Abstract
We have analyzed the unique epitope for the broadly neutralizing human monoclonal antibody (MAb) 2G12 on the gp120 surface glycoprotein of human immunodeficiency virus type 1 (HIV-1). Sequence analysis, focusing on the conservation of relevant residues across multiple HIV-1 isolates, refined the epitope that was defined previously by substitutional mutagenesis (A. Trkola, M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger, J. Virol. 70:1100-1108, 1996). In a biochemical study, we digested recombinant gp120 with various glycosidase enzymes of known specificities and showed that the 2G12 epitope is lost when gp120 is treated with mannosidases. Computational analyses were used to position the epitope in the context of the virion-associated envelope glycoprotein complex, to determine the variability of the surrounding surface, and to calculate the surface accessibility of possible glycan- and polypeptide-epitope components. Together, these analyses suggest that the 2G12 epitope is centered on the high-mannose and/or hybrid glycans of residues 295, 332, and 392, with peripheral glycans from 386 and 448 on either flank. The epitope is mannose dependent and composed primarily of carbohydrate, with probably no direct involvement of the gp120 polypeptide surface. It resides on a face orthogonal to the CD4 binding face, on a surface proximal to, but distinct from, that implicated in coreceptor binding. Its conservation amidst an otherwise highly variable gp120 surface suggests a functional role for the 2G12 binding site, perhaps related to the mannose-dependent attachment of HIV-1 to DC-SIGN or related lectins that facilitate virus entry into susceptible target cells.
Only a very few monoclonal antibodies (MAbs) are capable of neutralizing primary isolates of human immunodeficiency virus type 1 (HIV-1), and the polyclonal response is also weak (10, 20, 44, 46, 59, 68). Effective antibodies are scarce because HIV-1 has evolved various protective mechanisms to enable it to resist the binding of antibodies to its envelope glycoprotein (Env) complex (31-33, 43, 52, 58, 59, 62, 74, 75). Among the antibodies that can overcome these defenses is the human MAb 2G12 (68, 69). The 2G12 antibody recognizes a unique epitope on the surface glycoprotein gp120 that is not directly associated with the receptor-binding sites on this protein (45, 70). However, 2G12 is capable of inhibiting the interactions of HIV-1 with its cell surface binding sites and thereby neutralizing infectivity (24, 42, 67, 69, 70). The success of 2G12 at neutralizing HIV-1 in vitro is reinforced by its ability in passive immunization experiments, usually in combination with other antibodies, to protect macaques from simian-human immunodeficiency virus challenge (2, 37, 38).
The precise nature of the 2G12 epitope is uncertain. Antibody mapping studies using monomeric gp120 showed that 2G12 forms a unique competition group, in that no other MAb is able to prevent its binding to gp120, and vice versa (49). Moreover, a mutagenesis analysis revealed that the only amino acid substitutions in gp120 which disrupt the 2G12 epitope are at residues specifying sites for N-linked glycosylation in the C2, C3, C4, and V4 domains (see Fig. 1A) (69). The crystal structures of a gp120 fragment comprising the conserved core with truncations of the V1, V2, and V3 variable loops and of the gp41 interactive region have been obtained (31, 32). They showed that most of the predicted glycosylation sites thought be relevant to 2G12 binding are likely to be sufficiently proximal to one another to be within the footprint of an antibody epitope (74, 75). Furthermore, several of the relevant glycans are close to the receptor-binding sites on gp120 and probably play an important role in shielding these sites from antibody recognition (43, 74, 75). Thus, 2G12 may actually exploit the very glycan defenses that normally help protect HIV-1 from neutralizing antibodies (54). Because knowledge of neutralization epitopes might be exploitable for vaccine design, we have further analyzed the 2G12 epitope. Our results implicate a conserved patch of high-mannose and/or hybrid glycans as being involved in the formation of this epitope, with mannose residues as an essential component. There may be similarities between the 2G12 epitope and the mannose-dependent binding sites on gp120 for DC-SIGN, a lectin that facilitates HIV-1 entry by enhancing the presentation of virions to susceptible cells (3, 23, 25, 40, 61), and cyanovirin-N (CV-N), a cyanobacterial protein that inhibits HIV-1 infectivity (8, 19, 22).
FIG. 1.
(A) Carbohydrates on gp120 and their contribution to the 2G12 epitope as identified by substitutional mutagenesis. The schematic of CHO-expressed IIIB and JR-FL gp120 indicates N-linked glycosylation sites. The composition of the carbohydrates in IIIB gp120 was experimentally determined (35); the carbohydrate designations in the schematic of JR-FL gp120 are based on that study, assuming that glycans are processed similarly on the two Env glycoproteins. Two sites in JR-FL gp120 that are not present in IIIB gp120 are designated as being of unknown carbohydrate composition. Arrows indicate sites that were shown to be important for 2G12 binding in a substitutional mutagenesis study. Note that the sites at 392 and 397 were only deleted in combination (69). (B) Specificities of glycosidases. The schematic is derived from reference 28. The cleavage sites of some of the endo- and exoglycosidases used in this study are indicated on the structures of the three classes of carbohydrates: complex, hybrid, and high mannose. Note that the number and characteristics of the sugar residues in the outer branches can vary. Complex glycans generally have two to four outer branches and may have a fucose residue attached to the inner N-acetylglucosamine. Asterisks indicate enzymatic cleavages that affect 2G12 binding (see Fig. 4). Abbreviations: GlcNAc, N-acetylglucosamine; Man, mannose; Gal, galactose; S.A., sialic acid.
MATERIALS AND METHODS
Sequence analysis.
The sequences of isolates sensitive to neutralization by 2G12 (6, 44, 45) were obtained from the Los Alamos HIV Databank (http://hiv-web.lanl.gov/) and from D. Montefiori (D. Montefiori, personal communication), and their degree of conservation was analyzed. The sequences of 15 different isolates, BK132, HXBc2, JR-FL, QH0515, QH0692, PVO, TVO, 301593, 301657, 301660, 301727, 92BR030, 92RW009, 92RW021, and 92TH014, were analyzed for conservation. Each of these viruses is successfully neutralized by 2G12 (7, 68, 69). The sequence and structure of the gp120 core fragment for HXBc2 has been described previously (31, 32); the sequences for HIV-1 isolates PVO and TVO were obtained from D. Montefiori; database protein accession numbers for the other sequences were L03697, U63632, AF277061, AF277065, U08444, U04908, U04909, U04925, U08714, U88823, U08645 and U08801, respectively.
A variability criterion was devised, based upon whether a change would disrupt the binding of a hypothetical antibody. For example, a change of Lys to Glu conserves amino acid type, both being charged; however, such a substitution would disrupt the binding of an antibody that recognized the Glu residue, resulting in a variable classification. Residues were classified as variable only if such changes were present in at least 2 of the 15 sequences analyzed.
Deglycosylation of gp120.
HIV-1 JR-FL gp120, expressed and purified from Chinese hamster ovary (CHO) cells, was obtained from Progenics Pharmaceuticals, Tarrytown, N.Y. (67). The gp120 protein (1 μg) was incubated with various glycosidases for 16 h at 37°C in 100 μl of the appropriate buffer for each enzyme. Controls included untreated and mock-treated gp120 (digestion buffer, no enzyme). For each enzyme, an optimal digestion buffer recommended by the manufacturer was used in these studies. We selected the buffer for Endo F2 and Endo F3 to use for the mock-treated control, as it had the lowest pH and so was potentially the most disruptive to gp120 conformation; its composition was 50 mM sodium acetate, pH 4.5. All glycosidases were obtained from Calbiochem, La Jolla, Calif. The amounts of each enzyme used were as follows: NgF, 50 U; Endo D, 25 mU; Endo-β-galactosidase, 25 mU; Endo F1, 175 mU; Endo F2, 50 mU; Endo F3, 50 mU; Endo H, 50 mU; α2-3,6,8,9-neuraminidase, 62.5 mU; α1-2,3,6-mannosidase, 2,500 mU. Denaturation and reduction of gp120 was performed by boiling in 1% sodium dodecyl sulfate (SDS) and 50 mM dithiothreitol (DTT) (45).
