Abstract
The human immunodeficiency virus type 1 (HIV-1) capsid protein (CA) plays a crucial role in both assembly and maturation of the virion. Numerous recent studies have focused on either the soluble form of CA or the polymer end product of in vitro CA assembly. The CA polymer, in particular, has been used to study CA-CA interactions because it is a good model for the CA interactions within the virion core. However, analysis of the process of in vitro CA assembly can yield valuable insights into CA-CA interactions and the mechanism of core assembly. We describe here a method for the analysis of CA assembly kinetics wherein the progress of assembly is monitored by using turbidity. At pH 7.0 the addition of either of the isolated CA domains (i.e., the N or the C domain) to an assembly reaction caused a decrease in the assembly rate by competing for binding to the full-length CA protein. At pH 8.0 the addition of the isolated C domain had a similar inhibitory affect on CA assembly. However, at pH 8.0 the isolated N domain had no affect on the rate of CA assembly but, when mixed with the C domain, it alleviated the C-domain inhibition. These data provide biochemical evidence for a pH-sensitive homotypic N-domain interaction, as well as for an N- and C-domain interaction.
Retroviruses such as human immunodeficiency virus type 1 (HIV-1) assemble through the polymerization of ca. 1,500 molecules of the Gag polyprotein (39). HIV-1 Gag polyprotein consists of the matrix (MA), capsid (CA), p2, nucleocapsid (NC), p1, and p6 domains in that order (14, 23, 29). Approximately 11% of the Gag polyprotein molecules also consist of fusions with the protease, polymerase, and integrase proteins as a result of a −1 ribosomal frameshift (14, 23, 24). After assembly, the capsid buds from the cell, acquiring its membrane, and thereby releasing a spherical-immature virion (30). In the immature virion, the Gag polyprotein is radially arranged with the myristylated N terminus of the matrix domain proximal to the viral envelope (15, 42). Through a mechanism yet to be understood, budding triggers the activation of a virus-encoded protease that cleaves the Gag polyprotein, releasing the MA, CA, and NC proteins, as well as the p2, p1, and p6 peptides (14, 23). The individual proteins are now free to form new intersubunit interactions and indeed cleavage results in a profound morphological change in which the CA and NC proteins collapse to form a conical core, while the MA protein remains associated with the viral envelope (14, 23). There appears to be significant heterogeneity in the gross morphology of the core, although the local interactions between subunits are likely to be conserved (43). The diploid viral genome is contained in the center of this core in a complex with the NC protein (14, 23). Formation of the mature viral core is a critical step in the virus life cycle; mutations that block cleavage and subsequent maturation result in noninfectious particles (23). However, cleavage alone is not sufficient for function. Mutations that allow maturation and yet result in the formation of cores with aberrant morphology inhibit infectivity apparently by blocking the initiation of reverse transcription (9, 13, 32, 33, 38, 40, 41).
A number of studies have demonstrated that the CA protein alone or as part of a CA-NC fusion protein can be induced to polymerize into dimers, larger oligomers, and ultimately tubular polymers (6, 11, 20, 40). In fact, cones similar in size and shape to viral conical cores have been observed in small quantities in CA protein assembly reactions, indicating that in vitro-assembled CA was capable of forming bonding interactions similar to those formed in the virion (18, 27). Thus, in vitro-assembled CA polymers are thought to be good models for the viral core.
While the intact virus has proven refractory to structural studies, high-resolution structures of the MA, CA, and NC domains are available from crystallographic and nuclear magnetic resonance studies (8, 16, 17, 19, 22, 28, 44). The CA protein was composed of two structural domains, a 16.4-kDa N domain and a 9.3-kDa C domain. Cryoelectron microscopy and image analysis performed on tubes assembled in vitro from purified CA indicated that they are helical polymers with a surface lattice of hexameric rings arranged with approximate local p6 symmetry (27). At a lower radius (toward the interior of the tube), each subunit within the hexamer was connected to a subunit in an adjacent hexamer by inward projecting density. When the cryoelectron microscopy and crystallographic data were merged, the CA N domain packed best into the hexameric ring. In this structure, each N domain forms interactions with the adjacent N domain. Six cyclically arranged N domains thus comprise the hexameric turret seen in the image reconstructions. The inward projecting density that connects the hexamers was assigned to the C domain in the form of a dimer, a finding consistent with the observation that the C domain of HIV CA protein forms dimers in solution.
Typically, CA has been induced with salt to polymerize in vitro into long cylindrical structures. However, recent evidence suggests that the morphology of the assembled structures may be influenced by pH (12, 21). Gag constructs with most of MA deleted assemble into spherical polymers at pH 8.0 and assemble into cylindrical polymers at pH 6.0 (21). However, modeling of light-scattering data suggested that CA assembled into cylindrical polymers at pH 7.0 and into spherical polymers at pH 6.8 (12). Similarly, the CA protein itself undergoes changes concomitant with a change in pH, where the radius of CA protein increased by ca. 1.6 nm when the pH was increased from 6.6 to 7.0 (12). These changes in assembled morphology and CA protein indicate that CA protein interactions necessary for assembly are sensitive to pH.
Considerable effort has been put into developing in vitro assembly systems in which purified HIV structural domains can be induced to polymerize into biologically relevant structures. A common feature of all published protocols for the in vitro assembly of CA is a slow dialysis step in which a change in solvent conditions (typically, NaCl concentration) leads to polymerization (6, 11, 20, 40). The slow dialysis step precludes simple analysis of the assembly kinetics. Recently, Yu et al. used a dilution-based technique to monitor the kinetics of the Rous sarcoma virus Gag construct (ΔMBDΔPR) with turbidity (47). The ability to quantitatively monitor the rate of assembly can provide insight into the influence of mutations on assembly, as well as provide mechanistic insight into the pathway of assembly. We report here the development of a rapid dilution-induced technique for CA assembly and utilize this system to evaluate the effects of solvent conditions, protein concentration, and mutations on CA assembly. We also find evidence for an interaction between the N and C domains of CA in solution.
