Abstract
Feline calicivirus (FCV) nonstructural proteins are translated as part of a large polyprotein that undergoes autocatalytic processing by the virus-encoded 3C-like proteinase. In this study, we mapped three new cleavage sites (E46/A47, E331/D332, and E685/N686) recognized by the virus proteinase in the N-terminal part of the open reading frame 1 (ORF1) polyprotein to complete the processing map. Taken together with two sites we identified previously (E960/A961 and E1071/S1072), the FCV ORF1 polyprotein contains five cleavage sites that define the borders of six proteins with calculated molecular masses of 5.6, 32, 38.9, 30.1, 12.7, and 75.7 kDa, which we designated p5.6, p32, p39 (NTPase), p30, p13 (VPg), and p76 (Pro-Pol), respectively. Mutagenesis of the E to A in each of these cleavage sites in an infectious FCV cDNA clone was lethal for the virus, indicating that these cleavages are essential in a productive virus infection. Mutagenesis of two cleavage sites (E1345/T1346 and E1419/G1420) within the 75.7-kDa Pro-Pol protein previously mapped in bacterial expression studies was not lethal.
Feline calicivirus (FCV) is a common cause of upper respiratory tract disease and oral ulceration in cats (8, 14). FCV is classified in the genus Vesivirus of the family Caliciviridae. The family is comprised of a number of animal viruses (vesicular exanthema of swine virus, rabbit hemorrhagic disease virus [RHDV], canine calicivirus, and San Miguel sea lion virus) as well as human pathogens associated with acute gastroenteritis (Norwalk-like and Sapporo-like viruses) (11).
FCV virions are small (∼35 nm in diameter), nonenveloped, icosahedral particles that possess a linear, polyadenylated positive-sense single-stranded RNA genome of ∼7.7 kb. The genome has three major open reading frames (ORFs): ORF1 (nucleotides [nt] 20 to 5308), ORF2 (nt 5314 to 7317), and ORF3 (nt 7317 to 7634). The virus structural proteins, encoded by ORF2 and -3, are expressed from an abundant subgenomic RNA (3, 12, 24, 36). ORF2 encodes the capsid protein precursor (73 kDa), which is cleaved by the FCV ORF1-encoded proteinase into the 60-kDa mature capsid protein (VP1) and the 14.2-kDa leader of the capsid (LC) (3, 36, 37). ORF3 is located at the 3′ end of the subgenomic RNA and has a 1-nt overlap with ORF2. This small ORF encodes a minor structural protein designated VP2 (36).
FCV ORF1 encodes a large polyprotein that undergoes cotranslational proteolytic processing to yield several nonstructural proteins ranging in size from approximately 13 kDa to 96 kDa (2, 38). The 3C-like cysteine proteinase responsible for this processing as well as that of the capsid protein precursor has been mapped to the C-terminal part of the ORF1 polyprotein (37, 38). Four cleavage sites (E960/A961, E1071/S1072, E1345/T1346, and E1419/G1420) recognized by the proteinase at various efficiencies, were identified in this part of the ORF1 polyprotein in in vitro and bacterial expression studies (38). The E960/A961 and E1071/S1072 cleavage sites defined the borders of the 12.7-kDa VPg protein (36, 38). Efficient processing of the ORF1 polyprotein at the E1071/S1072 cleavage site led to release of the 75.7-kDa 3CD-like proteinase-polymerase (Pro-Pol) protein. The Pro-Pol protein accumulates in FCV-infected cells (38), and further proteolytic processing between the proteinase and polymerase domains of recombinant Pro-Pol was not required for polymerase activity in vitro (41). Two internal cleavage sites in the Pro-Pol protein, E1345/T1346 and E1419/G1420, were utilized less efficiently by the proteinase in the in vitro studies, and their role in virus replication was not clear (38).
The goal of the present work was to map the remaining cleavage sites in the FCV nonstructural polyprotein and to identify the cleavage events essential for virus growth. Our study demonstrates that the FCV 3C-like proteinase mediates at least three additional cleavages in the viral nonstructural polyprotein when analyzed in vitro at positions E46/A47, E331/D332, and E685/N686. Mutagenesis of individual nonstructural polyprotein cleavage sites in an infectious FCV cDNA clone demonstrated that the majority of cleavage events identified in in vitro mapping studies are critical for productive virus infection.
MATERIALS AND METHODS
Cells and virus.
Crandell-Rees feline kidney (CRFK) cells were maintained in Eagle's minimum essential medium containing amphotericin B (2.5 μg/ml), chlortetracycline (25 μg/ml), penicillin (250 U/ml), and streptomycin (250 μg/ml) and supplemented with 10% heat-inactivated fetal bovine serum. The source and molecular characterization of the Urbana (URB) strain of FCV (GenBank accession no. L40021) have been described previously (34).
Radiolabeling of virus-specific proteins.
CRFK monolayers (106 cells) were mock infected or infected with FCV at a multiplicity of infection (MOI) of 1 to 4 and incubated at 37°C. For radiolabeling of proteins, the cells were washed at 3.5 h postinfection with methionine- or cysteine-free growth medium and incubated in the same medium for 30 min. [35S]methionine (>1,000 Ci/mmol; Amersham) or [35S]cysteine (>800 Ci/mmol; ICN) was added to cells at a concentration of 100 μCi/ml, and the cells were incubated for an additional hour. The monolayer was washed with phosphate-buffered saline (PBS) before lysis in 500 μl of radioimmunoprecipitation assay buffer (31).
Plasmid construction.
Standard recombinant DNA methods were used for construction of plasmids (31). Construction of the plasmid pTMF-1 used for analysis of proteolytic processing of the FCV ORF1 polyprotein was described previously (37). The plasmid contained the entire ORF1 of the FCV genome, with the exception of the two C-terminal codons.
To express the ORF1 sequence fused at its N terminus to a glutathione S-transferase (GST)-TAG sequence (Novagen), plasmid pGSTFΔXm was constructed in two steps: first, plasmid pQ14, the full-length FCV cDNA clone (34), was cleaved with AspI, followed by incubation with the Klenow fragment of DNA polymerase I in the presence of dTTP and further treatment with EagI. The 4,547-bp AspI-EagI fragment was isolated and subcloned into SacI-EagI-digested pET-23a plasmid (Novagen). In a second step, the resulting intermediate plasmid p23FΔXm was digested with NheI and EagI, and the 4,591-bp NheI-EagI fragment was isolated and subcloned into SpeI-EagI-digested pET-41b plasmid (Novagen). The selected clone, pGSTFΔXm, encoded ORF1 polyprotein sequence from amino acid (aa) 2 to 1517 fused to an N-terminal GST-TAG sequence that was engineered immediately downstream from the T7 RNA polymerase promoter.
In order to analyze processing by the virus proteinase at the junction of the 38- and 30-kDa protein sequences, the corresponding regions of ORF1 polyprotein were amplified and subcloned into the pET-28b vector (Novagen) in two steps. First, a DNA fragment containing a truncated sequence of the Pro-Pol protein was amplified from plasmid pQ14 by using oligonucleotides 5′-acatataagcttTCTGGACCCGGCACC-3′ and 5′-agatagctcgagttaTTCTGAGGAAATGTTCAC-3′ (38). The sequence of the first oligonucleotide contained 15 nt corresponding to the beginning of the 3C-like proteinase gene (nt 3233 to 3247 [uppercase]) and included a HindIII site (underlined). The second oligonucleotide corresponded to the sequence complementary to nt 4037 to 4054 of the FCV genome (uppercase) and included an engineered termination codon (boldface) and an XhoI site (underlined). The purified PCR fragment was treated with HindIII and XhoI and ligated into HindIII-XhoI-linearized pET-28b. The resulting plasmid was designated pHP. The next step used oligonucleotides 5′-atacatatggctagcGTTTGTATTGTTGACGAG-3′ and 5′-tatataaaagtcgactgcttcaCCTCCGGCTCGAATCAC-3′ to amplify the DNA fragment corresponding to the 38-kDa-30-kDa protein junction sequence. The sequence of the first oligonucleotide contained 18 nt corresponding to nt 1589 to 1606 of the FCV genome (uppercase) and included an NheI site (underlined). The second oligonucleotide contained the sequence complementary to nt 2696 to 2712 (uppercase) and a SalGI site (underlined). The purified PCR-fragment was treated with NheI and SalGI and ligated into NheI-SalGI-linearized pHP. The resulting plasmid, designated pCAC, contained the 38-kDa-30-kDa protein junction sequence fused in frame at its C terminus to the 3C-like proteinase sequence and at its N terminus to the vector sequence. The chimeric sequence was placed under control of the T7 RNA polymerase promoter and downstream from the bacterial ribosome-binding site. The plasmid was verified by sequence analysis and employed in in vitro and bacterial expression studies.
In order to raise antisera specific to FCV proteins, selected regions of the virus genome were PCR amplified from plasmid pQ14 as template and cloned into the bacterial expression vector pET-28b (Novagen). The primer sequences and regions amplified are presented in Table 1. Purified PCR fragments from each region were treated either with BamHI and NotI or with HindIII and XhoI and ligated into the pET-28b vector linearized with the corresponding pair of enzymes. The resulting plasmids contained FCV ORF1 sequences fused to the vector sequence encoding an N-terminal six-histidine tag under the control of the T7 RNA polymerase promoter and placed downstream from the bacterial ribosome-binding site. All plasmids were verified by sequence analysis before expression in bacteria.
TABLE 1.