SDS-PAGE and Western blotting.
Glycosidase-treated gp120 (20 ng) was analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (6). Western blotting was performed following established protocols using the anti-gp120 V3 loop MAb PA-1 (1:5,000; final concentration, 0.2 μg/ml; Progenics) (67) and horseradish peroxidase-labeled anti-mouse immunoglobulin G (IgG) (1:5,000; final concentration, 0.2 μg/ml; Kirkegaard & Perry Laboratories, Gaithersburg, Md.). Luminometric detection of envelope glycoproteins was performed using the Renaissance Western Blot Chemiluminescence Reagent Plus system (NEN Life Science Products, Boston, Mass.).
ELISAs.
Enzyme-linked immunosorbent assays (ELISAs) were performed as described previously (48, 49). Glycosylated or deglycosylated gp120 (100 ng/ml) was captured onto the solid phase by using antibody D7324 to the C5 region (48, 49), and then bound gp120 was detected with either 2G12 or serum from an HIV-1-infected individual (coded as LSS). Denaturation of gp120 by boiling in the presence of SDS and DTT, followed by ELISA, was performed as described elsewhere (48). The IgG1b12 MAb was a gift from Dennis Burton (Scripps Research Institute, La Jolla, Calif.) (11).
Structure-based analyses.
The following structures were used: the crystal structure of core gp120 in complex with the N-terminal two domains of CD4 (D1D2) and the antigen-binding portion (Fab) of the human antibody 17b, as determined for the T-cell-line-adapted isolate HXBc2 (29, 30); a model (74) of the HXBc2 core extended by molecular dynamics to include the protein-proximal pentasaccharide structure consisting of the two N-acetylglycosamine and three mannose residues [GlcNAc-GlcNAc-Man-(Man)2] common to all N-linked sites (Fig. 1B); a model (33) further extended by using steric constraints to graft the nuclear magnetic resonance structure (13) of a V3 loop onto the gp120 core; and a trimeric model (33) obtained by optimization of quantifiable surface parameters. This trimeric model represents the orientation that gp120 most likely assumes in the functional viral spike (33).
Calculations of the accessibility of glycan and polypeptide components of the 2G12 epitope were made with the model containing the GlcNAc-GlcNAc-Man-(Man)2 moieties. This model probably underestimates the glycan contribution, since it only contains the central glycan core. All surface areas and distance measurements were made using the program GRASP (53).
RESULTS
Sequence analysis.
A substitutional mutagenesis study has implicated several N-linked glycans as being important components of the 2G12 epitope on gp120 (69). Amino acid substitutions that directly or indirectly affected the N-linked glycan sites at positions 295, 332, 386, 392, 397, and 448 were found to completely or partially reduce 2G12 binding (69) (Fig. 1 ). To gain insight into the conservation of the 2G12 epitope, the sequence variability of the above N-linked glycan sites was analyzed (Table 1). The Los Alamos Database (http://hiv-web-lanl.gov) was used, as it has been designed to represent the total sequence diversity of HIV-1. This database has 20 representative HIV-1 sequences from subtype A, 107 from subtype B, and 30 from subtype C (12). To make estimates of total variability for each residue, the total number of differences in subtypes A through C was divided by the total number of sequences (Table 1). Of the above six N-linked glycan sites, four were relatively well conserved, while two (Asn-295 and Asn-397) were moderately variable. The variability was subtype dependent; Asn-295, for example, was relatively conserved in subtype B but highly variable in subtypes A and C (12).
TABLE 1.
Variability of N-linked sites identified by substitutional mutagenesis as being part of the 2G12 epitope
| N-linked site | Glycan typea | Variability (%)
|
|||
|---|---|---|---|---|---|
| Clade A | Clade B | Clade C | Total | ||
| 295 | Mannose | 40 | 15 | 80 | 31 |
| 332 | Mannose | 25 | 14 | 13 | 15 |
| 386 | Mannose | 15 | 15 | 15 | 15 |
| 392 | Mannose | 5 | 11 | 16 | 11 |
| 397 | Complex | 45 | 26 | 37 | 31 |
| 448 | Mannose | 5 | 4 | 10 | 6 |
Mannose refers to either high-mannose or hybrid glycans.
The variability of the complete amino acid sequence was also analyzed among the different HIV-1 isolates that are efficiently neutralized by 2G12 and so must express the 2G12 epitope (7, 68, 69). We chose 15 isolates for which there was sequence information available (7, 68, 69). How the extent of sequence variability mapped onto the surface of gp120 is shown in Fig. 2. Despite our use of only 15 sequences, the variation appeared similar to that described previously when using sequences from the entire spectrum of primate immunodeficiency viruses (32), with much of the gp120 surface being variable. The only conserved surfaces were associated with either the CD4 or the coreceptor binding surfaces, or involved oligomer contact sites that were occluded within the functional viral spike (32, 44).
FIG. 2.
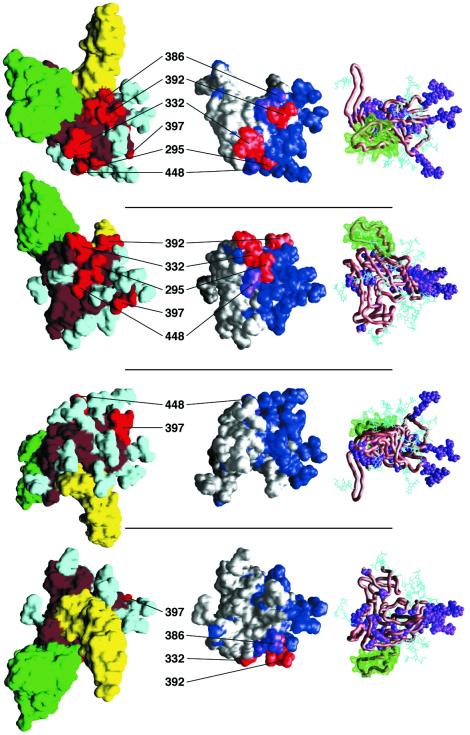
The 2G12 epitope. Four different orientations of an HIV-1 gp120 monomer are shown in three different representations. The top panel shows gp120 as viewed from the target cell membrane, looking towards the virus. Each subsequent panel shows a view rotated by 90°, with the bottom panel showing core gp120 oriented such that the viral membrane would be positioned above it and the target cell would be below it. The leftmost column depicts the solvent-accessible surface of gp120, colored according to the functionality of the underlying atoms. Red, residues and associated glycans identified by mutagenesis as being part of the 2G12 epitope; cyan, carbohydrate; brown, remaining gp120 surface. Shown for reference are the solvent-accessible surfaces of CD4 (yellow; N-terminal two domains) and the human neutralizing antibody 17b (green; variable [Fv] portion), as they are oriented in the core gp120-CD4-17b ternary crystal structure (31, 32). The rightmost column depicts a carbon-alpha worm of gp120 (brown), the molecular surface of the V3 loop as modeled into the gp120 core context (33) (green), the atoms of neutral mutants for 2G12 binding identified previously (69) (purple), and the bonds of modeled carbohydrate (74) (cyan). The middle column depicts the variability of strains that 2G12 neutralizes efficiently, mapped onto the solvent-accessible gp120 core surface. Conserved residues are shown in white, variable residues are in blue, and the mutationally identified 2G12 epitope is in red, for substitutions that decrease 2G12 binding by at least 90%, or in purple for substitutions that decrease binding by 60 to 90%. Selected residues are labeled to aid in orientation.
Among the N-linked sites implicated as probably being contributory to the 2G12 epitope (69), the sites at residues 332, 386, and 392 were conserved in all the test isolates, the site at residue 295 was conserved in all but isolate 92RW021, and the site at residue 448 was conserved in all but isolate QH0515 (Fig. 2). In contrast, the N-linked glycan site at residue 397 was variable in most strains; this site is located within the highly variable V4 loop region. The conservation analysis therefore indicates that the glycan at residue 397 is probably not part of the 2G12 epitope. Note that, in the published substitutional mutagenesis study, residue 397 was only analyzed in the context of a double substitution that simultaneously disrupted the glycan sites at both residues 392 and 397 (69). The observed loss of 2G12 binding to this 392-plus-397 double mutant is therefore likely to be due to the change at position 392 and not to the change at position 397.
