Abstract
The RNA polymerase of the influenza virus is responsible for the transcription and replication of the segmented RNA viral genome during infection of host cells. Polymerase function is known to be strictly dependent on interaction with its RNA promoter, but no attempts to investigate whether the virion RNA (vRNA) promoter stabilizes the polymerase have been reported previously. Here we tested whether the vRNA promoter protects the polymerase against heat inactivation. We prepared partially purified recombinant influenza A virus RNA polymerase, in the absence of influenza virus vRNA promoter sequences, by transient transfection of expression plasmids into human kidney 293T cells. The polymerase was found to be heat labile at 40°C in the absence of added vRNA. However, it was protected from heat inactivation if both the 5′ and 3′ strands of the vRNA promoter were present. By using the ability of vRNA to protect the enzyme against heat inactivation, we established a novel assay, in conjunction with a mutagenic approach, that was used to test the secondary structure requirement of the vRNA promoter for polymerase binding. Binding required a panhandle structure and the presence of local hairpin loop structures in both the 5′ and 3′ ends of vRNA, as suggested by the corkscrew model. The interaction of the vRNA promoter with the influenza virus RNA polymerase heterotrimeric complex is likely to favor a particular closed conformation of the complex, thereby ensuring the stability of the RNA polymerase within both the infected cell and the isolated virus.
Influenza virus is a negative-sense, segmented RNA virus with eight RNA segments coding for 10 genes. The three longest gene segments code for the three subunits PB1, PB2, and PA of the heterotrimeric influenza virus RNA polymerase complex, which is the enzyme responsible for transcription and replication of the viral genome in the infected cell. In addition to its role as an RNA-dependent RNA polymerase, which copies virion RNA (vRNA)→mRNA in transcription and vRNA→cRNA and cRNA→vRNA in replication, the complex is also an endonuclease, snatching capped RNAs from host cell pre-mRNA for use as primers for the synthesis of influenza virus-specific mRNA, and a poly(A) polymerase, catalyzing polyadenylation of influenza virus-specific mRNA (for reviews, see references 19, 24, and 31).
There has been significant progress in understanding the sequence and secondary structure properties of the vRNA promoter, which consists of 13 nucleotides (nt) at the 5′ end and 12 nt at the 3′ end of the vRNA, all of which are conserved in each RNA segment of every influenza A virus. These conserved nucleotides, along with an extra one to five segment-specific bases at the 5′ and 3′ ends of vRNA, form a partially double-stranded panhandle structure. Additional short hairpin loops, consisting of a 2-bp stem and a tetraloop, have been proposed either at the 5′ end (5′ hairpin loop model) or at both the 5′ and 3′ ends (corkscrew model) of the vRNA (4, 33).
The hairpin loop near the 5′ end of the vRNA promoter is probably required for binding to the polymerase complex, since whenever requirements for this hairpin loop in, e.g., endonuclease cleavage (21) or polyadenylation (33) have been critically examined, its presence has been found to be absolutely essential. The hairpin loop near the 3′ end of vRNA is also required for endonuclease cleavage (20) but not for ApG-primed transcription (8) or polyadenylation (33).
The influenza virus RNA polymerase binds vRNA and cRNA promoter sequences specifically (7-9, 32). The purpose of the present study was to test whether vRNA promoter binding stabilized the polymerase to heat inactivation, and if so, what the secondary structure requirements for this stabilization were. To investigate this problem, we required a source of recombinant influenza virus RNA polymerase prepared in the absence of influenza virus RNA, since active, nonrecombinant polymerase cannot readily be isolated from viral cores without contamination by residual influenza virus RNA (8). We therefore adapted a plasmid-based system to express influenza virus RNA polymerase transiently in human kidney 293T cells. We found that the vRNA promoter protected the isolated influenza virus RNA polymerase from heat inactivation. By utilizing this property of vRNA, we established a novel assay, in conjunction with a mutagenic approach, that was used to test the secondary structure requirement of the vRNA promoter for polymerase binding. Binding required a panhandle structure and the presence of local hairpin loop structures in both the 5′ and 3′ ends of vRNA, as suggested by the corkscrew model.
MATERIALS AND METHODS
Plasmids.
Full-length cDNA copies of the PB1, PB2, PA, and NP segments of influenza A/WSN/33 virus were cloned into the AgeI site of pcDNA3A, a variant of pcDNA3 (Invitrogen) modified by the insertion of an AgeI sequence into the HindIII site (further details are available from the authors of the present article), giving plasmids pcDNA-PB1 (abbreviated PB1), pcDNA-PB2 (abbreviated PB2), pcDNA-PA (abbreviated PA), and pcDNA-NP (abbreviated NP). These plasmids are under the control of a cytomegalovirus promoter/enhancer. The plasmid pPOLI-GFP (abbreviated vRNA; sequence available from the authors of the present article) (gift of Peter Palese) encodes the RNA of green fluorescent protein in negative sense flanked by the 5′ and 3′ ends of the vRNA promoter of segment 8 (slightly modified in their 5′ and 3′ noncoding regions) of influenza A/WSN/33 virus under the control of a truncated human RNA polymerase I promoter and a ribozyme terminator (5). The first 23 residues of an ApG-primed influenza virus RNA polymerase transcript of the vRNA promoter derived from pPOLI-GFP have the sequence AGCAAAAGCAGGGUGACAAAGAC.
Preparation of recombinant influenza virus RNA polymerase by transient transfection of 293T cells.
Exponentially growing 293T cells (usually 5 × 105 cells) were transfected with 1 μg of plasmids, as specified in the individual experiments; e.g., pcDNA-PB1, pcDNA-PB2, and pcDNA-PA were transfected with Lipofectamine 2000 (Gibco BRL) in suspension and plated onto 3.5-cm-diameter dishes in antibiotic-free Dulbecco's modified Eagle's medium containing 10% fetal calf serum. After 48 h (typically), cells were harvested, washed in phosphate-buffered saline, and lysed for 5 min at 0°C with 1 ml of buffer A (0.1% Nonidet NP-40, 10 mM Tris-HCl [pH 7.8], 10 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). Nuclei were isolated by centrifugation at 1,000 × g and lysed for 10 min at 0°C in 0.1 to 0.2 ml of buffer B (0.5% Nonidet NP-40, 50 mM Tris-HCl [pH 7.8], 0.2 M KCl, 0.2 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 25% glycerol). After a further centrifugation at 1,000 × g, the nuclear extract was either used directly or concentrated by precipitation with an equal volume of 3 M ammonium sulfate. The protein pellet was finally dissolved in 20 to 30 μl of buffer B and stored at −20°C. In some experiments, the transfection was scaled up seven times and plated on 9-cm-diameter dishes.
Preparation of RNA templates.