PCR primers used in construction of bacterial expression vectors
| Region | Sequencea | Nucleotide positions | Amino acid position | Expression vector | Calculated mol mass (kDa)b |
|---|---|---|---|---|---|
| A | atataggatccgATGTCTCAAACTCTG | 20-36 | 1-139 | p2AB | 19.1 |
| ttaatttgcggccgcctaTTCAGCCATAACCAACAG | |||||
| B4 | ggtagggatccaGCTTGTCCTTCTTGTGCC | 158-436 | 47-139 | p2B5 | 13.5 |
| ttaatttgcggccgcctaTTCAGCCATAACCAACAG | |||||
| B3 | ggtcgggatccgCAACGCTCTAGGTCGACACTC | 449-1012 | 144-331 | p2B3 | 25.3 |
| tttatatgcggccgcctaCTCTGATCTAAACCC | |||||
| B2 | ggtcggatccgGCTGCCAAACAAAAACTAAGG | 653-1195 | 212-392 | p2B2 | 23.5 |
| ttatatagcggccgcctaTTCAGAATTCCATGTTGC | |||||
| C | atataggatccgGTTTGTATTGTTGACGAG | 1589-2068 | 524-683 | p2C | 21.5 |
| tattaaagcggccgcctaTTCAAATGCCATGTTTTTC | |||||
| D3 | ggtcgggatccgGCCTCTAGCTCCATTGGG | 220-2713 | 728-898 | p3A3 | 22.1 |
| tattagcggccgctaACCTCCGGCTCGAATCAC | |||||
| D1 | ggtcgggatccgGGAAACTGTGTGCAATC | 2312-2899 | 765-960 | p3A1 | 25.1 |
| gtatatgcggccgcctaTTCAGATTTGGGTCTAAAC | |||||
| F | acatataagcttTCTGGACCCGGCACC | 3233-3595 | 1072-192 | p3C | 17.4 |
| gtatatctcgagctaATCTCCTGGGTGAG |
The nonviral sequences of the oligonucleotides are shown in lowercase. Restriction sites used for subcloning of corresponding PCR fragments are underlined. Sequences corresponding to a termination codon in the oligonucleotides complementary to the FCV genomic sequence are in boldface.
Calculated molecular masses are given for encoded recombinant proteins.
To express the FCV ORF1 sequences encoding p32 (aa 47 to 331), p39 (aa 332 to 685), and p30 (aa 686 to 960); the corresponding regions were PCR amplified from plasmid pQ14 as a template. The oligonucleotide pairs employed to amplify these regions were 5′-tataatacgcgtacctgcccgccatgGCTTGTCCTTCTTG-3′ and 5′-tataatgcggccgctgcagttatcaCTCTGATCTAAACCCTG-3′, 5′-tataatacgcgtaccaccatgGATGTGGCAAACTCATTC-3′ and 5′-tataatgcggccgctgcagttatcaCTCCGCTTCAAATGCCATG-3′, and 5′-tataatacgcgtacctgcccgccatgAATGGACATAGTGAGCAC-3′ and 5′-tataatgcggccgctgcagttatcaTTCAGATTTGGGTCTAAACC-3′, respectively. The sequence of the first oligonucleotide contained nucleotides corresponding to the beginning of the protein gene sequence (uppercase) and included an initiation codon (boldface) and an MluI site (underlined). The second oligonucleotide corresponded to the sequence complementary to the end of the protein gene sequence (uppercase) and contained a termination codon (boldface) and a NotI site (underlined). Purified PCR fragments were treated with MluI and NotI and ligated into the pCI vector (Promega) linearized with the corresponding enzymes. The resulting plasmids (p47/331, p332/685, and p686/960) contained p32, p39, and p30 gene sequences, respectively, starting with a methionine codon and placed under control of the T7 RNA polymerase promoter. The plasmids were verified by sequence analysis and employed in in vitro expression studies.
Bacterial expression of the recombinant proteins, purification, and production of specific antiserum.
The expression of recombinant proteins in E. coli BL21(DE3) cells was performed as previously described (37). The six-His-tagged recombinant proteins of the insoluble fraction were purified by the immobilized-metal affinity chromatography (IMAC) method with Ni-nitrilotriacetic (NTA) agarose (Qiagen). The purified proteins were dialyzed twice against a 1,000× volume of PBS and were used for immunization of guinea pigs as described previously (38). Production of Pro-Pol-specific serum was described previously (38).
In vitro translation assay.
One to 5 μg of plasmid DNA was used as the template in a coupled transcription and translation reaction (TNT T7 coupled reticulocyte lysate system; Promega). For radiolabeling of synthesized protein, [35S]methionine (>1,000 Ci/mmol; Amersham) or [35S]cysteine (>800 Ci/mmol; ICN) was used at a concentration of 1.5 mCi/ml.
Immunoprecipitation analysis.
Immunoprecipitations of viral proteins from infected-cell lysates and in vitro translation mixtures were performed as described previously (38) with region-specific antisera. The GST-TAG fusion proteins were immunoprecipitated with commercial rabbit polyclonal antiserum specific for the GST protein (Sigma).
Nucleotide and protein sequencing.
Nucleotide sequence analysis of DNA was performed with the Big Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer), and the sequencing products were resolved on an automated sequencer, ABI 310 (Applied Biosystems).
Direct N-terminal sequence analysis of the radiolabeled virus-specific proteins was conducted as follows. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Tricine running buffer (Invitrogen), transferred to a ProBlott membrane (Applied Biosystems), and located by autoradiography. The band of interest was excised and subjected to automated protein analysis. Edman degradation of protein was performed on an Applied Biosystems model 477A pulse liquid protein sequencer. A modified anilino-thiazolinone cycle (as described by the manufacturer) was used. The standard plumbing of the instrument was modified so that the cleaved amino acid derivative at each degradation cycle was delivered and collected by a fraction collector. The fractions were added to vials and counted for radioactivity in a Beckman scintillation counter.
Direct N-terminal sequence analysis of proteins expressed and purified from bacterial cells was conducted as described previously (37).
Site-directed mutagenesis of the FCV full-length cDNA clone.
The QuickChange site-directed mutagenesis kit (Stratagene) was utilized to introduce amino acid substitutions into the FCV ORF1 polyprotein coding sequence in the plasmid pQ14. Sense and antisense primers (Table 2) were employed in PCR mutagenesis, which was conducted according to the manufacturer's protocol. Clones were screened by sequence analysis, and plasmids containing the desired mutations in the cleavage site were subjected to complete genome sequence analysis. No other mutations elsewhere in the genome were found for plasmids pF3, pF10, pF22, pF36, pF65, pF71, and pF87. A single nucleotide substitution each was found in pF5 (nt 3988, G 224 T) and pF77 (nt 4873, C 224 A). The nucleotide substitutions were both located in the third position of a codon and did not affect the amino acid sequence of the encoded protein. The nucleotide substitution at position 4873 of pF77 was conserved with FCV strain CFI/68 (GenBank accession no. U13992).
TABLE 2.
Primers used to introduce cleavage site mutations into FCV full-length cDNA clone
| Localization | Sequencea | Amino acid change(s) | Full-length cDNA clone | Virus recovery |
|---|---|---|---|---|
| p5.6 | 1 5′-GTAAAAGAAATTTGAGACAAAGTCTCAAACTCTGAGC-3′ 37 | 1M→1K | pF10 | − |
| p5.6/p32 | 143 5′-CCTATAAGGGCAGCCGCTTGTCCTTCTTGTGCC-3′ 175 | 46EA→46AA | pF22 | − |
| p32/p39 | 998 5′-GGGTTTAGATCAGCCGATGTGGCAAACTCATTCTGG-3′ 1033 | 331ED→331AD | pF71 | − |
| p39/p30 | 2056 5′-CATGGCATTTGCCGCGGAGAATGGACATAG-3′ 2085 | 683EAEN→683AAEN | pF65 | + |
| p39/p30 | 2059 5′-GGCATTTGAAGCGGCCAATGGACATAGTGAGC-3′ 2090 | 683EAEN→683EAAN | pF36 | − |
| p30/VPg | 2880 5′-GGTTTAGACCCAAATCTGCCGCCAAAGGAAAAAC-3′ 2913 | 960EA→960AA | pF77 | − |
| VPg/Pro-Pol | 3215 5′-GTTAGCTTTGCTGAAGCCTCTGGACCCGGCACC-3′ 3247 | 1071ES→1071AS | pF87 | − |
| Pro-Pol | 4042 5′-CATTTCCTCAGCCACCTCTATGCTATC-3′ 4068 | 1345ET→1345AT | pF3 | + |
| Pro-Pol | 4262 5′-GAAAAGGTTGCTGCCGGAAAGCGAAGG-3′ 4288 | 1419EG→1419AG | pF5 | + |
The sequence of the sense primer is shown. The antisense primer (not shown) was the precise reverse and complementary sequence. The nucleotide positions of the beginning and the end of each of the primers in the FCV genome are indicated. Nucleotide substitutions are in bold, and cleavage site codons are underlined.
Recovery experiments.
Recovery of virus from plasmid DNA by using the MVA/T7 expression system was conducted by a protocol similar to that described previously (35). Briefly, confluent CRFK cell monolayers (2 × 106 cells) were infected with MVA/T7 at an MOI of 10 and incubated for 1.5 h at 37°C. Cells then were washed two times with Opti-MEM I (Invitrogen), and the plasmid transfection mixture was added. The transfection mixture was prepared as follows. One to 2 μg of plasmid DNA was diluted in 100 μl of Opti-MEM I, and 6 μl of Lipofectamine plus reagent (Invitrogen) was added, followed by incubation at room temperature for 15 min. The mixture was then combined with 100 μl of Opti-MEM I containing 4 μl of Lipofectamine (Invitrogen) and incubated at room temperature for an additional 15 min. Before addition to washed cells, the Lipofectamine-DNA solution was diluted to 1 ml with Opti-MEM I. Cells were incubated with the transfection mixture for 3 h at 37°C before the Lipofectamine-DNA-containing medium was replaced with 2 ml of maintenance medium. Following incubation for 24 h at 37°C, medium from the transfected cell monolayer was transferred to a fresh cell monolayer, and recovery of the virus was verified by plaque formation assay and immunofluorescent staining with serum raised in guinea pigs against FCV virions as described previously (34).