Treatment of gp120 with endo- and exoglycosidases.
We next used a biochemical technique to analyze the 2G12 epitope on gp120 from the HIV-1 JR-FL strain, expressed in CHO cells. The gp120 protein from the IIIB isolate, also expressed in CHO cells, has been shown experimentally to contain 13 complex glycans and 11 high-mannose and/or hybrid chains (35). JR-FL gp120 is predicted to have all of the complex glycan sites that are present in IIIB gp120, eight of the high-mannose and/or hybrid sites, and two additional N-linked sites that are absent from IIIB gp120 (Fig. 1A).
N-linked carbohydrates can be divided into three categories, termed high mannose, hybrid, and complex (Fig. 1B) (28). These groups share a common pentasaccharide core structure. High-mannose oligosaccharides usually have two to six mannose residues attached to this core. Complex carbohydrates can have different outer branches; bi-, tri-, and tetraantennary chains with a typical sialyllactosamine sequence are shown in Fig. 1B. Hybrid oligosaccharides contain elements of both high-mannose and complex carbohydrate structures. Sugars are transferred to asparagines in the rough endoplasmic reticulum and then trimmed to yield Man8GlcNAc2 oligosaccharide species. These high-mannose carbohydrates can be further modified during passage through the Golgi network to become complex-type carbohydrates (28). The theoretical molecular masses of the examples shown in Fig. 1B are 1.7 kDa for hybrid oligosaccharides, 2.0 kDa for high-mannose oligosaccharides, and 2.4 kDa for biantennary complex chains. The molecular masses of the equivalent tri- and tetraantennary complex chains are 3.1 and 3.8 kDa, respectively.
We treated monomeric JR-FL gp120 with a selection of endo- and exoglycosidases. These enzymes have different specificities for the three classes of carbohydrates, as indicated in Fig. 1B (28, 55, 65). N-glycosidase F (NgF) cleaves all three carbohydrate classes between the asparagine and the innermost N-acetylglucosamine residue. Endoglycosidases D, F1, F2, F3, and H cleave between the two core N-acetylglucosamine residues but they differ in their specificities: Endo F1 and Endo H cleave high-mannose and hybrid carbohydrates; Endo F2 cuts high-mannose and biantennary complex chains, but not hybrid chains; and Endo F3 cleaves bi- and triantennary complex chains. Endo D generally acts on Man3 and Man4 sugars. Endo-β-galactosidase cuts β1-4 galactosidase linkages in unbranched poly-N-acetyllactosamine chains. Neuraminidase removes terminal sialic acid residues from complex chains, and α-mannosidase cleaves terminal α-linked mannose residues.
The removal of carbohydrate chains from gp120 by glycosidase treatment was monitored by determining whether gp120 migrated with a lower molecular mass when analyzed by SDS-PAGE and Western blotting with the anti-V3 loop MAb, PA-1 (Fig. 3). A substantial reduction in the molecular mass of gp120, to ≈60 kDa, was caused by NgF treatment, indicating that most carbohydrates had been removed. No proteolytic degradation of HIV-1 gp120 was observed after NgF treatment, in contrast to what occurs with virion-derived gp120 from simian immunodeficiency virus (SIV) (39). Endo F2- or Endo F3-treated gp120s migrated diffusely with glycoprotein species running from around 80 to 110 kDa. The diffuseness is probably caused by heterogeneity in the complex carbohydrates, only some of which are cleaved by these enzymes. Thus, Endo F2 digests only biantennary complex chains and high-mannose chains, whereas Endo F3 cleaves bi- and triantennnary but not tetraantennary chains. A shift of 40 kDa corresponds to a predicted loss of 15 biantennary complex oligosaccharides plus 4 high-mannose carbohydrates in the case of Endo F2, and of 14 triantennary oligosaccharides in the case of Endo F3. Smaller changes in the mobility of HIV-1 gp120 were observed in response to treatment with Endo F1 or Endo H (both of ≈10 kDa, corresponding to five to seven hybrid or high-mannose chains), with α-mannosidase (≈10 kDa, corresponding to ≈55 mannose residues, each of 0.18 kDa), or with neuraminidase (≈10 kDa, corresponding to ≈30 sialic acid residues, each 0.32 kDa in size, on complex oligosaccharide chains).
FIG. 3.
Mobilities of glycosidase-treated gp120 on SDS-PAGE. CHO-expressed JR-FL gp120 was incubated with various endo- and exoglycosidases and then analyzed by SDS-PAGE and Western blotting. Control lanes included untreated and mock-treated gp120 (digestion buffer, no enzyme). Asterisks indicate enzymes that affect 2G12 binding (see Fig. 4).
The loss of ≈30 sialic acid residues is consistent with the removal of all the residues from 15 biantennary or 10 triantennary complex chains, assuming they are of the type depicted in Fig. 1B. Hence, many of the complex carbohydrate chains must be susceptible to neuraminidase modification. Similarly, mannosidase must affect most of the hybrid and high-mannose oligosaccharides, since the molecular mass loss is so substantial (Fig. 1B).
No changes in gp120 migration were observed after exposure of the glycoprotein to Endo D or Endo-β-galactosidase (Fig. 3). Hence, these particular glycosidases were probably unable to remove any glycan components from the gp120 surface, either because their function was blocked by the presence of terminal sugars (Endo D) or their sites of action were either absent (Endo-β-galactosidase) or inaccessible.
2G12 binding to deglycosylated gp120.
We next used an ELISA to assess whether 2G12 was able to bind to gp120 that had been exposed to the various glycosidases (Fig. 4). Treatment of gp120 with NgF, Endo F1, Endo H, or α-mannosidase completely destroyed its ability to bind 2G12. Conversely, Endo F2, Endo F3, neuraminidase, Endo D, or Endo-β-galactosidase treatment had no significant effect on 2G12 binding (Fig. 4A, left panel). Endo-β-galactosidase had a very modest apparent effect on 2G12 binding, but this was not reproducible in repeat experiments (data not shown). In contrast to what was observed with 2G12 binding, most of the above enzyme treatments had little effect on the gp120 binding of polyclonal antibodies from the serum of an HIV-1-infected person (code LSS), although NgF digestion of gp120 did significantly reduce the binding of the serum antibodies (Fig. 4A, right panel).
FIG. 4.
Mannosidase treatment of gp120 inhibits 2G12 binding. (A) 2G12 binding of glycosidase-treated gp120. Aliquots of the same glycosidase-treated gp120 preparations analyzed by SDS-PAGE (Fig. 3) were tested for 2G12 reactivity in an ELISA. Bound gp120 was detected with either 2G12 (left panel) or serum from an HIV-1-infected individual (LSS; right panel). Modest variations in the ELISA signals derived from different enzymatic digests probably reflect small variations in the amounts of gp120 captured from the individual reaction buffers, and they are not considered experimentally significant. The results shown are representative of three independent experiments with similar outcomes, except where indicated in the text. (B) Deglycosylation of gp120 does not significantly affect IgG1b12 binding. The experiment was similar to that shown in panel A but compared the binding of the IgG1b12 and 2G12 MAbs. (C) Denaturation and reduction of gp120 significantly decreases 2G12 binding. Treatment of gp120 to denature and reduce it was performed as described in Materials and Methods. The binding of MAbs or HIV-1+ serum antibody (LSS) binding were then measured.
To further assess whether the glycosidases that affected the binding of 2G12 did so through a specific effect on the 2G12 epitope or through a nonspecific perturbation of gp120 structure, we compared the ability of enzyme-treated gp120 to bind 2G12 and another neutralizing MAb to a discontinuous gp120 epitope, IgG1b12 (Fig. 4B). We observed that IgG1b12 binding was not significantly affected by treatment of gp120 with Endo F1, Endo F2, Endo F3, Endo H, or α-mannosidase, whereas Endo F1, Endo H, or α-mannosidase treatment destroyed the 2G12 epitope (Fig. 4B). In additional experiments, NgF-treated gp120 was found to bind IgG1b12 very poorly (data not shown).