Short 15- or 14-nt synthetic oligonucleotides at the 5′ end (5′ AGUAGAAACAAGGCC 3′) or 3′ end (5′ GGCCUGCUUUUGCU 3′), respectively, of wild-type influenza A virus vRNA were purchased from Dharmacon, unblocked by following the manufacturer's instructions, and used without further purification. These oligonucleotides contained the consensus sequence (underlined) plus an additional two residues. Mutant oligonucleotides (15 and 14 nt) corresponding to the 5′ and 3′ ends of vRNA, respectively, were obtained from our collection of synthetic RNAs, which had been synthesized and gel purified in 1993 to 1994 and stored at −20°C (8). Purity of the RNA was assessed, essentially as described previously, as >75% (8) by [γ-32P]ATP labeling with T4 polynucleotide kinase and analysis on 20% polyacrylamide gel electrophoresis (PAGE) in 7 M urea. The main contaminants were oligonucleotides one residue shorter than the desired sequence (results not shown). Nucleotides are numbered with prime numbers from the 5′ end of the 5′ strand of the promoter and without prime numbers from the 3′ end of the 3′ strand of the promoter (see Fig. 5 to 7).
FIG. 5.
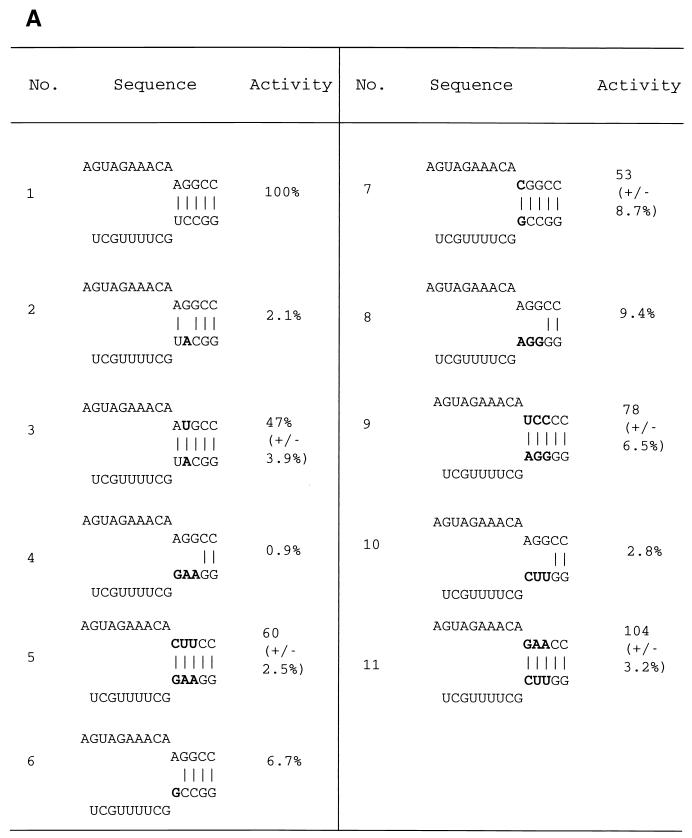
Base pairing between the conserved 5′ strand and 3′ strand of the vRNA promoter is required to protect the RNA polymerase from heat inactivation. (A) The transcriptional activity remaining after heat inactivation of the polymerase in the presence of structures 1 to 11. Mutations are shown in bold; base pairs are shown by vertical lines. For sequences 3, 5, 7, 9, and 11, activity was expressed as a percentage of wild-type (sequence 1) activity (means and standard deviations of three independent experiments), by quantitation of the major bands a and a1 to a5; activity values for sequences 2, 4, 6, 8, and 10 were from single measurements. (B) RNA polymerase, obtained by transfection of 293T cells with PB1, PB2, and PA, was heated for 15 min at 40°C in the presence of the indicated sequences (1 μM), transcription was evaluated with the [α-32P]GTP incorporation assay, and the polymerase was fractionated by 20% PAGE in 7 M urea. Lanes 1, 3, 7, 9, 11, 4, and 5 depict the results for the corresponding sequences shown in panel A. Bands b and c depict minor, 13-nt or 12-nt transcripts, shorter than the main 14-nt product, band a; the minor band, x, depicts a longer, 15-nt transcript, possibly formed by the nontemplated addition of an extra base. Bands a1 to a5 differ in gel mobility from one another because of their differing G content.
FIG. 7.
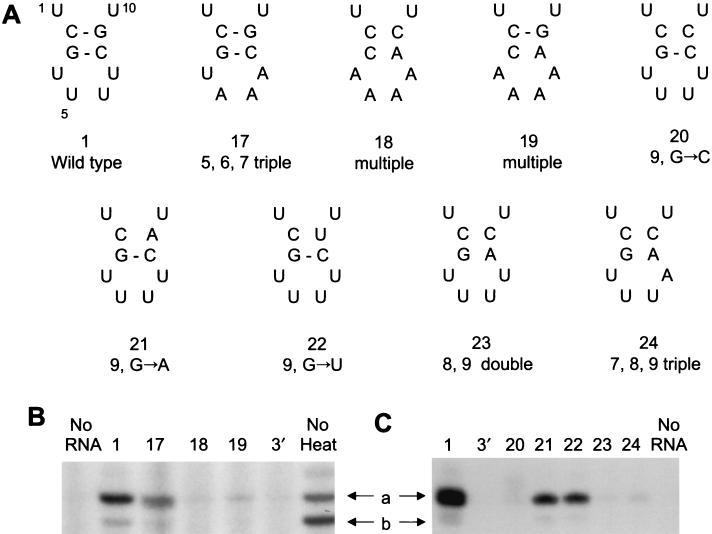
Base pairs in the stem of the 3′ hairpin loop of the promoter are required to protect the RNA polymerase from heat inactivation. (A) The wild-type vRNA (1) and mutants (17 to 24) tested. Only the 3′ hairpin loop (or potential loop) of a 14-nt 3′ strand is shown; all experiments used in addition an equimolar amount of the 15-nt wild-type 5′ strand of vRNA. Sequences are numbered from their 3′ ends. (B and C) RNA polymerase, reconstituted in vivo from PB1, PB2, and PA plasmids, was heated for 15 min at 40°C in the presence of the indicated sequences (1 μM), transcription was evaluated by the [α-32P]GTP incorporation assay with ApG and added wild-type vRNA sequences (2.5 μM), and the polymerase was fractionated by 22% PAGE in 7 M urea. See legend to Fig. 6 for explanation of additional symbols. A duplicate experiment gave similar results.
ApG-primed transcription, using an [α-32P]GTP incorporation assay, of short model vRNAs.
Transcription was performed as described before (8), except that reactions were performed with 2.5 μCi of [α-32P]GTP (>3,000 Ci/mmol; Amersham) in the presence of 1 mM ATP-0.5 mM CTP-0.1 μM GTP-0.5 mM UTP-1 mM ApG (Sigma) using up to 1.5 μl of recombinant RNA polymerase and 5′ (15 nt) and 3′ (14 nt) wild-type or mutant templates (up to 15 pmol of each) in a 3-μl reaction mixture. After 1 h at 30°C, formamide dyes (10 μl) were added and the mixture was heated at 95°C for 2 min. Fractionation was by either 18%, 20%, or 22% PAGE in 7 M urea. Gels were autoradiographed using Kodak BioMax Trans-screen HE intensifying screens and Biomax MS film. Quantitation was by phosphorimage analysis. When products had a differing G content, the relative molar yields were calculated by dividing the raw phosphorimage data by the number of G residues in the transcript. In the case of the base-paired as opposed to the mismatched sequences (Fig. 5A, sequences 1, 3, 5, 7, 9, and 11), the [α-32P]GTP incorporation assay was modified by the omission of wild-type vRNA, since preliminary experiments had shown that the addition of wild-type vRNA was unnecessary (data not shown). In the case of Fig. 6, the [α-32P]GTP incorporation assay was modified by the addition of a pseudo-wild-type panhandle structure derived from the specified mutant 15-nt 5′ and 14-nt 3′ vRNA oligonucleotides, which resulted in the formation of an A→U base pair mutation at positions 11 and 12′.