The presence of engineered mutations in the genomes of recovered viruses was confirmed by direct sequence analysis of reverse transcriptase (RT)-PCR products derived from viral RNA as described previously (37).
RESULTS
Generation and characterization of a panel of FCV ORF1 region-specific antisera.
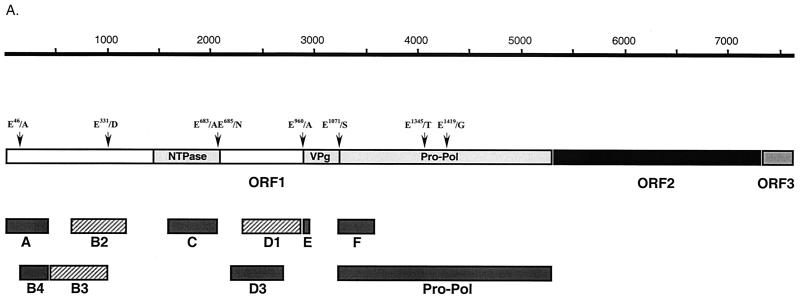
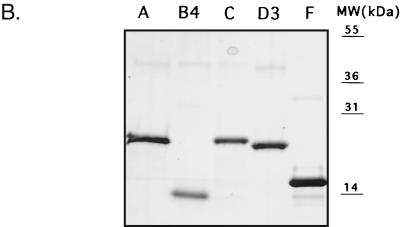
Our previous studies demonstrated that the 3C-like proteinase encoded in the C-terminal part of the FCV ORF1 was responsible for autocatalytic processing of the ORF1 polyprotein. We had directly mapped the cleavage sites bordering the proteinase as well as those for the virus VPg by N-terminal sequence analysis of cleaved proteins obtained by expression of the C-terminal part of ORF1 in bacteria (38). In order to map the remaining upstream polyprotein cleavage sites of FCV, we initially attempted expression of larger fragments in bacteria. However, the levels of expression were insufficient for direct sequence analysis. We then developed a strategy for mapping that involved the microsequencing of radioactively labeled proteins produced in in vitro translation reactions coupled with the generation of region-specific antisera to confirm the localization of the coding sequence in the genome. To generate a panel of sera specific for the FCV nonstructural proteins, several regions were chosen for expression in bacteria (Fig. 1A and Table 1) as six-histidine-tagged fusion proteins by using the pET-28b plasmid (Novagen). One exception was the N-terminal region of the VPg protein (designated as region E in Fig. 1A), from which a 20-aa peptide was synthesized to raise antisera (36). The recombinant plasmids were transformed into E. coli (BL21) cells, and protein expression was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). For plasmids p2AB (region A), p2B5 (region B4), p2C (region C), and p3C (region F), SDS-PAGE analysis of the IPTG-induced bacterial cells followed by Coomassie blue staining demonstrated efficient expression of proteins of the expected size (data not shown). Plasmid p3A3 (region D3, aa 728 to 898) also showed efficient expression, but the mobility of the recombinant protein was higher (observed mass, 18 to 19 kDa) than expected (22.1 kDa), although no sequence errors were present in the cDNA clone (Fig. 1B). Expression of plasmid p3A1 (region D1, aa 765 to 960) resulted in a small amount of protein (data not shown). Plasmid p2B3 (region B3, aa 144 to 331) and plasmid p2B2 (region B2, aa 212 to 392) could not be expressed to detectable levels (data not shown). Similar problems associated with efficiency of expression of these regions in bacteria were described for RHDV (43).
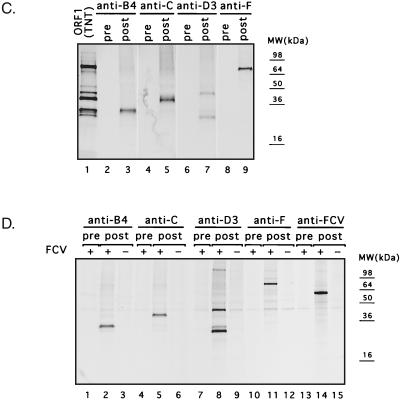
FIG. 1.
Localization and expression of FCV polyprotein fragments in E. coli. (A) Schematic diagram showing the organization of the FCV genome and location of cDNA fragments expressed in E. coli. The FCV ORF1 proteins, VPg and Pro-Pol, as well as the domain with amino acid homology to the picornavirus NTPase (nt 1445 to 2101) described by Neill (23) and the RHDV NTPase (19) are indicated as light gray boxes. The dipeptide cleavage sites previously identified (38) and the putative cleavage sites analyzed in the present study are indicated. The cDNA fragments efficiently expressed in E. coli are depicted as dark gray bars. The cDNA fragments corresponding to the B2, B3, and D1 regions that were found to be toxic for bacterial cells are shown as hatched gray bars. Bar E represents the peptide sequence corresponding to the beginning of the putative VPg. (B) Fusion proteins corresponding to regions A, B4, C, D3, and F were purified with Ni-NTA agarose. Aliquots were analyzed by SDS-PAGE followed by Coomassie blue staining. The expression levels of B2, B3, and D1 sequences were minimal. MW, molecular mass. (C) Immunoprecipitation of proteins obtained in coupled TNT reaction from pTMF1 (ORF1) with antisera raised against purified recombinant B4, C, D3, and F proteins. The radiolabeled in vitro translation products derived from pTMF1 (lane 1) were precipitated with either pre- or postimmunization sera raised against B4 protein (lanes 2 and 3), C protein (lanes 4 and 5), D3 protein (lanes 6 and 7), and F protein (lanes 8 and 9), subjected to SDS-PAGE, and visualized by autoradiography. (D) The same panel of antisera was analyzed for specificity by immunoprecipitation of radiolabeled proteins produced in FCV- or mock-infected CRFK cells. The CRFK cells were infected at an MOI of 1 to 4 with the FCV URB strain or mock infected. At 3.5 h postinfection, cells were metabolically labeled with [35S]methionine for 1 h. The radiolabeled proteins were immunoprecipitated from cell lysates, subjected to SDS-PAGE, and visualized by autoradiography. Radiolabeled proteins from FCV-infected CRFK cells that underwent immunoprecipitation with either pre- or postimmunization sera are shown as follows: lanes 1 and 2, anti-B4 sera; lanes 4 and 5, anti-C sera; lanes 7 and 8, anti-D3 sera; lanes 10 and 11, anti-F sera; and lanes 13 and 14, anti-FCV sera. As a control for cellular proteins that could be precipitated with the postimmunization sera, radiolabeled proteins precipitated from mock-infected CRFK cells with corresponding postimmunization sera are included as follows: lane 3, anti-B4 serum; lane 6, anti-C serum; lane 9, anti-D3 serum; lane 12, anti-F serum; and lane 15, anti-FCV serum.
Most of the expressed proteins were found in the insoluble fraction of the bacterial lysate. A general purification procedure included solubilization of the inclusion bodies in urea, binding of protein to an Ni-NTA agarose column, and consecutive washing and elution of proteins by changing the pH of the buffer. Eluted proteins were more than 90% pure (Fig. 1B) and were used to immunize guinea pigs. The specificity of the antisera against regions B4, C, D3, and F was examined by immunoprecipitation analysis of radiolabeled proteins synthesized in vitro from pTMF1 (ORF1 polyprotein) (Fig. 1C). The preimmunization sera did not recognize the FCV ORF1 TNT products (Fig. 1C, lanes 2, 4, 6, and 8, respectively), whereas the postimmunization sera demonstrated specific recognition of proteins in the TNT mixture (Fig. 1C, lanes 3, 5, 7, and 9, respectively).
The specificity of the new antisera was then examined by immunoprecipitation analysis of radiolabeled proteins from mock- and FCV-infected cells. The specificity of each antiserum was similar to that observed with the ORF1 TNT products (Fig. 1D), although there were more minor bands in the cell lysates. A minor band corresponding to an approximately 60-kDa protein was frequently observed in immunoprecipitation experiments with the FCV-infected cell lysates (Fig. 1D and Fig. 2), but was not observed in mock-infected cells (Fig. 1D, lanes 3, 6, 9, 12, and 15). This protein comigrated with the virus capsid protein that was precipitated by antisera raised against FCV virions (Fig. 1D, lane 14) and likely represents coprecipitating traces of the FCV capsid protein, which is abundant in infected cells.
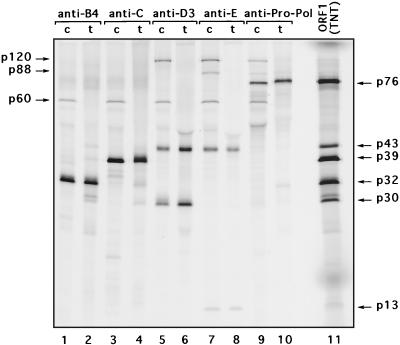
FIG. 2.
