Abstract
Successful implementation of the global poliomyelitis eradication program raises the problem of vaccination against poliomyelitis in the posteradication era. One of the options under consideration envisions completely stopping worldwide the use of the Sabin vaccine. This strategy is based on the assumption that the natural circulation of attenuated strains and their derivatives is strictly limited. Here, we report the characterization of a highly evolved derivative of the Sabin vaccine strain isolated in a case of paralytic poliomyelitis from a 7-month-old immunocompetent baby in an apparently adequately immunized population. Analysis of the genome of this isolate showed that it is a double (type 1-type 2-type 1) vaccine-derived recombinant. The number of mutations accumulated in both the type 1-derived and type 2-derived portions of the recombinant genome suggests that both had diverged from their vaccine predecessors ∼2 years before the onset of the illness. This fact, along with other recent observations, points to the possibility of long-term circulation of Sabin vaccine strain derivatives associated with an increase in their neurovirulence. Comparison of genomic sequences of this and other evolved vaccine-derived isolates reveals some general features of natural poliovirus evolution. They include a very high preponderance and nonrandom distribution of synonymous substitutions, conservation of secondary structures of important cis-acting elements of the genome, and an apparently adaptive character of most of the amino acid mutations, with only a few of them occurring in the antigenic determinants. Another interesting feature is a frequent occurrence of tripartite intertypic recombinants with either type 1 or type 3 homotypic genomic ends.
The use of highly efficacious poliovirus vaccines, the oral live vaccine made from the Sabin strains (oral poliovirus vaccine [OPV]) and the inactivated Salk vaccine, has resulted in a dramatic worldwide decrease in the circulation of wild-type polioviruses (73). The success has been so great that the global eradication of wild-type poliovirus (74) seems a realistic goal for the foreseeable future (3). Development of an optimal strategy for poliovirus control in the posteradication era is a big challenge, and currently there is no consensus in this regard (20, 34, 37, 56, 73). One of the suggested options (34, 73) includes complete and possibly synchronous worldwide cessation of vaccination, in particular by OPV, at some point after the world is certified to be free of the circulating wild-type polioviruses. The basic assumption underlying this strategy is that natural circulation of derivatives of Sabin strains is strictly limited in time. Therefore, such derivatives are believed to be unable to survive in nature long enough to evolve into highly transmissible neuropathogenic variants, even though the size of the susceptible population would increase very rapidly after the discontinuation of vaccination. This assumption is indeed supported by some evidence (reviewed in references 22 and 73), but the evidence is not unequivocal. Long-term (several years) persistence of vaccine derivatives in immunocompromised persons and the ability of the evolved variants to cause paralytic disease are well-established phenomena (5, 36, 44, 77). Recent outbreaks of poliomyelitis in Egypt (11), the Dominican Republic and Haiti (13), and the Philippines (12), caused by evolved derivatives of vaccine viruses of types 1 and 2, support the notion that there is a significant risk of prolonged circulation of the vaccine viruses in populations with a low immunity level, as well as their conversion into epidemic strains. Highly evolved Sabin vaccine derivatives have also been isolated from sewage even in the absence of apparent cases of paralytic poliomyelitis (60).
Prospective and retrospective characterization of Sabin vaccine-derived polioviruses may shed additional light on this important topic and provide valuable information for making a rational decision on vaccination policies. Here we report the characterization of the genome of a type 1 highly evolved vaccine-derived poliovirus isolated from a patient with paralytic disease. The data suggest that long-term survival and evolution of Sabin 1 poliovirus causing paralytic disease may occur even in apparently well-immunized populations. These data also provide some insights into the patterns of natural evolution of poliovirus.
MATERIALS AND METHODS
Virus isolation and characterization.
Virus isolation from stool samples was done in RD cells (human rhabdomyosarcoma cell line) by the standard methods (75). The viruses were typed in microneutralization tests with type-specific sera (75), and intratypic differentiation was performed with cross-absorbed polyclonal antisera (71).
Characterization of poliovirus RNA.
RNA was extracted from cell lysates with Trizol reagent (Life Technologies) and was reverse transcribed using random hexamer primers (Boehringer Ingelheim) with avian myeloblastosis virus reverse transcriptase (Promega) at 42°C for 1 h. PCR with Sabin strain-specific primers was performed as described previously (76). PCR-restriction fragment length polymorphism (RFLP) analysis was done as proposed by Balanant et al. (4) and Georgescu et al. (29). In short, DNA copies of segments of the VP1 (nucleotides [nt] 2402 to 2881; numbering of the Sabin 1 RNA), 2AB (nt 3617 to 4152), 2C (nt 4169 to 4965), and 3D (nt 6086 to 6376) genomic regions were amplified by PCR with appropriate primers; the samples were separately treated with restriction endonucleases HaeIII, HinfI, HpaII, DdeI, and RsaI; and fragments were analyzed by 3% agarose gel electrophoresis at 5 V/cm for 2 h and visualized by ethidium bromide staining. For determination of complete genome sequences, DNA copies of 11 genomic fragments were amplified by PCR (57, 64), and the PCR products were purified with the QIAquick system (Qiagen) and sequenced using an ABI Prism 310 Genetic Analyzer (Applied Biosystems) as specified by the manufacturer. Nucleotides at each position of the genome were determined at least twice by reading both complementary strands.
Comparative analysis of nucleotide sequences and proteins.
For comparative analysis, we used the genome sequences of Sabin 1 and Sabin 2 strains as determined by Toyoda et al. (70) and as corrected by direct sequencing of PCR products (57, 64). Multiple alignment of the sequences determined here and those of Sabin strains was carried out with the program CLUSTAL W, version 1.74 (67). The assessment of the degree of synonymous and nonsynonymous nucleotide divergence and estimation of genetic distances were performed according to the method of Li et al. (42). The following parameters were calculated: the overall number of mutations (Nt) and the percentage of mutated sites among all sites (Kt), the number of synonymous differences (Ns) and the percentage of mutated synonymous sites among all synonymous sites (Ks), and the number of nonsynonymous differences (Na) and the percentage of mutated nonsynonymous sites among all nonsynonymous sites (Ka) (26). To assess the similarity of determined nucleotide and deduced amino acid sequences to all of the picornavirus sequences available in GenBank, the BLAST search programs were used (http://www.ncbi.nlm.nih.gov/BLAST/). Prediction and comparative analysis of protein structures based on the resolved crystal structure of type 1 poliovirus Mahoney strain (33) were performed using the programs Swiss-Pdb Viewer, version 3.7b2 (31), and RasMol, Windows version 2.7.2.1 (7, 59).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper are available from the GenBank nucleotide sequence database under accession no. AF462418 and AF462419.
RESULTS
Case.