The control experiments show that the overall antigenic structure of gp120 was not substantially perturbed by the various glycosidases or by the moderately low pH buffers that were used in the digests. An exception was NgF treatment, which did significantly reduce the binding of HIV-1+ serum antibodies and the IgG1b12 MAb to gp120 (Fig. 4A and data not shown). This is perhaps not surprising, given that NgF converts each relatively hydrophobic N-linked site into a hydrophilic aspartic acid. This modification of so many carbohydrates could affect the structure of multiple antibody epitopes, including but not limited to that for IgG1b12, either directly or by affecting the overall conformation of gp120. However, Endo F1, Endo H, and α-mannosidase clearly destroy the 2G12 epitope without affecting the IgG1b12 epitope (Fig. 4B).
When gp120 was denatured by boiling in SDS and DTT, 2G12 was still able to bind, but with an approximately 500-fold reduction in affinity (Fig. 4C, left panel). In contrast, serum antibody binding was only reduced 10-fold by denaturation and reduction of gp120 (Fig. 4C, middle panel) (45). The binding of IgG1b12 was completely eliminated by gp120 denaturation and reduction (Fig. 4C, right panel).
Together, these findings imply that the 2G12 epitope is discontinuous in nature, or otherwise sensitive to gp120 conformation (48), and that it contains mannose residues. Moreover, consistent with our conservation analysis, the complex glycan at residue 397 is probably not involved in 2G12 binding.
Structure-based analysis of the 2G12 epitope.
Antibodies against almost all of the carbohydrate-free surface of the gp120 core have been characterized (48, 49, 74). Since 2G12 forms a unique competition group, these other ligands do not sterically compete with 2G12 binding (50). This restriction on the definition of the 2G12 epitope is graphically demonstrated in Fig. 2, which illustrates the binding to gp120 of the CD4 ligand and the 17b MAb.
The substitution mutagenesis data and the sequence-variation information were placed into the context of the gp120 structure (Fig. 2). This sequence- and structure-based analysis implicated a high-mannose and/or hybrid carbohydrate-rich region, centered upon the glycans of residues 295, 332, and 392, with the glycans at residues 386 and 448 flanking opposite ends (Fig. 2). The earlier mutagenesis study showed that residues 295, 332, and 392 were critical to 2G12 binding, in that substitutions at these positions caused a complete or substantial loss of 2G12 binding, whereas removal of the glycans from residues 386 or 448 had a significant, although incomplete, inhibitory effect (69). The central 295-332-392 glycan site (Asn plus pentasaccharide core) displays a solvent-accessible surface of 2,161 Å2, with glycan sites at 386 and 448 contributing 770 Å2 and 730 Å2, respectively.
The glycans at residues 386 and 392 are slightly separated from the rest of the 2G12 epitope by the V3 loop. Deletion of the V3 loop from gp120, either completely or partially (removal of residues 303 to 323), modestly reduces 2G12 binding (7, 69). Thus, the V3 loop is unlikely to be part of the 2G12 epitope, but its close proximity suggests that its removal could indirectly perturb the structure of the epitope.
Directly proximal to residues 295, 332, and 448 is the high-mannose or hybrid glycan of Asn-262 (in Fig. 2, this is the conserved carbohydrate directly left of residues 295 and 448 in the second panel from the top). Asn-262 is conserved in all primate immunodeficiency viruses. A substitution at residue 262 that eliminates this glycosylation site (262N/T) causes a substantial enhancement in 2G12 binding (69). Thus, while the glycan on residue 262 cannot form part of the 2G12 epitope, its absence probably affects the orientation and accessibility of the nearby glycans on residues 295 and 332. Substitutions at residue 262 are known to have a substantial impact on the overall folding of gp120 that affects multiple antibody epitopes (50). The other substitutions associated with an increase in 2G12 binding involve residues 88, 103, and 256 (69). The change at position 88 (88N/P) is likely to significantly distort the β-strand at the gp41-interactive region of gp120, whereas residues 103 and 256 are both buried within the gp120 structure; their substitution with larger residues (103Q/F and 256S/Y) probably adversely affects gp120 folding. Indeed, the 256S/Y substitution has a very similar phenotype to that of the 262N/T change, in that it significantly perturbs the structure of many antibody epitopes (J. P. Moore and J. Sodroski, unpublished data).
Because our biochemical analysis demonstrated the importance of the mannose residues on the glycans, we analyzed the mannoses of residues 295, 332, 386, 392, and 448 within the context of the functional envelope trimer (Fig. 5). These glycans form a surface that is proximal to the chemokine receptor binding surface, on a face orthogonal to the CD4 binding face. When amino acid substitution among different natural isolates is considered, much of this surface is seen to be relatively variable. The most conserved regions are the glycans at 332, 386, 392, and 448 that make up part of the 2G12 epitope, together with the base of the V3 loop and the glycan at residue 262, both of which are directly proximal to the 2G12 epitope. A small portion of this surface is also composed of polypeptide main chain (Fig. 5, rightmost column), which is generally conserved even with substantial side chain variation.
FIG. 5.
The 2G12 epitope in the context of the functional envelope trimer. Various representations of gp120 are displayed in the trimeric orientation that it mostly assumes in the functional, virion-associated Env complex. This orientation was determined by optimization of quantifiable surface parameters, as described previously (33). Three different views of the trimer are shown, each rotated by 90° about a horizontal axis. The top panel shows gp120 as viewed from the virus, the middle panel shows a side view with the virus membrane positioned above and the target cell below, and the bottom panel shows a view from the perspective of the target cell. (Note: the rightmost proteomer in the bottom panel corresponds in orientation to the top panel of gp120 monomers in Fig. 2). The left column depicts the solvent-accessible surface of gp120 colored according to functionality as follows: cyan, surface associated with carbohydrate; yellow, surface within 3 Å of CD4; green, surface associated with residues that are part of the CCR5-binding surface (64); brown, the remaining gp120 surface. The next column depicts a carbon-alpha worm trace of gp120 (brown), carbohydrate (cyan), and a carbon-alpha worm of CD4 (yellow). The third column depicts gp120 colored according to the sequence variability of the underlying residues, ranging from white (conserved) to dark blue (highly variable). The conservation scheme depicted here was described earlier in the structure analysis of core gp120 (31). Shown in red are the mannose residues of glycans 295, 332, and 392, which we have identified here as being critical for 2G12 binding; shown in olive-green are the mannose residues of 386 and 448, which also contribute to the epitope. The rightmost column depicts in purple the solvent-accessible surface associated with complex carbohydrates and, in green, the surface associated with main-chain atoms. Since main-chain atoms do not change upon amino acid variation, this portion is less subject to change upon side chain variation. Comparison of the leftmost and rightmost panels shows that much of the gp120 surface facing the cell is dominated by high-mannose and/or hybrid glycans. The figure was made using the program GRASP (53). (The left two columns were previously shown in reference 33 and are reproduced here as a visual aid for orienting the other panels.)
The mutational, sequence-variation, biochemical, and structure-based analyses all suggested that the 2G12 epitope was composed of high-mannose and/or hybrid glycans. We therefore investigated the extent to which the polypeptide surface of gp120 in this region was accessible to antibody binding (Table 2). We investigated a range of different radial distances from the mannoses of residues 295, 332, 386, 392, and 448. The average diameter of an antibody epitope is 15 to 20 Å, with a surface area of approximately 600 to 800 Å2 (18, 57). With a 5-Å radial cutoff, the total solvent-accessible surface surrounding the mannoses of residues 295, 332, 386, 392, and 448 is 3,403 Å2; at 10 Å, it is 6,014 Å2; and at 15 Å, it is 9,513 Å2. Thus, the actual 2G12 epitope must represent only a small portion of this total surface. We removed from consideration the gp120 surface associated with the binding of CD4 and MAb 17b, because 2G12 does not compete with these ligands for binding to gp120 (49).
TABLE 2.