FIG. 6.
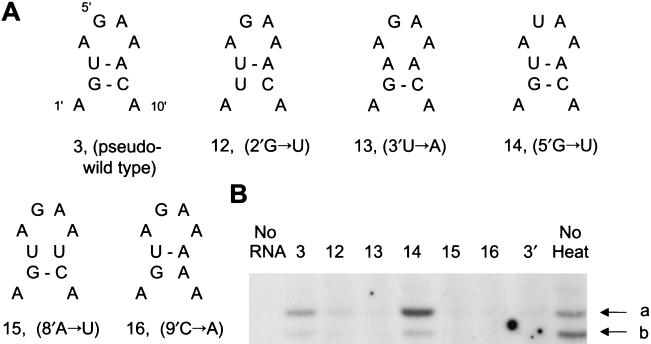
Base pairs in the stem of the 5′ hairpin loop of the promoter are required to protect the RNA polymerase from heat inactivation. (A) Pseudo-wild-type (3) (see Fig. 5A, sequence 3, for full structure) and point (12 to 16) mutants tested. Only the first 10 residues of the 15-nt 5′ hairpin loop are shown; for the full duplex structure, see Fig. 5A. Sequences are numbered from their 5′ ends (1′, 2′, 3′, etc.). (B) The RNA polymerase preparation, reconstituted in vivo from PB1, PB2, and PA plasmids, was heated for 15 min at 40°C in the presence of the indicated sequences (1 μM) and controls. Transcription was evaluated by the [α-32P]GTP incorporation assay with added pseudo-wild-type vRNA sequences (1.25 μM), and the polymerase was fractionated by 22% PAGE in 7 M urea. a, 14-nt transcription product; b, 13-nt product. Control lanes: No RNA, no added RNA; 3′, only the 3′ strand of the promoter was present; No heat, unheated enzyme. An independent experiment gave essentially similar results.
Heat inactivation of recombinant RNA polymerase.
The recombinant enzyme (in buffer B) was heated at 40°C for 15 min with a <20% volume of added oligonucleotides or deionized water and placed on ice. Heated extracts were assayed immediately for ApG-primed transcription by the [α-32P]GTP incorporation assay (see above).
RESULTS
Recombinant RNA polymerase isolated from 293T cells is transcriptionally active.
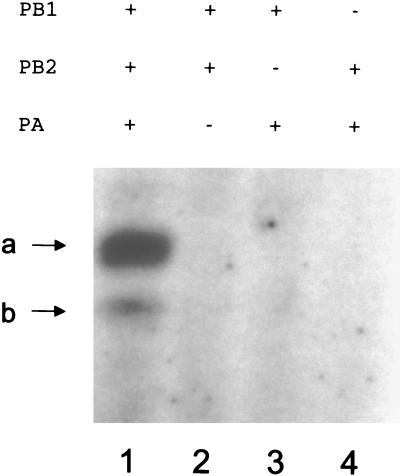
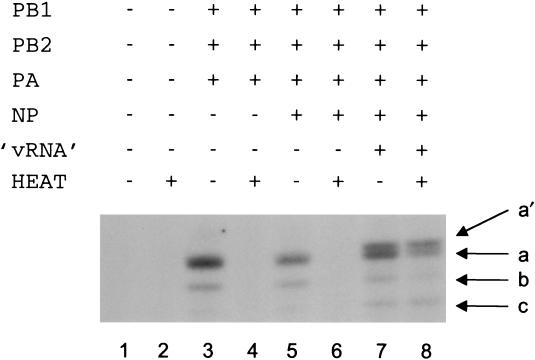
We had to prepare a source of recombinant RNA polymerase that was active in transcription in the absence of influenza virus vRNA promoter sequences before testing the heat stability of influenza virus RNA polymerase in the presence of the influenza virus vRNA promoter. To detect polymerase activity with the limited amounts of RNA polymerase present in transfected cells, we modified an existing transcription assay (8) by reducing the concentration of GTP to 0.1 μM, thereby improving its sensitivity. In this assay, a short synthetic 14-nt model vRNA 3′ end sequence, in the presence of a 15-nt 5′ end vRNA sequence, was transcribed in the presence of ApG, which resulted in the incorporation of [α-32P]GTP into a major 14-nt transcript, 5′ AGCAAAAGCAGGCC 3′ (Fig. 1, lane 1, band a). A minor, 13-nt transcript (Fig. 1, lane 1, band b) was also present, presumably transcribed from a 13-nt oligonucleotide (5′ GCCUGCUUUUGCU 3′) contaminating the 14-nt template (see Materials and Methods). No activity was detected if any of the three polymerase subunits was omitted from the transient transfection (Fig. 1, compare lanes 1 to 4). This result confirmed that all three polymerase subunits (PB1, PB2, and PA) were required for transcription.
FIG. 1.
ApG-primed transcription of 14-nt influenza virus vRNA model templates requires a RNA polymerase complex containing PB1, PB2, and PA. The results of in vitro transcription of a model 14-nt vRNA 3′ sequence in the presence of an equimolar concentration (2.5 μM) of a model 15-nt 5′ end sequence, evaluated using the [α-32P]GTP incorporation assay with different preparations of RNA polymerase prepared in parallel from the indicated plasmids (see Materials and Methods) and fractionated by 20% PAGE in 7 M urea, are shown. a, 14-nt product; b, 13-nt product (see text).
Recombinant RNA polymerase requires both the 5′ and 3′ strands of the vRNA promoter for ApG-dependent transcriptional activity.
