Abstract
De novo RNA synthesis by hepatitis C virus (HCV) nonstructural protein 5B (NS5B) RNA-dependent RNA polymerase has been investigated using short RNA templates. Various templates including those derived from the HCV genome were evaluated by examining the early steps of de novo RNA synthesis. NS5B was shown to be able to produce an initiation dinucleotide product from templates as short as 4-mer and from the 3′-terminal sequences of both plus and minus strands of the HCV RNA genome. GMP, GDP, and guanosine were able to act as an initiating nucleotide in de novo RNA synthesis, indicating that the triphosphate moiety is not absolutely required by an initiating nucleotide. Significant amounts of the initiation product accumulated in de novo synthesis, and elongation from the dinucleotide was observed when large amounts of dinucleotide were available. This result suggests that NS5B, a template, and incoming nucleotides are able to form an initiation complex that aborts frequently by releasing the dinucleotide product before transition to an elongation complex. The transition is rate limiting. Furthermore, we discovered that the secondary structure of a template was not essential for de novo initiation and that 3′-terminal bases of a template conferred specificity in selection of an initiation site. Initiation can occur at the +1, +2, or +3 position numbered from the 3′ end of a template depending on base composition. Pyrimidine bases at any of the three positions are able to serve as an initiation site, while purine bases at the +2 and +3 positions do not support initiation. This result implies that HCV possesses an intrinsic ability to ensure that de novo synthesis is initiated from the +1 position and to maintain the integrity of the 3′ end of its genome. This assay system should be an important tool for investigating the detailed mechanism of de novo initiation by HCV NS5B as well as other viral RNA polymerases.
Hepatitis C virus (HCV) is a single-stranded, positive-sense RNA virus. It encodes a single large polyprotein that is subsequently processed into at least 10 separate viral proteins by host as well as virally encoded proteases (17, 21). Of these proteins, nonstructural protein 5B (NS5B) is an RNA-dependent RNA polymerase (RdRp) that is responsible for replication of viral RNA genome. Because NS5B is unique to the virus, it is a potential target for developing antiviral drugs and is being subjected to intense research. Recently, several crystal structures of NS5B have been reported, revealing several unique structural features, including a fully encircled active site and a β-hairpin structure in the thumb subdomain protruding towards the active site (1, 2, 9). Efforts have been made to characterize NS5B RdRp functions and to understand the mechanistic differences between NS5B and cellular polymerases. This information would help to design direct and specific inhibitors for HCV replication.
HCV NS5B is believed to initiate RNA synthesis by a de novo mechanism. De novo initiation of RNA synthesis is common for most viral RdRps, with a few exceptions such as poliovirus 3Dpol, which uses a protein primer (7). Recently, several groups reported that recombinant NS5B isolated from Escherichia coli or the baculovirus/insect cell expression system was able to initiate de novo RNA synthesis (12, 14, 23). Most of these studies were focused on identifying essential features in templates for initiating de novo RNA synthesis. A recombinant NS5B from E. coli was shown to initiate de novo RNA synthesis and yield a full-length HCV RNA genome (14). A minimal 98 nucleotides (nt) from the 3′ end of the plus strand or 239 nt from the 3′ end of the minus strand HCV RNA genome was found to serve as a template for de novo RNA synthesis, indicating the presence of cis-acting sequences for HCV RNA synthesis (14). In line with this observation, shorter pieces of 3′-terminal HCV RNA were reported to be unable to direct de novo RNA synthesis (8, 14). Another study showed that a 21- or 22-nt template from the 3′ terminus of the HCV(+) RNA genome was replicated in the presence of Mn2+, while hardly any products were observed with Mg2+ as a cofactor (23). Non-HCV RNA templates were also tested, and a 25-nt RNA derived from the brome mosaic virus genome was shown to be able to initiate de novo synthesis by NS5B (8). Recently, it was reported that the deletion of 98 nt from the 3′ terminus of the plus strand HCV RNA enhanced RNA synthesis, while the deletion of 45 nt from the 3′ terminus of the minus strand did the opposite for replication (16). These studies led to the conclusion that efficient de novo initiation requires specific sequences and structural elements recognized by NS5B.
These previously reported investigations are mostly based on assays that detect template-length RNA products in the presence of a template and all four nucleoside triphosphates (NTPs). Interpretation of these results might not be straightforward, because the polymeric products were hard to characterize. Initiation of de novo RNA synthesis consists of several steps. First, NS5B recognizes and binds the 3′ terminus of the viral RNA genome to form an enzyme/template complex. Two incoming nucleotides complementary to 3′-terminal and penultimate bases then join the complex to form Watson-Crick base pairs with the template. In the enzyme/template/nucleotide tetrameric complex, the 3′-hydroxyl group of the initiating nucleotide initiates a nucleophilic attack onto the α-phosphate of the elongating nucleotide to generate the first phosphodiester bond, resulting in the formation of a dinucleotide initiation product. The next complementary nucleotide then enters into the complex to start the second round of nucleotidyl transfer reaction, where the dinucleotide is extended from the 3′-hydroxyl group to form a trinucleotide product. To understand the mechanism of de novo initiation by NS5B or other viral RdRps, kinetic and structural characterization of each step should be performed using an appropriate assay system.
A recent study has shown that NS5B can utilize 2- or 3-nt RNA primers to efficiently initiate RNA replication (22). It was proposed that di- or tri-nucleotides, generated either by polymerases or from abortive cellular RNA transcription, might act as a primer for RNA replication by NS5B. That study provided a well-defined assay to investigate the elementary steps of a primer-dependent polymerase reaction.
In this report, we show that RNA oligonucleotides are templates that can be efficiently utilized to examine the early steps of de novo initiation by HCV NS5B with Mg2+ as a cofactor. We used various short RNA templates, including those derived from the HCV genome, to examine various aspects of de novo initiation, including initial dinucleotide formation, elongation, and template requirements. Altered nucleotides and nucleosides were tested as an initiating substrate. The implications of our studies for understanding the replication mechanism of the HCV genome are discussed.
MATERIALS AND METHODS
Materials.
[α-33P]CTP, [γ-33P]GTP, and [α-33P]UTP (3,000 Ci/mmol) were purchased from Perkin-Elmer. Various nucleoside 5′ triphosphates were obtained from Gibco BRL. GDP, GMP, guanosine, N-2 methyl guanosine, inosine, and ApC were from Sigma. All RNA oligonucleotides were from Oligos Etc. (Wilsonville, Oreg.) and were purified by polyacrylamide gel electrophoresis. All other reagents were of the highest grade available from ICN, Sigma, Fisher, or Ambion.
Protein expression and purification.