MATERIALS AND METHODS
Protein expression and purification.
Plasmids for expression of wild type (WISP93-73) (19, 46) and M185A mutant HIV-1 CA (17) were obtained from W. Sundquist (University of Utah) and transformed into Escherichia coli BL21(DE3). A 5-liter culture was grown to an A600 of 0.8 at 37°C and induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After 4 h, the cells were collected by centrifugation and stored at −80°C. Cells were lysed by mixing thawed pellets with lysozyme to a final concentration of 12.5 mg/ml in 50 mM Tris-HCl (pH 8.0)-5 mM β-mercaptoethanol. Cellular debris larger than 100 s was removed by centrifugation, and the CA protein was precipitated from the supernatant with 25% saturated ammonium sulfate; a precipitate larger than 500 S was collected by centrifugation, resuspended in 50 mM Tris-HCl (pH 8.0)-5 mM β-mecaptoethanol, and then dialyzed into 25 mM Tris-HCl (pH 8.1). The crude CA was bound to a 40-ml Q-Sepharose column (Amersham Pharmacia Biotech) and eluted with 25 mM Tris-Cl (pH 8.1) and 75 mM NaCl. The protein was determined to be ca. 99% pure when analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Plasmids encoding the N and C domains of CA (representing CA residues 1 to 148 and CA residues 148 to 231, respectively) were obtained from Ming Luo (University of Alabama at Birmingham) and transformed into E. coli BL21(DE3). Induction of expression was done as for whole CA protein. The cells were lysed in 10 mM citrate buffer (pH 5.7). Cellular debris larger than 100S was removed by centrifugation and the C domain was bound to a 5-ml SP-Sepharose column (Amersham Pharmacia Biotech). The protein was eluted in 10 mM citrate buffer (pH 5.7) with a 0 to 500 mM NaCl gradient. The C domain eluted at ca. 300 mM NaCl and was ca. 99% pure as analyzed by SDS-PAGE. The N domain was prepared similarly to the C domain, except that it was loaded onto a Q-Sepharose column in 25 mM Tris-HCl (pH 8.0). The N domain eluted at ca. 100 mM NaCl and was also 99% pure by SDS-PAGE. Mass spectrometry measurements of the purified proteins revealed that the N-terminal methionines had been removed.
In vitro capsid assembly.
Purified CA and the C and N domains were dialyzed into 50 mM Na2HPO4 at pH 8.0 (Buffer D). CA was concentrated to ca. 1 mM by using a MACROSEP 10K centrifugal concentrator (Pall Filtron, East Hills, N.Y.). CA in Buffer D was triggered to assemble by the addition of 50 mM Na2HPO4-4 M NaCl at pH 8.0 (Buffer A). The final salt and protein concentrations were varied to test their effect on CA assembly. (During initial assembly experiments it was noticed that assembly in sodium phosphate buffers was more consistent than in Tris-HCl buffers. Therefore, all subsequent experiments were performed in sodium phosphate buffers.)
Thin-section transmission electron microscopy.
For thin-section electron microscopic analysis, CA protein was assembled at 400, 250, 120, and 38 μM CA protein at final NaCl concentrations of 1.0, 1.2, 1.6, and 2.25 M, respectively. After the assembly reactions were complete (1 h at 20°C), 50% glutaraldehyde was added to produce a final concentration of 1.0%. This was allowed to incubate overnight at room temperature. Assembled polymers were collected by centrifugation at 70,000 rpm for 30 min in a Beckman TLA100.3 rotor (conditions calculated to pellet any species whose sedimentation coefficient is larger than 24 S), and the buffer was removed. The pellet was washed with 2% OSO4 in buffer and water, stained en bloc for 30 min with a saturated aqueous solution of uranyl acetate, dehydrated in a graded acetone series, and then embedded in epoxy resin Embed-812 (Electron Microscopy Sciences). Thin sections (60 to 90 nm) were picked up on copper grids, stained for 20 min on drops of saturated uranyl acetate, rinsed with water, and touched to Whatman filter paper; sections were then stained for 10 min on drops of Reynolds' lead citrate in the presence of NaOH pellets (34), rinsed with water, and dried on Whatman filter paper. Electron micrographs were taken with a Hitachi H-7100 transmission electron microscope at an accelerating voltage of 75 kV and a magnification of ×40,000.
Kinetic analysis of assembly reactions.
For kinetic analysis, unless otherwise indicated, assembly reactions were performed at 38 μM CA protein and 2.25 M NaCl. After the addition of Buffer A to initiate assembly, the reaction was rapidly mixed and placed into a cuvette. Approximately 20 s elapsed between the time of addition and the first time point measured. The increase in optical density was monitored on a Beckman DU640 spectrometer at 350 nm every 20 s for 1 h. To estimate the rate of assembly, the increase in turbidity at early times was fit to a straight line by using Origin 6.0 software (Microcal Software, Inc.). To determine the effect of NaCl concentration on assembly, the volumes of Buffer A and Buffer D were varied to produce the desired final NaCl concentration. For the assembly reactions with combinations of CA and the separate domains, the species were mixed together in Buffer D prior to triggering assembly with Buffer A.
Calculation of the effective CA concentration.
To calculate the concentration of CA not incorporated into heterodimers (and hence free for assembly), the Kd values of the heterodimer and both homodimers were assumed to be equivalent. The concentrations of each species were then calculated by using the known Kd value (17), the C-domain and CA protein concentrations, and the statistical probability of forming each species. For example, in equimolar concentrations the probability of forming the CA homodimer, CA and C-domain heterodimer, and C-domain homodimer is 1:2:1, respectively. The concentration of effective CA (CA not in a heterodimer) is then equal to the total CA concentration minus the heterodimer CA concentration.
RESULTS
In vitro assembly by rapid dilution.
