37°C. All reagents utilized for cell and virus propagation were obtained from Invitrogen Australia unless otherwise stated.
Transfection of CRIB-1 cells.
CRIB-1 cells were seeded in six-well plates (Corning) at 5 × 105 cells/well 24 h prior to transfection and incubated at 37°C in an atmosphere of 5% CO2. The CRIB-1 monolayers were transfected with Lipofectamine (Invitrogen Australia) as described below. For each transfection, 1 to 2 μg of DNA was diluted to 100 μl in OptiMEM and mixed with 8 μl of Lipofectamine diluted to 100 μl in OptiMEM. This reaction mixture was incubated at room temperature for 45 min to allow lipid-DNA complexes to form. The reaction volume was increased to 1 ml with OptiMEM, and the mixture was added to monolayers which had been washed twice with OptiMEM. The transfected monolayers were incubated at 37°C with 5% CO2 for 16 to 18 h prior to the addition of 1 ml of OptiMEM containing 10% donor calf serum. At 24 h, the transfection liquid was removed and replaced with maintenance medium. Monolayers were monitored for the development of cytopathic effect (CPE) for up to 7 days posttransfection.
Construction of BoHV-1 infectious clone BAC.
Unless otherwise stated, all enzymes and reagents used for DNA cloning were obtained from Roche Molecular Biosystems.
BoHV-1 genomic DNA was purified from viral particles as follows. CRIB-1 cells were infected with BoHV-1 strain V155 at a multiplicity of infection (MOI) of 5, and the infection was allowed to proceed to completion. The cell culture supernatant was clarified by centrifugation at 5,000 × g for 10 min. The mature BoHV-1 virions were pelleted by centrifugation at 120,000 × g for 2 h, and the supernatant was discarded. BoHV-1 genomic DNA was recovered from the pelleted virus by using the Qiagen genomic DNA extraction kit essentially as described by the manufacturer. Viral pellets were resuspended in genomic DNA extraction buffer at a ratio of 1 to 65 of the starting supernatant volume. Following elution from the column, the BoHV-1 DNA was stored in aliquots at −20°C until required.
To facilitate the insertion of transgenes into the TK gene of BoHV-1 by homologous recombination, a deletion-and-insertion vector was constructed. This vector was constructed by using two segments of the TK gene, TKleft and TKright, as recombination arms. TKleft and TKright were amplified from purified BoHV-1 genomic DNA by PCR using standard procedures except for the inclusion of 5% (vol/vol) dimethyl sulfoxide and 10% (vol/vol) glycerol in the reaction mixture. Following amplification, the TKleft PCR product was digested with KpnI and SalI, gel purified, and ligated into pBluescript SK+ (Stratagene), which had also been digested with KpnI and SalI. The TKright PCR product was cloned into the plasmid containing the TKleft product following EcoRI and SpeI digestion by similar cloning procedures. The resultant plasmid was called pTKdel. The primers used for the PCR amplification of the TK targeting regions are shown in Table 1. NsiI sites were incorporated into the Tkleft5′ and Tkright3′ primers to allow the excision of the transgene product from the pTKdel vector for recombination experiments. Four unique restriction endonuclease sites are present between the two TK crossover regions to allow the insertion of transgenes for transfer to the BoHV-1 genome. These sites are SalI, HindIII, EcoRV, and EcoRI.
TABLE 1.
Oligonucleotides used to generate the PCR products described in this study
| Primer | Primer sequencea | Product (size in bp) and plasmid |
|---|---|---|
| TKleft5′ | 5′-GT GGTACCATGCAT CTGATACCCCTTCGCCCGCTACTG-3′ | Tkleft (301); pTKdel |
| KpnINsiI | ||
| TKleft3′ | 5′-TTTGC GTCGAC CCACTCCAGCGCGTCCCAG-3′ | |
| SalI | ||
| TKright5′ | 5′-AT GAATTC GCCGCGCTCGCAGACCCCA-3′ | TKright (337); pTKdel, TK-probe |
| EcoRI | ||
| TKright3′ | 5′-GGACTAGTCATGCAT CTCTAGCGCGAACTGACG-3′ | |
| SpeI NsiI | ||
| ChloramF | 5′-TCACTGGATATACCACCGTTGA-3′ | CAPr gene (402); CAPr-probe |
| ChloramR | 5′-TCACCGTAACACGCCACATCTT-3′ | |
| gE-KanF | 5′-GGGGAACGGCGCACGCGAGAGGGTTCGAAAAGGGCATTTGGCAATGCAAC-ATTTAAAT-ccacgttgtgtctcaaaatctctgatg-3′ | gE-Kanr (1,237) |
| SwaI | ||
| gE-KanR | 5′-TCGCGCTGCTACCACGGTGTAATCTGGTGCGGCCGGGGTCCGCGCTGGCG-ATTTAAAT-cggttgatgagagctttgttgtaggtg-3′ | |
| SwaI | ||
| BHV1.3 | 5′-GGG CAT TTG GCA ATG CAA C-3′ | gE-probe (845) |
| BHV1.6 | 5′-CGT CTC GTA TAT GCG GAT G-3′ | |
| Kanrfwd | 5′-GGT ATT AGA AGA ATA TCC TGA TTC-3′ | Kanr-probe (483) |
| Kanrrev | 5′-CTC ATC GAG CAT CAA ATG AAA CT-3′ |
Restriction enzyme sites are underlined in the oligonucleotide sequence. Sequences in lower case for gE-KanF and gE-KanR are specific for the kanamycin resistance cassette of EZ::TN 〈Kan-1〉 (Epicentre Technologies).
The BAC vector pBello-BAC II (31) (kindly provided by Robert Henry, Southern Cross University, Lismore, New South Wales, Australia) was digested with HindIII and gel purified. It was ligated into pTKdel which had been digested with HindIII and dephosphorylated. Following transformation into E. coli strain XL1-Blue cells, transformants were plated on selective agar containing 12.5 μg of chloramphenicol (CAP)/ml and 100 μg of ampicillin/ml. Insertion of the BAC vector was confirmed by excision with HindIII. The TK deletion fragment (TK-BAC), containing the BAC vector flanked by TKleft and TKright, was excised from pTKdel-BAC by digestion with NsiI and gel purified.