A cDNA clone encoding the HCV-1b CON1 strain was obtained from R. Bartenschlager and was used to make an NS5B expression construct with the C-terminal 21 amino acids deleted and with a His tag added at the C terminus as reported previously (4). The resulting plasmid was transformed into E. coli BL21(DE3) cells. Protein production was induced by 0.5 mM isopropyl-β-d-thiogalactopyranoside at 24°C when cells reached an optical density at 600 nm of 1.0. Soluble cell lysates were loaded onto a Ni-chelating Sepharose column (Amersham Pharmacia Biotech, Piscataway, N.J.). The bound protein was eluted with a buffer containing 70 to 300 mM imidazole. Fractions containing NS5B were pooled and loaded onto an SP column. NS5B was eluted with a linear gradient of 0 or 1 M NaCl. Fractions containing pure NS5B were pooled, concentrated, and stored at −80°C before use. The D318A/D319A double mutant was made using the QuikChange site-directed mutagenesis kit (Stratagene), and the mutant enzyme was purified by the same procedures described for the wild-type enzyme.
De novo initiation assay.
A standard reaction was carried out in 10 μl containing 50 mM HEPES (pH 7.3), 10 mM dithiothreitol, 5 mM MgCl2, 20 μM RNA template, 2.5 to 5 μM NS5B, 100 μM 33P-labeled nucleotide, and a 1 mM concentration of an initiating or elongating nucleotide as indicated. The reaction mixture was incubated at 30°C for 1 h and then quenched by the addition of 10-μl loading buffer (90% formamide, 0.025% bromophenol blue, and 0.025% xylene cyanol). The quenched reaction mixture was heated to 70°C for 2 to 5 min prior to loading 2 or 3 μl onto a denaturing 25% polyacrylamide-7 M urea-Tris-borate-EDTA (TBE) gel. Electrophoresis was performed in 1× TBE at 70 to 90 W. Gels were visualized and analyzed using a PhosphorImager.
RESULTS
Formation of initiation product.
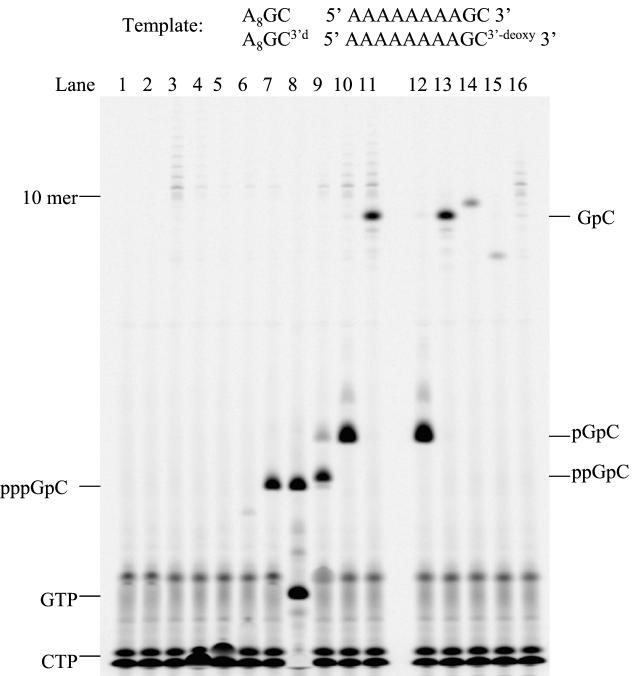
To examine the formation of a dinucleotide initiation product in de novo RNA synthesis by NS5B, we designed a 10-mer RNA template, 5′-AAAAAAAAGC-3′ (A8GC). In the presence of template, NS5B synthesized a dinucleotide product, pppGpC, using GTP as an initiating nucleotide and a radiolabeled elongating nucleotide, [α-33P]CTP. The product was resolved in a 25% acrylamide-7 M urea-TBE gel (Fig. 1, lane 7). Omitting either the template (Fig. 1, lane 2) or GTP (Fig. 1, lane 3) or using an initiating nucleotide other than GTP, including CTP, ATP, or UTP (Fig. 1, lanes 4 to 6), in the reaction gave no dinucleotide product. The results indicate that de novo initiation requires a template and a nucleotide complementary to the 3′-terminal base of the template. Replacing GTP and [α-33P]CTP with [γ-33P]GTP and CTP resulted in a similar radiolabeled band on the gel, confirming that the dinucleotide was produced by de novo synthesis with GTP as the initiating nucleotide and that the triphosphate of the initiating nucleotide was intact in the product (Fig. 1, lane 8). Use of 2′-deoxy-GTP instead of GTP gave no dinucleotide product (data not shown), indicating that NS5B, like other RNA polymerases, is highly selective for ribonucleotides (NTP) over 2′-deoxyribonucleotides (2′-deoxy-NTP). It was reported that the 5′ triphosphate group of an initiating nucleotide was not strictly required for de novo initiation by several RNA polymerases. Either GMP or GDP was shown to serve as an initiating nucleotide for de novo RNA synthesis by bovine viral diarrhea virus RdRp (6) and HCV NS5B (8). Guanosine or GMP has also been shown to be an initiator for transcriptional initiation by T7 RNA polymerase (13). The significance of the 5′ phosphate group of an initiating nucleotide in de novo initiation by NS5B was further explored. As shown in Fig. 1 (lanes 9 to 11), the reaction containing GDP, GMP, or guanosine as the only guanosine source produced a distinct band on the polyacrylamide gel that had a mobility different from that for the pppGpC band. The reaction with GDP produced a band (ppGpC) with a slightly lower mobility than the pppGpC band (Fig. 1, lane 9). The GMP-initiated reaction gave a more intense band (pGpC) with even lower mobility than the ppGpC band (Fig. 1, lane 10). Interestingly, the reaction initiated by guanosine gave a band that was well separated from the other dinucleotide bands (Fig. 1, lane 11). To confirm that this band represents GpC, several guanosine analogs were tested as an initiator. If the guanosine analogs can initiate dinucleotide synthesis, the resulting dinucleotides are expected to have mobilities similar but not identical to that of GpC due to the differences in charge and molecular weight. As shown in Fig. 1, guanosine analogs—N-2 methyl guanosine (lane 14) and inosine (lane 15)—yielded products with mobilities close to yet clearly distinct from those for the GpC band on the polyacrylamide gel electrophoresis gel. This experiment indirectly confirmed the identity of GpC. It is the first demonstration that guanosine or guanosine analogs can serve as an initiating nucleotide in de novo RNA synthesis catalyzed by NS5B. These results also illustrate that minor change in mass or charge of RNA products can be easily distinguished in our assay system.