A poliovirus type 1 strain (99/056-252-14; hereinafter abbreviated as isolate 14) was isolated in October 1999 in a children's hospital in the city of Perm (Russia) from stools of a 7-month-old nonvaccinated female on day 19 after the appearance of the first signs of flaccid paralysis. Isolate 14 exhibited non-Sabin-strain-like antigenic properties when assayed by intratypic differentiation in an enzyme-linked immunosorbent assay with cross-absorbed polyclonal antisera (71). The link between the isolate and poliomyelitis was supported by a high and increasing level of serum antibodies against type 1 poliovirus (1:1,024 and 1:2,048 on days 18 and 42, respectively, versus constant antibody titers of 1:8 and 1:64 against polioviruses of types 2 and 3, respectively). The baby was admitted to an orphanage at the age of 1 month and then, before presenting paralysis, spent a considerable part of her life in pediatric hospitals with such diagnoses as bronchitis, pneumonia, and sepsis. Judging by the immunogram determined in September 2000 (920, 130, and 89 mg/dl for immunoglobulin G [IgG], IgM, and IgA, respectively), the patient was not seriously immunocompromised.
Overall features of the genome of isolate 14.
PCR with Sabin strain-specific primers indicated the vaccine nature of isolate 14. PCR-RFLP analysis (data not shown) demonstrated that the HaeIII and HpaII restriction fragment patterns of the VP1 region were identical to those of the Sabin 1 strain. The HaeIII restriction pattern of the 2AB region was similar to that of Sabin 2, but there was an additional cutting site for DdeI. The HaeIII and RsaI restriction patterns of the 3D region were also similar to those of Sabin 2. Based on the results of serotyping and PCR with Sabin strain-specific primers, as well as PCR-RFLP analysis, we concluded that isolate 14 is a vaccine-derived poliovirus with an evolved recombinant genome.
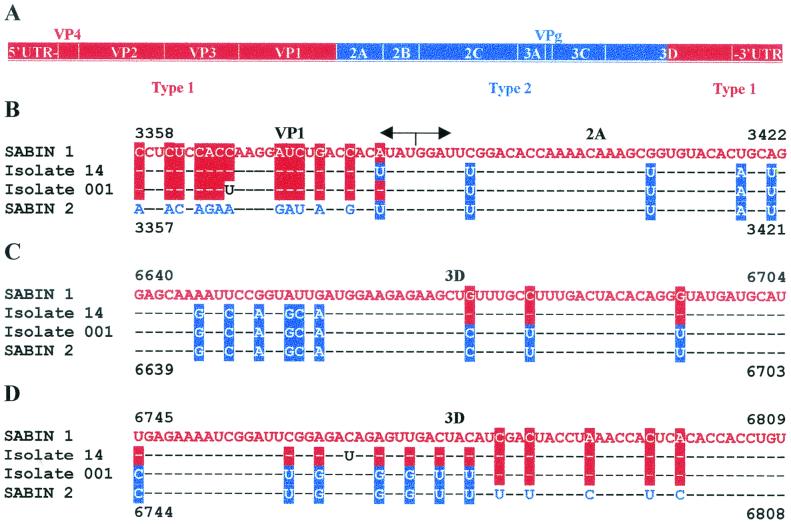
The full-length genome sequencing of isolate 14 revealed that it was a double recombinant between Sabin 1 and Sabin 2 viruses (type 1-type 2-type 1) (Table 1 and Fig. 1A ). The first crossover site was within the extreme 3′ end of the VP1 coding region, before or within its penultimate codon at nt 3380-3381 (Fig. 1B). The second crossover was in the middle of the 3D coding region (nt 6659 to 6672) (Fig. 1C).
TABLE 1.
Comparison of various regions of the genome of isolate 14 to corresponding regions of the Sabin 1 and Sabin 2 genomes
| Genomic region | Nucleotide identity (%) of isolate 14 RNA to:
|
|
|---|---|---|
| Sabin 1 RNA | Sabin 2 RNA | |
| 5′ UTR | 98.9 | 86.1 |
| VP4 | 98.1 | 77.8 |
| VP2 | 98.9 | 72.1 |
| VP3 | 99.0 | 74.9 |
| VP1 | 97.4 | 68.4 |
| 2A | 81.7 | 98.0 |
| 2B | 76.6 | 99.3 |
| 2C | 81.9 | 98.6 |
| 3A | 82.8 | 99.2 |
| VPg | 83.3 | 100 |
| 3C | 90.3 | 98.4 |
| 3D (nt 5987-6665) | 84.4 | 98.8 |
| 3D (nt 6666-7369) | 99.1 | 89.2 |
| 3′ UTR | 100 | 95.8 |
FIG. 1.
Isolates 14 and 001 are recombinants. (A) Schematic representation of the genomic structure of isolates 14 and 001. The segments of the Sabin 1 and Sabin 2 genomes are highlighted in red and blue, respectively. (B) The first (5′) crossover regions in the genomes of isolates 14 and 001. (C and D) The second (3′) crossover regions in isolates 14 (C) and 001 (D). Dashes correspond to nucleotides identical to those of the Sabin 1 genome.
A total of 58 (1.39%) and 44 (1.34%) nucleotide substitutions were identified in the isolate 14 genome parts derived from Sabin 1 and Sabin 2 parents, respectively. Mutations were found in all of the genomic regions, except the VPg coding sequence and the 3′ untranslated region (3′ UTR) (Tables 1 and 2).
TABLE 2.
Nucleotide and deduced amino acid differences between isolate 14 and appropriate Sabin strains
| Genomic region | Changesa |
|---|---|
| 5′ UTR | C334U, C471U, G505A, G506A, U525C, U575A, A600G, A714G |
| VP4 | G868U, C880U, C923U, A928U |
| VP2 | G1078A, U1141C, A1333G, G1351A, U1390C, C1414U, G1608A→S220N, C1705U, A1738G |
| VP3 | U1798C, A1944C→K60T, C2092U, C2185U, U2293C, A2410G, A2438C→M225L |
| VP1 | C2554U, G2645A→V56I, G2678A→V67I, A2734G, A2749G→I90M, G2776U→K99N, U2785C, A2795G→T106A, G2848A, A2912U→T145S, G2957A→V160I, A3043G, C3055U, G3058A, G3085A, C3109U, A3181U, C3187U, C3244U, A3271G, G3292A, G3313A, C3337U, G3349A |
| 2A | A3448G, A3460G, A3496G, C3535U, U3688C, U3709C, G3775A, G3802C, G3817A |
| 2B | U3877C, A3943U |
| 2C | U4159C, U4165C, G4195A, U4267C, G4300A, A4504G, C4510U, A4558G, G4594A, U4603C, G4606A, A4780G, C4894U, C4936U |
| 3A | A5280U→N57I, C5359U |
| 3C | A5458G, C5580U→A48V, C5587U, C5620U, G5674A, C5686U, C5704U, U5855C, A5905G |
| 3D | G6031U, U6080C→Y32H, A6175G, A6211G, C6241U, C6298U, U6448A, A6577G, C6766U, A6823G, C6922U, C7283U, A7330U |
Reversions to the Mahoney strain are underlined. Amino acid residues are separately numbered for each protein and indicated in boldface.