Comparative immunoprecipitation analysis of virus-specific proteins derived from ORF1. Shown are the results of SDS-PAGE analysis of the radiolabeled FCV ORF1 polyprotein cleavage products that were immunoprecipitated from FCV-infected cells (c) with ORF1 region-specific antisera (lanes 1, 3, 5, 7, and 9) or ORF1 in vitro translation mixture (t) (lanes 2, 4, 6, 8, and 10). Lane 11, radiolabeled TNT products derived from the ORF1 clone (pTMF-1).
Localization of the FCV nonstructural proteins by immunoprecipitation.
The ORF1 region-specific antisera were then used to localize the coding sequences for FCV nonstructural proteins in the genome. Figure 2 contains a direct comparison of immunoprecipitated radiolabeled proteins synthesized in FCV-infected cells or in vitro from the FCV ORF1 clone pTMF-1.
Antiserum to the B4 region (near the N terminus of ORF1) precipitated a protein of approximately 32 kDa from infected cells and from the in vitro translation mixture derived from pTMF-1 (Fig. 2, lanes 1 and 2), which we designated p32. The protein profile was similar for the anti-A region serum (data not shown).
Antiserum to the C region (corresponding to the 2C-like protein region) precipitated a protein with a molecular mass of approximately 38 to 39 kDa from both FCV-infected cells and the ORF1 TNT lysate (Fig. 2, lanes 3 and 4), which we designated p39. Faint bands corresponding to proteins with molecular masses of approximately 71 and 77 kDa were observed among proteins precipitated by anti-C and anti-B4 sera (Fig. 2, lanes 1 to 4).
Antiserum to the D3 region (raised against aa 728 to 898 of the ORF1 polyprotein) precipitated two major proteins with masses of approximately 43 and 30 kDa (Fig. 2, lanes 5 and 6), which we designated p43 and p30, respectively. In addition, a minor band corresponding to an approximately 120-kDa protein (designated p120) was observed in the FCV-infected cell lysate (Fig. 2, lane 5). In the lane corresponding to the proteins immunoprecipitated from the in vitro translation mixture (Fig. 2, lane 6), the 120-kDa band was detectable only after prolonged exposure of the gel (data not shown).
Further analysis showed that p43 was immunoprecipitated efficiently with antiserum to the region E peptide (corresponding to the VPg), whereas p30 was not (Fig. 2, lanes 7 and 8). The anti-Pro-Pol serum we characterized previously (38) precipitated the 76- to 78-kDa Pro-Pol precursor in both FCV-infected cells and the TNT lysate, but did not recognize the 43- and 30-kDa proteins. The anti-F serum (shown previously in Fig. 1) developed against the N terminus of Pro-Pol showed a similar result. We then deduced that p43 overlaps the VPg and contains sequences upstream of it. Given that the VPg sequence comprises the C-terminal part of p43, the N-terminal part of this protein should be a protein with a molecular mass of approximately 30 kDa (assuming that cleavage occurs at the E960/A961 site defined in our earlier bacterial expression studies). Thus, p30 would correspond to the protein located just upstream of the virus VPg.
The anti-VPg-peptide (anti-E) serum also precipitated a small 13- to 14-kDa protein and two larger proteins with molecular masses of 88 to 89 kDa (designated p88) and 120 kDa (p120) (Fig. 2, lanes 7 and 8), which were consistent with our previous study (36). These large-molecular-mass proteins were also recognized by anti-Pro-Pol serum (Fig. 2, lanes 9 and 10). It should be noted that detection of the two bands corresponding to these proteins precipitated from the in vitro translation mixture required a longer exposure of the gel. The major proteins precipitated by anti-Pro-Pol serum were a 76-kDa protein in the infected cell lysate and a 78-kDa protein in the ORF1 translation mixture (Fig. 2, lanes 9 and 10). The observed difference in protein size reflects the presence of a fused vector sequence at the C terminus of the Pro-Pol protein in the pTMF-1 vector (37, 38).
On the basis of these results, it was possible to propose the following organization of the FCV ORF1 polyprotein: NH2-p32-p39-p30-VPg-Pro-Pol.
Direct mapping of p32.
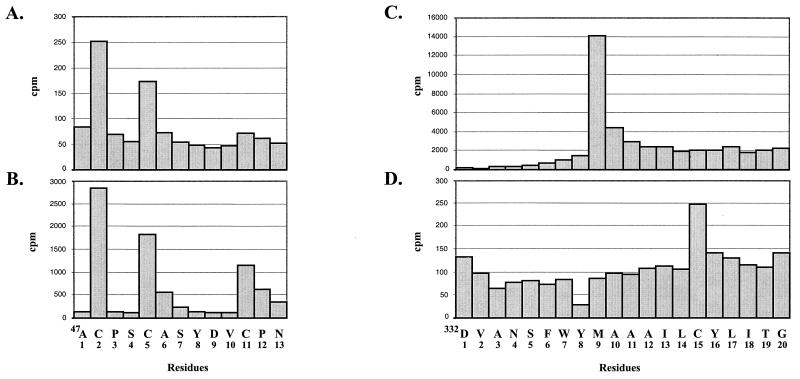
Microsequencing was performed by radiolabeling the proteins derived by in vitro translation of the pTMF-1 ORF1 clone in the presence of [35S]cysteine or [35S]methionine and sequencing the N terminus by Edman degradation. Mapping p32 in the pTMF-1 ORF1 translation products involved the radiolabeling of the products with [35S]cysteine. Direct microsequencing analysis indicated substantial yields of radioactivity for residues 2 and 5, and a third less-intense peak of radioactivity was found at position 11 (Fig. 3A). A computer search identified this pattern of cysteines as unique and was consistent with cleavage at aa 47 near the N terminus of the polyprotein. We compared the microsequencing profile of p32 derived from the ORF1 clone to that of the [35S]cysteine-radiolabeled p32 synthesized in FCV-infected cells. A plot of the radioactivity showed similar peaks (Fig. 3B).
FIG. 3.
Direct mapping of p32 and p39. Coupled transcription and translation of pTMF-1 were performed in the TNT system in the presence of [35S]cysteine. p32 was electrotransferred onto a polyvinylidene difluoride membrane following SDS-PAGE and subjected to microsequencing. The released radioactivity was determined for each cycle and is plotted against the residue number (A). To perform microsequence analysis of p32 synthesized in FCV-infected cells, the cells were labeled with [35S]cysteine, lysates were prepared, and radiolabeled protein was immunoprecipitated with anti-B4 serum. p32 was isolated and subjected to microsequence analysis as described above (B). For direct mapping of p39 protein, the coupled transcription and translation of pTMF-1 were performed in the presence of [35S]methionine or [35S]cysteine. Following gel separation, electrotransfer onto a polyvinylidene difluoride membrane, and microsequencing, the released radioactivity was determined for each cycle. Sequencing profiles for [35S]methionine and [35S]cysteine-labeled p39 are shown in panels C and D, respectively.
The localization of a predicted cleavage site (E/A) at aa 46 to 47 was evidence for initiation of translation at the first AUG of ORF1 polyprotein that would produce a small N-terminal protein of approximately 5.6 kDa. However, we were not able to detect the predicted 5.6-kDa protein in an immunoprecipitation analysis of radiolabeled FCV-infected cells by using either anti-A serum or serum raised in rabbits against a peptide corresponding to the first 20 aa of the ORF1 polyprotein (data not shown).
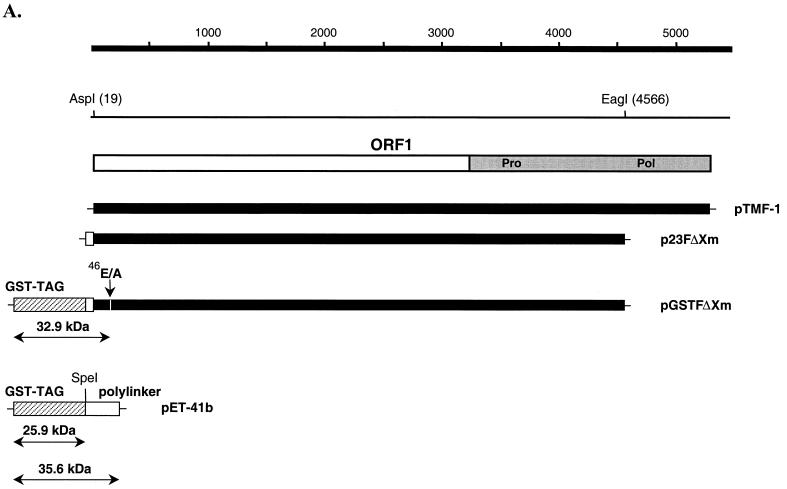
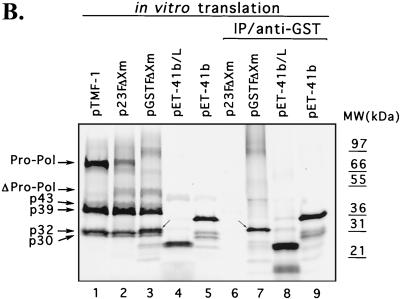
To confirm the synthesis of the N-terminal cleavage product in vitro, we constructed plasmid pGSTFΔXm, which encoded an ORF1 polyprotein sequence that was fused to a GST-TAG sequence at the N terminus and truncated in the polymerase region (Fig. 4A). The additional N-terminal sequence and the truncated polymerase region did not affect proteolytic processing activity. Cleavage products were produced from both plasmids p23FΔXm (ORF1 without the N-terminal GST-TAG) and pGSTFΔXm (ORF1 with the N-terminal GST-TAG) (Fig. 4B, lanes 1, 2, and 3). The 50-kDa band (not present in pTMF-1; Fig. 4B, lane 1) corresponded to the truncated Pro-Pol sequence encoded by these plasmids (Fig. 4B, lanes 2 and 3). The approximately 78-kDa protein (with a mobility similar to that of Pro-Pol) observed in translation products derived from p23FΔXm (Fig. 4B, lane 2) corresponded to the N-terminal part of the FCV ORF1 fused to the polylinker sequence of the pET-23a vector based on our previous studies with similar ORF1-encoded plasmids (38). In addition, this protein was not precipitated by anti-Pro-Pol serum (data not shown). In order to identify a potential fusion protein containing the GST-TAG sequence, an immunoprecipitation was performed with GST-specific antibodies. A positive control (plasmid pET-41b, either uncut or cut with SpeI) was used to demonstrate the specific binding of the GST antibodies. The GST antibodies precipitated a protein with a molecular mass of approximately 35 to 36 kDa from uncut plasmid pET-41b-derived translation products that represented the GST-TAG protein sequence fused to the entire polylinker sequence encoded by this vector (Fig. 4B, lane 9). The pET-41b control plasmid was cut with SpeI to exclude the polylinker sequence, and the GST antibodies precipitated an approximately 26-kDa protein from the translation products, consistent with the molecular mass of the GST-TAG protein alone (Fig. 4B, lane 8). The GST-specific antibodies did not react with TNT products derived from p23FΔXm, which did not contain the GST-TAG sequence (Fig. 4B, lane 6). The GST antibodies were then used to examine the TNT products derived from the plasmid pGSTFΔXm (Fig. 4B, lane 7). The observed protein with a molecular mass of approximately 32 to 33 kDa corresponded in size to a predicted fusion protein containing the GST polypeptide (25.9 kDa), polylinker sequence (1.6 kDa), and the small FCV ORF1 N-terminal protein (5.6 kDa), which would be produced by cleavage at E46/A47.