FIG. 1.
Formation of initiation products in NS5B de novo RNA synthesis using oligoribonucleotide 5′-A8GC-3′ as a template. Each reaction contained 20 μM RNA template, 2.5 μM NS5B, 100 μM [α-33P]CTP, and 1 mM concentration of one of the following initiating nucleotides or nucleosides unless specified: lane 1, GTP (no enzyme); lane 2, GTP (no template); lane 3, no initiating nucleotide; lane 4, CTP; lane 5, UTP; lane 6, ATP; lane 7, GTP; lane 8, 100 μM [γ-33P]GTP (initiating nucleotide) to replace [α-33P]CTP, and 1 mM CTP; lane 9, GDP; lane 10, GMP; lane 11, guanosine; lanes 12 to 15, A8GC3′d as an template and GMP (lane 12), guanosine (lane 13), N-2 methyl guanosine (lane 14), and inosine (lane 15) as an initiating nucleotide; and lane 16, GTP as an initiating nucleotide with NS5B D318A/D319A double mutant. The reaction products were resolved on a 25% polyacrylamide-7 M urea-TBE gel and were analyzed by a PhosphorImager.
Terminal transferase activity.
We noticed that in the above experiments a ladder of RNAs longer than 10-mer was also produced. It was highly visible in the reaction containing [α-33P]CTP (100 μM) as the sole nucleotide (Fig. 1, lane 3). Addition of 1 mM cold NTP to the reaction diminished the intensity of these long RNA bands significantly (Fig. 1, lanes 4 to 6). The reactions with GMP or guanosine as an initiating nucleotide also resulted in similar long RNA bands (Fig. 1, lanes 10 and 11). These long RNA products could be produced by a terminal transferase activity that incorporates [α-33P]CTP to the 3′ terminus of the template. Since any NTP can be a substrate for terminal transferases, the presence of cold NTPs will compete with [α-33P]CTP for incorporation and reduce the intensity of the bands. However, GMP and guanosine lack the 5′ triphosphate moiety and cannot compete with CTP for the terminal transferase activity. To find out whether the long RNA products were the result of terminal transferase activity, a new template (A8GC3′d) was designed that replaced the 3′-terminal cytidine with 3′ deoxycytidine. Due to the lack of the 3′-hydroxyl group for elongation, the new template should not be utilized by any terminal transferase activity. The previous experiments were repeated with this new template. Complete abolition of all the long RNA products was observed (Fig. 1, lanes 12 to 15), confirming that these products were the result of terminal transferase activity. There are contradicting reports about the origin of terminal transferase activity in NS5B assays. The activity was believed to be either an intrinsic property of NS5B (15) or a contaminant from protein purification (10). To distinguish between these two possibilities, we prepared the D318A/D319A double mutant of NS5B. The two aspartic acid residues are highly conserved in many polymerases and serve as catalytic residues in phosphodiester bond formation (18). Thus, replacement of both Asp residues with Ala should abolish all the nucleotidyl transfer activities of NS5B. As shown in Fig. 1, lane 16, the D318A/D319A double mutant produced no dinucleotide product but still gave long RNA products, suggesting that the long RNA products are most likely from a contaminating terminal transferase copurified with NS5B. This result also demonstrates that these two Asp residues are critical for de novo initiation.
Template specificity.
To comprehend template requirements for de novo initiation, various short templates were chemically synthesized and tested for their ability to generate RNA products. Products were then analyzed on a denaturing polyacrylamide-urea-TBE gel. None of the RNA templates has any secondary structures that may be implicated in the interaction with NS5B for initiating de novo RNA synthesis. Therefore, our studies were focused solely on the enzyme's intrinsic ability to initiate de novo synthesis from a simple RNA template.
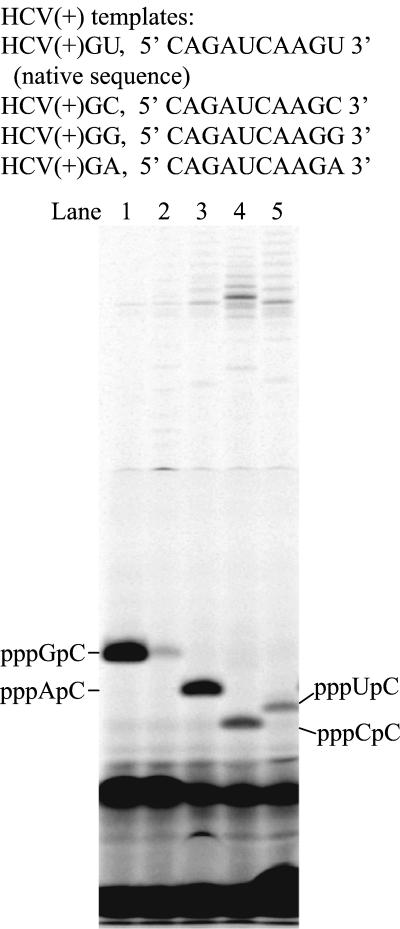
The HCV(+)GU template (10 nt, 5′-CAGAUCAAGU-3′) is derived from the 3′ terminus of the plus strand HCV RNA genome (Fig. 2). Templates with different 3′-terminal bases were also chemically synthesized and denoted as HCV(+)GC, HCV(+)GG, and HCV(+)GA, respectively. It was reported that short templates derived from the 3′-terminal HCV genome failed to generate products in de novo synthesis in the presence of Mg2+ (23). However, in our assay, all four HCV(+) templates were found to generate expected initiation products, pppApC, pppGpC, pppCpC, and pppUpC (Fig. 2). The relative activity of NS5B to utilize these templates are in the order of A8GC > HCV(+)GU > HCV(+)GG > HCV(+)GA > HCV(+)GC.
FIG. 2.
Formation of initiation products in NS5B de novo RNA synthesis using A8GC (lane 1) or HCV(+) templates (lanes 2 to 5). Each reaction contained 2.5 μM NS5B, 100 μM [α-33P]CTP, 20 μM specified RNA template, and 1 mM concentration of a nucleotide as follows: lane 1, A8GC and GTP; lane 2, HCV(+)GC and GTP; lane 3, HCV(+)GU and ATP; lane 4, HCV(+)GG and CTP; and lane 5, HCV(+)GA and UTP. The reaction products were resolved on a 25% polyacrylamide-7 M urea-TBE gel and were subjected to PhosphorImager analysis.
Elongation from initiation product.