5′ UTR.
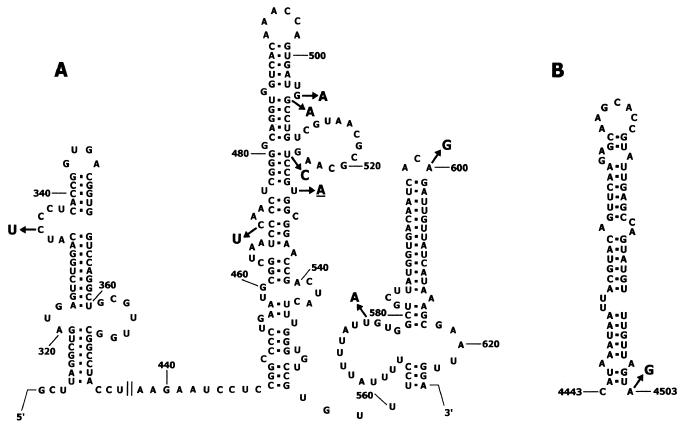
Among eight substitutions in the 5′ UTR, all but one (A714G) mapped to the internal ribosome entry site (IRES), known to have a highly conserved secondary structure (55, 61). Two mutations (G506A and U525C) are expected to stabilize the stem-loop structure of domain V by changing GU base pairs to AU and GC base pairs, respectively (Fig. 2A). The remaining five mutations were at positions supposed to be single stranded, i.e., the loops and bulges of domains IV, V, and VI, and hence unlikely to affect the secondary structure stability of the IRES (Fig. 2A).
FIG. 2.
Mutations within cis-acting elements of the isolate 14 and 001 RNA. (A) Mutations in the IRES. The structure of hairpin-loop elements of the Sabin 1 IRES is given according to the work of Pilipenko et al. (55). The single change found in isolate 001 is underlined. (B) Mutation in CRE. The structure of Sabin 2 CRE is given according to the work of Rieder et al. (58). Position 4503 corresponds to position 4504 in the RNA of isolate 14.
Nucleotide substitutions in the coding sequence.
The overwhelming majority (81 out of 94 [86.2%]) of mutations in the coding region of the isolate 14 RNA were silent (synonymous) substitutions. The distribution of these substitutions was not entirely random. The highest proportion of synonymous substitutions (Ks values in the range of 6.7 to 7.9) was found in the VP4, VP1, and 2A coding regions, whereas the VP2, VP3, and 3A regions turned out to be the least variable (Ks values in the range of 1.1 to 2.8) (Table 3). As already mentioned, there were no mutations in the VPg coding region, the smallest in the poliovirus genome.
TABLE 3.
Proportions of synonymous and nonsynonymous substitutions accumulated in different regions of the isolate 14 RNAa
| Genomic region | Ks | Ka | Ka/Ks |
|---|---|---|---|
| VP4 | 7.9 | 0 | |
| VP2 | 2.8 | 0.2 | 0.07 |
| VP3 | 2.1 | 0.3 | 0.14 |
| VP1 | 6.7 | 1.1 | 0.16 |
| 2A | 7.2 | 0 | |
| 2B | 3.3 | 0 | |
| 2C | 4.2 | 0 | |
| 3A | 1.1 | 0.5 | 0.45 |
| 3C | 4.4 | 0.3 | 0.07 |
| 3D | 3.8 | 0.1 | 0.03 |
| Full coding part of genome | 4.1 | 0.3 | 0.07 |
The RNA segments of the isolate 14 genome with coordinates nt 743 to 3380 and 6666 to 7369 were compared with the relevant segments of the Sabin 1 RNA, whereas the segment nt 3381 to 6665 was compared with the homologous segment of the Sabin 2 RNA.
The 2C coding region is known to contain a conserved stem-loop structure serving as an essential cis-acting replicative element (CRE) (30) involved in the uridylylation of VPg (54, 58). The substitution A4504G should strengthen the CRE stem-loop by forming an additional GC base pair in the base of the stem (Fig. 2B).
Amino acid substitutions.
Among 13 amino acid substitutions found in the isolate, 10 were located in the capsid proteins (Table 2). The individual capsid proteins were affected to different extents: there were one, two, and seven mutations in VP2, VP3, and VP1, respectively. Both of the VP3 mutations (Lys60Thr and Met225Leu) and two of the VP1 mutations (Ile90Met and Thr106Ala) were reversions to the parental Mahoney strain.
The structure of two known antigenic sites was altered. The substitutions Lys99Asn in VP1 and Lys60Thr in VP3 should affect antigenic sites 1 and 3, respectively (53). The former is at the top of the VP1 B-C loop (residues 96 to 104), while the latter is in the VP3 hairpin knob (residues 58 to 60) of beta-strand B (33). Two additional mutations in VP1, Ile90Met and Thr106Ala, were located in beta-strands B and C, respectively. The two latter residues appear to be involved in hydrophobic interactions with each other.
The mutation Val160Ile of VP1 (located at the protomer interface) as well as the already-mentioned substitution Lys60Thr in VP3 may affect receptor recognition by the virus (6, 16, 17, 32). The former may also influence the stability of the capsid (17).
Amino acids at positions 225 in VP3 and 56 in VP1 appear to be involved in hydrophobic interactions between VP1 and VP3 within a protomer, whereas residues at positions 67 and 145 of VP1 take part in contacts with VP2 and VP1, respectively, of a neighboring protomer. The substitutions at these positions involved similar amino acid residues in each case and hence are unlikely to cause any significant biological consequences.
Missense mutations were also present in nonstructural proteins 3A, 3C, and 3D of isolate 14 (Table 2). Residue 57 in 3A lies near a critical hydrophobic sequence that appears to be required for membrane association of the poliovirus RNA replication complex (69). The functional significance of the Asn→Ile substitution at this position, as well as of the amino acid changes in 3C (Ala48Val) and 3D (Tyr32His) proteins, is difficult to predict.
The age of isolate 14.
The approximately linear rate of fixation of mutations in naturally evolving polioviruses (5, 26, 36, 44) made it possible to estimate the time of divergence of the isolate from its progenitor attenuated Sabin strain. However, similar to previously published observations (26), different segments of isolate 14 RNA were evolving asynchronously. A reliable estimate of the viral age is possible, if calculations are based on accumulation of synonymous mutations in the VP1 coding region (see Discussion). The reported value for the rate of VP1 evolution of Sabin 1 was 3.28 × 10−2 (26) or 2.85 × 10−2 (calculated from the data in reference 5) synonymous substitutions per synonymous site per year. As the Ks value for VP1 of isolate 14 was 6.7 × 10−2 (Table 3), the age of isolate 14 can be calculated to be 2.04 or 2.35 years, respectively.
A slightly evolved poliovirus isolate with a similar location of the crossover sites.