FIG. 4.
Detection of the N-terminal cleavage product of the ORF1 polyprotein fused to a GST-TAG sequence. (A) Localization of clones pTMF-1 (ORF1 clone), p23FΔXm, pGSTFΔXm, on the FCV ORF1 map. Plasmids were engineered as described in Materials and Methods. The GST-TAG sequence in pGSTFΔXm is shown as hatched gray bar. The polylinker sequence originating from plasmid pET-23a (Novagen), which was used to construct plasmid p23FΔXm, is indicated by an open bar. (B) SDS-PAGE analysis of the products obtained in TNT reactions and immunoprecipitation (IP) analysis with the GST-specific antibodies. Radiolabeled TNT translation products derived from the indicated plasmid are shown as follows: lane 1, pTMF-1; lane 2, p23FΔXm; lane 3, pGSTFΔXm; lane 4, pET-41b linearized (L) with SpeI; lane 5, pET-41b. The radiolabeled in vitro translation products derived from p23FΔXm, pGSTFΔXm, pET-41b linearized with SpeI, and pET-41b were subjected to immunoprecipitation analysis with anti-GST antibodies (Sigma) and analyzed on the same gel: lane 6, p23FΔXm; lane 7, pGSTFΔXm; lane 8, pET-41b linearized with SpeI; and lane 9, pET-41b. Small arrows indicate the protein band that corresponds to the GST-TAG-polylinker-p5.6 fusion protein. MW, molecular mass.
Indirect evidence for the importance of the cleavage of this small protein from the N terminus of the polyprotein was obtained when the sequence of the cleavage site between the predicted 5.6-kDa protein and p32 was mutagenized (E46/A47 into A46/A47) in the FCV full-length cDNA clone (pQ14), so that it could not be cleaved by the virus-encoded 3C-like proteinase. Following verification of the mutagenized genome sequence, the resulting plasmid, pF22 (Table 2), was analyzed in transfection experiments. Transfection of the parental pQ14 plasmid DNA into MVA/T7-infected CRFK cells resulted in intracellular transcription of the genome and subsequent production of infectious FCV. However, virus could not be recovered from cells transfected with plasmid pF22, indicating that this mutation was lethal for the virus. In addition, mutagenesis of the first AUG codon in ORF1 into AAG was lethal (plasmid pF10), suggesting that synthesis of the extreme N terminus of the polyprotein is essential.
Direct mapping of p39.
The p39 protein was labeled separately with [35S]methionine and [35S]cysteine in a TNT reaction of pTMF-1. The microsequencing profiles of the [35S]methionine and [35S]cysteine-labeled 38-kDa protein demonstrated that residue 9 is a methionine (Fig. 3C) and residue 15 is a cysteine (Fig. 3D). A computer search indicated that this pattern was unique and corresponded to cleavage at position 331. Of interest, the region near the sequence of this E331/D332 cleavage site in the FCV polyprotein shared similarity with the region surrounding the Q399/G400 cleavage site described for Southampton virus (SHV). The FCV protein shares sequence relatedness with the SHV and RHDV NTPases (19, 27). Substitution of E331 for A331, which would abrogate the E331/D332 cleavage site, prevented recovery of viable virus when MVA/T7-infected CRFK cells were transfected with the corresponding plasmid, pF71 (Table 2).
Mapping of p43 and p30.
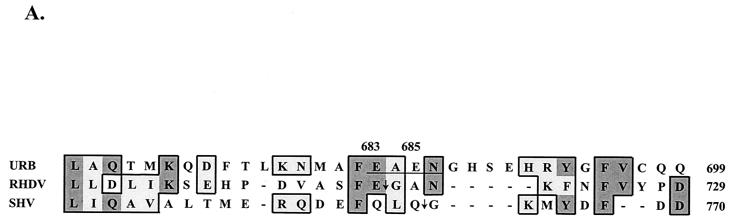
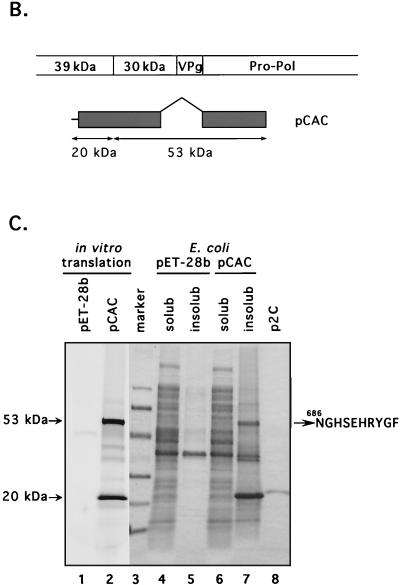
We were unable to map the N-terminal sequence of p43 and p30 definitively by the microsequencing technique. The localization of this protein on the map of ORF1 polyprotein was predicted on the basis of immunoprecipitation data (see above). The FCV ORF1 polyprotein sequence corresponding to the putative border between p39 and p30 was aligned with the corresponding regions from the SHV and RHDV polyproteins (Fig. 5A). The analogous location in the FCV ORF1 polyprotein showed two adjacent dipeptide sequences with potential for recognition by the virus 3C-like proteinase. In order to identify the correct N-terminal border of this protein, we first expressed large fragments of ORF1 overlapping the sequences of p39, p30, VPg, and the 3C-like proteinase sequence in E. coli (data not shown). However, expression levels were low. Further analysis of plasmids encoding p30 suggested a toxic effect from the C terminus (data not shown). To overcome this problem, we constructed a plasmid (pCAC) encoding the C-terminal part of p39 and the N-terminal sequence of p30 fused to the complete sequence of the 3C-like proteinase (Fig. 5B). The deletion of the C-terminal part of p30 in pCAC plasmid allowed efficient expression in vitro and in bacteria of two major proteins of approximately 53 and 20 kDa (Fig. 5C, lanes 2 and 7). Analysis of the 53-kDa protein expressed in bacteria identified the sequence NGHSEHRYGF consistent with cleavage at aa 685 to 686 of ORF1. Sites E685/N686 and E960/A961 (38) defined the borders of a protein with a calculated molecular mass of 30.1 kDa that mapped just upstream of VPg.
FIG. 5.
Analysis of the cleavage site between p39 and p30. (A) An alignment of the deduced amino acid sequences of the putative junction of the FCV p39 and p30 with the corresponding regions of RHDV (GenBank accession no. M67473) and SHV (GenBank accession no. L07418) was created with the Multalin computer program (5) with the following parameters: gap weight, 5.0; gap length weight, 0. The numbers given next to the protein sequences correspond to numbering systems for the virus polyproteins. Identical and similar amino acids of calicivirus sequences are shaded in dark and light tones, respectively. The sequence of the putative N-terminal cleavage site of the FCV p39 is underlined. Arrows are used to indicate the corresponding cleavage sites in sequences of the RHDV and SHV polyproteins. (B) Schematic representation of the pCAC plasmid, which was engineered to contain the p39-p30 junction and an active 3C-like proteinase. The entire VPg sequence and C-terminus sequence of p30 were deleted. (C) SDS-PAGE analysis of proteins encoded in pCAC when expressed in vitro and in E. coli. Lanes: 1 and 2, in vitro TNT translation products derived from pET-28b (vector) and pCAC, respectively (autoradiography); 3, electrophoresis calibration kit protein markers (Pharmacia) corresponding to molecular masses of 94, 67, 43, 30, 20, and 14 kDa (from top to bottom of gel); 4 and 5, soluble (solub) and insoluble (insolub) fractions of induced bacterial cells transformed with pET-28b, respectively; 6 and 7, soluble and insoluble fractions of induced bacterial cells transformed with pCAC; 8, purified 21.6-kDa recombinant protein encoded by plasmid p2C. Proteins in lanes 3 to 8 were detected by Coomassie blue staining. Arrows point to the two major pCAC proteins expressed in both in vitro translation reactions and in bacteria. The N-terminal sequence of the 53-kDa protein (NGHSEHRYGF) is shown.