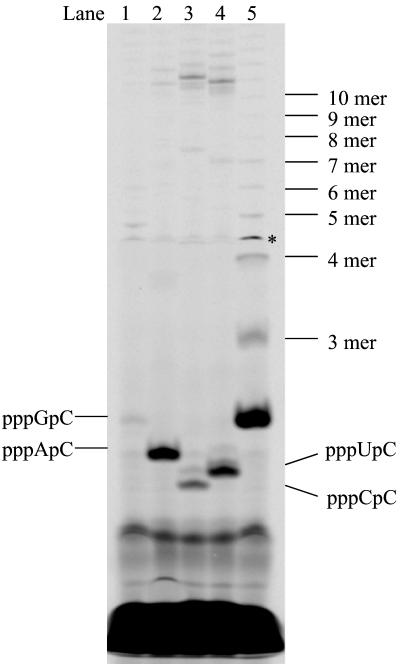
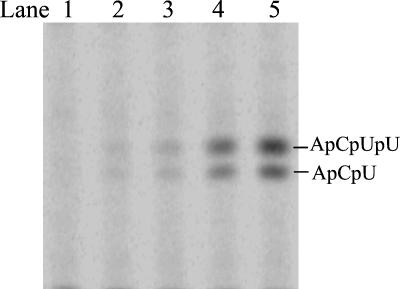
The above 10-nt RNA templates were used to further investigate the elongation steps after dinucleotide formation. Addition of UTP in the dinucleotide formation reactions is expected to give RNA products of 10-mer and 4-mer from A8GC and the HCV(+) templates, respectively. Using A8GC as a template in the elongation condition, a ladder of RNA products longer than 3-mer was observed (Fig. 3, lane 5). The products shorter than 10-mer were likely from the elongation reaction, and the ones longer than 10-mer were the result of the terminal transferase activity that was described above. Interestingly, the initial de novo synthesis product, pppGpC, accumulated significantly even in the presence of excess amounts of UTP. Use of a higher concentration of the template had little effect on the result, indicating that the template concentration was not the limiting factor. The elongation intermediates accumulated evenly on the gel, indicating that NS5B incorporates nucleotides at a uniform rate during elongation. However, when the HCV(+) templates were used, no extended products from dinucleotides were observed and the dinucleotides accumulated at a lower level than that found with A8GC (Fig. 3, lane 1 to 4). These reactions appeared to abort after the formation of dinucleotides. One explanation for the lack of elongation might be that, once the dinucleotide product is formed, it dissociates from the enzyme/template complex. The elongation requires a higher concentration of dinucleotide to proceed. To find out whether the amount of dinucleotide limits further elongation, an exogenous dinucleotide, ApC, was used as a primer for HCV(+)GU. As shown in Fig. 4, elongated products ApCpU and ApCpUpU started to appear when the ApC concentration reached 0.1 mM and kept increasing as more ApC was supplied, confirming that elongation was limited by the amount of dinucleotide formed.
FIG. 3.
Elongation from initiation products in NS5B de novo RNA synthesis. Each reaction contained 2.5 μM NS5B, 100 μM [α-33P]CTP, 2 mM UTP, 20 μM RNA template, and 1 mM concentration of an initiating nucleotide as indicated. Lane 1, HCV(+)GC template and GTP; lane 2, HCV(+)GU template and ATP; lane 3, HCV(+)GG template and CTP; lane 4, HCV(+)GA template and UTP; and lane 5, A8GC template and GTP. The reaction products were resolved on a 25% polyacrylamide-7 M urea-TBE gel and were analyzed using a PhosphorImager. Asterisk denotes gel artifact.
FIG. 4.
Elongation in NS5B de novo synthesis with HCV(+)GU as a template and ApC as a primer. Each reaction contained 2.5 μM NS5B, 100 μM [α-33P]UTP, 20 μM HCV(+)GU template, and one of the following concentrations of ApC: lane 1, none; lane 2, 50 μM; lane 3, 100 μM; lane 4, 500 μM; and lane 5, 1 mM. The reaction products were resolved on a 25% polyacrylamide-7 M urea-TBE gel and were subjected to PhosphorImager analysis.
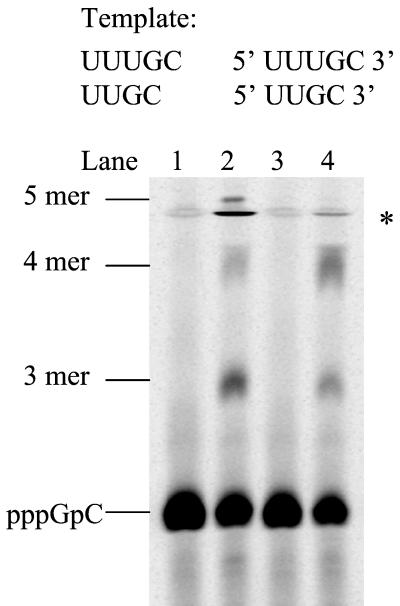
Minimal template length.
To find out if there was a minimal requirement of template length for initiation of de novo synthesis, 5-mer and 4-mer RNA templates were chemically synthesized and tested for de novo initiation. As shown in Fig. 5, in the presence of GTP and [α-33P]CTP, both UUUGC and UUGC were able to serve as a template and generated large amounts of the dinucleotide product, pppGpC. Addition of ATP into the reactions resulted in elongation intermediates and full-length products, as well as the accumulation of predominant amounts of pppGpC. There was no significant difference in the amount of products formed using either of the templates. The amount of dinucleotide generated from both templates was comparable to that from A8GC, indicating that a template as short as 4 nt is able to form a productive complex with the polymerase. In the dinucleotide-primed replication assay, the minimal template length for NS5B was reported to be 5 nt and a significant reduction in the polymerase activity was observed with a 4-nt template (22). The discrepancy between the two observations may reflect the template sequence differences or the utilization of a dinucleotide as a primer in the latter assay.
FIG. 5.
Minimal length requirement of an RNA template in NS5B de novo synthesis. Each reaction contained 2.5 μM NS5B, 100 μM [α-33P]CTP, 1 mM GTP, and 1 mM concentrations of the RNA template and nucleotide as follows: lane 1, template UUUGC only; lane 2, UUUGC and ATP; lane 3, template UUGC only; and lane 4, UUGC and ATP. The reaction products were resolved on a 25% polyacrylamide-7 M urea-TBE gel and were analyzed using a PhosphorImager. Asterisk denotes gel artifact.