To promote homologous recombination between the BAC-TK fragment and BoHV-1 genomic DNA, purified BoHV-1 DNA was digested with NsiI and dephosphorylated with bacterial alkaline phosphatase (Pharmacia). The BAC-TK fragment and NsiI-digested BoHV-1 genomic DNA were cotransfected into CRIB-1 cells as described above. After 18 to 24 h, the transfection mixture was removed and replaced with complete Hanks' minimal essential medium containing 2 mM N,N′-hexamethylene-bis-acetamide (ICN Pharmaceuticals) to promote viral gene transcription (28).
The resultant viral supernatants were passaged once in CRIB-1 cells. Insertion of the BAC vector into the BoHV-1 genome was confirmed by a PCR assay specific for the CAP resistance gene using the primer pair ChloramF and ChloramR (Table 1). PCR templates were prepared by incubation of 10 μl of viral supernatant with 10 μl of lysis buffer (10 mM Tris-HCl [pH 8.0] containing 0.45% [vol/vol] Triton X-100 and 0.45% [vol/vol] Tween 20) with 2 μl of 10-mg/ml proteinase K followed by incubation at 60°C for 2 h. The proteinase K was inactivated at 95°C for 15 min. PCRs were performed with 1 to 2 μl of this preparation as a template. Following PCR detection of the CAP resistance gene within the BoHV-1 genome, bulk genomic DNA was recovered from virus particles as described above.
To facilitate transformation and growth in a bacterial host, the purified BoHV-1 genomic DNA was circularized by standard ligation procedures. Aliquots of the ligation mixtures were electroporated into E. coli DH10B cells (1.5 kV, 100 Ω, 25 μF; Electroporator II; Invitrogen, San Diego, Calif.). Following electroporation, DH10B cells were recovered in 960 μl of SOC broth (Invitrogen) and incubated at 37°C for 5 to 6 h with gentle shaking. Aliquots of the electroporation mix were plated onto Luria-Bertani (LB) plates containing 12.5 μg of CAP per ml. Colonies were allowed to develop for 24 to 48 h at 37°C.
CAP-resistant colonies were inoculated into 5 ml of LB broth containing 12.5 μg of CAP per ml and grown at 37°C for 16 h. BAC DNA was recovered by a standard alkaline lysis method (1). The HindIII digestion profiles of these BAC clones were compared with the HindIII profile of BoHV-1 genomic DNA. BAC clones with HindIII profiles similar to those of genomic DNA were transfected into CRIB-1 cells as described above. The transfections were monitored daily for the development of CPE considered typical of BoHV-1.
Stable propagation of the BoHV-1 BAC clone in DH10B cells.
To determine the stability of the BAC clone in the bacterial host, DH10B cells containing a BAC carrying the BoHV-1 genome were grown in serial culture for 1, 5, 10, and 20 days. BAC DNAs were isolated as previously described and characterized by restriction endonuclease digestion and field inversion gel electrophoresis (FIGE). DNA samples from each time point were also transfected into CRIB-1 cells to confirm that the BAC DNA was still infectious.
Recombination of BoHV-1 BAC in DH10B.
Modifications of the BACs containing the complete BoHV-1 genome were performed using GET recombination in DH10B cells. GET cloning facilitates homologous recombination reactions between the BAC and linear transgenes (16). The plasmid pGETrec (kindly provided by Panos Ioannou, Murdoch Institute, Melbourne, Australia) was transformed into DH10B cells harboring pBACBHV37. DH10B cells containing both plasmids were selected on agar containing 12.5 μg of CAP per ml and 100 μg of ampicillin per ml. Electrocompetent cells of double-resistant DH10B cells were prepared using 0.2% (wt/vol) arabinose induction to enable homologous recombination as previously described (16).
To demonstrate that GET recombination could be utilized to modify the BoHV-1 genome, the gene encoding gE was targeted for deletion. A minimal kanamycin resistance cassette (Kanr) was amplified by PCR from the transposon EZ::TN<KAN-1> (Epicentre Technologies). The PCR primers gE-KanF and gE-KanR utilized included regions of 50-bp homology to the 5′ (base numbers 121671 to 121720 of AJ004801) and 3′ (base numbers 123371 to 123420 of AJ004801) ends of the gE gene and are shown in Table 1. The resultant product, gE-Kanr, was 1,300 bp in length and was recovered following agarose gel separation by using a gel extraction kit according to the manufacturer's instructions (Qiagen). Approximately 200 ng of the gel-purified PCR product was electroporated into the electrocompetent DH10B(pBACBHV37, pGETrec) as previously described. Recombinant colonies were identified by plating on LB plates containing 12.5 μg of CAP per ml and 50 μg of kanamycin per ml.
Southern blotting.
Viral genomic DNA for Southern blotting was purified as described below. Cell monolayers were infected with BoHV-1 strain V155 at an MOI of 5, and the infection was allowed to proceed until approximately 40% of the cells showed a “rounding up” type of morphology. Following removal of the growth medium, the monolayer was gently washed twice with phosphate-buffered saline at 0°C. Cell lysis buffer at 0°C (10 mM sodium phosphate, pH 7.3, containing 1% [vol/vol] Nonidet P-40) was added to each flask (4 ml per 175 cm2), and the flasks were rocked so that the lysis buffer contacted the entire monolayer. The lysis buffer was removed and placed on ice, and another 4 ml was added to each flask with gentle rocking; then it was removed and added to the initial lysis solution. The lysates were clarified at 4,300 × g for 10 min at 4°C. The supernatant was collected and centrifuged at 112,700 × g for 100 min at 5°C. The viral pellet was resuspended in 500 μl of G2 buffer (Qiagen). Proteinase K (25 μl at a concentration of 10 mg/ml) was added to the resuspended viral pellet followed by incubation at 50°C for 1 h. Genomic viral DNA was recovered by using genomic tip 20/G (Qiagen) as follows. The genomic tip 20/G was equilibrated with 1 ml of QBT buffer. The material treated with proteinase K was diluted with an equal volume of QBT buffer and loaded onto the genomic tip (660 μl per tip). Tips were washed twice with 7.5 ml of QC buffer. Viral DNA was eluted from the tip with 2 × 1.5 ml of QF buffer at 50°C. The DNA was precipitated, washed once with 75% ethanol, and resuspended in 50 μl of 10 mM Tris-HCl, pH 8.5.