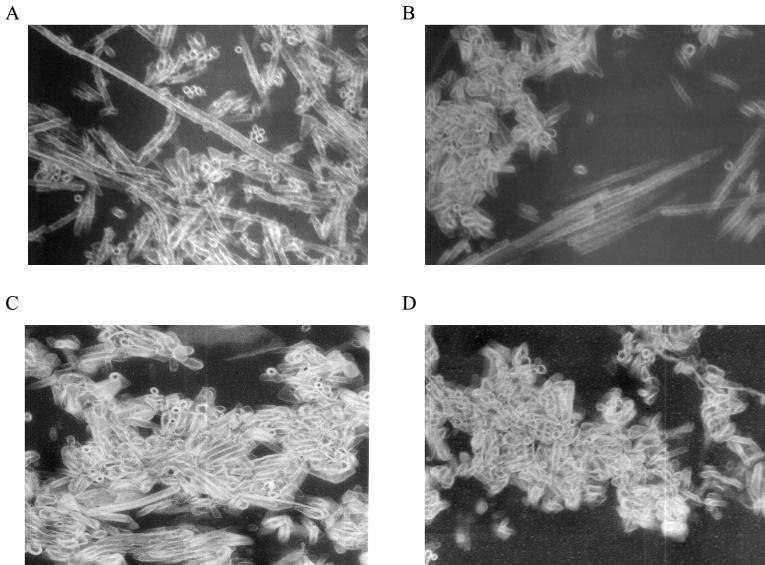
Purified HIV-1 capsid protein was rapidly diluted into 4 M NaCl such that the final concentrations of CA and NaCl were 400 μM and 1 M, respectively. These final conditions are comparable to those previously used in assembly by dialysis experiments (6, 11, 20, 40). The reaction was allowed to proceed at 20°C for 1 h, at which point the reaction was fixed with 1% glutaraldehyde, and the polymerized material was collected by centrifugation, embedded, thin sectioned, and observed by transmission electron microscopy (Fig. 1A). Long tubular polymers (100 to >1,000 nm) were observed, as well as end-on views of these tubes, that appeared roughly cylindrical, with an average diameter of ca. 33 nm. To determine the effect of CA protein and salt concentration on morphology, CA was assembled at 250, 120, and 38 μM at final NaCl concentrations of 1.2, 1.6, and 2.25 M, respectively. The polymerized material was analyzed by thin-section electron microscopy as described above (Fig. 1). The polymers formed at 250 μM CA and 1.2 M NaCl were shorter on average than those formed at 400 μM CA and 1 M NaCl; however, the end-on views of these cylinders were similar in diameter to the cylinders formed at a higher CA concentration, indicating that the tube morphology was probably the same (Fig. 1A and B). However, lower CA protein concentrations and higher NaCl concentrations (120 and 38 μM CA protein and 1.6 and 2.25 M NaCl, respectively) yielded similar decreases in polymer length while maintaining the same tube diameter. Despite the differences in length, the gross morphology of the polymers was similar. These tubes, formed by dilution, are comparable in diameter, as well as in ultrastructural morphology, to those formed during in vitro assembly by dialysis (6, 11, 18, 20, 27, 40). These CA polymers are generally thought to represent good models for biophysical and structural studies.
FIG. 1.
CA was assembled at final concentrations of 400 (A), 250 (B), 120 (C), and 38 (D) μM and final NaCl concentrations of 1, 1.2, 1.6, and 2.25 M, respectively. After completion of assembly (∼1 h), the CA polymers were pelleted (>24S) and prepared for thin section electron microscopy as described in Materials and Methods.
The formation of large cylindrical polymers during assembly is accompanied by an increase in light scattering that can be easily detected as an increase in turbidity. A variety of salt and CA concentrations were surveyed to identify conditions under which the rate of assembly would be amenable to routine laboratory analysis. In an initial series of survey experiments, assembly was performed by using small volumes in glass capillary tubes, and the turbidity was monitored visually. The rate of assembly was observed to increase with increasing salt and protein concentrations. Based on these survey experiments, assembly reactions were then performed within a narrow range of protein and salt concentrations, and the change in turbidity over time was monitored in a spectrophotometer.
The rate of assembly is dependent upon salt and protein concentrations.
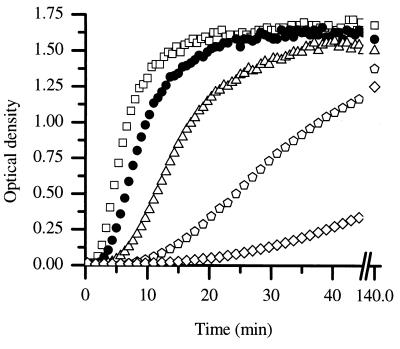
To evaluate the effect of ionic strength on the kinetics of assembly, CA was diluted to a final protein concentration of 38 μM at final salt concentrations ranging from 1.8 to 2.4 M, and the increase in turbidity was monitored over time (Fig. 2). The rate of increase in turbidity was sharply dependent upon the salt concentration; increasing the salt concentration caused an increase in the rate of polymerization. There are two limiting explanations for this type of salt dependence: either salt can increase the rate of assembly directly, for example, through charge shielding of unfavorable interactions or salt can induce a conformational change within the CA protein subunit, resulting in a higher fraction of active protein. Although there was a pronounced effect of salt on the rate of assembly, at longer time points (140 min) the final amount of turbidity achieved was relatively independent of the salt concentration. This suggests that salt increases the rate of assembly rather than the fraction of CA protein capable of assembly. A final salt concentration of 2.25 M was chosen as the standard condition.
FIG. 2.
CA was assembled at 38 μM at final NaCl concentrations of 2.4 M (□), 2.25 M (•), 2.10 M (▵), 1.95 M ( ), and 1.80 M (⋄).
), and 1.80 M (⋄).