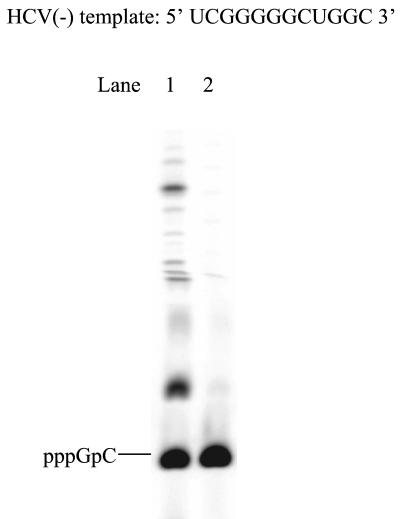
HCV minus strand as a template.
De novo synthesis was further examined by using a template derived from the last 12 nt of the HCV(−) RNA genome, 5′-UCGGGGGCUGGC-3′ (HCV[−] template). After incubation of the template with [α-33P]CTP, GTP, and NS5B, a ladder of RNA products was observed, starting from the dinucleotide pppGpC (Fig. 6, lane 1). This reaction produced RNAs longer than 4-mer because GTP was capable of base-pairing with U on the template. Products exceeding template length might be from terminal transferase activity or a template-switching event. The dinucleotide product, pppGpC, was synthesized at a level comparable to that found with A8GC (Fig. 6, lane 2). Considering that A8GC is a much better template than HCV(+)GU (Fig. 3), then HCV(−) should be a more efficient template than HCV(+)GU. This result is consistent with those from the reported experiments in dinucleotide-primed RNA synthesis, where the 14-nt RNA derived from the 3′ terminus of the HCV minus strand was more efficient as a template than that from the 3′ terminus of the HCV plus strand (22). A recent study on in vitro replication of the 3′-terminal region of the minus and plus strand viral RNA reported the same observation (16).
FIG. 6.
De novo initiation by NS5B using a template derived from the HCV minus strand genome. Each reaction contained 5 μM NS5B, 100 μM [α-33P]CTP, 1 mM GTP, and 20 μM concentration of the HCV(−) template (lane 1). For comparison, the same reaction was run with A8GC as a template (lane 2). The reaction products were resolved on a 25% polyacrylamide-7 M urea-TBE gel and were analyzed using a PhosphorImager.
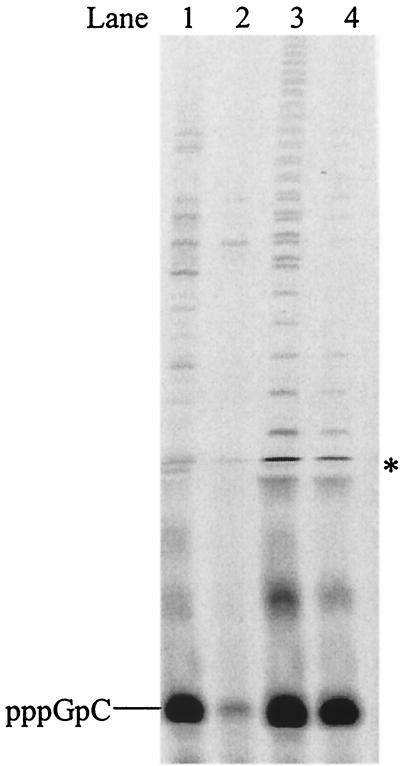
Effect of Mn2+ on de novo initiation and elongation.
It is known that the presence of Mn2+ in a polymerase reaction can increase polymerase activity as well as the frequency of misincorporation. To find out the effect of Mn2+ on de novo synthesis of NS5B, experiments were performed on the HCV(+)GU template that previously did not show any elongation products in the presence of Mg2+. In this case, GTP was used as an initiating nucleotide to form the “Wobble” base pair with the 3′-terminal U in the template to assess the effect of Mn2+ on misincorporation. Under the elongation conditions specified for Fig. 7 with Mn2+ as a cofactor, HCV(+)GU gave elongated products longer than 4-mer as well as significant accumulation of pppGpC (Fig. 7, lane 1), whereas it generated only pppGpC in much lower quantity in the presence of Mg2+ (Fig. 7, lane 2). This experiment clearly shows that Mn2+ increased both polymerase activity and misincorporation frequency. The rate of elongation for A8GC3′d also increased significantly in the presence of Mn2+ from that in the presence of Mg2+ (Fig. 7, lanes 3 and 4).
FIG. 7.
Effect of Mn2+ on NS5B de novo RNA synthesis activity. Each reaction contained 5 μM NS5B, 100 μM [α-33P]CTP, 1 mM GTP, 1 mM UTP, 5 mM concentration of a metal cofactor, and 20 μM concentration of an RNA template as listed. Lane 1, Mn2+ and the HCV(−) template; lane 2, Mg2+ and the HCV(−) template; lane 3, Mn2+ and A8GC3′d template; and lane 4, Mg2+ and A8GC3′d template. The reaction products were resolved on a 25% polyacrylamide-7 M urea-TBE gel and were analyzed using a PhosphorImager. Asterisk denotes gel artifact.
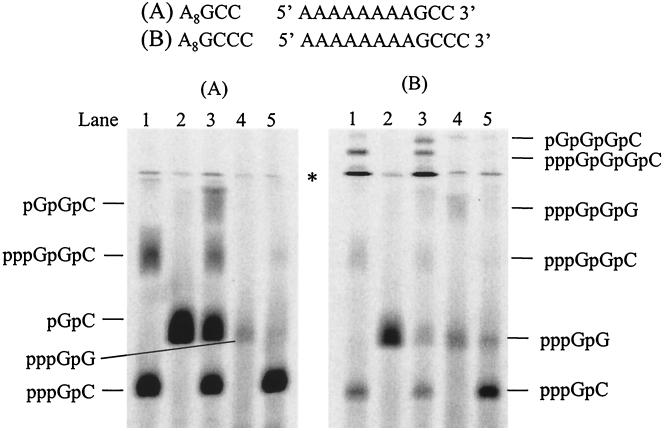
Selection of initiation site on a template.