Restriction enzyme-digested DNA samples were electrophoresed on a 1% gel in 0.5× Tris-borate-EDTA buffer for 13.5 h by use of a FIGE apparatus (Bio-Rad) at 5°C. The switch time ramp was 0.1 to 2 s (linear shape) with a forward voltage of 180 V and a reverse voltage of 120 V. The DNA fragments were transferred to a Hybond-N noncharged membrane (Amersham) by capillary action, and the DNA was fixed to the membrane by application of UV light (1). Probes were labeled with digoxigenin-11-dUTP (Roche Molecular Biosystems). Probes were synthesized by PCR using a reaction mix containing the following: Taq polymerase buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl [pH 8.5]); 1.25 mM each dATP, dCTP, dGTP, and dTTP; 1.25 μM DIG-II-dUTP; 1 U of Taq polymerase (Roche Molecular Biosystems); and 5 ng of template DNA in a final volume of 20 μl. PCR conditions were 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by final incubation at 72°C for 7 min. Following gel purification, PCR probes were hybridized to membranes for 12 to 16 h at 55°C (DIG System User's Guide for Filter Hybridization; Roche Molecular Biosystems). Hybridizations were carried out in rotating bottles in a hybridization oven (Hybaid). Membranes were washed in dishes on a shaking platform at room temperature.
Replication kinetics.
The replication kinetics of the various rBoHV-1 strains were determined by standard virologic techniques. Briefly, one 50% tissue culture infective dose (TCID50) of virus was allowed to infect 105 CRIB-1 cells plated in 24-well plates for 90 min at 37°C in a 5% CO2 atmosphere. Any extracellular virus was then inactivated by addition of sodium citrate solution (40 mM sodium citrate, 10 mM KCl, 135 mM NaCl [pH 3.0]), the cell layers were then washed twice with phosphate-buffered saline, 1 ml of maintenance medium was added, and the mixture was incubated at 37°C in a 5% CO2 atmosphere. Viral supernatants and cell pellets were collected at 2, 4, 6, 12, 24, 48, and 72 h postinfection (p.i.) and frozen at −70°C until required. The TCID50 of each supernatant from each time point was then determined in triplicate. Following one freeze-thaw cycle, the TCID50 of intracellular virus was also determined for each time point in triplicate.
RESULTS
Construction and analysis of a BAC containing the BoHV-1 genome.
To promote the insertion of foreign DNA into the BoHV-1 genome, we have used a method called restriction enzyme-mediated recombination (REMR), which will be reported in detail elsewhere. Briefly, REMR utilizes a unique NsiI restriction site (nucleotide position 63980 of the Cooper strain genomic sequence) to promote homologous recombination between BoHV-1 DNA and transgene DNA molecules. The NsiI site is located within the TK gene, which has previously been shown to be nonessential for in vitro and in vivo growth of the BoHV-1 strain V155 used in this study (26). Following NsiI digestion and dephosphorylation, if the BoHV-1 DNA is transfected into CRIB-1 cells, the two genomic segments do not produce infectious BoHV-1. To exploit this, a transfer vector, pTK-del, which contains the BoHV-1 genomic sequences immediately upstream and downstream of the NsiI site was constructed (Fig. 1). Following excision, the insert of pTK-del was cotransfected into CRIB-1 cells with NsiI-dephosphorylated genomic DNA; homologous recombination between the three molecules facilitated the reconstitution of infectious BoHV-1 (data not shown). After cloning into pTK-del the BAC vector was excised by NsiI digestion and gel purified (Fig. 1). The BAC vector was transfected into CRIB-1 cells along with NsiI-digested and -dephosphorylated BoHV-1 genomic DNA. Typically, between 5 and 7 days following transfection, a CPE considered typical of BoHV-1 was evident. Following one passage of the resultant virus, PCR amplification of the CAP resistance gene from pBelloBacII confirmed that the BAC vector had been inserted into the BoHV-1 genome (data not shown).
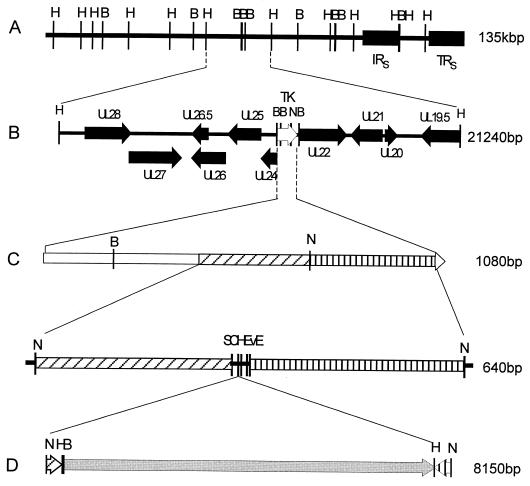
FIG. 1.