Our collection of poliovirus isolates from patients with vaccine-associated paralytic poliomyelitis (VAPP) contained another Sabin 1-derived isolate (RUS-1161-96-001, abbreviated as 001), which, according to the RFLP analysis, was a recombinant between Sabin 1 and Sabin 2 viruses with crossover sites located similarly to those in isolate 14 (data not shown). The strain was isolated in December 1996 in the Ryazan region (Russia) from a stool sample of a 10-month-old female on the 7th day after the first signs of paralysis (or on the 23rd day after receiving the first dose of OPV). We thought it would be of interest to compare the RNA sequences of this isolate and isolate 14.
The full genome of strain 001 was sequenced, and the isolate turned out to be also a vaccine-derived double (type 1-type 2-type 1) recombinant (Fig. 1A). Its first crossover site was mapped to the VP1/2A border at nt positions 3383 to 3390, which is in the immediate vicinity of the first crossover site in the isolate 14 RNA (Fig. 1B). The second crossover in the 001 genome occurred also in the 3D region (positions 6779-6780 [Fig. 1D]) at an ∼100-nt distance from the second crossover site of isolate 14.
Altogether, 10 mutations were fixed in the RNA of isolate 001 RNA compared to that of its Sabin progenitor strains. Among three mutations in the 5′ UTR, one (U529A) was within the IRES (Fig. 2A), while the other two (U698C and C739U) mapped to the hypervariable region. There were also two mutations in VP3 (U1834C and G2110A), four mutations in VP1 (G2502A, U2881C, U3346C, and C3367U), and one mutation in 3D (C7363U). All these mutations except one resulted in synonymous substitutions. The only deduced amino acid change converts Ser8 in VP1 to Asn.
DISCUSSION
The pattern of mutation fixation in isolate 14.
There are two major mechanisms ensuring fixation of mutations in an evolving viral population, selective pressure and random sampling from heterogeneous (quasispecies) populations (19, 26). These mechanisms are not mutually exclusive, and both appeared to contribute to the genetic makeup of isolate 14.
The substitution U525C in domain V of the 5′ UTR most likely conferred a selective advantage. It restored the stability of a base pair between nt 480 and 525 (AU in Mahoney, GU in Sabin-1, and GC in isolate 14). The destabilization of this base pair in the Sabin 1 RNA appears to be associated with a decrease in neurovirulence (15, 35) and translational template activity both in vitro (62, 63) and in vivo, i.e., in human neuroblastoma cells (40). Although the true reversion G480A, restoring the original base pair found in the wild-type Mahoney strain, was most frequently observed in Sabin 1-derived isolates (5, 27, 41, 49, 52), the pseudoreversion U525C was also encountered (41, 49). The latter was shown elsewhere to restore the translational template activity of the viral RNA (49).
The C334U mutation in the 5′ UTR may also have some biological consequences. This mutation decreases the length of a C-rich sequence in loop b of domain IV, a site known to bind poly(C)-binding protein 2 involved in control of viral translation (8, 25). The U residue at position 334 was also observed in the genomes of another highly divergent Sabin 1-related strain (5) as well as in wild-type 1 polioviruses (39).
It can be assumed with reasonable confidence that several amino acid changes in capsid proteins are also of an adaptive character. Substitutions for the VP3 residue Lys60 and the VP1 residue Lys99, common findings among vaccine-derived polioviruses (5, 27, 36, 45, 48; our unpublished data) may facilitate virus escape from antibodies (53, 72). Amino acid residue Val160 was changed to Ile in VP1 in a poliovirus mutant resistant to a soluble receptor (17), and hence, its mutation as well as that of Lys60 of VP3 (6, 32) may modulate virus interaction with the receptor. The Val160Ile mutation may affect virion stability (17). Some other mutations in capsid proteins, e.g., Met225Leu in VP3 or Val56Ile, Val67Ile, Ile90Met, and Thr106Ala in VP1, may also have a selective advantage, because they were not infrequently found in other evolving lineages of Sabin 1 (5, 27, 36, 45, 48; our unpublished data).
Importantly, some of the above amino acid substitutions (Lys60Thr and Met225Leu in VP3 and Ile90Met and Thr106Ala in VP1) were in fact direct reversions to the parental Mahoney strain. Moreover, the back mutations at positions 225 in VP3 and 106 in VP1 were previously reported to be associated with increased neurovirulence (9). Thus, some nucleotide substitutions in the 5′ UTR as well as many nonsynonymous mutations in the coding region of isolate 14 could be of an adaptive character.
The overwhelming majority of the mutations in the coding region of isolate 14 RNA were, however, synonymous. In principle, some synonymous substitutions may well affect biological properties of a virus (for a more detailed discussion of this point, see reference 26), and the occurrence of such silent mutations as C880U, U1141C, G1351A, U1798C, C2092U, and U2293C in another Sabin 1 derivative (5) may hint at their possible biological relevance. Nevertheless, the majority of synonymous substitutions are expected to be phenotypically neutral. Since, in line with the results of other studies (5, 26, 27), synonymous mutations in the genome of isolate 14 significantly outnumber nonsynonymous ones (the Ka/Ks ratio is low [Table 3]), our data buttress the previously expressed notion that a random sampling mechanism provides a major quantitative contribution to the natural evolution of poliovirus RNA (2, 26). The fact that the sets of silent mutations that accumulated in different highly evolved derivatives of Sabin 1 (5, 26, 27, 36; this study) overlap to only a small extent provides additional evidence in support of this notion.
While considering the character of changes in evolving vaccine-derived polioviruses, it is perhaps appropriate to mention not only mutations that usually accumulate in vaccine-derived strains but also those that could be expected to accumulate but have not been observed. In this respect, it is interesting that antigenic site 2 and the VP1 part of site 3 appeared to be highly conserved. The reason for this stability is yet to be elucidated, but the prevalence of changes in antigenic site 1 and the VP3 part of site 3 should be taken into account in epidemiological studies.
Also, it seems that nonsynonymous nucleotide substitutions are underrepresented in the genomes of isolate 14 as well as of other highly evolved Sabin poliovirus derivatives. Indeed, the proportion of synonymous substitutions should roughly correspond to one-third of all the theoretically possible mutations (i.e., in the third codon positions). Although changes caused by nonsynonymous mutations may often have adverse effects, many such changes should involve amino acids with similar physical and/or chemical properties and therefore are not expected to be selected against. Nevertheless, the proportion of nonsynonymous mutations is surprisingly low (Table 3), suggesting that the amino acid composition of the virus-specific proteins is perhaps more finely adjusted to perform their specific functions than is generally appreciated. This may constitute a significant evolutionary constraint.
Recombination in poliovirus evolution.