As described above, de novo synthesis was initiated efficiently from the very 3′-terminal base of A8GC to produce an initiation product. However, no product was observed when A8GC was incubated with [α-33P]CTP and UTP, indicating that initiation from the +2 position did not occur on this template (Fig. 1, lane 3). Initiation from the +3 position was ruled out since incubation of the template with [α-33P]UTP did not yield any products (data not shown). To further examine the requirements for an initiation site in de novo synthesis by NS5B, a series of RNA templates with changes on the 3′ end were designed. In the reaction that used A8GCC as a template, NS5B produced two major products in the presence of GTP and [α-33P]CTP (Fig. 8A, lane 1). The lower band was identified as pppGpC generated from penultimate initiation, and the upper band was assigned as 3-nt RNA, pppGpGpC, that was produced from terminal initiation. When GMP was included as the only guanosine species in the reaction, pGpC was observed as the sole product, confirming that initiation can occur from the penultimate position (Fig. 8A, lane 2). When both GMP and GTP were supplied as the initiating nucleotides, four products were observed, pppGpC, pGpC, pppGpGpC, and pGpGpC, which were initiated from both the penultimate and terminal positions (Fig. 8A, lane 3). Initiation efficiency from the terminal and penultimate positions was further compared using [γ-33P]GTP as an initiating nucleotide (Fig. 8A, lanes 4 and 5). The dinucleotide product from the terminal initiation (pppGpG) was significantly less than that from the initiation of the penultimate position (pppGpC), partially due to the different concentrations of elongating nucleotide (Fig. 8A, 100 μM GTP in lane 4 versus 1 mM CTP in lane 5). Quantitative analysis showed that the product ratio was 1:40. Considering that the elongation nucleotide concentration in lane 5 was 10 times higher than that in lane 4, it seems that the internal initiation is preferred for this template. As the products from the internal and the terminal initiation are different, more detailed quantitative analysis and kinetic experiments are needed to address the question. The results of the above experiments are summarized in Table 1.
FIG. 8.
Selective initiation from +1, +2, and +3 positions of an RNA template by NS5B. Each reaction contained 2.5 μM NS5B, 20 μM concentration of an RNA template (template A8GCC for panel A and template A8GCCC for panel B), and 100 μM and 1 mM concentrations, respectively, of the following [α-33P] nucleotide and unlabeled nucleotide(s): for panel A, lane 1, [α-33P]CTP and GTP; lane 2, [α-33P]CTP and GMP; lane 3, [α-33P]CTP, GTP and GMP; lane 4, [γ-33P]GTP; and lane 5, [γ-33P]GTP and CTP; and for panel B, lane 1, [α-33P]CTP and GTP; lane 2, [α-33P]CTP and GMP; lane 3, [α-33P]CTP, GTP, and GMP; lane 4, [γ-33P]GTP, and lane 5, [γ-33P]GTP and CTP. The reaction products were resolved on a 25% polyacrylamide-7 M urea-TBE gel and were subjected to PhosphorImager analysis. Asterisk denotes gel artifact.
TABLE 1.
Summary of de novo initiation sites on the different templates
| Template | Products with selective initiation at
|
||
|---|---|---|---|
| +1a | +2 | +3 | |
| 5′AAAAAAAAGC3′ | 5′GC3′ | None | None |
| AAAAAAAAGCC | GGC/GG | GC | None |
| AAAAAAAAGCCC | GGGC/GGG/GG | GGC/GG | GC |
| CAGAUCAAGC | GC | None | None |
| CAGAUCAAGU | AC | None | None |
| CAGAUCAAGG | CC | None | None |
| CAGAUCAAGA | UC | None | None |
| CAGAUCAAGUA | UA (minimal) | AC | None |
| CAGAUCAAGUAG | None | None | AC |
Under the conditions of initiation product formation.
The same experiments were performed on template A8GCCC. In the presence of GTP and [α-33P]CTP, the enzyme yielded three products, pppGpC, pppGpGpC, and pppGpGpGpC (Fig. 8B, lane 1). Total activity appeared to be much less than that from A8GCC. Notably, the trinucleotide product that was initiated from the penultimate position was barely observed. Nevertheless, the result clearly shows that de novo synthesis can be initiated from the +3 position. This was confirmed by using GMP as an initiating nucleotide with pGpC as the sole product (Fig. 8B, lane 2). In the presence of both GTP and GMP as the initiating nucleotides, four RNA products initiated from +1, +2, and +3 positions were observed. Among them, two products that initiated from the penultimate and +3 positions, respectively, were hardly visible (Fig. 8B, lane 3). Experiments using [γ-33P]GTP as an initiating nucleotide were performed to compare the initiation efficiency from the terminal, +2, and +3 positions (Fig. 8B, lanes 4 and 5). Similar to the results using template A8GCC, initiation products pppGpC and pppGpG were again observed. It seems that internal initiation was preferred for this template; however, more detailed kinetic analysis is needed to draw this conclusion. The results of alternative initiation with A8GCCC as a template are summarized in Table 1.
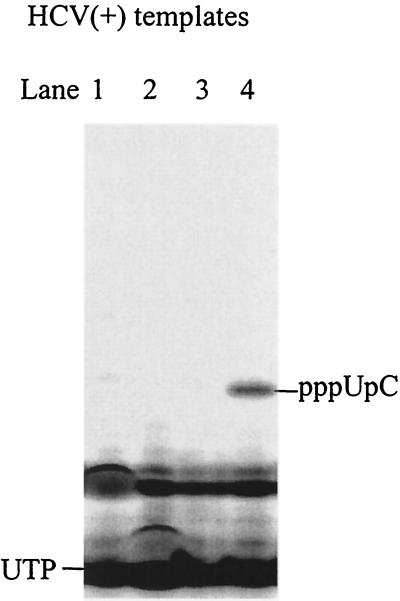
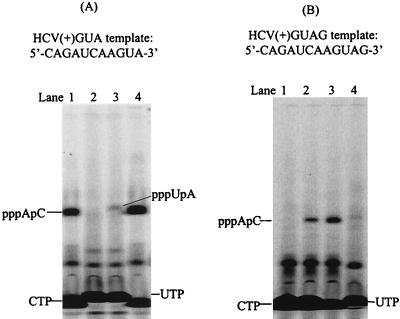
To find out whether de novo synthesis can be initiated from the +2 or +3 positions of the HCV(+) templates, reactions were performed in the presence of [α-33P]UTP and CTP that were complementary to the +3 and +2 bases of the templates. No product was observed, except from HCV(+)GA, which produced a dinucleotide, pppUpC, that was initiated from the +1 position (Fig. 9). This clearly demonstrates that +2 or +3 initiation does not occur from the viral templates. A new template, HCV(+)GUA, was designed by addition of a purine nucleotide at the 3′ end of HCV(+)GU. Interestingly, de novo initiation from HCV(+)GUA in the presence of [α-33P]CTP, ATP, and UTP resulted in a single dinucleotide product on the gel (Fig. 10A, lane 1). This dinucleotide product was likely pppApC that was initiated from the +2 position. The identity of the dinucleotide was confirmed by a reaction in the presence of [α-33P]CTP and ATP, where a dinucleotide with the same mobility on the gel was produced (Fig. 10A, lane 4). To see whether initiation at the +1 position for this template can occur, a reaction was performed in the presence of [α-33P]UTP and ATP. The reaction gave a dinucleotide product that corresponds to pppUpA, but the yield was much lower than that from the penultimate position, pppApC (Fig. 10A, lane 3). Finally, a new template, HCV(+)GUAG, was designed to introduce an additional G at the 3′ end of the template to determine which G site is preferred for de novo initiation. In the presence of [α-33P]CTP and UTP, no product was observed, indicating that +1 or +4 initiation did not occur as shown in Fig. 10B, lane 1. However, a single dinucleotide product was generated in the presence of [α-33P]CTP, UTP, and ATP (Fig. 10B, lane 2). It was identified to be pppApC that was initiated from the +3 position, as the same product was observed without UTP in the reaction (Fig. 10B, lane 3). The +2 initiation was examined by incubating with [α-33P]UTP and ATP, and no apparent product was detected (Fig. 10B, lane 4). These results are summarized in Table 1.