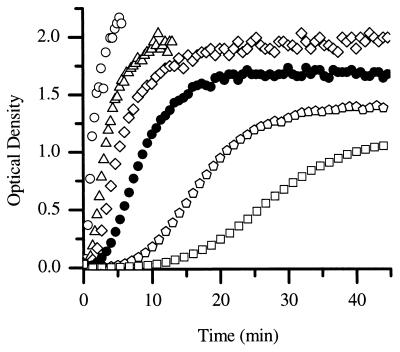
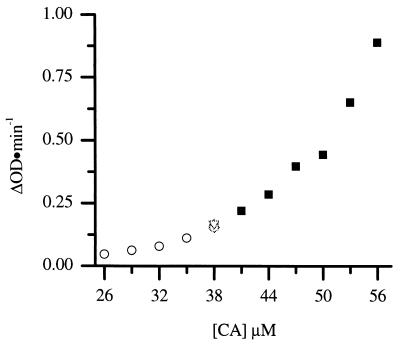
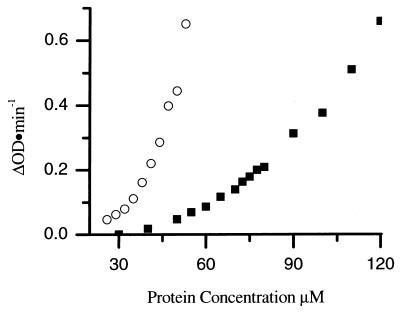
The concentration dependence of the kinetics of assembly reactions can provide information about the number of protein subunits involved in the rate-limiting step of assembly. Therefore, the kinetics of CA assembly was examined as a function of protein concentration over the range from 26 to 56 μM (Fig. 3). The rate of assembly increased with increasing CA protein concentration. The final amount of turbidity achieved was proportional to the initial CA protein concentration, indicating that turbidity is a sensitive measure of the amount of protein polymerized, and therefore the rate of polymerization is proportional to the change in turbidity with time. The initial assembly rates were determined by drawing tangents to the initial region of the progress curves, and a plot of the rate versus protein concentration was generated (Fig. 4) (31). A 2-fold change in the protein concentration (from 27 to 55 μM) results in a 16-fold increase in the rate of assembly, indicating that the rate of assembly is highly concentration dependent. High-order dependence on protein concentrations is typical of nucleated polymerization processes and implies cooperative assembly (31, 35). These data also demonstrate the reproducibility of the measurements; data from two independent sets of experiments, as well as datum points obtained at 38 μM CA in five different series of experiments, lie on the same curve (Fig. 4).
FIG. 3.
CA protein was assembled at final concentrations of 2.25 M NaCl and 56 μM (○), 50 μM (▵), 44 μM (⋄), 38 μM (•), 32 μM ( ), and 26 μM (□) CA.
), and 26 μM (□) CA.
FIG. 4.
Rate of CA protein assembled at a final concentration of 2.25 M NaCl. The results were assembled during two trials (denoted by circles and squares, respectively). The superimposed symbols at 38 μM represent the rate of CA assembly collected during five different assembly experiments.
C-domain dimer interactions are important for assembly.
The assembled tubular polymers are composed of hexamers of the N-terminal CA domain held together by dimerization of the C domain (27). If the interactions present in the final structure are required during assembly, mutations that alter these interactions will have an effect on the rate of assembly. One such mutation is M185A, which lies within the C domain. C-domain dimerization is driven by the parallel packing of helix 2 of the C domain across a dyad axis to form a hydrophobic core (17). Substitution of alanine for methionine in helix 2 of the C domain decreases the extent of dimerization. In vivo, this results in the formation of noninfectious virions (17). If dimerization plays a substantial role in assembly, the M185A mutant would be expected to assemble more slowly than the wild-type protein. To determine the effect the mutation has on assembly rates, CA protein carrying the M185A mutation was assembled at different protein concentrations. The rates for these assembly reactions were determined and compared to the wild-type CA protein rates (Fig. 5). At equivalent protein concentrations, the rate of assembly of M185A was reduced ∼10-fold from that of the wild type. Presumably, these decreased rates reflect the decreased ability of the C domain of M185A to dimerize. These data demonstrate that dimerization is important in assembly and that changes in subunit-subunit interactions in solution manifest themselves by changing the assembly rate.
FIG. 5.
Rates for wild-type CA (○) and mutant M185A CA (▪) assembled with a final NaCl concentration of 2.25 M.
Inhibition of in vitro assembly.
Based on the importance of C-terminal dimerization in assembly, it would be predicted that molecules that disrupt dimerization would also alter assembly rates. It has been demonstrated that the isolated C domain forms dimers as readily as does the intact CA protein (17). Therefore, we predicted that the addition of isolated C domain to assembly reactions would inhibit polymer formation through the formation of a mixed heterodimer composed of one full-length CA protein molecule and one C domain. This heterodimer would either be unable to become incorporated into growing cylinders or would act as a cap to prevent further cylinder elongation. In either case, the result would be a decreased rate of assembly. To test this hypothesis, purified recombinant C domain was included in assembly reactions and the effect on the rate of CA assembly was determined (Fig. 6A).
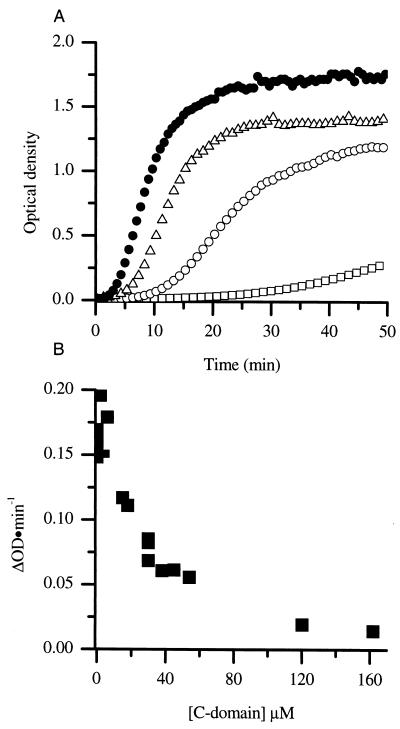
FIG. 6.