Poliovirus isolates from VAPP patients are very often represented by intertypic recombinants (23, 24, 29, 43). Several lines of indirect evidence suggest that the recombination events generating the genome of isolate 14 took place not long after its divergence from the Sabin vaccine strains. Indeed, (i) intertypic recombinants are known to accumulate very early after OPV vaccination (10, 18, 47), (ii) the crossover sites in the isolate 14 RNA are located strikingly similarly to those in the very young isolate 001 (Fig. 1), and (iii) the genomic regions of isolate 14 derived from Sabin 1 and Sabin 2 exhibited roughly similar divergence from their respective predecessors (Table 1).
It is widely accepted that recombination helps RNA viruses prevent accumulation of adverse mutations, which are readily generated by the error-prone viral RNA polymerases. This genome purification may be especially important if random sampling from a heterogeneous population is a significant contributor to viral evolution, as appears to be the case (see above), and if sequential bottlenecking events result in a significant decrease in viral fitness (the so-called Muller's ratchet [14]). Several factors may favor the accumulation of intertypic recombinants in recipients of OPV. Genetic determinants of attenuation usually decrease virus fitness. Although certain of these determinants may be similarly located in the genomes of all three Sabin strains (46), it is very likely that there are also type-specific attenuating mutations. If so, intertypic recombination may be instrumental in getting rid of these fitness-decreasing lesions. For example, a Tyr→His mutation at position 73 of 3D polymerase, acquired upon derivation of Sabin 1 from its Mahoney progenitor and proposed to contribute to the attenuated phenotype of the former (15, 50, 51, 65), has been reversed in the genomes of isolates 14 and 001 as a result of replacement by the Sabin 2-derived segment.
The relationship between recombination and the incidence of VAPP cases is an intriguing but still open question. On the one hand, some attenuating mutations may be efficiently eliminated by recombination events (see above), thereby increasing viral neurovirulence. On the other, the apparently long-term circulation of isolate 14 without any known adverse manifestations may suggest that the recombination itself did not result in a significant immediate increase in neurovirulence. At the present state of our knowledge, it would perhaps be safe to state that recombination may or may not contribute to some increase in neurovirulence. But if this increase is real, it can result in an overt disease only in hosts with increased susceptibility or in combination with other mutations.
The crossover sites are distributed nonrandomly in the recombinants generated in both cultured cells (68) and human gut (18). The bias may be due to the existence of preferred sites (hot spots) for recombination and/or selective advantages and/or disadvantages of certain genomic arrangements (1). Interestingly, the crossovers in the VP1/2A region of isolates 14 and 001 occurred at nearly identical sites (nt 3380-3381 and nt 3383 to 3390, respectively [Fig. 1]). Moreover, in another Sabin 1-Sabin 2-Sabin 1 recombinant isolated from sewage in Russia in 1998, the first crossover region was mapped again between nt 3383 and 3390 (unpublished data). It is tempting to assume that there are some still-unknown structural features in poliovirus RNA facilitating its recombination in this particular region.
In addition to the three recombinants mentioned above, a fourth type 1 recombinant isolated from a VAPP patient in 1998 in Russia had a similar tripartite arrangement of the genome (unpublished data). This fact may suggest that Sabin vaccine-derived recombinants with the type 1-type 2-type 1 arrangement have a selective advantage. For example, the virus may prefer homotypic 5′- and 3′-terminal regions. Indeed, in our collection of intertypic poliovirus recombinants, all 4 of the type 1 and 9 out of 16 of the type 3 recombinants have tripartite genomes with homotypic termini (unpublished data). Type 3 recombinants of this kind have been described by others also (10, 18, 21, 28, 38, 47, 66). Interestingly, no similar type 2 tripartite recombinants have thus far been observed.
Apparently uneven rates of evolution of different parts of the poliovirus genome.
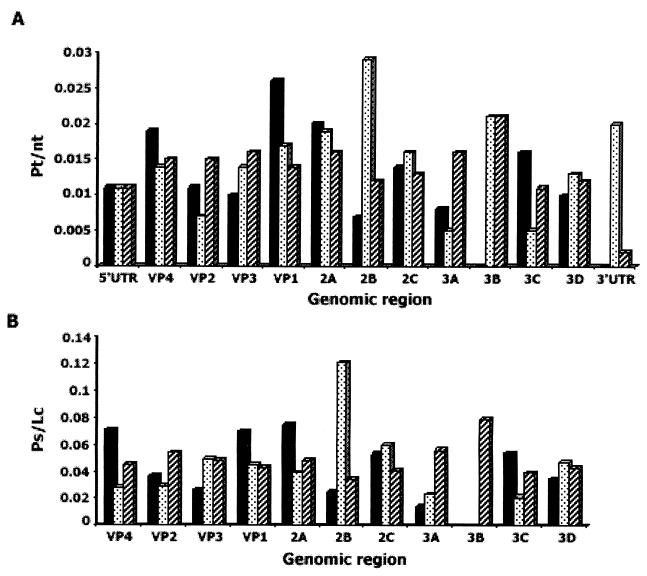
An intriguing feature of the evolution of the poliovirus genome is the apparently different rates of evolution of its different parts (5, 26). This is true when a single lineage is considered (Table 3) as well as when different lineages are compared with each other (Fig. 3).
FIG. 3.
Uneven rates of evolution of different parts of the poliovirus genome of type 1 vaccine-derived strains. The specific mutability parameters for total (A) and synonymous (B) mutations were calculated because the lineages were of different ages. The total number of appropriate mutations (compared to the Sabin vaccine) in the whole genome was taken as 100%, and the proportion of these mutations (Pt and Ps for total and synonymous substitutions, respectively) in a given region of the genome was divided by the number of nucleotides (nt) or codons (Lc) in this region, respectively. The data for isolates 14 (black bars), 1-IIs (27) (dotted bars), and IS2 (5) (hatched bars) are presented.
There are obvious constraints limiting amino acid substitutions in certain areas of capsid proteins (e.g., those involved in the interaction with receptors or active centers of viral enzymes). Understandably, replicative and translational cis-acting elements in the viral RNA would accept certain nucleotide changes rather reluctantly. Previously we discussed some other possible grounds for uneven evolution of different parts of the poliovirus genome (for example, see reference 26). However, it should be admitted that all the above considerations can hardly explain why, say, the 2B coding region in an isolate described by Georgescu et al. (27) was accumulating synonymous mutations extremely efficiently while this region was over five times more stable in our isolate 14 (Fig. 3).
It seems warranted to consider the following hypothetical explanation of this puzzling phenomenon. In addition to other mechanisms, apparently different rates of evolution of different regions of a given genome may be a consequence of homotypic recombination between different coexisting lineages, which may or may not originate from the same predecessor(s). The ease with which heterotypic recombination occurs suggests that homotypic recombination takes place even more commonly. If it is assumed that there are lineages unable to mix freely in an organism (for example, due to spatial separation [28]), they may evolve with different apparent rates because the rate of mutation fixation depends on several variables, such as the duration and frequency of reproduction cycles, the size of population, etc. These variables may not be the same for different pockets of viral reproduction. Admittedly, any experimental support for this hypothesis is lacking at the moment. It is theoretically possible, though not very likely, that the homotypic donor of genomic segments may also be represented by superinfecting virus. Currently, the reliable identification of recombination events is feasible only if they involve heterotypic or otherwise poorly related partners. Recombination between homotypic partners may manifest itself only in apparently unequal degrees of divergence of different genomic regions.