FIG. 9.
Selective initiation by NS5B from the +2 or +3 position of the HCV(+) templates. Each reaction contained 2.5 μM NS5B, 100 μM [α-33P]UTP, 1 mM CTP, and 20 μM concentration of one of the following RNA templates: lane 1, HCV(+)GC; lane 2, HCV(+)GU; lane 3, HCV(+)GG; and lane 4, HCV(+)GA. The reaction products were resolved on a 25% polyacrylamide-7 M urea-TBE gel and were analyzed using a PhosphorImager.
FIG. 10.
Selective initiation using HCV(+)GUA (A) or HCV(+)GUAG (B) as a template. Each reaction contained 2.5 μM NS5B and a 20 μM concentration of the specified RNA template and the following nucleotides: for panel A, lane 1, 100 μM [α-33P]CTP, 1 mM ATP, and 1 mM UTP; lane 2, 100 μM [α-33P]UTP and 1 mM ATP (no enzyme control); lane 3, 100 μM [α-33P]UTP and 1 mM ATP; and lane 4, 100 μM [α-33P]CTP and 1 mM ATP; and for panel B, lane 1, 100 μM [α-33P]CTP and 1 mM UTP; lane 2, 100 μM [α-33P]CTP, 1 mM UTP, and 1 mM ATP; lane 3, 100 μM [α-33P]CTP and 1 mM ATP; and lane 4, 100 μM [α-33P]UTP and 1 mM ATP. The reaction products were resolved on a 25% polyacrylamide-7 M urea-TBE gel and were analyzed using a PhosphorImager.
DISCUSSION
In this paper, we describe an assay using short RNA templates to investigate the early steps of de novo RNA synthesis catalyzed by HCV NS5B. The use of short oligonucleotide RNA templates provides opportunities to explore various features of de novo initiation. With this assay system, incorporations of various nucleotide/nucleoside analogs into the de novo initiation products were directly detected and confirmed, as a small change in charge and mass of RNA products was easily resolved using a 25% denaturing gel. The assay utilized Mg2+ as the metal ion cofactor instead of Mn2+, and thus the results are more physiologically relevant.
De novo RNA synthesis starts with the formation of an initiation product, a dinucleotide, from the first nucleotidyl transfer reaction. The formation of dinucleotides in the de novo RNA synthesis catalyzed by NS5B was examined using various templates in the presence of two NTPs that are complementary to the 3′-terminal and penultimate bases of a template. All the templates tested, including those derived from the 3′ termini of both HCV plus and minus strands of the RNA genome, were able to serve as a template for dinucleotide formation. However, elongation of the dinucleotides was inefficient. The HCV(+) templates did not yield any products except the dinucleotides under the elongation conditions. This may explain the earlier failure to observe de novo RNA synthesis from short templates derived from the 3′ termini of the HCV RNA genome, in which only fully elongated products were examined (8, 14). Elongation products as well as a large accumulation of dinucleotide products were observed from A8GC, HCV(−), UUUGC, and UUGC templates. Higher concentrations of elongating nucleotides or RNA templates showed little effect on the results, implying that the concentration of nucleotides or a template is not a limiting factor. Product distribution from these templates suggests that NS5B RdRp may exist in at least two states, an initiation complex that accumulates dinucleotides and an elongation complex that incorporates NTP onto the dinucleotides at a uniform rate. It is possible that short RNA templates might not form a productive elongation complex efficiently. However, gel analysis of the products from de novo replication of the poly(C) template (∼300 nt) showed a similar accumulation of dinucleotide products (data not shown), suggesting that this phenomenon is intrinsic to NS5B RdRp. The results imply that the transition from initiation to elongation is a rate-limiting step. We note that the amount of dinucleotide accumulated from the A8GC, HCV(−), UUUGC, or UUGC template is much larger than that from the HCV(+) templates. Several lines of evidence support the hypothesis that large amounts of dinucleotide are needed to drive the reaction to the further elongation steps. Elongation products from one of the HCV(+) templates began to appear when an appropriate dinucleotide was supplied to the elongation reaction in excess of 100 μM. The presence of Mn2+ also resulted in more elongation products, possibly by increasing dinucleotide formation. These results indicate that dinucleotides may dissociate rapidly from the polymerase/template complex. The dissociation and accumulation of dinucleotides are similar to a process known as abortive initiation. It has been described for T7 RNA polymerase in which an unstable initiation complex releases 2- to 8-nt RNA transcripts before transition to a stable elongation complex (20). Recently, it has been reported that NS5B can utilize di- or trinucleotides as a primer for RNA replication (22). This has led to a proposal that NS5B may use short abortive RNA transcripts as a primer generated either from abortive cellular transcription or from de novo synthesis by NS5B. Our study demonstrates that such short abortive RNA transcripts are intrinsic to the mechanism of de novo initiation by NS5B. Furthermore, it implies that a conformational change may be required for the transition from the initiation complex to the elongation complex. A recent study on T7 RNA polymerase has suggested that a conformational change of the specificity loop of the polymerase may be accompanied by the transition of the unstable initiation complex to the stable elongation complex (20). Crystal structures of NS5B revealed a unique β-loop in the thumb subdomain protruding into the active site (2, 9). This β-loop has been proposed to position and stabilize the 3′ terminus of a template and dinucleotide primer (5, 22). Conceivably, the transition from initiation complex to elongation complex could be triggered by flipping of the β-loop. A similar mechanism has been proposed for φ6 RNA polymerase in which the C-terminal domain locks in the initiation complex and is displaced away upon further elongation (3). The role of the β-loop in the mechanism of de novo synthesis of NS5B is presently under investigation using site-directed mutagenesis and kinetic analysis. It should be noted that the abortive initiation observed here is an intrinsic property of NS5B polymerase alone. It is possible that, in the replication complex in vivo, HCV may have a mechanism to ensure a smooth and efficient transition from the initiation complex to the elongation complex.