Schematic representation of the construction of an infectious clone for BoHV-1. (A) Combined HindIII and BamHI restriction map of the bovine herpesvirus genome (135,307 bp; GenBank accession number AJ004801) derived from the complete genome sequence of the BoHV-1 Cooper strain. The internal repeat sequence (IRS) and terminal repeat sequence (TRS) are illustrated. (B) BoHV-1 HindIII genomic fragment containing the TK gene (white arrow). Open reading frames (black arrows) surrounding the TK gene are illustrated. The unique NsiI (N) site of the BoHV-1 genome used to promote homologous recombination is illustrated. (C) TK gene of BoHV-1. TK gene-specific crossover regions immediately upstream (301 bp, forward hatch) and downstream (337 bp, vertical hatch) of the unique NsiI site were amplified by PCR and cloned to the recombination transfer, vector pTKdel. The transfer vector contains five unique restriction enzyme sites (including ClaI and EcoRV) for insertion of transgenes to be transferred to the BoHV-1 genome. The completed transgenes can be excised from the transfer vector by NsiI digestion or amplified by PCR. (D) The BAC vector, pBelloBACII, was cloned into the HindIII site of pTKdel. The resultant transgene was excised with NsiI prior to transfer into the BoHV-1 genome by homologous recombination. Abbreviations for the illustrated restriction enzyme sites: B, BamHI; E, EcoRI; H, HindIII; N, NsiI; S, SalI.
To facilitate propagation in E. coli strain DH10B, viral genomic DNA was purified from virions and circularized by standard ligation protocols. This ligation process utilizes single-base 3′ overhangs on the termini of the virus genome which are required for virus circularization prior to replication in vivo (8). The circularized genomes were electroporated into DH10B cells and plated onto CAP-selective agar plates. CAP-resistant colonies were screened by a gE-specific PCR, which identified six clones as putatively positive BAC clones (data not shown). Plasmid DNAs were prepared from overnight cultures of the six colonies. The HindIII digestion profiles of these six BAC clones identified two of them, pBACBHV27 and pBACBHV37, with profiles that could not be distinguished from that of native BoHV-1 DNA. Plasmid DNA from both of these clones was transfected into CRIB-1 cells. A CPE considered typical of BoHV-1 was observed in the cells transfected with both of these clones after 4 days (data not shown). Neither the plaque size nor the morphology produced by virus derived from the BAC clones differed from those of the wild-type virus. All subsequent experiments were conducted with the BAC clone pBACBHV37.
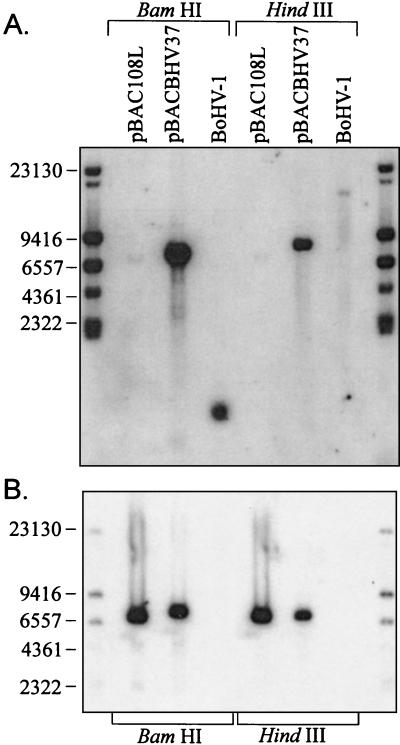
The insertion of the BAC vector into the BoHV-1 genome was demonstrated by Southern blotting (Fig. 2). The CAP-specific PCR product was digoxigenin (DIG) labeled and hybridized to pBACBHV37 DNA and BoHV-1 genomic DNA which had been digested with HindIII and BamHI. Figure 2B illustrates specific hybridization of the CAP probe to the positive control BAC vector, pBac108L, which also carries the CAP resistance gene. The CAP probe also binds specifically to a 7.5-kbp band in the HindIII-digested pBACBHV37 DNA which corresponds to the excised pBeloBACII vector (Fig. 2B). The hybridizing fragment in the BamHI digest is slightly larger than the HindIII fragment, as the BAC vector is inserted into a 969-bp BamHI genomic fragment. As the vector contains a BamHI site, digestion with this enzyme releases the BAC vector plus 500 bp of BoHV-1 genomic DNA resulting in the hybridization pattern observed. As expected, there was no hybridization of the CAP probe to DNA isolated from wild-type BoHV-1 virions (Fig. 2B). The slight variation in the sizes of the hybridized bands is due to the difference in size of the control vector pBac108L (6,812 bp) and the BAC vector inserted into the BoHV-1 genome pBelloBacII (7,507 bp).
FIG. 2.
Southern blotting analysis of the clone pBACBHV37 illustrating the insertion of the BAC vector, pBelloBACII, into the genome of the BoHV-1 strain V155. DNA was digested with either BamHI or HindIII and separated by FIGE in a 1% agarose gel. DNA fragments were transferred onto nylon membranes by Southern blotting. Duplicate membranes were hybridized with DIG-labeled probes specific for either TK of BoHV-1 or the CAP resistance gene (CAPr) of the BAC vector. (A) Southern blot hybridized with a DIG-labeled probe specific for the TK gene of BoHV-1. (B) Southern blot hybridized with a DIG-labeled probe specific for the CAPr gene. Standard molecular sizes (in base pairs) are given.
The specific insertion of the BAC vector into the NsiI site was demonstrated by stripping the membrane and reprobing it with a TK gene-specific probe (TKright) (Table 1). The TK probe did not bind to the BAC vector DNA on the membrane (Fig. 2A). The TK gene is located on a 21.2-kbp HindIII genomic fragment (Fig. 1B). This fragment is evident in the HindIII-digested BoHV-1 DNA (Fig. 2A). The relatively low intensity of this band is probably due to inefficient transfer of such a high-molecular-weight fragment. As expected, in BoHV-1 DNA digested with BamHI, the TK probe bound specifically to a 1,000-bp band (Fig. 1 and 2A). Insertion of the BAC vector (7,507 bp) into the 21.2-kbp HindIII genomic fragment at the NsiI site increases the size to 28.2 kbp and introduces two HindIII sites and one BamHI site (Fig. 1). As a result, the TK-specific band detected by the probe increased in size to 9 kbp, due to the incorporation of the BAC vector in BamHI-digested pBACBHV37 DNA (Fig. 1 and 2A). In HindIII-digested pBACBHV37 DNA, the TK-specific band decreases to approximately 9 kbp due to the HindIII sites introduced during insertion of the BAC vector (Fig. 1 and 2A).