Obviously, variable levels of mutation fixation in different genomic regions introduce some ambiguity in the determination of the age of a lineage by comparing its genome sequence with that of its progenitor. However, taking into account that the major quantitative contribution to the accumulated mutations is coming from the random sampling mechanism, the estimates based on the accumulation of synonymous substitutions in sufficiently large genomic segments exhibiting a relatively high level of divergence (as is the case, for example, with the VP1 coding region) are expected to be reasonably adequate.
Implications for the program of poliomyelitis eradication.
Accepting that the isolate 14 was ∼2 years old, whereas its last host was only 7 months old, we have to conclude that the Sabin-derived virus evolved for a relatively long period before the birth of the patient and during her life before the onset of paralytic disease. The presence of antibodies to all three poliovirus serotypes in the nonimmunized patient, the ultimate host of isolate 14, suggests that contact infections could readily occur and indeed have occurred in this case. The intermediate host or hosts of the virus are unknown, and they did not appear to develop paralytic disease.
In assessing the significance of the long-term evolution of a Sabin-derived virus and its conversion into a seemingly neuropathogenic variant, it is important to stress that the proportion of the nonimmune population in the city of Perm (as well as in the majority of other regions of Russia) is likely to be relatively low. Indeed, in addition to routine OPV immunization, National Immunization Days have been carried out each year from 1996 to 2000. A serological survey of children in Perm kindergartens (i.e., approximately 3 to 7 years old) performed by the local Center of Government Sanitary and Epidemiological Surveillance in 1998-1999 revealed that less than 6% of children had no antibodies to a given poliovirus serotype.
Admittedly, such long-term persistence and evolution of Sabin vaccine-derived polioviruses in a presumably well-immunized population are quite a rare phenomenon. Neither similarly evolved Sabin virus derivatives nor VAPP cases have been registered in the Perm region since at least 1997. However, the reported data indicate that vaccine-derived viruses may make their way through narrow breaches and evolve into transmissible pathogens even in adequately immunized populations. This fact adds an additional dimension to the accumulating information about long-term circulation of vaccine-derived polioviruses in areas with inadequate immunization (11-13). Collectively, these data point to a high risk associated with the strategies that involve stopping polio vaccinations after the eradication of naturally circulating wild polioviruses.
Acknowledgments
We thank A. V. Alekseevsky and S. A. Spirin for help in analysis of the structure of mutant viral proteins. We also express our gratitude to virologists from the Government Center for Sanitary and Epidemiological Surveillance of Perm for providing us with clinical and epidemiological information. The logistical help provided by G. Y. Lipskaya is highly appreciated.
This work was supported by EU grant Copernicus, a grant from the U.S. Defense Advanced Research Project Agency, and the Polio Eradication Partnership through the European Office of the World Health Organization.
REFERENCES
- 1.Agol, V. I. 1997. Recombination and other genomic rearrangements in picornaviruses. Semin. Virol. 8:1-9. [Google Scholar]
- 2.Agol, V. I., G. A. Belov, E. A. Cherkasova, G. V. Gavrilin, M. S. Kolesnikova, L. I. Romanova, and E. A. Tolskaya. 2001. Some problems of molecular biology of poliovirus infection relevant to pathogenesis, viral spread and evolution, p. 43-50. In F. Brown (ed.), Progress in polio eradication: vaccine strategies for the end game, vol. 105. S. Karger, Basel, Switzerland. [PubMed]
- 3.Aylward, R. B., H. F. Hull, S. L. Cochi, R. W. Sutter, J. M. Olive, and B. Melgaard. 2000. Disease eradication as a public health strategy: a case study of poliomyelitis eradication. Bull. W. H. O. 78:285-297. [PMC free article] [PubMed] [Google Scholar]
- 4.Balanant, J., S. Guillot, A. Candrea, F. Delpeyroux, and R. Crainic. 1991. The natural genomic variability of poliovirus analyzed by a restriction fragment length polymorphism assay. Virology 184:645-654. [DOI] [PubMed] [Google Scholar]
- 5.Bellmunt, A., G. May, R. Zell, P. Pring-Akerblom, W. Verhagen, and A. Heim. 1999. Evolution of poliovirus type I during 5.5 years of prolonged enteral replication in an immunodeficient patient. Virology 265:178-184. [DOI] [PubMed] [Google Scholar]
- 6.Belnap, D. M., D. J. Filman, B. L. Trus, N. Cheng, F. P. Booy, J. F. Conway, S. Curry, C. N. Hiremath, S. K. Tsang, A. C. Steven, and J. M. Hogle. 2000. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 74:1342-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein, H. J. 2000. Recent changes to RasMol, recombining the variants. Trends Biochem. Sci. 25:453-455. [DOI] [PubMed] [Google Scholar]
- 8.Blyn, L. B., K. M. Swiderek, O. Richards, D. C. Stahl, B. L. Semler, and E. Ehrenfeld. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 93:11115-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouchard, M. J., D. H. Lam, and V. R. Racaniello. 1995. Determinants of attenuation and temperature sensitivity in the type 1 poliovirus Sabin vaccine. J. Virol. 69:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cammack, N., A. Phillips, G. Dunn, V. Patel, and P. D. Minor. 1988. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology 167:507-514. [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2001. Circulation of a type 2 vaccine-derived poliovirus—Egypt, 1982-1993. Morb. Mortal. Wkly. Rep. 50:41-42, 51. [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 2001. Public Health dispatch: acute flaccid paralysis associated with circulating vaccine-derived polioviruses—Philippines, 2001. Morb. Mortal. Wkly. Rep. 50:874-875. [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 2001. Update: outbreak of poliomyelitis—Dominican Republic and Haiti, 2000-2001. Morb. Mortal. Wkly. Rep. 50:855-856. [PubMed] [Google Scholar]
- 14.Chao, L. 1990. Fitness of RNA virus decreased by Muller's ratchet. Nature 348:454-455. [DOI] [PubMed] [Google Scholar]
- 15.Christodoulou, C., F. Colbere-Garapin, A. Macadam, L. F. Taffs, S. Marsden, P. Minor, and F. Horaud. 1990. Mapping of mutations associated with neurovirulence in monkeys infected with Sabin 1 poliovirus revertants selected at high temperature. J. Virol. 64:4922-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colston, E., and V. R. Racaniello. 1994. Soluble receptor-resistant poliovirus mutants identify surface and internal capsid residues that control interaction with the cell receptor. EMBO J. 13:5855-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colston, E. M., and V. R. Racaniello. 1995. Poliovirus variants selected on mutant receptor-expressing cells identify capsid residues that expand receptor recognition. J. Virol. 69:4823-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuervo, N. S., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, R. Crainic, and F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domingo, E., C. Escarmis, C. Menendez-Arias, and J. J. Holland. 1999. Viral quasispecies and fitness variations, p. 141-161. In E. Domingo, R. Webster, and J. J. Holland (ed.), Origin and evolution of viruses. Academic Press, San Diego, Calif.