(A) Assembly of CA protein at 38 μM (•) and in the presence of C domain at 18 μM (▵), 54 μM (○), and 162 μM (□) (A). (B) Rates of CA assembled at 38 μM in the presence of various concentrations of C domain.
Assembly reactions were performed at a fixed CA concentration of 38 μM in the presence of various concentrations of C domain ranging from 18 to 162 μM. The C domain was added to the CA protein prior to the addition of NaCl to allow equilibration to occur. After the addition of NaCl to a final concentration of 2.25 M, the rate and extent of polymerization were monitored by turbidity. The inclusion of the C domain in assembly reactions resulted in a decrease in the apparent rate and extent of assembly of CA (Fig. 6A). Even at longer times (120 min) there was a decrease in the extent of polymerization. In control experiments, the addition of unrelated proteins such as T4 lysozyme at similar concentrations had little effect on assembly (not shown). The effect of added C domain on the rate of assembly was evaluated by plotting the rate of assembly versus C-domain concentration (Fig. 6B).
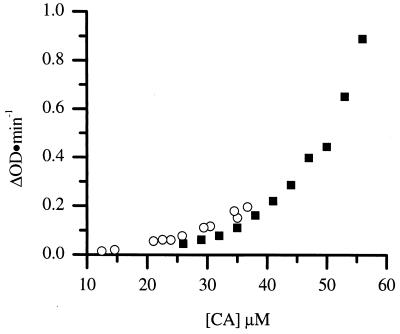
The heterodimer inhibition model predicts that the fraction of full-length CA protein bound in heterodimers is unavailable for assembly. By taking advantage of the fact that the association constant for dimerization of the isolated C domain is identical to that of the intact protein (17), it is possible to calculate the concentration of heterodimers for any given mixture of intact CA and C domain. This corresponds to a reduction in effective concentration of the full-length CA. This calculation was performed for the CA and C-domain mixtures used in the assembly experiments, and the experimentally determined rate of assembly was plotted versus the calculated concentration of biologically active CA in homodimers (Fig. 7). The experimentally determined rate of assembly in the presence of C domain falls, within reasonable agreement, on the same curve observed for CA in the absence of inhibitor. This suggests the mechanism of inhibition is via the formation of inactive heterodimers.
FIG. 7.
The rates of CA assembled at a final 2.25 M NaCl concentration are shown (▪). CA protein was assembled at 38 μM with a final 2.25 M NaCl concentration in the presence of C domain at different concentrations, and the biologically active CA concentration was calculated and plotted versus the respective rates (○).
Role of the N-terminal domain.
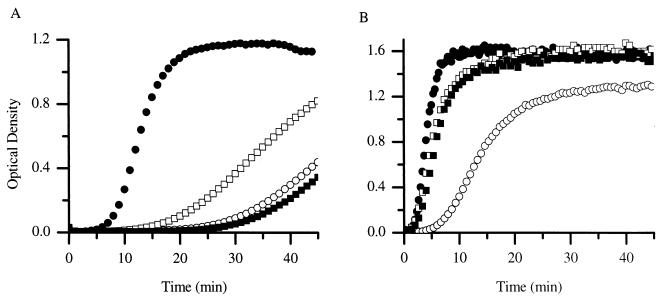
The structure of the CA cylinder obtained by Li et al. (27) suggests that interactions between the N domains to form hexameric clusters are required for assembly. In experiments analogous to those described for the C domain, the N domain and CA were mixed together prior to assembly to determine whether the N domain would affect CA assembly. The addition of N-terminal domain at concentrations ranging from 20 to 160 μM at pH 8.0 had no apparent effect on CA assembly (Fig. 8; only the data for 45 μM are shown). Although this is somewhat puzzling, it is consistent with the earlier reports that there appear to be no strong interactions between N domains in solution.
FIG. 8.
The CA protein (38 μM) was assembled in the absence of both domains (•), in the presence of N domain at 45 μM (□), in the presence of the C domain at 45 μM (○), and in the presence of both N and C domains at 45 mM each (▪) at pH 7.0 (A) and pH 8.0 (B).
In recent studies, Ehrlich et al. have shown that the hydrodynamic radius of CA decreases when the pH is decreased from 8.0 to 6.6, suggesting that the CA dimer became more compact (12), and Gross et al. have demonstrated that epitopes on CA become inaccessible at a pH below 6.8 (21). Gross et al. have also reported, based on circular dichroism spectra, that there is no significant change in subunit secondary structure over this pH range (21). One possible mechanism by which the dimer could become more compact and epitopes masked is via the formation of a new inter- or intrasubunit interaction(s). pH has also been reported to affect the form of the polymer assembled (12, 21). To examine the effect of pH, assembly reactions were performed over a range of pH values from 7.0 to 8.0. The assembly rate of CA protein changed little from pH 7.25 to 8.0 (data not shown). However, the assembly rate of CA protein at pH 7.0 was significantly decreased from the rates observed at a pH of >7.0. As assayed by thin-section electron microscopy, the products of the assembly reaction at pH 7.0 were very similar in morphology and quantity to the assembly products observed at pH 8.0 (data not shown). This pH dependence is consistent with earlier reports on the pH dependence of assembly of HIV capsid protein (6, 12, 20, 21).
pH affects on N- and C-domain interactions with CA protein.
The effect of added N and C domain was examined at pH 7.0. C domain was added to a final concentration of 45 μM, and a marked inhibition of assembly was observed. The inhibitory effect of the C domain was slightly greater at pH 7.0 (sixfold) than at pH 8.0 (fourfold).