Stable propagation of the BoHV-1 genome as a BAC.
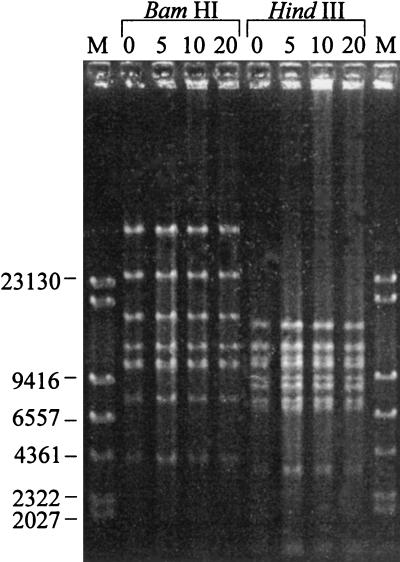
The stable propagation of pBACBHV37 in E. coli DH10B was demonstrated by serial culture for 5, 10, and 20 days. As E. coli has a generation time of approximately 20 min, and given that the stationary growth phase is reached at 12 h, each day represents about 36 generations. Following culture, the restriction endonuclease pattern of BAC DNA purified from cells at each time point was examined by FIGE. The restriction endonuclease patterns following HindIII and BamHI digestion of the purified DNA are illustrated in Fig. 3. There were no distinguishable differences in the profiles of the infectious clone pBACBHV37 after multiple passages in the bacterial host (Fig. 3). Further, BAC DNA isolated at each time point was infectious following transfection into CRIB-1 cells. The recovered virus produced a CPE which was typical of BoHV-1 and pBACBHV37 (data not shown).
FIG. 3.
Stability of the BAC pBACBHV37 over multiple generations in E. coli DH10B. DH10B cells containing pBACBHV37 were serially grown for 5, 10, and 20 days. BAC DNA was prepared from these cultures, and the restriction enzyme profiles were compared by FIGE with 1% agarose. DNA fragments were stained with ethidium bromide and photographed. Standard molecular sizes (in base pairs) are given.
Replication kinetics of BoHV-1 rescued from a BAC.
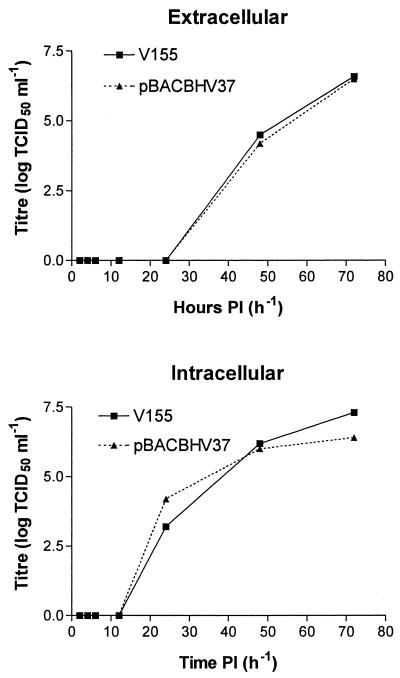
Replication kinetics of the infectious clone was compared to that of wild-type BoHV-1 to determine if incorporating the BAC vector into the BoHV-1 genome has any effect on viral replication in vitro, as the insertion of the vector essentially deletes TK activity. CRIB-1 cells were infected with BoHV-1 and with pBACBHV37-derived virus at equivalent MOIs. The titers of viral supernatant were determined at 2, 4, 6, 12, 24, 48, and 72 h p.i. The results of these titrations are illustrated in Fig. 4. On the basis of these titrations, the insertion of the BAC vector into the BoHV-1 genome did not have any effect on the replication kinetics of BoHV-1. There was no difference in the amount of virus recovered following infection with either pBACBHV37 or BoHV-1 (Fig. 4).
FIG. 4.
Replication kinetics of infectious BoHV-1 rescued from pBACBHV37 compared to wild-type BoHV-1 strain V155. Following recovery of infectious BoHV-1 from pBACBHV37, DNA and viral stock was amplified and titrated. CRIB-1 monolayers were infected with one TCID50 of either the BoHV-1 strain V155 or pBACBHV37. The culture supernatants of each infection were recovered at 2, 4, 6, 12, 24, 48, and 72 h p.i. (PI), and the TCID50 was determined for each. The titrations were done in triplicate, and averaged titers are illustrated. TCID50s were determined for both extracellular and intracellular viruses.
Specific recombination of the BoHV-1 infectious clone.
The efficient and specific modification of BAC in DH10B cells has recently become possible through the development of GET cloning (16). To determine if this procedure was adaptable to the plasmid pBACBHV37, the gene encoding gE was targeted for deletion. A kanamycin resistance cassette (Kanr) was amplified by PCR. To facilitate the specific deletion of the gE gene, 50-bp sequences of homology to the gE gene 5′ and 3′ regions were incorporated into the Kanr PCR product. The purified Kanr PCR product was electroporated into DH10B cells containing pBACBHV37 and pGETrec, which had been induced with arabinose.