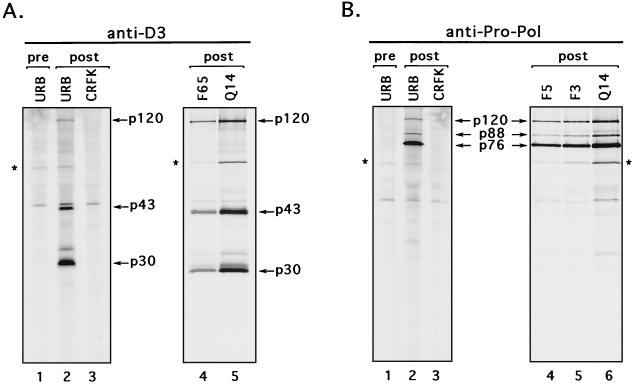
Further evidence for the utilization of the E685/N686 site was obtained in mutagenesis experiments. When this cleavage site (E685/N686) was changed into A685/N686 in the full-length cDNA clone (plasmid pF36), we could not detect virus replication. However, changing the upstream E683/A684 site into A683/A684 (plasmid pF65) did not affect virus viability, because transfection of the MVA/T7-infected CRFK cells with plasmid pF61 led to the production of infectious FCV particles. The recovered virus demonstrated growth rate and plaque size characteristics similar to those of the wild-type virus (data not shown). Immunoprecipitation analysis with anti-D3 serum showed identical protein processing for viruses generated from the parental clone and pF65 plasmid as well as for wild-type virus (Fig. 6A, lanes 2, 4, and 5). The 2-nt substitutions introduced into the E683/A684 dipeptide sequence were verified in the RNA genome of the virus recovered from the plasmid pF65.
FIG. 6.
Comparative analysis of virus polyprotein processing products in CRFK cells infected with cleavage site mutants. The CRFK cells were infected at an MOI of 1 to 4 with FCV URB or with viruses recovered from plasmids pQ14 (Q14), pF65 (F65), pF3 (F3), and pF5 (F5). A mock-infected CRFK control was included. At 3.5 h postinfection, cells were metabolically labeled with [35S]methionine for 1 h. (A) The radiolabeled proteins were immunoprecipitated from cell lysates with anti-D3 (specific for p30) sera (lane 1, preimmunization serum; lanes 2, 3, 4, and 5, postimmunization serum). (B) The radiolabeled proteins were also immunoprecipitated from cell lysates with anti-Pro-Pol sera (lane 1, preimmunization serum; lanes 2, 3, 4, 5, and 6, postimmunization serum). Asterisks denote a 60-kDa protein coprecipitated from virus-infected cells.
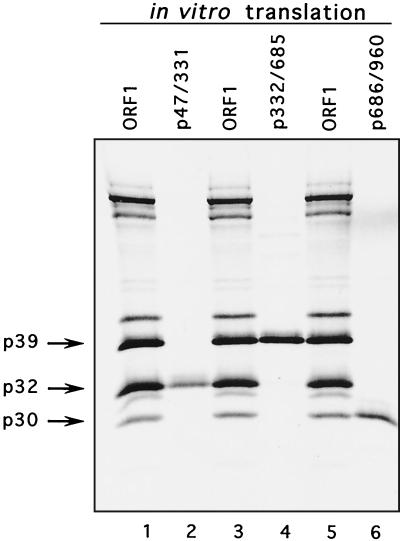
The ORF1 sequences bordered by cleavage sites E46/A47 and E331/D332, E331/D332 and E685/N686, E685/N686 and E960/A961 were subcloned into the pCI vector under control of the T7 RNA polymerase promoter in order to compare the mobility of proteins expressed individually with those produced by cleavage of the ORF1 polyprotein. SDS-PAGE analysis of the in vitro translation products derived from the corresponding plasmids p47/331, p332/685, and p686/960 demonstrated that the proteins encoded by these plasmids had the same apparent mobility as products of the autocatalytic processing of the ORF1 polyprotein, p32, p39, and p30, respectively (Fig. 7, lanes 2, 4, and 6, respectively). The introduction of a methionine residue at the N termini of the proteins did not appear to affect mobility. This experiment provided additional support for the newly defined borders in our proposed cleavage map.
FIG. 7.
Comparison of p39, p32, and p30 synthesized in vitro with products produced by cleavage of the FCV ORF1 polyprotein synthesized in vitro. Lanes: 1, 3, and 5, radiolabeled TNT products derived from ORF1 clone (pTMF-1); 2, 4, and 6, radiolabeled TNT products produced by in vitro translation of cDNA clones encoding the fragments of the FCV ORF1 corresponding to aa 47 to 331, 332 to 685, and 686 to 960, respectively.
Processing of the p120 precursor.
Analysis of the ORF1 in vitro translation products and radiolabeled FCV-infected cell lysates (Fig. 2) showed the presence of an approximately 120-kDa protein (p120) that was immunoprecipitated by three different sera derived from the C-terminal part of ORF1 (regions D3, E, and Pro-Pol). Thus, p120 was likely a precursor for other intermediate and mature proteins (p30-p13-p76) observed previously in studies of the C-terminal part of ORF1. These include the p43 precursor comprised of p30 and p13 and the p88 precursor comprised of p13 and p76 (36, 38). Although p13 has been identified as the putative VPg and p76 as a bifunctional proteinase-polymerase complex, the functions of the precursors to these proteins, if any, are not clear. A strategy was designed to examine the role of proteolytic processing and the subsequent generation of various precursors in virus replication. Site-specific mutations were introduced into four cleavage sites found previously in the p120 sequence. Virus could not be recovered from cells transfected with the plasmid pF77, which contained a mutated cleavage site between p30 and VPg (E960/A961 was changed into A960/A961). In addition, substitution of E1071 for A1071 in the E1071/S1072 cleavage site between the VPg and 3C-like proteinase prevented recovery of viable virus when MVA/T7-infected CRFK cells were transfected with the corresponding plasmid, pF87. However, we were able to recover virus with growth characteristics similar to the wild-type virus (data not shown), when we mutagenized two internal cleavage sites in the Pro-Pol sequence, E1345/T1346 and E1419/G1420, described earlier (38), into A1345/T1346 (pF3) and A1419/G1420 (pF5), respectively. The presence of engineered mutations in the genomes of recovered viruses was confirmed by direct sequence analysis of RT-PCR products derived from viral RNA. Antiserum prepared against Pro-Pol (38) was used to immunoprecipitate radiolabeled proteins from CRFK cells infected with viruses recovered from the plasmids pF3 and pF5. The protein profiles were similar to that of virus recovered from the parental plasmid (pQ14) control and to that of wild-type virus (Fig. 6B, lanes 2, 4, 5, and 6).
DISCUSSION
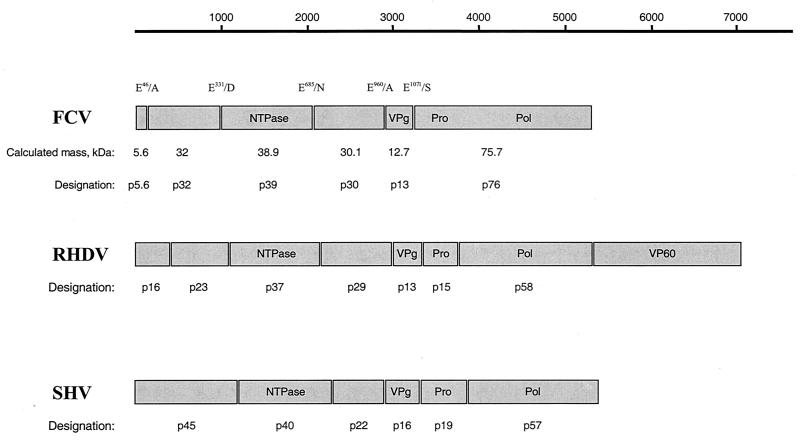
The goals of the present study were to complete the FCV ORF1 cleavage map and to identify cleavage sites in the ORF1 polyprotein essential for growth of the virus. We found that the FCV 3C-like proteinase processes the N-terminal half of the ORF1 polyprotein at cleavage sites E46/A47, E331/D332, and E685/N686 to yield three proteins with calculated masses of 5.6, 32, and 38.9 kDa, which we designated p5.6, p32, and p39, respectively. In addition, cleavage at E685/N686 defines the N-terminal border of a 30.1-kDa protein that we designated p30. Taken together with our previous data (38), the proposed FCV ORF1 polyprotein gene order is p5.6-p32-p39 (NTPase)-p30-VPg-Pro-Pol (Fig. 8). Mutagenesis studies of an infectious cDNA clone of FCV demonstrated that cleavage between each of these gene products (with the exception of Pro-Pol) is critical for the growth of the virus.
FIG. 8.
Comparison of the genetic map of FCV ORF1 with those of other caliciviruses, SHV and RHDV. A schematic representation of proteins encoded by ORF1 of the FCV, RHDV, and SHV genomes is shown. Protein coding sequences are drawn as boxes, with the molecular mass of the encoded protein shown above. The locations and designations of the proteins encoded by RHDV and SHV ORF1 are adapted from studies by Meyers et al. (22) and Liu et al. (18).