Whereas the addition of the N domain had little affect on the assembly of CA protein at pH 8.0 (Fig. 8B), the N domain drastically decreased the rate of assembly at pH 7.0 (Fig. 8A). This finding indicated that the N-domain interaction with CA was sharply pH dependent. Because both the N and the C domains inhibited at pH 7.0 when added separately, their combined effect on assembly was examined. The mixture of N and C domains caused a small decrease in the rate from that observed when the domains were added alone (Fig. 8A).
However, at pH 8.0 a surprising result was obtained when both the C domain and the N domains were combined with CA prior to assembly. The N and C domains were combined at a ratio of 1:1 with CA at pH 8.0, and then assembly was triggered by increasing the salt concentration. Interestingly, inclusion of the N domain partially alleviated the C-domain inhibition. In control experiments, it was demonstrated that 1:1 mixtures of N- and C-terminal domains in the absence of CA displayed no change in turbidity upon addition of salt (data not shown). These data strongly suggest an interaction between the N and C domains in solution during CA protein assembly.
Mechanism of inhibition.
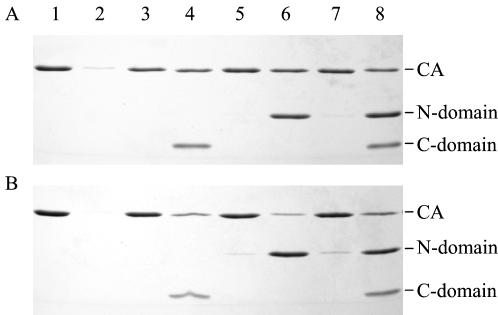
A prediction based on the heterodimer model for inhibiting assembly is that neither the C nor the N domain would be incorporated into the assembled polymers. To test for domain incorporation into cylinders, CA was assembled (38 μM) in the presence of C and N domains (both at 45 μM), and after 1 h the polymerized material (any species > 24S) was collected by centrifugation. The pellet was resuspended in a volume of buffer equivalent to the starting volume, and the pellet and supernatant fractions were analyzed by SDS-PAGE. This experiment was performed at both pH 7.0 and pH 8.0 (Fig. 9A and B, respectively). At both pH values examined, the amount of CA protein in the pellet fraction was decreased in the presence of the C domain compared to the assembly reactions without any added C domain. This result is consistent with the observation that the rate and extent of assembly was decreased by the presence of the C domain. The C domain remains primarily in the supernatant, suggesting that it is not efficiently incorporated into the assembling polymer, a result consistent with inhibition by mixed dimer formation.
FIG. 9.
The CA protein was assembled at a concentration of 38 μM, with a final 2.25 M NaCl concentration, in the absence of N and C domains (lanes 1 and 2), in the presence of C domain (lanes 3 and 4), in the presence of N domain (lanes 5 and 6), and in the presence of both N and C domains (lanes 7 and 8) at pH 7.0 (A) and pH 8.0 (B). CA polymers of >24S were pelleted and resuspended in a volume equivalent to that of the supernatant. Equal volumes of the pelleted fraction (lanes 1, 3, 5, and 7) and the supernatant fraction (lanes 2, 4, 6, and 8) were analyzed by SDS-PAGE on a 15% reducing gel.
In the case of the added N domain, the amount of CA in the pellet was decreased when the experiment was performed at pH 7.0 but not when it was performed at pH 8.0. These results are also consistent with the kinetic observations based on turbidity. A small amount of N domain was found associated with the assembled CA at pH 8.0, but the bulk remained in the soluble fraction. At pH 7.0, there was no evidence of N domain associated with the polymerized CA.
In the mixtures of the N and C domains, there is essentially no effect on the partitioning of the domains between the supernatant or pellet fractions at either pH. This result suggested that the mechanism of rescue of inhibition at pH 8.0 is unlikely to be complementation of C domain by N domain and incorporation into the growing polymer.
DISCUSSION
We demonstrated here that the direct dilution of the CA protein into a high-salt solution results in the polymerization of tubes reminiscent of the cores found in mature virions. Although the average length of these tubes varies with protein and salt concentration, the diameter is relatively invariant, suggesting conserved local interactions. Structural rearrangement after Gag cleavage is a conserved feature of the retrovirus life cycle. However, the morphology of the core formed is variable. While in the case of HIV and the lentiviruses in general the core is conical, in the case of Mason-Pfizer monkey virus, the core is cylindrical and in the case of murine leukemia virus and Rous sarcoma virus, the core is polyhedral (26, 30, 45). Despite this variable morphology, the basic fold of the capsid protein is conserved across the retroviruses, and the subunit interfaces observed in the crystal structure are generally conserved across species (25). Thus, protein subunits which form similar local interactions at the intersubunit interface can display different (although related) gross morphologies. This situation is similar to that in which icosahedral viral particles can be induced to assemble into particles of different T numbers (hepatitis B, P22) (10, 49), overall symmetry (simian virus 40) (37), or even sheets and spirals, and is formally similar to the mechanism by which icosahedral symmetry is generated from chemically identical subunits in such virus particles. In the cases examined to date, the fundamental interactions in the different quasi-equivalent positions are preserved, but the disposition of the subunits within the lattice is altered through hinge-bending type motions (7, 36). Thus, local interactions can be largely conserved, whereas global morphology can be profoundly altered. It is likely that global morphology is dictated by factors such as assembly kinetics, effector molecules, and long-range interactions (charge-charge repulsion) rather than through local interactions. However, the mechanism of viral form determination remains obscure in general, and the reason conical tubes are favored over straight tubes in the case of HIV is particularly difficult to explain. It should be noted, however, that in the case of HIV there appears to be considerable particle-to-particle variability in core morphology, which may in part reflect the difficulty of controlling these long-range interactions.