- 20.Dove, A. W., and V. R. Racaniello. 1997. The polio eradication effort: should vaccine eradication be next? Science 277:779-780. [DOI] [PubMed] [Google Scholar]
- 21.Driesel, G., S. Diedrich, U. Kunkel, and E. Schreier. 1995. Vaccine-associated cases of poliomyelitis over a 30-year period in East Germany. Eur. J. Epidemiol. 11:647-654. [DOI] [PubMed] [Google Scholar]
- 22.Fine, P. E., and I. A. Carneiro. 1999. Transmissibility and persistence of oral polio vaccine viruses: implications for the global poliomyelitis eradication initiative. Am. J. Epidemiol. 150:1001-1021. [DOI] [PubMed] [Google Scholar]
- 23.Friedrich, F., E. F. Da-Silva, and H. G. Schatzmayr. 1996. Type 2 poliovirus recombinants isolated from vaccine-associated cases and from healthy contacts in Brazil. Acta Virol. 40:27-33. [PubMed] [Google Scholar]
- 24.Furione, M., S. Guillot, D. Otelea, J. Balanant, A. Candrea, and R. Crainic. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology 196:199-208. [DOI] [PubMed] [Google Scholar]
- 25.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gavrilin, G. V., E. A. Cherkasova, G. Y. Lipskaya, O. M. Kew, and V. I. Agol. 2000. Evolution of circulating wild poliovirus and of vaccine-derived poliovirus in an immunodeficient patient: a unifying model. J. Virol. 74:7381-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Georgescu, M. M., J. Balanant, A. Macadam, D. Otelea, M. Combiescu, A. A. Combiescu, R. Crainic, and F. Delpeyroux. 1997. Evolution of the Sabin type 1 poliovirus in humans: characterization of strains isolated from patients with vaccine-associated paralytic poliomyelitis. J. Virol. 71:7758-7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Georgescu, M. M., J. Balanant, S. Ozden, and R. Crainic. 1997. Random selection: a model for poliovirus infection of the central nervous system. J. Gen. Virol. 78:1819-1828. [DOI] [PubMed] [Google Scholar]
- 29.Georgescu, M. M., F. Delpeyroux, M. Tardy-Panit, J. Balanant, M. Combiescu, A. A. Combiescu, S. Guillot, and R. Crainic. 1994. High diversity of poliovirus strains isolated from the central nervous system from patients with vaccine-associated paralytic poliomyelitis. J. Virol. 68:8089-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 32.He, Y., V. D. Bowman, S. Mueller, C. M. Bator, J. Bella, X. Peng, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2000. Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. USA 97:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 A resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 34.Hull, H. F., and R. B. Aylward. 1997. Ending polio immunization. Science 277:780.. [DOI] [PubMed] [Google Scholar]
- 35.Kawamura, N., M. Kohara, S. Abe, T. Komatsu, K. Tago, M. Arita, and A. Nomoto. 1989. Determinants in the 5′ noncoding region of poliovirus Sabin 1 RNA that influence the attenuation phenotype. J. Virol. 63:1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kew, O. M., R. W. Sutter, B. K. Nottay, M. J. McDonough, D. R. Prevots, L. Quick, and M. A. Pallansch. 1998. Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J. Clin. Microbiol. 36:2893-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimman, T. F., M. P. Koopmans, and H. G. van der Avoort. 1998. Ending polio immunization. Science 279:788-789. [DOI] [PubMed] [Google Scholar]
- 38.King, A. M. Q. 1988. Genetic recombination in positive strand RNA viruses, p. 149-165. In E. Domingo, J. J. Holland, and P. Ahlquist (ed.), RNA Genetics. CRC Press, Inc., Boca Raton, Fla.
- 39.Kinnunen, L., T. Poyry, and T. Hovi. 1992. Genetic diversity and rapid evolution of poliovirus in human hosts. Curr. Top. Microbiol. Immunol. 176:49-61. [DOI] [PubMed] [Google Scholar]
- 40.La Monica, N., and V. R. Racaniello. 1989. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol. 63:2357-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, J., L. B. Zhang, T. Yoneyama, H. Yoshida, H. Shimizu, K. Yoshii, M. Hara, T. Nomura, H. Yoshikura, T. Miyamura, and A. Hagiwara. 1996. Genetic basis of the neurovirulence of type 1 polioviruses isolated from vaccine-associated paralytic patients. Arch. Virol. 141:1047-1054. [DOI] [PubMed] [Google Scholar]
- 42.Li, W. H., C. I. Wu, and C. C. Luo. 1985. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 2:150-174. [DOI] [PubMed] [Google Scholar]
- 43.Lipskaya, G. Y., A. R. Muzychenko, O. K. Kutitova, S. V. Maslova, M. Equestre, S. G. Drozdov, R. Perez Bercoff, and V. I. Agol. 1991. Frequent isolation of intertypic poliovirus recombinants with serotype 2 specificity from vaccine-associated polio cases. J. Med. Virol. 35:290-296. [DOI] [PubMed] [Google Scholar]
- 44.Martin, J., G. Dunn, R. Hull, V. Patel, and P. D. Minor. 2000. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J. Virol. 74:3001-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuura, K., M. Ishikura, H. Yoshida, T. Nakayama, S. Hasegawa, S. Ando, H. Horie, T. Miyamura, and T. Kitamura. 2000. Assessment of poliovirus eradication in Japan: genomic analysis of polioviruses isolated from river water and sewage in Toyama Prefecture. Appl. Environ. Microbiol. 66:5087-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minor, P. D. 1992. The molecular biology of poliovaccines. J. Gen. Virol. 73:3065-3077. [DOI] [PubMed] [Google Scholar]
- 47.Minor, P. D., A. John, M. Ferguson, and J. P. Icenogle. 1986. Antigenic and molecular evolution of the vaccine strain of type 3 poliovirus during the period of excretion by a primary vaccinee. J. Gen. Virol. 67:693-706. [DOI] [PubMed] [Google Scholar]
- 48.Mulders, M. N., J. H. Reimerink, M. Stenvik, I. Alaeddinoglu, H. G. van der Avoort, T. Hovi, and M. P. Koopmans. 1999. A Sabin vaccine-derived field isolate of poliovirus type 1 displaying aberrant phenotypic and genetic features, including a deletion in antigenic site 1. J. Gen. Virol. 80:907-916. [DOI] [PubMed] [Google Scholar]
- 49.Muzychenko, A. R., G. Lipskaya, S. V. Maslova, Y. V. Svitkin, E. V. Pilipenko, B. K. Nottay, O. M. Kew, and V. I. Agol. 1991. Coupled mutations in the 5′-untranslated region of the Sabin poliovirus strains during in vivo passages: structural and functional implications. Virus Res. 21:111-122. [DOI] [PubMed] [Google Scholar]
- 50.Nomoto, A., N. Kawamura, M. Kohara, and M. Arita. 1989. Expression of attenuation phenotype of poliovirus type 1, p. 297-305. In B. L. Semler and E. Ehrenfeld (ed.), Molecular aspects of picornavirus infection and detection. American Society for Microbiology, Washington, D.C.