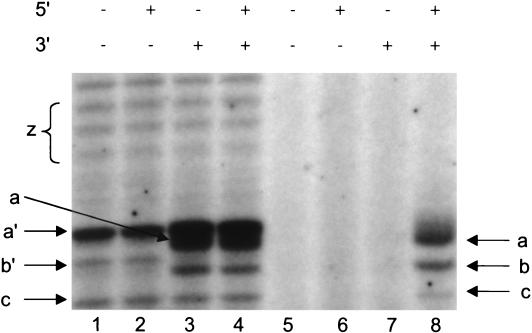
Evidence that both the 5′ and 3′ ends of the influenza virus vRNA promoter are required for transcription by influenza virus RNA polymerase has been derived mainly from early in vitro studies with RNA polymerase isolated from virus (7, 8). More recent studies of recombinant RNA polymerase isolated from baculovirus (13) confirmed these findings. Nevertheless, before studying heat inactivation of influenza virus RNA polymerase isolated from nuclear extracts of transiently transfected 293T cells, it was necessary to confirm the vRNA promoter requirements for the ApG-dependent [α-32P]GTP incorporation assay that we proposed to use to monitor heat inactivation. Figure 2 shows the effect of adding the 15 nt from the 5′ end, the 14 nt from the 3′ end, or oligonucleotides from both the 5′ and 3′ ends of the vRNA promoter (see Materials and Methods) to an influenza virus RNA polymerase (prepared in the presence of NP) with or without endogenous influenza virus-like vRNA derived from the plasmid pPOLI-GFP. In the case of nuclear extracts prepared from 293T cells cotransfected with pPOLI-GFP, in addition to transfection with expression plasmids for PB1, PB2, PA, and NP, we found that the RNA polymerase transcribed the endogenous vRNA without needing any added vRNA 5′ or 3′ end nucleotides (Fig. 2, lane 1). This result confirmed that the 5′ and 3′ ends of the influenza virus vRNA promoter were supplied by the endogenous RNA transcribed by the host Pol I from the transfected plasmid, pPOLI-GFP.
FIG. 2.
Both 5′ and 3′ vRNA promoter arms are needed to initiate transcription by RNA polymerase prepared without endogenous influenza virus-like RNA. ApG-primed in vitro transcription using the [α-2P] GTP incorporation assay was performed in the presence of short oligonucleotides (0.2 μM) corresponding to the 5′ end (15 nt), 3′ end (14 nt), or both ends of wild-type vRNA as shown, and the polymerase was fractionated by 20% PAGE in 7 M urea. Lanes 1 to 4, RNA polymerase prepared from PB1, PB2, PA, NP, and pPOLI-GFP; lanes 5 to 8, RNA polymerase prepared from PB1, PB2, PA, and NP. a′, 14-nt product AGCAAAAGCAGGGU, b′, 13-nt product AGCAAAAGCAGGG derived from endogenous pPOLI-GFP; z, longer transcription products, estimated as 17 to 19 nt in length, derived from endogenous pPOLI-GFP; a, 14-nt major end product derived from the added 14-nt 3′ end of wild-type vRNA; b, 13-nt products derived from added vRNA template (see text); c, 12-nt product AGCAAAAGCAGG, present in transcripts derived from both pPOLI-GFP and the added 14-nt 3′ end of wild-type vRNA.
A major 14-nt partial transcription product, AGCAAAAGCAGGGU (Fig. 2, lane 1, band a′), was present in good yield under our reaction conditions. We deduced the sequence of this product, firstly because it was present at the 14-nt position on the gel and migrated only slightly slower than a previously characterized (7) 14-nt product (Fig. 2, lane 1, band a), AGCAAAAGCAGGCC, derived from a known added 14-nt vRNA template (see Materials and Methods and Fig. 2, lane 8). Secondly, the 14-nt partial transcription product has the expected sequence derived from the plasmid pPOLI-GFP (see Materials and Methods). Thirdly, the 14-nt partial transcription product migrates marginally more slowly than band a, despite its similar length, because of its higher G content (five versus four G residues), which is consistent with the greater retardation effect of the presence of G residues in dense denaturing polyacrylamide gels (35). Fourthly, it accumulates in high yields, because the next, or 15th, residue to be incorporated is a G residue, after which a G is not incorporated until residue 21 (see Materials and Methods for the sequence derived from the vRNA promoter of pPOLI-GFP). The reaction conditions, in which GTP is present at low concentrations (0.1 μM), should, therefore, favor its accumulation. However, other unknown factors may be responsible for the high yield of this reaction product.
The addition of the 14-nt 3′ end of the influenza virus vRNA promoter catalyzed the synthesis of a 14-nt transcription product (a) (Fig. 2, lane 3) that migrated slightly faster than the 14-nt product (a′) derived from pPOLI-GFP (see above). No significant improvement in the yield of band a was observed if the 5′ end of vRNA was present in addition (Fig. 2, lane 4). This confirmed that the synthesis of the 14-nt product a in lanes 3 and 4 had occurred through the copying of the added 3′ end in trans by the RNA polymerase, which was bound to the endogenous vRNA promoter, as suggested before (30).
In contrast, influenza virus RNA polymerase prepared from nuclear extracts of 293T cells transfected with expression plasmids for PB1, PB2, PA, and NP but in the absence of pPOLI-GFP transcribed an added 3′ end vRNA template only in the presence of an added 5′ end of vRNA (Fig. 2, lane 8). Neither the 5′ nor the 3′ end alone of vRNA stimulated transcription (Fig. 2, lanes 6 and 7). However, when used at a 10-fold-higher concentration (2.5 μM), weak (<10%) activity was reproducibly detected with the 3′ end of the vRNA promoter alone (data not shown; see Discussion).
Overall, we conclude that both the 5′ and 3′ ends of the vRNA promoter are required for significant transcriptional activity in an ApG-dependent assay using a mammalian recombinant influenza virus RNA polymerase prepared in the absence of endogenous influenza virus-like vRNA promoter sequences.
Influenza virus vRNA promoter sequences protect the RNA polymerase from heat inactivation.
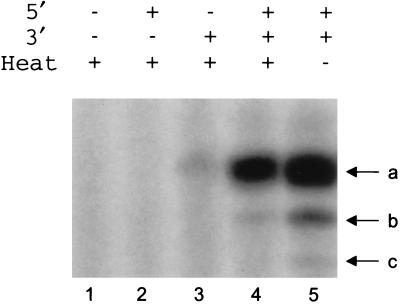
Preliminary experiments with an RNA polymerase that expressed endogenous influenza virus-like vRNA, prepared by transfection of 293T cells with the PB1, PB2, and PA expression plasmids plus an NP-expressing plasmid and pPOLI-GFP, showed that transcription activity was stable for 15 min at 40°C but became unstable at 45°C and higher temperatures (results not shown). We then tested if the heat stability of the RNA polymerase was a function of the presence of NP alone or of NP and influenza virus-like vRNA together. We prepared nuclear extracts of transiently transfected 293T cells expressing PB1, PB2, and PA and either NP alone or NP with influenza virus-like vRNA derived from the plasmid pPOLI-GFP. This plasmid contains the promoter and 5′ and 3′ noncoding regions of influenza A/WSN/33 virus segment 8, controlling the expression of a negative-sense green fluorescent protein. After 15 min of heat treatment at 40°C, the activity of the polymerase was evaluated with the [α-32P]GTP incorporation assay in the presence of added 15- and 14-nt oligonucleotides derived from the 5′ and 3′ ends, respectively, of the influenza virus vRNA promoter (see Materials and Methods). Only the RNA polymerase reconstituted in the presence of influenza virus-like vRNA derived from the plasmid pPOLI-GFP was heat stable (Fig. 3; compare lanes 7 and 8), thus giving rise to the major 14-nt transcript (band a) and minor transcripts b and c (see legend to Fig. 3). In addition, the previously detected 14-nt partial product, band a′ (5′ AGCAAAAGCAGGGT 3′), derived from pPOL1-GFP (see Fig. 2), accumulated preferentially. RNA polymerase, derived either from PB1, PB2, and PA or from PB1, PB2, PA, and NP, was heat labile (Fig. 3, lanes 4 and 6). Unheated controls (Fig. 3, lanes 3, 5, and 7) showed the expected major 14-nt transcription product. Thus, it was likely that the vRNA promoter was critical in protecting the polymerase against heat inactivation, although the influence of other nucleotides within the 5′ and 3′ noncoding regions, and the possibility that the NP cooperated with vRNA, could not be excluded at this stage.