Antibody accessibility of the gp120 polypeptide surface containing the 2G12 epitopea
| Radius of probe (Å) | Glycan-protein surface area ratio when radial distance from mannose epitope (Å) is:
|
||
|---|---|---|---|
| 5.0 | 10.0 | 15.0 | |
| 1.4 | 6.8 | 3.1 | 2.3 |
| 2.5 | 9.9 | 4.1 | 3.2 |
| 5.0 | 9.1 | 5.0 | 4.7 |
| 10.0 | 13.9 | 10.2 | 9.0 |
Surface accessibility is dependent on the size of the probe, with water, for example, being able to penetrate into small crevices that are not accessible to bulky aromatic side chains. To analyze the degree to which the gp120 polypeptide surface was accessible beneath overlying glycan residues, the 2G12 epitope was analyzed with spherical probes of different radii, ranging from 1.4 Å, the radius of a water molecule, to 10 Å, the radius of an antibody epitope. Because the precise extent of the 2G12 epitope is not known, a range of boundaries extending from the known 2G12 mannose binding site was considered. At each radial boundary distance (columns) and probe radius (rows), the ratio of glycan surface area to polypeptide surface area was calculated for different potential epitopes. The actual 2G12 paratope is likely to be a mixture of different probe radii, with the 2G12 epitope boundary within the range of radial distances presented here.
We next investigated a variety of probe radii. A 1.4-Å probe, the radius of a water molecule, provides a rough approximation of the penetration of a side chain to the protein surface. A radius of between 2.5 and 5 Å approximates the penetration of a β-hairpin turn, for example, from a CDR loop of an antibody. A radius of 10 Å approximates the reach of an entire antibody-combining region (18, 57). We found that even in the extreme of a 15 Å radial distance with a 1.4-Å probe, the glycan-to-protein surface area ratio was 2.3. This corresponds to a surface that is 70% carbohydrate. At the other extreme, a 5 Å radial distance with a 10-Å probe, the glycan-to-protein surface area ratio was 13.9, which corresponds to a surface of 93% carbohydrate. These results probably underestimate the true amount of the glycan component of the 2G12 epitope, since they were derived from a model that contains only the protein-proximal pentasaccharide, and most glycans would be twice this size. On the other hand, glycans are also flexible and may permit better penetration in an induced-fit scenario than would be predicted by the rigid model used in our probe analysis. Nonetheless, these results demonstrate that the 2G12 epitope is primarily composed of high-mannose and/or hybrid glycans.
DISCUSSION
Our experiments indicate that mannose residues in N-linked high-mannose and/or hybrid glycan chains are essential for 2G12 binding to recombinant gp120. Thus, exo-mannosidase treatment is sufficient to destroy the 2G12 epitope, without affecting the discontinuous epitope for the neutralizing MAb IgG1b12 (Fig. 4). Endo F1 and Endo H treatment also abolished 2G12 binding to CHO cell-expressed gp120, whereas Endo F2 had no effect, indicating that the mannose residues of hybrid, rather than high-mannose, carbohydrates may be involved in the 2G12 epitope (Fig. 1). This conclusion can be drawn because Endo F2 is able to remove mannose residues from high-mannose chains but not from hybrid chains (28, 55, 65); moreover, Endo F2 does successfully digest gp120 (Fig. 3) while leaving the 2G12 epitope intact (Fig. 4). However, the efficiency of Endo F2 at cleaving high-mannose carbohydrates is at least 20-fold lower than its cleavage of complex chains (65). It is therefore possible that, in addition to hybrid chain carbohydrates, high-mannose chains resistant to Endo F2 treatment could also be involved in 2G12 binding. Endo H and Endo D treatment of JR-FL gp120 produced from Drosophila melanogaster cells has been shown to remove at most only 90% of the high-mannose carbohydrate (30), although SIV gp160 has been reported to be completely sensitive to Endo H (14). Thus, while we expect most high-mannose glycans on HIV-1 gp120 to be sensitive to Endo H, Endo F1, or Endo F2, the observed mobility change of only 10 kDa after Endo F1 and Endo H treatment suggests that part of the high-mannose and hybrid carbohydrates on HIV-1 JR-FL gp120 are not, in fact, accessible to endoglycosidases.
N-linked glycans are added onto proteins during synthesis as predominately mannose, preformed oligosaccharides; only through later modification in the Golgi apparatus do these oligosaccharides lose their terminal mannose sugars. The precise characterization of N-linked glycans has been carried out only on recombinant, monomeric gp120, so it is possible that the glycans on the native, trimeric Env complex might be modified differently. However, since 2G12 neutralizes HIV-1 virions derived from human cells (69), the MAb must be able to recognize the native Env complex. The recognition of terminally linked mannose residues by 2G12 is not, therefore, an artifact of our use of recombinant gp120 expressed in CHO cells; the critical mannose residues must be exposed for 2G12 binding on the surfaces of both the monomeric gp120 molecule and the native, trimeric Env complex.
Mannosidase treatment reduces the infectivity of SIV virions (39). This observation further confirms that terminal mannoses are present on the functionally relevant, trimeric Env complex. Indeed, the oligomerization of Env late in its biosynthesis may decrease the accessibility of gp120 to the glycan-modifying enzymes in the Golgi apparatus and thereby increase the retention of high-mannose glycans on the native Env complex. Consistent with this view, an analysis of N-linked oligosaccharides on gp120 derived from chronically virus-infected, human H9 cells showed that more than 80% of the gp120 glycans are of the high-mannose or hybrid variety (41).
In contrast to the inhibitory effect of mannosidases, neuraminidase treatment increased the infectivity of SIV (39). One explanation for this might be that the complex, sialic acid-containing carbohydrates of the variable loops are involved in shielding conserved functional regions of gp120 on the native Env complex (43, 75); another explanation is that alterations in the electrostatic properties of virions caused by neuraminidase treatment might increase their binding to the cell surface. Regardless of the precise explanation, the results obtained using neuraminidase show that it is not glycosidase treatment in general that decreases virion infectivity but rather the specific activity of the particular glycosidases that are used.
A mutagenesis study on LAI gp120 revealed that the 2G12 epitope is destroyed by amino acid substitutions that affect several different N-linked glycan residues in the C2, C3, C4, and V4 regions of gp120 (Fig. 1A) (69). Most of these glycans consist of high-mannose and/or hybrid chains, not complex chains (35, 69). An exception is the complex glycan at residues 397, but our data indicate that this residue is probably not involved in 2G12 binding. In general, complex carbohydrates are present on the variable loops of gp120 and their positions often differ among HIV-1 isolates (12, 35, 36). In contrast, gp120 glycans of a high-mannose or hybrid character that are located in the less-variable regions of the protein are usually conserved among divergent HIV-1 isolates and may play an important structural role by facilitating the correct folding of gp120 (12, 35).
The sequence analysis of isolates sensitive to 2G12 neutralization proved to be unexpectedly powerful in defining the 2G12 epitope. Such variational analysis works well in the context of the dense information provided by the highly variable HIV-1 genome. A similar but less detailed analysis helped to define some features of the epitope for the broadly neutralizing anti-gp41 MAb 2F5 (68). The variational analysis of amino acids that are conserved in the 2G12-sensitive isolates, but highly variable in HIV-1 otherwise (31), defines eight amino acids, including that at position 295. These residues are scattered across the surface, but only position 295 is a site of N-linked glycosylation. Thus, even in the absence of any substitutional mutagenesis data, the sensitivity of the 2G12 epitope to deglycosylation would have been sufficient for the variational analysis to locate this epitope on gp120. However, a limitation of the variational analysis method of epitope definition is that there needs to be significant natural sequence variation within the epitope; it is thus most useful for defining the less-conserved gp120 epitopes.