The salt-induced assembly process requires specific intersubunit interactions, as witnessed by the fact that neither the isolated N or C domains nor a mixture of the two domains displays any increase in turbidity. Turbidity has long been used to monitor the polymerization of viral particles, and for rods whose length is large relative to the wavelength of light the amount of turbidity observed is proportional to the total number of subunits assembled (3). Thus, the rate of change in turbidity is proportional to the rate of assembly. In the present study we confirmed that the rate of assembly is dependent upon the concentration of both protein and salt. Examination of the end point of the assembly reactions suggests that the effect of salt is to increase the rate of assembly rather than increase the fraction of protein competent for assembly. In the cryoelectron microscopic image reconstruction of CA tubes obtained by Li et al., the N-terminal domains pack through the interaction of helices I and II (27). It is possible that significant charge-charge repulsion between adjacent N domains needs to be overcome to allow the N-terminal domain to form the hexameric clusters which comprise the outer surface of the tube. The ability to quantitatively evaluate the effect of mutations on the rate of assembly should allow for testing of this hypothesis.
The rate of assembly was highly dependent on the concentration of CA, suggesting that the rate-limiting step in assembly is the self-association of several CA molecules. A contrasting case is one wherein a rate-limiting conformational change in the protein subunit is required prior to continued assembly, such as is seen in the polymerization of bacterial flagellin (2). This does not appear to be the case for the assembly of CA into tubes. Furthermore, the high-order dependence of the rate of assembly on protein concentration suggests that several molecules of CA, rather than simply a dimer, are involved in the rate-limiting step (31, 35). However, it is clear that dimerization plays an important role in promoting the assembly of tubes. The assembly kinetics of the M185A mutant, which has been reported to block dimer formation (17), are substantially reduced relative to wild-type protein, and it has previously been observed that RNA-induced dimerization of a Gag Δp6 fusion protein (5) or dimerization through the introduction of a coiled-coil domain in place of the NC domain (1, 48) promotes assembly. It thus seems likely that dimers form a basic assembly building block. Consistent with the idea that dimers represent a basic building block for assembly is the observation that the addition of the isolated C domain to an assembly reaction can inhibit polymerization and that it likely does so through the formation of inactive mixed heterodimers. Based on the fact that assembly and inhibition proceed at both pH 8.0 and pH 7.0, we conclude that the C-domain homologous interaction is relatively pH independent. This interaction constitutes one of the fundamental interactions driving tube formation and, as such, represents a potential therapeutic target.
If the final structure of the tubes is similar to those analyzed by Li et al. (27), additional interactions involving the N domain are necessary. These interactions must be sufficiently stable to promote higher-order assembly and persist in the tubes. However, isolated N domains do not show any tendency to polymerize into hexamers in solution even under conditions which favor assembly (J. Lanman and P. E. Prevelige, Jr., unpublished data). Furthermore, the N domain generally packs as dimers in crystals, both as the isolated domain and as part of the intact CA protein. It is possible that the failure to find N-domain interactions in solution is due to the fact that removal of the C domain alters the N-domain conformation and obviates the interaction. Consistent with previous data, our kinetic experiments failed to provide evidence of N-domain interactions at pH 8.0. Given the highly concentration-dependent nature of the assembly kinetics, strong-stable interactions between N domains would be expected to be manifest as a decrease in the assembly rate when isolated N domain was added to the assembly reaction. However, at pH 7.0 we do find evidence of a homologous N-domain interaction. The addition of N domain to an assembly reaction results in a decrease in the rate of assembly. Although in principle the added N domain could interact with either the N or the C domain of the CA and inhibit assembly, the observation that a mixture of N domain and C domain further inhibits assembly suggests that this is a homotypic interaction. If the N domain interacted with the C domain in a mixture of the two, some fraction of each domain would be associated with one another and the effect would be to decrease the amount of inhibition observed. This is, in fact, precisely the result seen when both domains are mixed at pH 8.0. At this pH, at which the N domain alone is ineffective, it is capable of mitigating the inhibitory effect of the C domain. This suggests that there is an interaction between the N and C domains at pH 8.0. If this interaction in fact exists, why did the N domain not inhibit assembly at pH 8? One possible explanation is that the isolated N domain cannot effectively compete with the N domain of the intact CA for the C-domain binding site because of steric or effective concentration factors. If there is in fact an N-domain-C-domain interaction at pH 8.0, why then were the two domains not found in the assembled polymer? It must be that these species cannot successfully be incorporated due to steric or conformational effects.
Thus, we propose that there are three interactions that lead to assembly: the well-documented C-domain homotypic dimerization, a low-pH-dependent N-domain homotypic interaction previously not observed in solution, and a high-pH-dependent N-domain-C-domain interaction. Others have published data supporting the existence of a pH-dependent alteration in subunit-subunit interactions (12, 21), and there is both genetic evidence from Rous sarcoma virus (4) and calorimetric (I. Protassevitch and P. E. Prevelige, Jr., unpublished data) evidence from HIV for an N-domain-C-domain interaction. It is also likely that the notion of localizing the interactions to lie within or between domains, while convenient, is overly simplified and that the true footprint of these interactions involves contributions from both domains. These data suggest that an N-domain-C-domain interaction is involved in the assembly in both Rous sarcoma virus and HIV and that this interaction could be formed in other retroviruses.
Acknowledgments
We are grateful to W. Sundquist for the CA and M185A plasmid constructs and to Leigh Millican at the UAB Miscroscopy Core Facility for assistance with electron microscopy.
This work was supported by NIH grants AI44626 (P.E.P.), NIH/NCI/NRSA T32 CA09467 (J.L.), and AI43230 (M.S.).