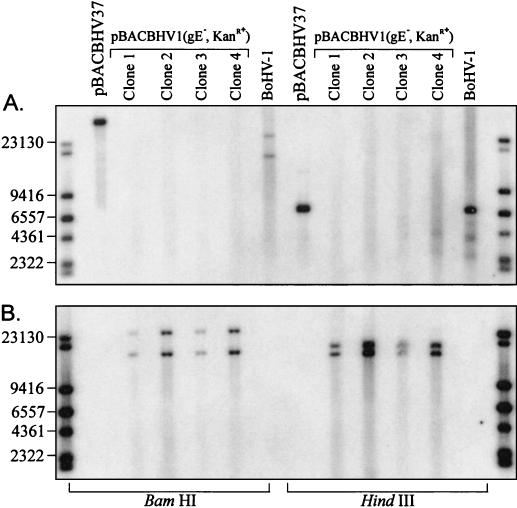
Following electroporation, the cells were allowed to recover and were plated onto CAP- and kanamycin-selective agar plates. BAC DNA was recovered from six of the double-resistant colonies and transfected into CRIB-1 cells. Five of the six colonies transfected produced a CPE considered typical of BoHV-1. The restriction enzyme profiles of these clones were examined following HindIII and BamHI digestion and FIGE. To determine if Kanr had replaced the gE gene, Southern blotting was performed. The membrane was first probed with a gE gene-specific probe. It was evident that the fragment which normally contains the gE gene had been significantly altered (Fig. 5A). It was apparent that the gE gene had been deleted via GET recombination. Following removal of the gE probe, the blot was probed with a Kanr-specific probe as illustrated in Fig. 5B. The Kanr probe did not bind to any fragments in either BoHV-1 DNA or pBACBHV37. However, following GET-mediated recombination, the Kanr probe specifically bound to two DNA fragments in both the BamHI and HindIII digests (Fig. 5). The DNA utilized for the construction of this Southern blot was isolated from virus-infected cells. The gE gene is located within the unique short (US) region of the BoHV-1 genome, which is flanked by the internal and terminal genomic repeat sequences. As the US region may switch orientations during virus replication, the relative locations of the restriction enzyme sites change and hence the digestion pattern is also altered, giving rise to two genomic isomers which harbor the gE gene on fragments of different sizes.
FIG. 5.
Southern blot analysis of four BoHV-1 infectious BAC clones following the replacement of the gene coding for glycoprotein E (gE) with a kanamycin resistance gene (Kanr). Viral genomic DNA was isolated from cells infected with modified virus. DNA was digested with either BamHI or HindIII, resolved on a 1% agarose gel by FIGE, and transferred onto nylon membranes. Blots were hybridized with a DIG-labeled probe specific for either gE or Kanr. Standard molecular sizes (in base pairs) are given. (A) Southern blot hybridized with a probe specific for the gE gene. (B) Southern blot hybridized with a probe specific for the Kanr gene.
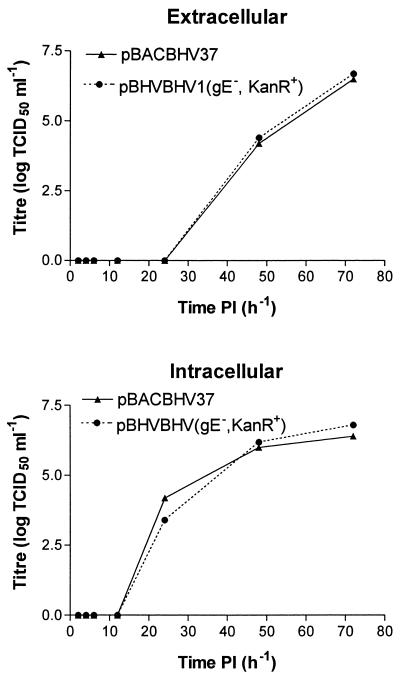
To determine if the deletion of the gE gene from pBACBHV37 has any effect on the replication, the kinetics of virus rescued from pBACBHV1 (gE− Kanr+) was compared to that of virus rescued from pBACBHV37. Following infection at the same MOI, samples of the supernatants were collected at 2, 4, 6, 12, 24, 48, and 72 h p.i. and the viral titers were determined. The results of these titrations are illustrated in Fig. 6. On the basis of these titrations, there was no difference in the amounts of virus recovered following transfection of pBACBHV37 or pBACBHV1 (gE− Kanr+) after in vitro recovery of virus. The replication kinetics of the negative viruses were determined by using TCID50 calculations, as the rBoHV-1 strains negative for the gE gene did not produce visible plaques under agarose overlays.
FIG. 6.
Comparison of the replication kinetics of BoHV-1 virus rescued from pBACBHV37 with that of rescued virus following the deletion of the gE gene from the BAC clone. Following recovery of infectious BoHV-1 from pBACBHV37 or pBACBHV1 (gE−, KanR+), the viral stock was amplified and titrated. CRIB-1 monolayers were infected with one TCID50 of virus from either pBACBHV37 or pBACBHV1 (gE−, KanR+). The viral supernatants were sampled at 2, 4, 6, 12, 24, 48, and 72 h p.i. (PI), and the TCID50 was determined for each. These samples were titrated. The titrations were done in triplicate, and the averaged titers are illustrated. TCID50s were determined for both extracellular and intracellular viruses.
DISCUSSION
Here we have described the construction of the first infectious clone for BoHV-1. This infectious clone contains the complete genome of BoHV-1 strain V155. We have demonstrated that when maintained as a BAC, the BoHV-1 clone is stable through multiple generations of the host strain E. coli DH10B. The stability was evaluated by restriction enzyme analysis and also through the reconstitution of infectious BoHV-1 following transfection into bovine cells.
Further, the recently described GET mutagenesis system (16) has been successfully used to delete the gene coding for gE. While the gE gene has been deleted from other strains of BoHV-1 (20, 30), this is the first report describing the deletion of gE from the 1.2b subtype. Previous attempts in our laboratory using established homologous recombination protocols failed to identify any gE-negative mutants despite extensive screening (unpublished data). As a result, we had suspected that gE is an essential gene in BoHV-1 subtype 1.2b, which represents a significant divergence from conventional paradigms of herpesvirus biology.