Our analysis identifies N-linked glycans at positions 295, 332, 392, 386, and 448 as making up the 2G12 epitope. Clearly, some of these glycans (e.g., those at positions 386 and 448) can be modified and not completely eliminate 2G12 binding. It is generally true of all protein-protein interactions that modification of the interactive surface can often be tolerated except at special hot spots of thermodynamic energy (16, 60). However, N-linked glycans differ in several respects from the typical amino acid side chain, and these differences should be noted. Thus, N-linked glycans are larger, with an average molecular weight more than 20 times that of a typical amino acid side chain. They also contain more structure and they affect a greater volume of their surrounding environment. Our surface analysis showed that 2G12 must bind to only a small portion of the N-linked glycans that we identified as contributing to the epitope—the total surface area of a single pentasaccharide is roughly the same size as a typical antibody footprint. In our analysis, we did not specify the precise details of how 2G12 recognizes glycans. Rather, we identified glycans on the surface of gp120 that form a mannose-rich structure which 2G12 recognizes. This structure may be affected by alterations of any of the glycan sites we have identified, but compensatory changes may also occur to restore the epitope. Thus, in the context of natural HIV-1 sequences, some of the glycans that we have identified here could perhaps be absent without necessarily eliminating 2G12 binding. Moreover, other, 2G12-like antibodies might exist that recognize broadly similar glycan-dependent epitopes without necessarily competing for 2G12 binding to gp120; we suspect, however, that such glycan-dependent antibodies will be rare.
Although 2G12 is broadly reactive with many HIV-1 isolates, it is not pan-reactive (68). For example, analysis of 2G12 neutralization resistance identified subtype C viruses that were 2G12 resistant (9). We analyzed the subset of those resistant viruses for which sequence information was available (isolates DU151, DU179, and DU422) (9). All of them lacked glycan 295. While the diversity represented by database sequences probably does not reflect the frequency of viral populations, it nonetheless shows that many HIV-1 isolates will naturally lack some of the N-linked glycans required for the formation of the 2G12 epitope (12). Indeed, glycan 295 is poorly conserved among subtype C strains (Table 1), suggesting that most subtype C isolates will be resistant to 2G12 neutralization.
The 2G12 epitope that we have identified on gp120 contains, or is directly proximal to, seven of the eight high-mannose or hybrid sites that are conserved between the JR-FL and HXBc2 isolates. Indeed, the site is the only conserved, exposed surface on the gp120 trimer that does not interact with the known cellular receptors, CD4 and a chemokine receptor (33). Nonetheless, 2G12 is able to interfere with the binding of gp120 to CCR5 (67) and with the attachment of HIV-1 virions to cells (70). The positioning of its epitope on the gp120 moieties of the native Env trimer suggests that this inhibition is an indirect, steric effect manifested by the sheer bulk of an antibody molecule located physically close to the receptor-binding sites. Such interference is particularly relevant in the context of the physically crowded virion-receptor complex on the cell surface.
Terminal mannose residues are rarely found on mammalian cell surface or serum glycoproteins (reviewed in references 66, 71, and 73). Indeed, the presence of a terminal mannose results in binding of proteins to hepatic lectin receptors and their rapid clearance from the plasma (34). Virions expressing HIV-1 envelope glycoproteins are very rapidly removed from plasma after their infusion into macaques, with a half-life measured in minutes (27). Thus, the presence of terminal high-mannose residues on gp120 glycans represents a paradox: these residues appear to be highly conserved and so presumably have a relevant function, yet their presence should be detrimental to the viral life cycle by accelerating the rate of virion clearance. No doubt, overall, HIV-1 finds the terminal mannose residues to be advantageous in its battle with the human immune system.
What could be an evolutionarily conserved function for the terminal mannose residues of gp120? One explanation is that HIV-1 is known to use the mannose components of its gp120 N-linked glycans to bind to the cell surface receptors DC-SIGN and DC-SIGNR (3, 25, 61). These dendritic cell and macrophage receptors augment the efficiency of both vertical and horizontal HIV-1 transmission by enhancing the presentation of virions to both macrophages and T cells (reviewed in references 3 and 61). Structural and biochemical analyses of DC-SIGN and DC-SIGNR show that these proteins bind to the central, protein-proximal mannose residues of high-mannose glycans (23, 40). The rate-limiting step for retroviral infection is known to be the initial stage of virus-cell attachment (15, 51, 56, 63, 72), so the use of DC-SIGN as a high-affinity attachment site provides a significant advantage to HIV-1 (3, 25, 61). The high-mannose and/or hybrid sugars that form and surround the 2G12 epitope are a possible component of the binding site for DC-SIGN and related proteins. Of note is that the location of these high-mannose sugars on a surface distal from the viral membrane (Fig. 5), facing outwards from the virus, is optimal for cell surface binding.
There is precedent for the glycan residues of gp120 being the target of an antiviral compound. The cyanobacterial protein CV-N is an inhibitor of HIV-1 entry that acts by binding to gp120 (8, 19, 22). The binding site for CV-N on gp120 comprises exclusively mannose residues on N-linked glycans, specifically Manα1-2Manα moieties presented on Man8 or Man9 high-mannose structures (4, 5). CV-N has high-affinity and low-affinity binding sites, each of which recognizes the mannose moieties of a single N-linked glycan (4). CV-N can block 2G12 binding to gp120, but it does not inhibit the gp120 binding of other neutralizing or nonneutralizing MAbs (22). The converse competition does not occur, however, in that 2G12 does not inhibit CV-N binding to gp120, most probably because there are multiple binding sites for CV-N, only some of which are occluded by 2G12 (Table 3) (4, 22). Overall, however, there are clear similarities in the gp120 binding sites of 2G12 and CV-N and probably also in the mechanisms of action of these infection inhibitors. However, 2G12 does not inhibit DC-SIGN binding to gp120 (Table 3). This is, again, probably a result of the relatively promiscuous binding of DC-SIGN to gp120, in that DC-SIGN can probably recognize any of the exposed high-mannose glycans, whereas 2G12 is more selective in its interactions. The converse competition between DC-SIGN and 2G12 has not been reported (Table 3). Any partial overlap that does occur between the DC-SIGN and 2G12 binding sites could help explain why unusually low concentrations of 2G12 are able to protect some macaques from vaginal challenge with SHIV-89.6P, albeit inconsistently (38). Much greater concentrations of other anti-Env MAbs are required to achieve the same degree of protection (58).
TABLE 3.
Competition between reagents that bind mannose residues on gp120
T. B. Geijtenbeek, and Y. van Kooyk, personal communication.
Derived from reference 19.
X, autologous cross-competition; YES, heterologous competition occurs; NO, no competition; ?, no information available.
Our principal conclusion is that the 2G12 MAb recognizes an epitope that is dependent on the presence of mannose residues on N-linked glycans. In all probability, the epitope is completely composed of sugars, with no involvement of the gp120 peptide backbone. This will need to be confirmed by crystallographic analysis of the 2G12-gp120 complex. However, for several reasons, we believe that 2G12 is not a conventional anti-carbohydrate antibody (26). Firstly, 2G12 is specific for HIV-1 gp120 and does not, for example, recognize SIV gp120 expressed in the same cells (unpublished data). Secondly, denaturation of gp120 with SDS and DTT causes at least a 500-fold reduction in 2G12 binding, yet the mannose residues are still present on denatured gp120. Thirdly, the N-linked moieties that 2G12 recognizes are present on many extracellular, host proteins. For 2G12 to avoid being self-reactive, it cannot bind with high affinity to just a single N-linked moiety. Furthermore, given the flexibility of N-linked attachments, a binding site involving even two or three moieties would probably not provide enough specificity. Hence, we think that 2G12 recognizes a discontinuous structure that comprises the mannose elements of several individual glycan chains, folded into proximity. Based on our analyses, up to five individual N-linked glycans could be involved in forming the 2G12 epitope.
If we are correct that the 2G12 epitope is a discontinuous structure comprising only carbohydrate residues, it may be very difficult to exploit this information for HIV-1 vaccine development. Common N-linked glycans are rarely immunogenic, and sera from HIV-1-infected individuals do not compete with 2G12 binding to gp120 (65). In addition, raising anticarbohydrate antibodies of broad specificity could cause problems from the perspective of autoimmunity. On the other hand, if the 2G12 epitope is indeed a discontinuous structure unique to gp120, perhaps that structure could be appropriately immunogenic in the context of a vaccine antigen if it can be further defined and then appropriately presented. After all, the structure was immunogenic in the individual whose immune system made 2G12, and the resulting antibody does recognize and neutralize a broad range of HIV-1 isolates (68, 69).