FIG. 3.
Heat stability of the influenza virus RNA polymerase. The results of assays of the heat stability of the RNA polymerase are shown. RNA polymerase, isolated from 293T cells transfected with the plasmids as indicated, was preincubated for 15 min at 40°C (even-numbered lanes) or not preincubated (odd-numbered lanes), evaluated by the [α-32P]GTP incorporation assay in the presence of added 5′ and 3′ ends (each at 5 μM) of the vRNA promoter, and fractionated by 20% PAGE in 7 M urea. a′, 14-nt partial product AGCAAAAGCAGGGT, derived from the RNA of plasmid pPOLI-GFP; a, 14-nt product, derived from the added 14-nt 3′ end of wild-type vRNA; b, 13-nt minor transcript, probably derived from a contaminating 13-nt vRNA template; c, 12-nt product (see legend to Fig. 2). The nuclear extracts (see Materials and Methods) used in lanes 1 and 2 were prepared from mock-transfected (no added plasmids) 293T cells.
Next we tested whether the addition of the 5′ and 3′ ends of the vRNA promoter to the RNA polymerase prior to heat treatment would give the same protective effect in vitro. Figure 4 shows that when the 15- and 14-nt oligonucleotides (see Materials and Methods), corresponding to the 5′ and 3′ ends, respectively, of the vRNA promoter, were added to the RNA polymerase (reconstituted without influenza virus-like vRNA or NP) prior to heat treatment, the enzyme was protected against heat inactivation when evaluated in the [α-32P]GTP incorporation assay. Significant protection required the presence of both oligonucleotides (Fig. 4, lane 4), although a very minor level of protection was reproducibly observed with the 3′ end alone (see lane 3 and Discussion). No protection was seen with the 5′ end alone (Fig. 4, lane 2). Since the 15- and 14-nt vRNA promoter oligonucleotides contain only the consensus promoter sequences plus two additional base pairs, this suggests that the segment 8-specific 5′ and 3′ noncoding regions present in pPOLI-GFP and the NP were unlikely to have contributed to the heat stability of the polymerase observed in vivo (Fig. 3, lane 8). Overall, we conclude that both the 5′ and 3′ ends of vRNA are required to protect the influenza virus RNA polymerase against heat inactivation.
FIG. 4.
Both the 5′ and 3′ ends of the vRNA promoter are needed to obtain significant protection of the RNA polymerase from heat inactivation. RNA polymerase, obtained by transfection with PB1, PB2, and PA, was heated for 15 min at 40°C in the presence of the indicated oligonucleotides (5 μM), and transcription was subsequently evaluated with the [α-32P]GTP incorporation assay with ApG in the presence of both 5′ and 3′ vRNA oligonucleotides (2.5 μM). Lane 1, no added RNA; lane 2, 5′ vRNA strand (15 nt); lane 3, 3′ vRNA strand (14 nt); lane 4, 5′ plus 3′ strand; lane 5, unheated control; a, 14-nt product; b, 13-nt minor product; c, 12-nt minor product.
Base pairing between the 5′ and 3′ strands of the vRNA promoter is needed to protect the RNA polymerase from heat inactivation.
By introducing single or multiple mutations between residues 10 and 14 of the 3′ end of the vRNA promoter, thereby disrupting one to three base pairs of the panhandle, we tested whether both the 5′ and 3′ ends of the vRNA promoter were needed to protect the influenza virus RNA polymerase from heat inactivation. Then, using a previously established methodology (8), we complemented the mismatched base(s) by mutating the appropriate base(s) of the 5′ end of the vRNA promoter to reform alternative base pairs. Figure 5A shows the wild type and 10 mutant panhandle structures tested. Overall, the activity levels of the mismatched constructs remaining after heat inactivation, as evaluated by the [α-32P]GTP incorporation assay, ranged from 1 to 9% of the wild-type activity level (Fig. 5A). A potential complication is the need to ensure that the ApG-dependent transcription assay can effectively measure the polymerase activity of mutants, which are known from previous work to be inactive or only minimally active in transcription (7, 8). Thus, we performed control experiments in which we showed that, prior to testing by the [α-32P]GTP incorporation assay, wild-type vRNA promoter sequences efficiently displaced these mismatched constructs from the polymerase. These results (data not shown) revealed that the wild-type promoter used in the ApG transcription assay can displace any potential binding of these mismatched promoter sequences to the polymerase. This confirms that the low activities observed after heat inactivation in the presence of the mismatched panhandle constructs (Fig. 5A, sequences 2, 4, 6, 8, and 10) were valid results and were not an artifact of the testing procedure. Significantly, in the case of mutants 3, 5, 7, 9, and 11, for which we complemented mismatched base pairs with either single, double, or triple base pairs, we were able to recover activity levels ranging from 47 to 104% (Fig. 5A) of the wild-type activity level. Figure 5B shows a typical result for those mutants (lanes 1, 3, 5, 7, 9, and 11). It may be noted (Fig. 5B) that because of their differing sequences, the mutant transcripts (bands a1 to a5), although all were 14 nt in length, differed slightly from one another and from the wild-type transcript (band a) in their gel mobility, as expected from previous work (7, 8). Further control experiments (data not shown) confirmed that in the case of the base pair mutants which provided significant protection against heat inactivation (Fig. 5B, mutants 3, 5, 7, 9, and 11), the results obtained were independent of the addition of wild-type vRNA promoter sequences. This confirmed that the vRNA promoters for these particular mutants remained bound to the polymerase and, under our experimental conditions, were not significantly displaced by the inclusion of wild-type vRNA promoter sequences in the ApG-dependent transcription assay which was used as a readout of polymerase activity.
The fact that disruption of even one base pair of the panhandle interfered with its ability to protect the polymerase in the heat inactivation assay, coupled with the fact that alternative base pairs partially or completely restored protection, argues strongly in favor of a role for the panhandle in polymerase protection.
Hairpin loops in both the 5′ and 3′ strands of the vRNA promoter are needed to protect the RNA polymerase from heat inactivation.
As the panhandle structure protected the RNA polymerase from heat inactivation, it was of interest to establish whether protection required a panhandle with both a 5′ and a 3′ hairpin loop, as in the corkscrew model, or whether only a 5′ hairpin loop was needed, as in the 5′ hairpin loop model (33).