The mature proteins identified in this study in in vitro experiments were all detected in FCV-infected cells, with the exception of the predicted protein, p5.6, encoded at the extreme N terminus of ORF1. The antibodies developed against region A, which include the p5.6 coding region, were unable to efficiently precipitate this protein. Thus, two indirect approaches were used to prove its existence. First, mutagenesis of the first AUG of ORF1 (which would be the initiating methionine of p5.6) and the cleavage site between p5.6 and p32 in the infectious FCV cDNA clone prevented virus recovery, indicating that its synthesis and release from the N terminus are important. It should be noted that the second AUG of ORF1 is located well within p32, and the direct mapping of the N terminus of p32 indicates that upstream protein sequence (presumably as the C-terminal part of p5.6) would be needed to form the E46/A47 cleavage site. The second approach involved the incorporation of a GST tag at the N terminus of ORF1. Evidence for the production of a GST-p5.6 fusion protein was found by immunoprecipitation analysis with anti-GST antibodies. Further studies are in progress to identify and localize p5.6 in infected cells.
The next protein in the linear arrangement of the FCV ORF1 polyprotein map was p32 (aa 47 to 331), which was observed both in in vitro translation products derived from an ORF1 clone and in virus-infected cells. It is likely that this protein is an analog for the RHDV 23-kDa protein and the 45-kDa N-terminal protein of SHV (Fig. 8) (17, 43). The amino acid similarity of these proteins ranges from 25 to 31% and represents one of the most divergent regions of polyprotein sequence within and between Caliciviridae genera (6, 9, 28, 29). It was suggested that the RHDV 23-kDa protein might have functional similarity with the 2B protein of picornaviruses that alone or as a part of its precursor, 2BC, is involved in membrane modifications in virus-infected cells that include rearrangements and vesicle formation (4, 40), change in membrane permeability (1, 13), and disassembly of the Golgi complex (32).
The p39 (immediately following p32) contains the predicted NTP-binding domain conserved in picornavirus 2C proteins (10, 23). Evidence for NTP-hydrolysis activity of this protein has been reported for RHDV and SHV (19, 27). Comparison of the amino acid sequence of the putative FCV NTPase with those of RHDV and SHV shows similarities of 47 and 35%, respectively. In addition, the FCV p39 shares a structural feature with the corresponding RHDV and SHV proteins, which is the presence of an N-terminal cluster of hydrophobic amino acids. Of interest, transmembrane helices (33) are predicted for aa 4 to 22 and 35 to 57 of the FCV p39. Based on their location in the protein sequence, it is possible that this protein is anchored to intracellular membranes via its N terminus. The picornavirus 2C proteins also have a conserved region near the N terminus, which is a putative amphipathic helix (26). Certain mutations in the helix sequence affect proteolytic processing of virus proteins and subsequent virus replication (26). In addition, deletions in this sequence in the poliovirus 2C protein significantly lowered its ability to bind to membranes (7). The finding of the 2C-like p39 protein in FCV membranous replication complexes (Green et al., unpublished observations) suggests that this calicivirus protein is involved in virus genome replication and might be associated with membranes.
The p30 (aa 686 to 960) is located between p39 and the VPg. The function of p30 is unknown, and it maps to the second most variable nonstructural protein region among caliciviruses. Computer analysis predicts an amphipathic helix in the C-terminal part of the protein (aa 920 to 939) (30), which might be responsible for the association of the protein with intracellular membranes. The protein can be detected in its mature form as well as in the form of p43 precursor (with VPg) in infected cells and in FCV membranous replication complexes (Green et al., unpublished). It is possible, by analogy with the picornaviruses, that p30 (as part of the p43 precursor) serves as a membrane anchor for the VPg protein. The release of the VPg from the p43 precursor might be dependent on several factors, including RNA synthesis initiation, as suggested for picornaviruses (16, 25, 39). Our mutagenesis experiments with plasmid pF77 indicated that cleavage between p30 and VPg was essential for a productive virus infection.
The final protein encoded in the FCV ORF1 is the 76-kDa Pro-Pol (aa 1072 to 1763). Pro-Pol has been described previously as a stable precursor to the FCV proteinase and polymerase that were observed in in vitro translation studies and in infected cells (38). Although stable complexes of the proteinase and polymerase were observed in in vitro studies of RHDV and SHV, cleavage events leading to release of mature proteins were identified for these two viruses only when the complexes were expressed to high levels in bacterial systems (18, 42). The internal cleavage sites we previously described for the FCV Pro-Pol in bacterial expression studies (E1345/T1346 and E1419/G1420) would be predicted to inactivate the polymerase by truncation of conserved functional domains (15, 38, 41). Inactivation of these two putative cleavage sites by mutagenesis in the present study showed no detectable effect on viral replication. Wei and colleagues (41) demonstrated that bacterially expressed recombinant FCV Pro-Pol with an inactivated proteinase exhibited properties expected of a replicative polymerase. The possibility has not been ruled out that the release of individual FCV polymerase and proteinase proteins requires the presence of certain cellular factors and that the in vitro approaches used thus far have not allowed the identification of an “authentic” cleavage site between these two proteins. The anti-Pro-Pol serum precipitates some unidentified minor proteins from virus-infected cells (38) (Fig. 2, lane 9), and it is possible that these proteins could represent further processing of the Pro-Pol protein.
The mapping and mutagenesis data defined the borders of the “mature” virus proteins encoded in ORF1. However, it should be noted that evidence was found for several stable precursor proteins. First, 77- and 71-kDa precursors derived from the N terminus were detected. These proteins are likely processed into the p5.6, p32, and p39 proteins or the p32 and p39 proteins, respectively. More stable potential precursors were observed for proteins encoded by the C-terminal part of ORF1 polyprotein. The p120 (calculated molecular mass, 118.5 kDa) precursor corresponding to p30-VPg-Pro-Pol could be detected in virus-infected cells as early as 2 to 3 h postinfection (38). It is likely that this large precursor protein undergoes further processing in at least two different ways. Data from immunoprecipitation analysis indicates that the 118.5-kDa protein can be cleaved into either p30 and the p88 precursor (VPg-Pro-Pol) or into the p43 precursor (p30-VPg) and p76 (Pro-Pol).
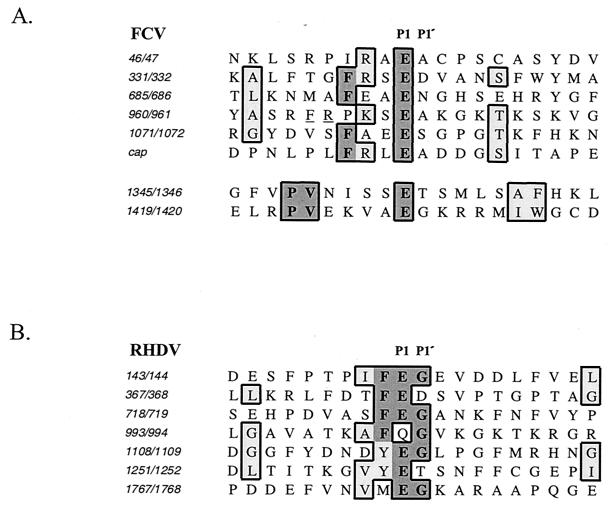
The completion of an FCV cleavage map that was verified by mutagenesis of the infectious FCV clone allowed a comparative analysis of the cleavage sites within the FCV coding sequence (Fig. 9A). The FCV 3C-like proteinase exhibits strict specificity for pairs of amino acids with E in the P1 position. The mechanism of cleavage site recognition is not known for caliciviruses. It was suggested that the RHDV proteinase could have a minor preference for a large, hydrophobic amino acid in the P2 position (42). Six of the seven mapped RHDV cleavage sites have a phenylalanine or tyrosine residue in the P2 position (Fig. 9B). When amino acids surrounding the cleavage sites recognized by the FCV proteinase were compared, we found that all of the six essential cleavage site sequences contained a hydrophobic residue at the P4 position, four of these being phenylalanine (Fig. 9A). In addition, four out of six cleavage sites had positively charged amino acid residues (arginine or lysine) in the P3 position. Of interest, the site between p30 and VPg has phenylalanine and a positively charged residue next to the cleavage site sequence, but these are more distant in positions P6 and P5. It will be interesting to examine whether the location of these amino acids relative to the cleavage site influences the efficiency of cleavage by the proteinase. A lower efficiency of the cleavage of the p43 precursor might be associated with the control of the release of p30 and VPg. The internal cleavage sites, E1345/T1346 and E1419/G1420, found in the Pro-Pol protein, which were not essential for infectivity, do not have the phenylalanine residues in adjacent positions. Instead, their common feature is the presence of a proline-valine pair in the P7 and P6 positions (Fig. 9A).
FIG. 9.
Comparison of cleavage sites identified in the polyproteins encoded by ORF1 of FCV and RHDV. (A) Alignment of the amino acid sequences flanking the cleavage sites of the FCV ORF1 polyprotein and the sequence of the FCV capsid precursor cleavage site. An alignment of sequences adjacent to the internal cleavage sites of the 75.7-kDa protein (Pro-Pol) is shown separately. Identical and similar amino acids are shaded in dark and light tones, respectively. The arginine and phenylalanine residues at positions −5 and −6 of the E960/A961 cleavage site are underlined. (B) Alignment of the amino acid sequences flanking cleavage sites of the RHDV ORF1 polyprotein. Studies by Meyers et al. provided the RHDV sequence (21) and cleavage site (22) data.
The sequence similarity in the cleavage sites of the capsid precursors of canine calicivirus and FCV may, in part, explain why the FCV proteinase was successful in processing the canine calicivirus protein (20). The E157/S158 cleavage site of the capsid precursor of canine calicivirus has phenylalanine and arginine residues present at the P4 and P3 positions, respectively.