There are very few conserved neutralization epitopes on gp120, yet there is a great need to exploit these limited weaknesses in the otherwise efficient defenses present on gp120 (47). HIV-1 sequence analysis demonstrates that gp120 glycans are often conserved. Moreover, a loss of five glycosylation sites when SIV was cultured in vitro was reversed when the virus replicated in macaques (21). This confirms the functional requirement to preserve gp120 glycans, probably to help resist the humoral immune response (54, 74). Unusual approaches to raising 2G12-like antibodies that focus on carbohydrate chemistry should, therefore, now be explored. For example, a synthetic structure containing clustered mannosyl structures on a peptide scaffold, resembling the recently synthesized trimeric Lewis(y) conjugate (29), could be considered. Alternatively, peptide mimeotopes of carbohydrate antigens might be a useful technique (1, 17). Our results also suggest that vaccine design strategies intended to deglycosylate gp120, and thereby uncover hidden neutralization epitopes, should focus on the complex carbohydrates and perhaps leave the high-mannose-containing structures intact, in the hope that 2G12-like antibodies might somehow be induced.
Acknowledgments
We thank Paul Maddon and Bill Olson for gifts of JR-FL gp120 and the PA-1 MAb and Dennis Burton for the IgG1b12 MAb. We are grateful to Larry Shapiro for helpful comments, to Bette Korber and Brian Foley for suggestions on protein sequence analysis, and to David Montefiori for sequences of 2G12-sensitive and -resistant subtype C isolates.
This work was supported by NIH grants AI36082, AI39420, and AI45463. J.P.M. is a Stavros S. Niarchos Scholar. P.D.K. was a recipient of a Burroughs Wellcome Career Development award. The Department of Microbiology and Immunology at the Weill Medical College gratefully acknowledges the support of the William Randolph Hearst Foundation.
REFERENCES
- 1.Agadjanyan, M., P. Luo, M. A. Westerink, L. A. Carey, W. Hutchins, Z. Steplewski, D. B. Weiner, and T. Kieber-Emmons. 1997. Peptide mimicry of carbohydrate epitopes on human immunodeficiency virus. Nat. Biotechnol. 15:547-551. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. [DOI] [PubMed] [Google Scholar]
- 3.Baribaud, F., S. Pohlmann, and R. W. Doms. 2001. The role of DC-SIGN and DC-SIGNR in HIV and SIV attachment, infection, and transmission. Virology 286:1-6. [DOI] [PubMed] [Google Scholar]
- 4.Bewley, C. A. 2001. Solution structure of a cyanovirin-N:Manα1-2Manα complex: structural basis for high-affinity carbohydrate-mediated binding to gp120. Structure 9:931-940. [DOI] [PubMed] [Google Scholar]
- 5.Bewley, C. A., and S. Otero-Quinten. 2001. The potent anti-HIV protein cyanovirin-N contains two novel carbohydrate binding sites that selectively bind to Man8D1D3 and Man9 with nanomolar affinity: implications for binding to HIV envelope glycoprotein gp120. J. Am. Chem. Soc. 123:3892-3902. [DOI] [PubMed] [Google Scholar]
- 6.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant HIV-1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binley, J. M., R. Wyatt, E. Desjardins, P. D. Kwong, W. Hendrickson, J. P. Moore, and J. Sodroski. 1998. Analysis of the interaction of antibodies with a conserved, enzymatically deglycosylated core of the HIV type 1 envelope glycoprotein. AIDS Res. Hum. Retrovir. 14:191-198. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, M. R., K. R. Gustafson, J. B. McMahon, R. H. Shoemaker, B. R. O'Keefe, T. Mori, R. J. Gulakowski, L. Wu, M. I. Rivera, C. M. Laurencot, M. J. Currens, J. H. Cardellina II, R. W. Buckheit, Jr., P. L. Nara, L. K. Pannell, R. C. Sowder II, and L. E. Henderson. 1997. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob. Agents Chemother. 7:1521-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bures, R., L. Morris, C. Williamson, G. Ramjee, M. Deers, S. A. Fiscus, A. Abdool-Karim, and D. C. Montefiori. 2002. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J. Virol. 76:2233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton, D. R., and D. C. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11(Suppl. A):587-598. [PubMed] [Google Scholar]
- 11.Burton, D. R., J. Pyati, R. Koduri, G. B. Thornton, L. S. W. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, M. Lamacchia, E. Garratty, E. R. Stiehm, Y. J. Bryson, J. P. Moore, D. D. Ho, and C. F. Barbas III. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 12.Carr, J. K., B. T. Foley, T. Leitner, M. Salminen, B. Korber, and F. McCutchan. 1998. Reference sequences representing the principal genetic diversity of HIV-1 in the pandemic. In B. Korber, C. L. Kuiken, B. Foley, B. Hahn, F. McCutchan, J. W. Mellors, and J. Sodroski (ed.), Human retroviruses and AIDS 1998: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 13.Catasti, P., J. D. Fontenot, E. M. Bradbury, and G. Gupta. 1995. Local and global structural properties of the HIV-MN V3 loop. J. Biol. Chem. 270:2224-2232. [DOI] [PubMed] [Google Scholar]
- 14.Chen, B., G. Zhou, M. Kim, R. E. Hussey, B. Elly, J. J. Skehel, E. L. Reinherz, S. C. Harrison, and D. C. Wiley. 2000. Expression, purification, and characterization of gp160e, the soluble, trimeric ectodomain of the simian immunodeficiency virus envelope glycoprotein, gp160. J. Biol. Chem. 275:34946-34953. [DOI] [PubMed] [Google Scholar]
- 15.Chuck, A. S., and B. O. Palsson. 1996. Consistent and high rates of gene transfer can be obtained using flow-through transduction over a wide range of retroviral titers. Hum. Gene Ther. 7:743-750. [DOI] [PubMed] [Google Scholar]
- 16.Clackson, T., and J. A. Wells. 1991. A hot spot of binding energy in a hormone-receptor interface. Science 267:383-386. [DOI] [PubMed] [Google Scholar]
- 17.Cunto-Amnesty, G., T. K. Dam, P. Luo, B. Monzavi-Karbassi, C. F. Brewer, T. C. Van Cott, and T. Kieber-Emmon. 2001. Directing the immune response to carbohydrate antigens. J. Biol. Chem. 276:30490-30498. [DOI] [PubMed] [Google Scholar]
- 18.Davies, D. R., and G. H. Cohen. 1996. Interactions of protein antigens with antibodies. Proc. Natl. Acad. Sci. USA 93:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dey, B., D. L. Lemer, P. Lusso, M. R. Boyd, J. H. Elder, and E. A. Berger. 2000. Multiple antiviral activities of cyanovirin-N: blocking of human immunodeficiency virus type 1 gp120 interaction with CD4 and coreceptor and inhibition of diverse enveloped viruses. J. Virol. 74:4562-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Souza, M. P., D. Livnat, J. A. Bradac, S. Bridges, et al. 1997. Evaluation of monoclonal antibodies to HIV-1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 21.Edmonson, P., M. Murphey-Corb, L. N. Martin, C. Delahunty, J. Heeney, H. Kornfeld, P. R. Donahue, G. H. Learn, L. Hood, and J. I. Mullins. 1998. Evolution of a simian immunodeficiency virus pathogen. J. Virol. 72:405-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esser, M. T., T. Mori, I. Mondor, Q. J. Sattentau, B. Dey, E. A. Berger, M. R. Boyd, and J. D. Lifson. 1999. Cyanovirin-N binds to gp120 to interfere with CD-dependent human immunodeficiency virus type 1 virion binding, fusion, and infectivity but does not affect the CD4 binding site on gp120 or soluble CD4-induced conformational changes in gp120. J. Virol. 73:4360-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feinberg, H., D. A. Mitchell, K. Drickamer, and W. I. Weis. 2001. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 294:2163-2166. [DOI] [PubMed] [Google Scholar]
- 24.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 26.Hansen, J. E., H. Clausen, C. Nielsen, L. S. Teglbjaerg, L. L. Hansen, C. M. Nielsen, E. Dabelsteen, L. Mathiesen, S. I. Hakomori, and J. O. Nielsen. 1990. Inhibition of human immunodeficiency virus (HIV) infection in vitro by anticarbohydrate monoclonal antibodies: peripheral glycosylation of HIV envelope glycoprotein gp120 may be a target for virus neutralization. J. Virol. 64:2833-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Igarashi, T., C. Brown, A. Azadegan, N. Haigwood, D. Dimitrov, M. A. Martin, and R. Shibata. 1999. Human immunodeficiency virus type 1 neutralizing antibodies accelerate clearance of cell-free virions from blood plasma. Nat. Med. 5:211-216. [DOI] [PubMed] [Google Scholar]