To address this question, we initially used mutant 15-nt-long oligonucleotides of the 5′ strand of vRNA with point mutations within the 5′ hairpin loop. These were tested in the presence of 14-nt 3′-strand oligonucleotides with wild-type 3′ hairpin loop sequences. Four mutants were designed for disruption of base pairs in the stem of the 5′ hairpin loop (Fig. 6 mutants 12, 13, 15, and 16 at positions 2′, 3′, 8′, and 9′, respectively). However, one mutant, mutant 14, had a G→U mutation at position 5′ of the hairpin loop. All 5′-strand mutants had an additional 12′ G→U panhandle mutation, because they had been originally designed for other experiments. However, this additional 12′ G→U mutation in the panhandle did not interfere in the experimental approach, because the 5′ mutants were complemented with a 14-nt mutant 3′ strand containing an A→U base pair mutation at position 11. This mutant 3′ strand restored base pairing with the 5′ mutant strand in the panhandle, giving rise to an 11, 12′ A→U base pair that was known to be 47% as effective as wild-type vRNA (see Fig. 5A, sequence 3) in protecting the RNA polymerase against heat inactivation. We used the heat inactivation assay, in the presence of added pseudo-wild-type vRNA promoter (see Materials and Methods) to provide an active vRNA template for mutants that were known to be inactive in transcription, for testing the remaining polymerase activity in the [α-32P]GTP incorporation assay (7). Figure 6 shows that only mutant 14 significantly protected against heat inactivation. All of the other mutants, i.e., mutants 12, 13, 15, and 16, failed to give a convincing level of protection above that of a control with the 3′ strand of the vRNA promoter alone (Fig. 6, lane labeled 3′). These results suggest that base pairs in the stem of the 5′ hairpin loop are important in protection against heat inactivation of the influenza virus RNA polymerase.
To establish if the hairpin loop in the 3′ end of the vRNA promoter is required to protect against heat inactivation, we tested point and multiple mutations of the 14-nt 3′ end of vRNA (Fig. 7) in the presence of the 15-nt wild-type 5′ strand of vRNA to evaluate activity in the subsequent ApG-primed [α-32P]GTP incorporation assay, again in the presence of wild-type vRNA promoter sequences (see above). Initially we selected mutants 18 and 19 (see Fig. 7A) because these mutants, with multiple sequence changes from wild-type vRNA, are known to inhibit ApG-primed transcription (8). Mutant 17, with a triple mutation in the hairpin loop, was selected as a potential positive control, since it is known to retain full transcriptional activity (8). The results (Fig. 7B) suggest that the 3′ hairpin loop was required to protect against heat inactivation. If both base pairs in the stem of the loop were disrupted (as for mutant 18), the activity level was no higher than that of a control (Fig. 7B, 3′) in which inactivation was carried out in the presence of the 3′ strand of the promoter alone. If only one base pair was retained (as for mutant 19), slight residual activity was apparent (Fig. 7B). However, when both base pairs of the stem were present (as for mutant 17), significant protection was observed.
To confirm the hypothesis that the 3′ hairpin loop was required to protect against heat inactivation, five further point and multiple mutants were selected. These differed from the mutants selected above in that they were all known to be as active, or nearly as active, as the wild type in ApG-primed transcription (8). The results (Fig. 7C) suggest that none of the mutants efficiently protected against heat inactivation, except mutants 21 and 22, which showed partial protection. Interestingly, these two mutants retained one of the two base pairs of the 3′ hairpin loop, possibly suggesting that some secondary structure resembling the 3′ hairpin loop was forming. Mutant 20 (9, G→C) (Fig. 7C), however, showed no evidence of significant protection.
Overall, we conclude that both the 5′ and 3′ hairpin loops of vRNA appear to be important if vRNA is to efficiently protect the influenza virus RNA polymerase against heat inactivation.
DISCUSSION
The RNA polymerase of influenza virus is tightly bound to the viral RNA promoter and packaged with NP into eight ribonucleoprotein complexes. In the infected cell, however, the RNA polymerase is present both as free or partially assembled polymerase, in transit from its site of synthesis in the cytosol to the nucleus, and as bound polymerase, complexed to the vRNA or the cRNA promoter in the nucleus. However, most of the time the polymerase would be expected to be bound to either the influenza virus vRNA or cRNA promoter. The purpose of this study was to test whether the vRNA promoter was involved in stabilizing the enzyme, and if so, what sequence and secondary structure features of this promoter were required.
To address these questions, it was first necessary to isolate recombinant RNA polymerase in the absence of influenza virus promoter sequences. A plasmid-based, transient expression system in human kidney 293T cells was devised. This system was a modification of the 12-plasmid transfection procedure devised to rescue novel influenza viruses (5, 27). Although other influenza virus RNA polymerase expression systems are described in the literature, they all had significant disadvantages for our purposes. Baculovirus (13) or Pichia (16) expression vectors were not used, because insect or yeast cells may not express an RNA polymerase with the correct posttranslational modifications (e.g., phosphorylation) present in naturally infected avian or mammalian cells. Such modifications may be required for polymerase binding to host factors such as RNA polymerase II or hCLE, which might regulate influenza virus replication (6, 15). Vaccinia virus expression in HeLa cells was not attempted, because RNA polymerase isolated by this approach supported only limited transcription (12). However, a recently described affinity method for partially purifying vaccinia virus-expressed influenza virus RNA polymerase from HeLa cells looks more promising (22). Finally, transfection of plasmids into vaccinia virus T7 polymerase-infected COS-1 cells was inappropriate, because active RNA polymerase could not be isolated in the absence of endogenous influenza virus-like vRNA (28).
Characterization of the recombinant RNA polymerase.
We initially demonstrated that the isolated recombinant RNA polymerase is transcriptionally active by using ApG as a primer in the presence of two short synthetic oligonucleotides containing the 5′ and 3′ ends of the vRNA promoter (Fig. 1). Transcriptional activity was dependent on the presence of all three subunits of the RNA polymerase, which is a result consistent with previous plasmid reconstitution experiments (29) but inconsistent with other studies which suggested that all three subunits are not required (18, 25, 26). Perhaps all three subunits are needed for efficient synthesis but residual activity, below our level of detection, can occur with short model vRNA templates in the absence of one or more subunits. Although NP is essential for in vivo replication of influenza virus RNA (2, 14), the presence of NP was not required for the synthesis of short 14-nt transcripts (Fig. 1), confirming previous studies with short RNA templates (12, 13, 16, 22). Recombinant RNA polymerase prepared by our method also required, as expected (7, 8, 13), both the 5′ and 3′ ends of the vRNA promoter for significant transcriptional activity (Fig. 2). Moreover, the recombinant RNA polymerase had the other expected properties of an authentic RNA polymerase, e.g., priming with capped (globin mRNA) primers, endonuclease activity, and poly(A) polymerase activity (data not shown). Interestingly, we were able to directly demonstrate (Fig. 2) that the RNA polymerase, which bound to an endogenous vRNA promoter, copied an added 3′ end of the vRNA promoter in trans, thus confirming earlier suggestions that this could occur (30).
Influenza virus vRNA promoter in a corkscrew configuration is needed to protect the RNA polymerase from heat inactivation.