Template specificity studies indicate that the de novo polymerase activity is template dependent. It is still not well understood how the nucleotide sequence of RNA templates affects the activity. In vitro, two key interactions may affect the de novo polymerase activity: (i) interaction between the RNA template and polymerase and (ii) interaction between NTP and the polymerase/template complex. It has been suggested that a certain RNA secondary structure is required to initiate de novo synthesis. However, the fact that templates used in this study are free of secondary structures and still exhibit various de novo activities indicates that secondary structure is not a primary determinant for de novo initiation. The secondary structure could play a role in vivo for efficient elongation. It seems that the primary sequence of HCV templates encodes the basic information required for the de novo RNA polymerization as well. For example, much higher de novo initiation activity was observed from the HCV(−) template than from HCV(+)GU. This result coincides with the observation that the plus strand RNA is more abundant than the minus strand RNA in cells harboring the HCV RNA replicon (11).
The primary determinant of template specificity has been thought to be the initiation site in which initiating NTP interacts with the 3′-terminal base of the template in the polymerase/template complex. NS5B has been reported to show a higher activity when templates have cytidine at their 3′ terminus (23). This is the case for templates A8GC, HCV(−), and UUUGC; however, the HCV(+)GC templates show the contrary results. Changes of the 3′-terminal base of the HCV(+) template affected dinucleotide formation significantly, with the native 3′-terminal end GU giving the highest yield of dinucleotide formation, followed by GG, GA, and GC ends. This result indicates that the very 3′-terminal base is not the sole determinant for template specificity and that the specificity collectively exists in the initiation site involving the 3′-terminal base of template, incoming NTP, and the active site structure of a polymerase.
Experiments using various artificial templates show that NS5B can initiate RNA synthesis at the +1, +2, or +3 position from the 3′ terminus (Table 1). Like most other polymerases, NS5B seems to have a strong preference for pyrimidine bases as the initiation site on a template for de novo RNA synthesis. Initiation from pyrimidine bases was observed for all three positions, while initiation from purine bases was observed only at the +1 position. Initiation from pyrimidine bases showed much higher activity than that from purine bases. Notably, the 3′ terminus of both the plus and minus strands of the HCV RNA genome starts with a pyrimidine followed by two purine bases. Such arrangement ensures that NS5B starts replication only from the very 3′-terminal pyrimidine base. Templates with the addition of extra purine nucleotide(s) A or AG at the 3′ end failed to initiate de novo synthesis. These results are consistent with the observation that any changes of the +2 or +3 base as well as replacement of terminal U with purine bases at the 3′ end of the HCV genome appear to be detrimental to HCV replication in cell culture (W. Zhong and Z. Hong, unpublished results). It is conceivable that HCV has evolved with constraints on the 3′-terminal sequence to maintain the integrity and stability of its genome, especially the last three terminal bases in the order of purine, purine, and pyrimidine. Evidently, the 3′ X region is highly conserved among all the isolates of HCV (19). One possible function of the conserved sequence may be to ensure de novo synthesis from the very 3′-terminal end.
It has been reported that T7 RNA polymerase can utilize guanosine or GMP as an initiating nucleotide (13). Recently, GMP and GDP were shown to function as initiating nucleotides in de novo synthesis by HCV NS5B (8). Our assay not only confirms that HCV NS5B can utilize GMP and GDP as an initiator but also demonstrates for the first time that guanosine can serve as an initiator in de novo synthesis. Our experiments clearly show the incorporations of GDP, GMP, and guanosine into the products at the initiation site. The dinucleotide products, pppGpC, ppGpC, pGpC, and GpC, are well resolved on a denaturing gel. Interestingly, GMP gave the highest yield of dinucleotide product. Detailed kinetics using simple nucleosides as an initiating nucleoside are presently under investigation. This result suggests that 5′ phosphate groups, especially the β- or γ-phosphate motif of an initiating nucleotide, are not critical for productive formation of NS5B/template/nucleotides. It also implies that 5′ diphosphate, 5′ monophosphate nucleotides, or simple nucleosides may function as an initiating nucleotide in de novo RNA synthesis in vivo. Further in vivo experiments are needed to investigate the possible heterogeneity of the 5′ end of the viral RNA genome.
Acknowledgments
We thank R. Bartenschlager for supplying the cDNA clone of HCV NS5B and thank W. Zhong, N. Yao, R. Hamatake, C. Cameron, M. Walker, and T. Appleby for helpful discussions and J. Lau for support.
REFERENCES
- 1.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold Des. 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 2.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 4.Ferrari, E., J. Wright-Minogue, J. W. Fang, B. M. Baroudy, J. Y. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 6.Kao, C. C., A. M. Del Vecchio, and W. Zhong. 1999. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology 253:1-7. [DOI] [PubMed] [Google Scholar]
- 7.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 8.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 10.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 12.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin, C. T., and J. E. Coleman. 1989. T7 RNA polymerase does not interact with the 5′-phosphate of the initiating nucleotide. Biochemistry 28:2760-2762. [DOI] [PubMed] [Google Scholar]
- 14.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranjith-Kumar, C. T., J. Gajewski, L. Gutshall, D. Maley, R. T. Sarisky, and C. C. Kao. 2001. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis. J. Virol. 75:8615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reigadas, S., M. Ventura, L. Sarih-Cottin, M. Castroviejo, S. Litvak, and T. Astier-Gin. 2001. HCV RNA-dependent RNA polymerase replicates in vitro the 3′ terminal region of the minus-strand viral RNA more efficiently than the 3′ terminal region of the plus RNA. Eur J. Biochem. 268:5857-5867. [DOI] [PubMed] [Google Scholar]
- 17.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p. 931-960. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 18.Steitz, T. A. 1998. A mechanism for all polymerases. Nature 391:231-232. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka, T., N. Kato, M. J. Cho, K. Sugiyama, and K. Shimotohno. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Temiakov, D., P. E. Mentesana, K. Ma, A. Mustaev, S. Borukhov, and W. T. McAllister. 2000. The specificity loop of T7 RNA polymerase interacts first with the promoter and then with the elongating transcript, suggesting a mechanism for promoter clearance. Proc. Natl. Acad. Sci. USA 97:14109-14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, J. Z. 2001. Internally located signal peptides direct hepatitis C virus polyprotein processing in the ER membrane. IUBMB Life 51:19-23. [DOI] [PubMed] [Google Scholar]
- 22.Zhong, W., E. Ferrari, C. A. Lesburg, D. Maag, S. K. Ghosh, C. E. Cameron, J. Y. Lau, and Z. Hong. 2000. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:9134-9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong, W., A. S. Uss, E. Ferrari, J. Y. N. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]