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asakura, S. 1968. A kinetic study of in vitro polymerization of flagellin. J. Mol. Biol. 35:237-239. [DOI] [PubMed] [Google Scholar]
- 3.Berne, B. J. 1974. Interpretation of the light scattering from long rods. J. Mol. Biol. 89:755-758. [DOI] [PubMed] [Google Scholar]
- 4.Bowzard, J. B., J. W. Wills, and R. C. Craven. 2001. Second-site suppressors of Rous sarcoma virus Ca mutations: evidence for interdomain interactions. J. Virol. 75:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caspar, D. L. D., and A. Klug. 1962. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 27:1-24. [DOI] [PubMed] [Google Scholar]
- 8.De Guzman, R. N., Z. R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384-388. [DOI] [PubMed] [Google Scholar]
- 9.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earnshaw, W., and J. King. 1978. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J. Mol. Biol. 126:721-747. [DOI] [PubMed] [Google Scholar]
- 11.Ehrlich, L. S., B. E. Agresta, and C. A. Carter. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrlich, L. S., T. Liu, S. Scarlata, B. Chu, and C. A. Carter. 2001. HIV-1 capsid protein forms spherical (immature-like) and tubular (mature-like) particles in vitro: structure switching by pH-induced conformational changes. Biophys. J. 81:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzon, T., B. Leschonsky, K. Bieler, C. Paulus, J. Schroder, H. Wolf, and R. Wagner. 2000. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology 268:294-307. [DOI] [PubMed] [Google Scholar]
- 14.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 15.Fuller, S. D., T. Wilk, B. E. Gowen, H. G. Krausslich, and V. M. Vogt. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7:729-738. [DOI] [PubMed] [Google Scholar]
- 16.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 17.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 18.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 19.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 20.Gross, I., H. Hohenberg, and H. G. Kraüsslich. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249:592-600. [DOI] [PubMed] [Google Scholar]
- 21.Gross, I., H. Hohenberg, T. Wilk, K. Wiegers, M. Grattinger, B. Muller, S. Fuller, and H. G. Krausslich. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter, E. 1994. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin. Virol. 5:71-83. [Google Scholar]
- 24.Jacks, T., M. D. Power, F. R. Masiarz, P. A. Luciw, P. J. Barr, and H. E. Varmus. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280-283. [DOI] [PubMed] [Google Scholar]
- 25.Kingston, R. L., T. Fitzon-Ostendorp, E. Z. Eisenmesser, G. W. Schatz, V. M. Vogt, C. B. Post, and M. G. Rossmann. 2000. Structure and self-association of the Rous sarcoma virus capsid protein. Struct. Fold Des. 8:617-628. [DOI] [PubMed] [Google Scholar]
- 26.Kingston, R. L., N. H. Olson, and V. M. Vogt. 2001. The organization of mature Rous sarcoma virus as studied by cryoelectron microscopy. J. Struct. Biol. 136:67-80. [DOI] [PubMed] [Google Scholar]
- 27.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 28.Massiah, M. A., M. R. Starich, C. Paschall, M. F. Summers, A. M. Christensen, and W. I. Sundquist. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 244:198-223. [DOI] [PubMed] [Google Scholar]
- 29.Mervis, R. J., N. Ahmad, E. P. Lillehoj, M. G. Raum, F. H. Salazar, H. W. Chan, and S. Venkatesan. 1988. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J. Virol. 62:3993-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nermut, M. V., and D. J. Hockley. 1996. Comparative morphology and structural classification of retroviruses. Curr. Top. Microbiol. Immunol. 214:1-24. [DOI] [PubMed] [Google Scholar]
- 31.Prevelige, P. E., Jr., D. Thomas, and J. King. 1993. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys. J. 64:824-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reicin, A. S., A. Ohagen, L. Yin, S. Hoglund, and S. P. Goff. 1996. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J. Virol. 70:8645-8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reicin, A. S., S. Paik, R. D. Berkowitz, J. Luban, I. Lowy, and S. P. Goff. 1995. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J. Virol. 69:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds, E. S. 1963. Use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romberg, L., M. Simon, and H. P. Erickson. 2001. Polymerization of Ftsz, a bacterial homolog of tubulin. is assembly cooperative? J. Biol. Chem. 276:11743-11753. [DOI] [PubMed] [Google Scholar]
- 36.Rossmann, M. G. 1984. Constraints on the assembly of spherical virus particles. Virology 134:1-11. [DOI] [PubMed] [Google Scholar]
- 37.Salunke, D. M., D. L. Caspar, and R. L. Garcea. 1989. Polymorphism in the assembly of polyomavirus capsid protein VP1. Biophys. J. 56:887-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang, S., T. Murakami, B. E. Agresta, S. Campbell, E. O. Freed, and J. G. Levin. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 75:9357-9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogt, V. M., and M. N. Simon. 1999. Mass determination of rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 73:7050-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, C. T., and E. Barklis. 1993. Assembly, processing, and infectivity of human immunodeficiency virus type 1 gag mutants. J. Virol. 67:4264-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilk, T., I. Gross, B. E. Gowen, T. Rutten, F. de Haas, R. Welker, H. G. Krausslich, P. Boulanger, and S. D. Fuller. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilk, T., R. Welker, T. Rutten, D. Thomas, H. G. Krausslich, and S. D. Fuller. 2001. R—the key to HIV capsid assembly. Virus Res. 77:103-105. [Google Scholar]
- 44.Worthylake, D. K., H. Wang, S. Yoo, W. I. Sundquist, and C. P. Hill. 1999. Structures of the HIV-1 capsid protein dimerization domain at 2.6 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 55:85-92. [DOI] [PubMed] [Google Scholar]
- 45.Yeager, M., E. M. Wilson-Kubalek, S. G. Weiner, P. O. Brown, and A. Rein. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. USA 95:7299-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoo, S., D. G. Myszka, C. Yeh, M. McMurray, C. P. Hill, and W. I. Sundquist. 1997. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 269:780-795. [DOI] [PubMed] [Google Scholar]
- 47.Yu, F., S. M. Joshi, Y. M. Ma, R. L. Kingston, M. N. Simon, and V. M. Vogt. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zlotnick, A., N. Cheng, J. F. Conway, F. P. Booy, A. C. Steven, S. J. Stahl, and P. T. Wingfield. 1996. Dimorphism of hepatitis B virus capsids is strongly influenced by the C terminus of the capsid protein. Biochemistry 35:7412-7421. [DOI] [PubMed] [Google Scholar]