Having isolated recombinant RNA polymerase, we investigated its stability with respect to heat. The RNA polymerase was essentially stable during 15 min of heat treatment at 40°C in the presence of endogenously synthesized influenza virus vRNA sequences and NP. In the absence of such influenza virus-like vRNA sequences, the polymerase was completely inactivated (Fig. 3). Protection against heat inactivation was restored when the vRNA promoter was added prior to heat treatment. Protection by the vRNA promoter required both the 5′ and 3′ ends of the vRNA promoter, although a minor level of protection reproducibly occurred in the presence of the 3′ end of the vRNA promoter alone (Fig. 4). In high concentrations, the presence of the 3′ end of the vRNA promoter alone also stimulated the synthesis of low yields of transcripts in the ApG-dependent transcription assay. Because the RNA polymerase enzyme preparation was impure, we could not determine whether this low-level activity of the 3′ end of the vRNA promoter is an intrinsic property of the RNA polymerase itself or whether it results from the presence of cryptic influenza virus 5′ vRNA or influenza virus-mimicking vRNA species, e.g., small nuclear RNAs, that might contaminate the RNA polymerase preparation. Contamination with 5′ vRNA promoter sequences would seem to be the more likely scenario, however, since all presently available evidence suggests that both the 5′ and 3′ ends of the vRNA promoter are needed for efficient promoter activity, although the presence of the 5′ end of the vRNA promoter alone is sufficient for cap binding (1). Contamination with 5′ vRNA promoter sequences is certainly possible, because the 3′ noncoding regions of the mRNA for PB1, PB2, and PA contain antisense 5′ vRNA promoter sequences derived from their pcDNA3 expression plasmids (see Materials and Methods). If these expression plasmids were transcribed by the host RNA polymerase from a cryptic promoter in antisense orientation, albeit inefficiently, this transcription would result in the generation of 5′ vRNA influenza virus promoter sequences, which could explain why we observed low-level transcription in the presence of an added 3′ end of vRNA alone.
Moreover, 5′- and 3′-end promoter mutations that disrupted base pairs between the 5′ and 3′ ends of the vRNA promoter, thus preventing or interfering in the stability of the panhandle, abolished protection (Fig. 5). However, complementary mutations, which reformed alternative base pairs in the panhandle, restored protection partially, or in some cases completely, even although different base pairs were now present (Fig. 5). Overall, it was clear that significant protection against heat inactivation required both the 5′ and 3′ ends of the vRNA promoter, held together in a panhandle configuration. Other results (data not shown) suggested that influenza virus cRNA promoter sequences also protected the RNA polymerase against heat inactivation. Again, both the 5′ and 3′ strands of the cRNA promoter were required for significant protection.
Next, we investigated whether the local hairpin loops in the 5′ and 3′ arms of the vRNA promoter had to be present in vRNA to protect the RNA polymerase against heat inactivation. As expected, given the importance of the 5′ hairpin loop for binding, endonuclease cleavage, transcription, and polyadenylation (7-9, 17, 21, 30, 34), mutations which disrupted either of the base pairs in the stem of the 5′ hairpin loop failed to protect the RNA polymerase, whereas a mutation in the hairpin loop itself still protected against heat inactivation (Fig. 6).
The role of the 3′ hairpin loop has been more controversial than that of the 5′ hairpin loop, but recent evidence has confirmed its importance for endonuclease cleavage (20), despite earlier indications that it was not required for ApG-primed transcription and polyadenylation (8, 33). Here we found that mutations of the stem of the 3′ hairpin loop of the promoter convincingly showed that this secondary structure feature was required for efficient protection of the polymerase against heat inactivation. All mutants which disrupted both base pairs of the stem failed to confer protection. By contrast, a triple mutation at positions 5, 6, and 7 within the hairpin loop itself was tolerated (Fig. 7). However, a detailed study of three position 9 mutants showed that partial protection occurred in two of the mutants when only one of the base pairs of the stem was disrupted. This result suggests that the role of the 3′ hairpin loop may be somewhat less critical than that of the 5′ hairpin loop. Some degree of relaxation of secondary structure is still compatible with partial protection against heat inactivation of the polymerase. Our heat inactivation data are thus consistent with the hypothesis that the 3′ hairpin loop is a transient structure which has to melt before transcription can be initiated (20). Thus, the 3′ hairpin loop may aid binding to the polymerase but its presence is not as critical as that of the 5′ hairpin loop, which must remain bound to the template for correct polyadenylation (7, 8, 30, 33, 34, 37).
It would be interesting to extend the present studies on heat inactivation of the influenza virus RNA polymerase by investigating whether the vRNA secondary structure requirements for conferring protection against heat inactivation would differ if an activity of the polymerase other than ApG-dependent transcription were used as a readout assay. Thus, an endonuclease cleavage assay (21) or mRNA poly(A) assay (34) might give different results from those of the ApG-dependent transcription assay used in this study, e.g., for mutations which disrupt the stem of the 3′ hairpin loop of vRNA, as discussed above. However, to investigate this fully would require a detailed study that is beyond the scope of this paper.
General implications.
It is well known that the vRNA promoter binds strongly to the polymerase. Indeed, progress has been made in localizing the amino acid residues in PB1 involved in the various interactions (10, 11, 23). However, to our knowledge, there have been no previous experimental data showing that the vRNA promoter stabilizes the RNA polymerase and protects it against heat inactivation. In the absence of the vRNA promoter, we envisage a polymerase structure that is open and labile to heat inactivation. In the presence of the promoter, the polymerase structure might be closed and more stable. Since the RNA polymerase has to survive inside the infected cell throughout the infection, a polymerase complex with the maximum possible half-life could have evolved to give the virus a selective advantage. It would be interesting to test if avian influenza viruses, which replicate at the temperatures found in birds (i.e., higher than those found in mammals), have RNA polymerases which are more heat stable. It would also be instructive to test whether pathogenic human influenza viruses have polymerases that exhibit greater heat stability than less pathogenic strains. Such a property of human viruses could allow replication at the higher temperatures found in the lung, thus facilitating the development of pneumonia, whereas the less pathogenic influenza viruses, because of their heat lability, might be confined to the lower temperatures of the upper respiratory tract.
We have not studied the mechanism of heat inactivation of the RNA polymerase. One possibility, however, is that the PA subunit is bound more weakly than the other subunits to the polymerase complex and dissociates when subjected to heating, leaving an inactive, dimeric complex. This hypothesis is supported by previous observations that PA is less stably associated with PB1 and PB2 in a recombinant polymerase complex, formed in the absence of influenza virus vRNA, than in a complex isolated from influenza virus-infected cells (3, 36).
In conclusion, we have found that the presence of both the 5′ and 3′ hairpin loops of vRNA in the corkscrew model (4) appears to be important if vRNA is to efficiently protect the influenza virus RNA polymerase against heat inactivation. This result is further evidence of the importance of the panhandle and the 5′ and 3′ hairpin loops of the vRNA promoter in providing the necessary secondary structure to ensure tight binding of the promoter to the polymerase as a prelude to endonuclease cleavage, transcription, and polyadenylation.
Acknowledgments
G.G.B. and J.L.S. were supported by MRC program grant G9523972 to G.G.B.
We thank Peter Palese for the pPOLI-GFP, Ervin Fodor and Mandy Crow for helpful discussions, and Alice Taylor for DNA sequencing.