In the study described here, the application of a mutagenesis system which was independent of viral replication allowed for the rapid production of a gE-negative mutant. The subsequent transfection into bovine cells confirmed that gE is a nonessential gene in the strain of BoHV-1 used. Experiments with BoHV-1 gE-negative mutants recovered from the mutated infectious clone demonstrated that the virus did not form visible plaques under agarose overlays. DNA sequence analysis across the junctions of recombination indicated that the recombination was specific and had not altered any of the control elements for the genes either upstream (glycoprotein I) or downstream (tegument protein) from gE (data not shown), indicating that the lack of plaque formation observed was due to deletion of gE. Our initial experiments using conventional homologous recombination techniques aimed to replace the gE gene with a cassette expressing green fluorescent protein with mutants to be identified by visualization of rBoHV-1 through fluorescent plaques. As the BAC-derived gE-negative mutant was unable to form plaques, this is the most likely explanation for the inability to identify any fluorescent plaques. This result was unexpected, as previously reported rBoHV-1 strains negative for gE were able to form plaques under overlays, although the plaques were smaller than those of the parent virus (20, 30). However, all of these rBoHV-1 were constructed from subtype 1.1 viruses, not the subtype 1.2 virus used in this study. The as-yet-undefined genetic differences between the subtypes are the most likely causes for the lack of plaque formation observed.
Further application of the techniques described in this study will greatly aid the elucidation of the genetic elements responsible for the BoHV-1 subtypes. By specifically replacing or swapping genes and regulatory sequences from different subtypes, the roles that individual genes or genomic segments play in viral pathogenesis and/or biotypes could be specifically evaluated. To fulfill this goal, an infectious clone of a type 1.1 virus is currently under construction.
The example discussed above highlights the powerful approach of BAC-based methods for the manipulation and characterization of herpesvirus genomes. Schumacher et al. (23) have also demonstrated the power of these approaches through manipulation of MDV-1, which had previously proven very difficult to modify by conventional approaches. These studies also illustrated how BAC manipulation can be used to delete essential genes, which is not possible through conventional approaches without complementing cell lines or helper viruses.
An important and attractive feature of BAC-based manipulation of the BoHV-1 genome is the ease with which gene-targeting constructs can be generated. It has been demonstrated here and by Schumacher et al. (23) that successful recombination can occur with crossover regions of only 50 bp. Consequently, the crossover regions can be incorporated into PCR primers to allow rapid and simple generation of insertion and deletion constructs for any region of the genome for which the sequence is known. With an estimated 80 genes and a completed genome sequence available for BoHV-1, it is now possible to construct a full library of gene deletions for analysis.
Since gene deletions can now be constructed in a replication-independent environment, those genes which are essential for replication and those that severely limit replication can be differentiated, whereas conventional technologies do not permit this. The application of BAC technologies to herpesvirus biology (reviewed in reference 4) will enable a more complete understanding of these complex DNA viruses.
In conclusion, the complete genome of BoHV-1 has been cloned and can be stably propagated in E. coli strain DH10B as a BAC. In addition, the recently described GET recombination has been applied to demonstrate that specific changes can be made to the BoHV-1 genome in DH10B cells while maintaining the infectivity of the clone. Through application of this technology, significant advances in determining the roles that virus-encoded factors play in the different diseases involving BoHV-1 will be possible. Further, by simplifying the construction of recombinant BoHV-1, the more efficient utilization of BoHV-1 as a vaccine vector is now possible. The application of these technologies to BoHV-1 and other herpesviruses should lead to the development of safer and more efficient vaccines and potentially more-useful gene therapy vectors.
Acknowledgments
We are indebted to Donna Mahony for construction of figures and critical reading of the manuscript and to Panos Ioannou, Murdoch Institute, Melbourne, Australia, for providing the plasmid pGETrec.
This work was supported by grant Flot.203 from Meat and Livestock Australia.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmer, J. A. Smith, and K. Struhl. 2001. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 2.Bello, L. J., J. C. Whitbeck, and W. C. Lawrence. 1992. Bovine herpesvirus 1 as a live virus vector for expression of foreign genes. Virology 190:666-673. [DOI] [PubMed] [Google Scholar]
- 3.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brune, W., M. Messerle, and U. H. Koszinowski. 2000. Forward with BACs: new tools for herpesvirus genomics. Trends Genet. 16:254-259. [DOI] [PubMed] [Google Scholar]
- 5.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engels, M., F. Steck, and R. Wyler. 1981. Comparison of the genomes of infectious bovine rhinotracheitis and infectious pustular vulvovaginitis virus strains by restriction endonuclease analysis. Arch. Virol. 67:169-174. [DOI] [PubMed] [Google Scholar]
- 7.Flores, E. F., and R. O. Donis. 1995. Isolation of a mutant CRIB-1 cell line resistant to bovine viral diarrhea virus infection due to block of viral entry. Virology 208:565-575. [DOI] [PubMed] [Google Scholar]