- 51.Omata, T., M. Kohara, S. Kuge, T. Komatsu, S. Abe, B. L. Semler, A. Kameda, H. Itoh, M. Arita, E. Wimmer, and A. Nomoto. 1986. Genetic analysis of the attenuation phenotype of poliovirus type 1. J. Virol. 58:348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otelea, D., S. Guillot, M. Furione, A. A. Combiescu, J. Balanant, A. Candrea, and R. Crainic. 1993. Genomic modifications in naturally occurring neurovirulent revertants of Sabin 1 polioviruses. Dev. Biol. Stand. 78:33-38. [PubMed] [Google Scholar]
- 53.Page, G. S., A. G. Mosser, J. M. Hogle, D. J. Filman, R. R. Rueckert, and M. Chow. 1988. Three-dimensional structure of poliovirus serotype 1 neutralizing determinants. J. Virol. 62:1781-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pilipenko, E. V., V. M. Blinov, L. I. Romanova, A. N. Sinyakov, S. V. Maslova, and V. I. Agol. 1989. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology 168:201-209. [DOI] [PubMed] [Google Scholar]
- 56.Racaniello, V. R. 2000. It is too early to stop polio vaccination. Bull. W. H. O. 78:359-360. [PMC free article] [PubMed] [Google Scholar]
- 57.Rezapkin, G. V., L. P. Norwood, R. E. Taffs, E. M. Dragunsky, I. S. Levenbook, and K. M. Chumakov. 1995. Microevolution of type 3 Sabin strain of poliovirus in cell cultures and its implications for oral poliovirus vaccine quality control. Virology 211:377-384. [DOI] [PubMed] [Google Scholar]
- 58.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374.. [DOI] [PubMed] [Google Scholar]
- 60.Shulman, L. M., Y. Manor, R. Handsher, F. Delpeyroux, M. J. McDonough, T. Halmut, I. Silberstein, J. Alfandari, J. Quay, T. Fisher, J. Robinov, O. M. Kew, R. Crainic, and E. Mendelson. 2000. Molecular and antigenic characterization of a highly evolved derivative of the type 2 oral poliovaccine strain isolated from sewage in Israel. J. Clin. Microbiol. 38:3729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skinner, M. A., V. R. Racaniello, G. Dunn, J. Cooper, P. D. Minor, and J. W. Almond. 1989. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J. Mol. Biol. 207:379-392. [DOI] [PubMed] [Google Scholar]
- 62.Svitkin, Y. V., S. V. Maslova, and V. I. Agol. 1985. The genomes of attenuated and virulent poliovirus strains differ in their in vitro translation efficiencies. Virology 147:243-252. [DOI] [PubMed] [Google Scholar]
- 63.Svitkin, Y. V., T. V. Pestova, S. V. Maslova, and V. I. Agol. 1988. Point mutations modify the response of poliovirus RNA to a translation initiation factor: a comparison of neurovirulent and attenuated strains. Virology 166:394-404. [DOI] [PubMed] [Google Scholar]
- 64.Taffs, R. E., K. M. Chumakov, G. V. Rezapkin, Z. Lu, M. Douthitt, E. M. Dragunsky, and I. S. Levenbook. 1995. Genetic stability and mutant selection in Sabin 2 strain of oral poliovirus vaccine grown under different cell culture conditions. Virology 209:366-373. [DOI] [PubMed] [Google Scholar]
- 65.Tardy-Panit, M., B. Blondel, A. Martin, F. Tekaia, F. Horaud, and F. Delpeyroux. 1993. A mutation in the RNA polymerase of poliovirus type 1 contributes to attenuation in mice. J. Virol. 67:4630-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tatem, J. M., C. Weeks-Levy, A. Georgiu, S. J. DiMichele, E. J. Gorgacz, V. R. Racaniello, F. R. Cano, and S. J. Mento. 1992. A mutation present in the amino terminus of Sabin 3 poliovirus VP1 protein is attenuating. J. Virol. 66:3194-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tolskaya, E. A., L. I. Romanova, V. M. Blinov, E. G. Viktorova, A. N. Sinyakov, M. S. Kolesnikova, and V. I. Agol. 1987. Studies on the recombination between RNA genomes of poliovirus: the primary structure and nonrandom distribution of crossover regions in the genomes of intertypic poliovirus recombinants. Virology 161:54-61. [DOI] [PubMed] [Google Scholar]
- 69.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 70.Toyoda, H., M. Kohara, Y. Kataoka, T. Suganuma, T. Omata, N. Imura, and A. Nomoto. 1984. Complete nucleotide sequences of all three poliovirus serotype genomes. Implication for genetic relationship, gene function and antigenic determinants. J. Mol. Biol. 174:561-585. [DOI] [PubMed] [Google Scholar]
- 71.van der Avoort, H. G., B. P. Hull, T. Hovi, M. A. Pallansch, O. M. Kew, R. Crainic, D. J. Wood, M. N. Mulders, and A. M. van Loon. 1995. Comparative study of five methods for intratypic differentiation of polioviruses. J. Clin. Microbiol. 33:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wiegers, K., H. Uhlig, and R. Dernick. 1988. Evidence for a complex structure of neutralization antigenic site I of poliovirus type 1 Mahoney. J. Virol. 62:1845-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wood, D. J., R. W. Sutter, and W. R. Dowdle. 2000. Stopping poliovirus vaccination after eradication: issues and challenges. Bull. W. H. O. 78:347-357. [PMC free article] [PubMed] [Google Scholar]
- 74.World Health Assembly. 1988. Global eradication of poliomyelitis by the year 2000. Resolution WHA41.28. World Health Organization, Geneva, Switzerland.
- 75.World Health Organization. 1997. Manual for the virologic investigation of poliomyelitis. WHO/EPI/GEN/97.1. World Health Organization, Geneva, Switzerland.
- 76.Yang, C. F., L. De, B. P. Holloway, M. A. Pallansch, and O. M. Kew. 1991. Detection and identification of vaccine-related polioviruses by the polymerase chain reaction. Virus Res. 20:159-179. [DOI] [PubMed] [Google Scholar]
- 77.Yoneyama, T., A. Hagiwara, M. Hara, and H. Shimojo. 1982. Alteration in oligonucleotide fingerprint patterns of the viral genome in poliovirus type 2 isolated from paralytic patients. Infect. Immun. 37:46-53. [DOI] [PMC free article] [PubMed] [Google Scholar]