REFERENCES
- 1.Cianci, C., L. Tiley, and M. Krystal. 1994. Differential activation of the influenza virus polymerase via template binding. J. Virol. 69: 3995-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De la Luna, S., J. Martin, A. Portela, and J. Ortin. 1993. Influenza virus naked RNA can be expressed upon transfection into cells co-expressing the three subunits of the polymerase and the nucleoprotein from SV40 recombinant viruses. J. Gen. Virol. 74:535-539. [DOI] [PubMed] [Google Scholar]
- 3.Detjen, B. M., C. St. Angelo, M. G. Katze, and R. M. Krug. 1987. The three influenza virus polymerase (P) proteins not associated with viral nucleocapsids in the infected cell are in the form of a complex. J. Virol. 61:16-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flick, R., G. Neumann, E. Hoffmann, E. Neumeier, and G. Hobom. 1996. Promoter elements in the influenza vRNA terminal structure. RNA 2:1046-1057. [PMC free article] [PubMed] [Google Scholar]
- 5.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. García-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fodor, E., A. Mikulasova, L. J. Mingay, L. L. M. Poon, and G. G. Brownlee. 2000. Messenger RNAs that are not synthesized by RNA polymerase II can be 3′ end cleaved and polyadenylated. EMBO Rep. 1:513-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1995. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J. Virol. 69:4012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fodor, E., B. L. Seong, and G. G. Brownlee. 1993. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J. Gen. Virol. 74:1327-1333. [DOI] [PubMed] [Google Scholar]
- 10.González, S., and J. Ortín. 1999. Characterization of influenza virus PB1 protein binding to viral RNA: two separate regions of the protein contribute to the interaction domain. J. Virol. 73:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González, S., and J. Ortín. 1999. Distinct regions of influenza virus PB1 polymerase subunit recognize vRNA and cRNA templates. EMBO J. 18:3767-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagen, M., T. D. Y. Chung, J. A. Butcher, and M. Krystal. 1994. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J. Virol. 68:1509-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honda, A., A. Endo, K. Mizumoto, and A. Ishihama. 2001. Differential roles of vRNA and cRNA in functional modulation of the influenza virus RNA polymerase. J. Biol. Chem. 276:31179-31185. [DOI] [PubMed] [Google Scholar]
- 14.Huang, T. S., P. Palese, and M. Krystal. 1990. Determination of influenza viral proteins required for genome replication. J. Virol. 64:5669-5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huarte, M., J. J. Sanz-Ezquerro, F. Roncol, J. Ortin, and A. Nieto. 2001. PA subunit from influenza virus polymerase complex interacts with a cellular protein with homology to a family of transcriptional activators. J. Virol. 75:8597-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang J.-S., K. Yamada, A. Honda, K. Nakade, and A. Ishihama. 2000. Expression of functional influenza virus RNA polymerase in the methylotrophic yeast Pichia pastoris. J. Virol. 74:4074-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, H. J., E. Fodor, G. G. Brownlee, and B. L. Seong. 1997. Mutational analysis of the RNA-fork model of the influenza A virus vRNA promoter in vivo. J. Gen. Virol. 78:353-357. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, M., T. Toyoda, and A. Ishihama. 1996. Influenza virus PB1 protein is the minimal and essential subunit of RNA polymerase. Arch. Virol. 141:525-539. [DOI] [PubMed] [Google Scholar]
- 19.Lamb, R. A., and R. M. Krug. 1996. Orthomyxoviridae: the viruses and their replication, p. 1353-1395. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 20.Leahy, M. B., H. C. Dobbyn, and G. G. Brownlee. 2001. A hairpin loop structure in the 3′ arm of the influenza A virus virion RNA promoter is required for endonuclease activity. J. Virol. 75:7042-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leahy, M. B., D. C. Pritlove, L. M. Poon, and G. G. Brownlee. 2001. Mutagenic analysis of the 5′ arm of the influenza A virus virion RNA promoter defines the sequence requirements for endonuclease activity. J. Virol. 75:134-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, M. T. M., K. Bishop, L. Medcalf, D. Elton, P. Digard, and L. Tiley. 2002. Definition of the minimal viral components required for the initiation of unprimed synthesis by influenza virus RNA polymerase. Nucleic Acids Res. 30:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, M. L., B. C. Ramirez, and R. M. Krug. 1998. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 17:5844-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikulasova, A., E. Vareckova, and E. Fodor. 2000. Transcription and replication of the influenza A virus genome. Acta Virol. 44:273-282. [PubMed] [Google Scholar]
- 25.Nakagawa, Y., N. Kimura, T. Toyoda, K. Mizumoto, A. Ishihama, K. Oda, and S. Nakada. 1995. The RNA polymerase PB2 subunit is not required for replication of the influenza virus genome but is involved in capped mRNA synthesis. J. Virol. 69:728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawa, Y., K. Oda, and S. Nakada. 1996. The PB1 subunit alone can catalyze cRNA synthesis, and the PA subunit in addition to the PB1 subunit is required for viral RNA synthesis in replication of the influenza viral genome. J. Virol. 70:6390-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortega, J., J. Martín-Benito, T. Zürcher, J. M. Valpuesta, J. L. Carrascosa, and J. Ortín. 2000. Ultrastructural and functional analyses of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J. Virol. 74:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perales, B., and J. Ortín. 1997. The influenza A virus PB2 polymerase subunit is required for the replication of viral RNA. J. Virol. 71:1381-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poon, L. L., D. C. Pritlove, J. Sharps, and G. G. Brownlee. 1998. The RNA polymerase of influenza virus, bound to the 5′ end of virion RNA, acts in cis to polyadenylate mRNA. J. Virol. 72:8214-8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Portela, A., T. Zürcher, A. Nieto, and J. Ortín. 1999. Replication of orthomyxoviruses. Adv. Virus Res. 54:319-348. [DOI] [PubMed] [Google Scholar]
- 32.Pritlove, D. C., E. Fodor, B. L. Seong, and G. G. Brownlee. 1995. In vitro transcription and polymerase binding studies of the termini of influenza A virus cRNA: evidence for a cRNA panhandle. J. Gen. Virol. 76:2205-2213. [DOI] [PubMed] [Google Scholar]
- 33.Pritlove, D. C., L. L. Poon, L. J. Devenish, M. B. Leahy, and G. G. Brownlee. 1999. A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J. Virol. 73:2109-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pritlove, D. C., L. L. Poon, E. Fodor, J. Sharps, and G. G. Brownlee. 1998. Polyadenylation of influenza virus mRNA transcribed in vitro from model virion RNA templates: requirement for 5′ conserved sequences. J. Virol. 72:1280-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simoncsits, A., G. G. Brownlee, R. S. Brown, J. R. Rubin, and H. Guilley. 1977. New rapid gel sequencing method for RNA. Nature 269:833-836. [DOI] [PubMed] [Google Scholar]
- 36.St. Angelo, C., G. E. Smith, M. D. Summers, and R. M. Krug. 1987. Two of the three influenza viral polymerase proteins expressed by using baculovirus vectors form a complex in insect cells. J. Virol. 61:361-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tiley, L. S., M. Hagen, J. T. Matthews, and M. Krystal. 1994. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J. Virol. 68:5108-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]