- 8.Hammerschmidt, W., H. Ludwig, and H. J. Buhk. 1988. Specificity of cleavage in replicative-form DNA of bovine herpesvirus 1. J. Virol. 62:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horsburgh, B. C., M. M. Hubinette, and F. Tufaro. 1999. Genetic manipulation of herpes simplex virus using bacterial artificial chromosomes. Methods Enzymol. 306:337-352. [DOI] [PubMed] [Google Scholar]
- 10.Kaashoek, M. J., F. A. Rijsewijk, R. C. Ruuls, G. M. Keil, E. Thiry, P. P. Pastoret, and J. T. Van Oirschot. 1998. Virulence, immunogenicity and reactivation of bovine herpesvirus 1 mutants with a deletion in the gC, gG, gI, gE, or in both the gI and gE gene. Vaccine 16:802-809. [DOI] [PubMed] [Google Scholar]
- 11.Kit, S., M. Kit, and S. McConnell. 1986. Intramuscular and intravaginal vaccination of pregnant cows with thymidine kinase-negative, temperature-resistant infectious bovine rhinotracheitis virus (bovine herpesvirus 1). Vaccine 4:55-61. [DOI] [PubMed] [Google Scholar]
- 12.Kuhnle, G., A. Heinze, J. Schmitt, K. Giesow, G. Taylor, I. Morrison, F. A. Rijsewijk, J. T. van Oirschot, and G. M. Keil. 1998. The class II membrane glycoprotein G of bovine respiratory syncytial virus, expressed from a synthetic open reading frame, is incorporated into virions of recombinant bovine herpesvirus 1. J. Virol. 72:3804-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metzler, A. E., H. Matile, U. Gassmann, M. Engels, and R. Wyler. 1985. European isolates of bovine herpesvirus 1: a comparison of restriction endonuclease sites, polypeptides, and reactivity with monoclonal antibodies. Arch. Virol. 85:57-69. [DOI] [PubMed] [Google Scholar]
- 15.Nadin-Davis, S. A., C. Lutze-Wallace, and X. Zhong. 1996. Bovine herpesvirus 1 isolates contain variable copy numbers of GC-rich tandem repeats in the gI non-coding regions of their genomes. Virus Genes 13:263-268. [DOI] [PubMed] [Google Scholar]
- 16.Narayanan, K., R. Williamson, Y. Zhang, A. F. Stewart, and P. A. Ioannou. 1999. Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther. 6:442-447. [DOI] [PubMed] [Google Scholar]
- 17.Raggo, C., D. R. Fitzpatrick, L. A. Babiuk, and X. Liang. 1996. Expression of bovine interleukin-1 beta in a bovine herpesvirus-1 vector: in vitro analysis. Virology 221:78-86. [DOI] [PubMed] [Google Scholar]
- 18.Raggo, C., M. Habermehl, L. A. Babiuk, and P. Griebel. 2000. The in vivo effects of recombinant bovine herpesvirus-1 expressing bovine interferon-gamma. J. Gen. Virol. 81:2665-2673. [DOI] [PubMed] [Google Scholar]
- 19.Rijsewijk, F. A., M. J. Kaashoek, J. P. Langeveld, R. Meloen, J. Judek, K. Bienkowska-Szewczyk, M. A. Maris-Veldhuis, and J. T. van Oirschot. 1999. Epitopes on glycoprotein C of bovine herpesvirus-1 (BHV-1) that allow differentiation between BHV-1.1 and BHV-1.2 strains. J. Gen. Virol. 80:1477-1483. [DOI] [PubMed] [Google Scholar]
- 20.Rijsewijk, F. A., S. B. Verschuren, J. Madic, R. C. Ruuls, P. Renaud, and J. T. van Oirschot. 1999. Spontaneous BHV1 recombinants in which the gI/gE/US9 region is replaced by a duplication/inversion of the US1.5/US2 region. Arch. Virol. 144:1527-1537. [DOI] [PubMed] [Google Scholar]
- 21.Roizman, B. 1996. The function of herpes simplex virus genes: a primer for genetic engineering of novel vectors. Proc. Natl. Acad. Sci. USA 93:11307-11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitt, J., P. Becher, H. J. Thiel, and G. M. Keil. 1999. Expression of bovine viral diarrhoea virus glycoprotein E2 by bovine herpesvirus-1 from a synthetic ORF and incorporation of E2 into recombinant virions. J. Gen. Virol. 80:2839-2848. [DOI] [PubMed] [Google Scholar]
- 23.Schumacher, D., B. K. Tischer, W. Fuchs, and N. Osterrieder. 2000. Reconstitution of Marek's disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. J. Virol. 74:11088-11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, G. A., P. L. Young, and K. C. Reed. 1995. Emergence of a new bovine herpesvirus 1 strain in Australian feedlots. Arch. Virol. 140:599-603. [DOI] [PubMed] [Google Scholar]
- 26.Smith, G. A., P. L. Young, B. J. Rodwell, M. A. Kelly, G. J. Storie, C. A. Farrah, and J. S. Mattick. 1994. Development and trial of a bovine herpesvirus 1 thymidine kinase deletion virus as a vaccine. Aust. Vet. J. 71:65-70. [DOI] [PubMed] [Google Scholar]
- 27.Snowden, W. A. 1964. Infectious rhinotracheitis and infectious pustular vulvovaginitis in Australian cattle. Aust. Vet. J. 40:277-288. [Google Scholar]
- 28.Stavropoulos, T. A., and C. A. Strathdee. 1998. An enhanced packaging system for helper-dependent herpes simplex virus vectors. J. Virol. 72:7137-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor, G., F. A. Rijsewijk, L. H. Thomas, S. G. Wyld, R. M. Gaddum, R. S. Cook, W. I. Morrison, E. Hensen, J. T. van Oirschot, and G. Keil. 1998. Resistance to bovine respiratory syncytial virus (BRSV) induced in calves by a recombinant bovine herpesvirus-1 expressing the attachment glycoprotein of BRSV. J. Gen. Virol. 79:1759-1767. [DOI] [PubMed] [Google Scholar]
- 30.van Engelenburg, F. A., M. J. Kaashoek, F. A. Rijsewijk, L. van den Burg, A. Moerman, A. L. Gielkens, and J. T. van Oirschot. 1994. A glycoprotein E deletion mutant of bovine herpesvirus 1 is avirulent in calves. J. Gen. Virol. 75:2311-2318. [DOI] [PubMed] [Google Scholar]
- 31.Woo, S. S., J. Jiang, B. S. Gill, A. H. Paterson, and R. A. Wing. 1994. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 22:4922-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yates, W. D. G. 1982. A review of infectious bovine rhinotracheitis, shipping fever pneumonia and viral-bacterial synergism in respiratory disease of cattle. Can. J. Comp. Med. 46:225-263. [PMC free article] [PubMed] [Google Scholar]
- 33.Young, P. L., and G. A. Smith. 1995. Genetically altered herpesviruses as vaccines. Vet. Microbiol. 46:175-179. [DOI] [PubMed] [Google Scholar]