- 28.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631-664. [DOI] [PubMed] [Google Scholar]
- 29.Kudryashov, V., P. W. Glunz, L. J. Williams, S. Hinterman, S. J. Danishevsky, and K. O. Lloyd. 2001. Towards optimized carbohydrate-based anticancer vaccines: epitope clustering, carrier structure, and adjuvant all influence antibody responses to Lewis(y) conjugates in mice. Proc. Natl. Acad. Sci. USA 98:3264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwong, P. D., R. Wyatt, E. Desjardins, J. Robinson, J. S. Culp, B. D. Hellmig, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1999. Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1). J. Biol. Chem. 274:4115-4123. [DOI] [PubMed] [Google Scholar]
- 31.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 32.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwong, P., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, S. J., S. Evers, D. Roeder, A. F. Parlow, J. Risteli, L. Risteli, Y. C. Lee, T. Feizi, H. Langen, and M. C. Nussenzweig. 2002. Mannose receptor-mediated regulation of serum glycoprotein homeostasis. Science 295:1898-1901. [DOI] [PubMed] [Google Scholar]
- 35.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 36.Li, Y., L. Luo, N. Rasool, and C. Y. Kang. 1993. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J. Virol. 67:584-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. R. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 39.Means, R. E., and R. C. Desrosiers. 2000. Resistance of native, oligomeric envelope on simian immunodeficiency virus to digestion by glycosidases. J. Virol. 74:11181-11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell, D. A., A. J. Fadden, and K. Drickamer. 2001. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. J. Biol. Chem. 276:28939-28945. [DOI] [PubMed] [Google Scholar]
- 41.Mizuochi, T., T. J. Matthews, M. Kato, J. Hamako, K. Titani, J. Solomon, and T. Feizi. 1990. Diversity of oligosaccharide structures on the envelope glycoprotein gp120 of human immunodeficiency virus 1 from the lymphoblastoid cell line H9. J. Biol. Chem. 265:8519-8524. [PubMed] [Google Scholar]
- 42.Mondor, I., M. Moulard, S. Ugolini, P.-J. Klasse, J. Hoxie, A. Amara, T. Delaunay, R. Wyatt, J. Sodroski, and Q. Sattentau. 1998. Interactions among HIV gp120, CD4, and CXCR4: dependence on CD4 expression level, gp120 viral origin, conservation of the gp120 COOH- and NH2-termini and V1/V2 and V3 variable loops, and sensitivity to neutralizing antibodies. Virology 248:394-405. [DOI] [PubMed] [Google Scholar]
- 43.Moore, J. P., and J. M. Binley. 1998. Envelope's letters boxed into shape. Nature 393:630-631. [DOI] [PubMed] [Google Scholar]
- 44.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120 and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore, J. P., and D. D. Ho. 1993. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J. Virol. 67:863-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore, J. P., and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9(Suppl. A):S117-S136. [PubMed] [Google Scholar]
- 47.Moore, J. P., P. W. H. I. Parren, and D. R. Burton. 2001. Genetic subtypes, humoral immunity and human immunodeficiency virus type 1 vaccine development. J. Virol. 75:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore, J. P., Q. J. Sattentau, R. Wyatt, and J. Sodroski. 1994. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J. Virol. 68:469-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore, J. P., R. L. Willey, G. K. Lewis, J. Robinson, and J. Sodroski. 1994. Immunological evidence for interactions between the first, second and fifth conserved domains of the gp120 surface glycoprotein of human immunodeficiency virus type 1. J. Virol. 68:6836-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morgan, J. R., J. M. LeDoux, R. G. Snow, R. G. Tompkins, and M. L. Yarmush. 1995. Retrovirus infection: effect of time and target cell number. J. Virol. 69:6994-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Myzska, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insight from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11:281-296. [DOI] [PubMed] [Google Scholar]
- 54.Olofsson, S., and J. E. Hansen. 1998. Host cell glycosylation of viral glycoproteins—a battlefield for host defence and viral resistance. Scand. J. Infect. Dis. 30:435-440. [DOI] [PubMed] [Google Scholar]
- 55.O'Neill, R. A. 1996. Enzymatic release of oligosaccharides from glycoproteins for chromatographic and electrophoretic analysis. J. Chromatogr. A 720:201-215. [DOI] [PubMed] [Google Scholar]
- 56.Orloff, G. M., S. L. Orloff, M. S. Kennedy, P. J. Maddon, and J. S. McDougal. 1991. Penetration of CD4 T cells by HIV-1. The CD4 receptor does not internalize with HIV, and CD4-related signal transduction events are not required for entry. J. Immunol. 146:2578-2587. [PubMed] [Google Scholar]
- 57.Padlan, E. A. 1996. X-ray crystallography of antibodies. Adv. Protein Chem. 49:57-133. [DOI] [PubMed] [Google Scholar]
- 58.Parren, P. W. H. I., and D. R. Burton. 2001. The antiviral activity of antibodies in vitro and in vivo. Adv. Immunol. 77:195-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parren, P. W. H. I., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13(Suppl. A):S137-S162. [PubMed] [Google Scholar]
- 60.Pearce, K. H., Jr., M. H. Ultsch, R. F. Kelley, A. M. de Vos, and J. A. Wells. 1996. Structural and mutational analysis of affinity-inert contact residues at the growth hormone-receptor interface. Biochemistry 35:10300-10307. [DOI] [PubMed] [Google Scholar]
- 61.Pohlmann, S., F. Baribaud, and R. W. Doms. 2001. DC-SIGN and DC-SIGNR: helping hands for HIV. Trends Immunol. 22:643-646. [DOI] [PubMed] [Google Scholar]
- 62.Poignard, P., E. Ollman Saphire, P. W. H. I. Parren, and D. R. Burton. 2001. gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 63.Poret, C. D., K. V. Lukacs, G. Box, Y. Takeuchi, and M. K. L. Collins. 1998. Cationic liposomes enhance the rate of transduction by a recombinant retroviral vector in vitro and in vivo. J. Virol. 72:4832-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 65.Tarentino, A. L., and T. H. Plummer, Jr. 1994. Enzymatic deglycosylation of asparagine-linked glycans: purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol. 230:44-57. [DOI] [PubMed] [Google Scholar]
- 66.Taylor, M. E. 1993. Recognition of complex carbohydrates by the macrophage mannose receptor. Biochem. Soc. Trans. 21:468-473. [DOI] [PubMed] [Google Scholar]
- 67.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 68.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas, I. I. I., D. R. Burton, D. D. Ho, and J. P. Moore. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4 IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ugolini, S., I. Mondor, P. W. H. I. Parren, D. R. Burton, S. A. Tilley, P. J. Klasse, and Q. J. Sattentau. 1997. Inhibition of attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J. Exp. Med. 186:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vasta, G. R., M. Quesenberry, H. Ahmed, and N. O'Leary. 1999. C-type lectins and galectins mediate innate and adaptive immune functions: their roles in the complement activation pathway. Dev. Comp. Immunol. 23:401-420. [DOI] [PubMed] [Google Scholar]
- 72.Wang, H., R. Paul, R. E. Burgeson, D. R. Keene, and D. Kabat. 1991. Plasma membrane receptors for ecotropic murine retroviruses require a limiting accessory factor. J. Virol. 65:6468-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weis, W. I., M. E. Taylor, and K. Drickamer. 1998. The C-type lectin superfamily in the immune system. Immunol. Rev. 163:19-34. [DOI] [PubMed] [Google Scholar]
- 74.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 75.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]