Proteolytic processing of the nonstructural polyprotein by the virus-encoded cysteine proteinase is a feature common to all caliciviruses. The finding that proteolytic processing at most of the mapped cleavage sites in the FCV nonstructural polyprotein is essential for virus growth indicates the importance of accurate proteolytic processing in virus replication. Knowledge of the structural features of the cleavage sites that facilitate recognition and cleavage by the proteinase, as well as the structural characteristics of the proteinase itself, should lead to a better understanding of virus replication.
Acknowledgments
We thank John Shannon from Biomolecular Research Facility, University of Virginia, for assistance with the protein sequence analysis. We thank Jose Valdesuso and Tanaji Mitra for their dedicated technical support. We extend our appreciation to Albert Z. Kapikian and Robert M. Chanock, LID, NIAID, NIH, for continuing support.
REFERENCES
- 1.Aldabe, R., A. Barco, and L. Carrasco. 1996. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 271:23134-23137. [DOI] [PubMed] [Google Scholar]
- 2.Carter, M. J. 1989. Feline calicivirus protein synthesis investigated by Western blotting. Arch. Virol. 108:69-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter, M. J., I. D. Milton, P. C. Turner, J. Meanger, M. Bennett, and R. M. Gaskell. 1992. Identification and sequence determination of the capsid protein gene of feline calicivirus. Arch. Virol. 122:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 5.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dingle, K. E., P. R. Lambden, E. O. Caul, and I. N. Clarke. 1995. Human enteric Caliciviridae: the complete genome sequence and expression of virus-like particles from a genetic group II small round structured virus. J. Gen. Virol. 76:2349-2355. [DOI] [PubMed] [Google Scholar]
- 7.Echeverri, A. C., and A. Dasgupta. 1995. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology 208:540-553. [DOI] [PubMed] [Google Scholar]
- 8.Gaskell, R. M. 1985. Viral-induced upper respiratory tract diseases, p. 257-270. In E. A. Chandler, C. J. Gaskell, and A. D. R. Hilbery (ed.), Feline medicine and therapeutics. Blackwell Scientific Publications, Oxford, United Kingdom.
- 9.Glenn, M., A. D. Radford, P. C. Turner, M. Carter, D. Lowery, D. A. DeSilver, J. Meanger, C. Baulch-Brown, M. Bennett, and R. M. Gaskell. 1999. Nucleotide sequence of UK and Australian isolates of feline calicivirus (FCV) and phylogenetic analysis of FCVs. Vet. Microbiol. 67:175-193. [DOI] [PubMed] [Google Scholar]
- 10.Gorbalenya, A. E., E. V. Koonin, and Y. I. Wolf. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 262:145-148. [DOI] [PubMed] [Google Scholar]
- 11.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H. J. Thiel. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181:S322-S330. [DOI] [PubMed] [Google Scholar]
- 12.Herbert, T. P., I. Brierley, and T. D. Brown. 1996. Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from a single, functionally bicistronic, subgenomic mRNA. J. Gen. Virol. 77:123-127. [DOI] [PubMed] [Google Scholar]
- 13.Jecht, M., C. Probst, and V. Gauss-Muller. 1998. Membrane permeability induced by hepatitis A virus proteins 2B and 2BC and proteolytic processing of HAV 2BC. Virology 252:218-227. [DOI] [PubMed] [Google Scholar]
- 14.Kahn, D. E., and E. A. Hoover. 1976. Infectious respiratory diseases of cats. Vet. Clin. N. Am. 6:399-413. [DOI] [PubMed] [Google Scholar]
- 15.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn, R. J., and E. Wimmer. 1987. The replication of picornaviruses, p. 17-51. In D. J. Rowlands, M. A. Mayo, and B. W. J. Mahy (ed.), The molecular biology of the positive strand RNA viruses. Academic Press, London, United Kingdom.
- 17.Liu, B., I. N. Clarke, and P. R. Lambden. 1996. Polyprotein processing in Southampton virus: identification of 3C-like protease cleavage sites by in vitro mutagenesis. J. Virol. 70:2605-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, B. L., G. J. Viljoen, I. N. Clarke, and P. R. Lambden. 1999. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J. Gen. Virol. 80:291-296. [DOI] [PubMed] [Google Scholar]
- 19.Marin, M. S., R. Casais, J. M. Martin Alonso, and F. Parra. 2000. ATP binding and ATPase activities associated with recombinant rabbit hemorrhagic disease virus 2C-like polypeptide. J. Virol. 74:10846-10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuura, Y., Y. Tohya, M. Onuma, F. Roerink, M. Mochizuki, and T. Sugimura. 2000. Expression and processing of the canine calicivirus capsid precursor. J. Gen. Virol. 81:195-199. [DOI] [PubMed] [Google Scholar]
- 21.Meyers, G., C. Wirblich, and H.-J. Thiel. 1991. Rabbit hemorrhagic disease virus—molecular cloning and nucleotide sequencing of a calicivirus genome. Virology 184:664-676. [DOI] [PubMed] [Google Scholar]
- 22.Meyers, G., C. Wirblich, H. J. Thiel, and J. O. Thumfart. 2000. Rabbit hemorrhagic disease virus: genome organization and polyprotein processing of a calicivirus studied after transient expression of cDNA constructs. Virology 276:349-363. [DOI] [PubMed] [Google Scholar]
- 23.Neill, J. D. 1990. Nucleotide sequence of a region of the feline calicivirus genome which encodes picornavirus-like RNA-dependent RNA polymerase, cysteine protease and 2C polypeptides. Virus Res. 17:145-160. [DOI] [PubMed] [Google Scholar]
- 24.Neill, J. D., I. M. Reardon, and R. L. Heinrikson. 1991. Nucleotide sequence and expression of the capsid protein gene of feline calicivirus. J. Virol. 65:5440-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nomoto, A., B. Detjen, R. Pozzatti, and E. Wimmer. 1977. The location of the polio genome protein in viral RNAs and its implication for RNA synthesis. Nature 268:208-213. [DOI] [PubMed] [Google Scholar]
- 26.Paul, A. V., A. Molla, and E. Wimmer. 1994. Studies of a putative amphipathic helix in the N-terminus of poliovirus protein 2C. Virology 199:188-199. [DOI] [PubMed] [Google Scholar]
- 27.Pfister, T., and E. Wimmer. 2001. Polypeptide p41 of a Norwalk-like virus is a nucleic acid-independent nucleoside triphosphatase. J. Virol. 75:1611-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pletneva, M. A., S. V. Sosnovtsev, and K. Y. Green. 2001. The genome of Hawaii virus and its relationship with other members of the Caliciviridae. Virus Genes 23:5-16. [DOI] [PubMed] [Google Scholar]
- 29.Rinehart-Kim, J. E., W. M. Zhong, X. Jiang, A. W. Smith, and D. O. Matson. 1999. Complete nucleotide sequence and genomic organization of a primate calicivirus, Pan-1. Arch. Virol. 144:199-208. [DOI] [PubMed] [Google Scholar]
- 30.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol 232:584-599. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sandoval, I. V., and L. Carrasco. 1997. Poliovirus infection and expression of the poliovirus protein 2B provoke the disassembly of the Golgi complex, the organelle target for the antipoliovirus drug Ro-090179. J. Virol. 71:4679-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 34.Sosnovtsev, S., and K. Y. Green. 1995. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VPg for infectivity. Virology 210:383-390. [DOI] [PubMed] [Google Scholar]
- 35.Sosnovtsev, S., S. Sosnovtseva, and K. Y. Green. 1996. Recovery of feline calicivirus from plasmid DNA containing a full-length copy of the genome, p. 125-130. In D. Chasey, R. M. Gaskell, and I. N. Clarke (ed.), The 1st International Symposium on Caliciviruses. European Society for Veterinary Virology and Central Veterinary Laboratory, Reading, United Kingdom.
- 36.Sosnovtsev, S. V., and K. Y. Green. 2000. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology 277:193-203. [DOI] [PubMed] [Google Scholar]
- 37.Sosnovtsev, S. V., S. A. Sosnovtseva, and K. Y. Green. 1998. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 72:3051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sosnovtseva, S. A., S. V. Sosnovtsev, and K. Y. Green. 1999. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J. Virol. 73:6626-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takegami, T., B. L. Semler, C. W. Anderson, and E. Wimmer. 1983. Membrane fractions active in poliovirus RNA replication contain VPg precursor polypeptides. Virology 128:33-47. [DOI] [PubMed] [Google Scholar]
- 40.Teterina, N. L., K. Bienz, D. Egger, A. E. Gorbalenya, and E. Ehrenfeld. 1997. Induction of intracellular membrane rearrangements by HAV proteins 2C and 2BC. Virology 237:66-77. [DOI] [PubMed] [Google Scholar]
- 41.Wei, L., J. S. Huhn, A. Mory, H. B. Pathak, S. V. Sosnovtsev, K. Y. Green, and C. E. Cameron. 2001. Proteinase-polymerase precursor as the active form of feline calicivirus RNA-dependent RNA polymerase. J. Virol. 75:1211-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wirblich, C., M. Sibilia, M. B. Boniotti, C. Rossi, H.-J. Thiel, and G. Meyers. 1995. 3C-like protease of rabbit hemorrhagic disease virus: identification of cleavage sites in the ORF1 polyprotein and analysis of cleavage specificity. J. Virol. 69:7159-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wirblich, C., H.-J. Thiel, and G. Meyers. 1996. Genetic map of the calicivirus rabbit hemorrhagic disease virus as deduced from in vitro translation studies. J. Virol. 70:7974-7983. [DOI] [PMC free article] [PubMed] [Google Scholar]